Abstract
Background
The successful sequencing of SARS-CoV-2 cleared the way for the use of omics technologies and integrative biology research for combating the COVID-19 pandemic. Currently, many research groups have slowed down their respective projects to concentrate efforts in the study of the biology of SARS-CoV-2. In this bibliometric analysis and mini-review, we aimed to describe how computational methods or omics approaches were used during the first months of the COVID-19 pandemic.
Methods
We analyzed bibliometric data from Scopus, BioRxiv, and MedRxiv (dated June 19th, 2020) using quantitative and knowledge mapping approaches. We complemented our analysis with a manual process of carefully reading the selected articles to identify either the omics or bioinformatic tools used and their purpose.
Results
From a total of 184 articles, we found that metagenomics and transcriptomics were the main sources of data to perform phylogenetic analysis aimed at corroborating zoonotic transmission, identifying the animal origin and taxonomic allocation of SARS-CoV-2. Protein sequence analysis, immunoinformatics and molecular docking were used to give insights about SARS-CoV-2 targets for drug and vaccine development. Most of the publications were from China and USA. However, China, Italy and India covered the top 10 most cited papers on this topic.
Conclusion
We found an abundance of publications using omics and bioinformatics approaches to establish the taxonomy and animal origin of SARS-CoV-2. We encourage the growing community of researchers to explore other lesser-known aspects of COVID-19 such as virus-host interactions and host response.
Keywords: Omics sciences, COVID-19, SARS-CoV-2, VOSviewer, Bibliometrics
Highlights
-
•
China, USA, UK, and India lead SARS-CoV-2 research using bioinformatics.
-
•
Genome, proteome, and metagenome sequence analysis helped to identify virus nature.
-
•
Molecular docking is a widely used approach in identifying therapeutic targets.
1. Introduction
The single-stranded RNA virus SARS-COV-2 is the causative agent of the COVID-19 disease that impacts the lower respiratory tract, with the potential to spread and injure other tissues [1]. Since the first step in SARS-CoV-2 infection is the binding to host cell receptor angiotensin-converting enzyme 2 (ACE2) [2,3], it has been a focus of interest to identify the pattern of expression of this receptor across different cells and tissues. To the date, we know that there is a variety of host cells expressing it in different tissues, such as lung AT2, nasal epithelial cells, tongue keratinocytes, liver cholangiocyte, colon colonocytes, esophagus keratinocytes, ileum and rectum enterochromaffin cells, stomach epithelial cells, and kidney proximal tubules [[4], [5], [6]]. Accordingly, there is a broad set of symptoms that ranges from asymptomatic infections, mild respiratory symptoms, severe pneumonia, acute kidney injury [7], digestive and circulatory system affection, to fatality [1].
It is becoming common knowledge that the surface-anchored spike protein mediates coronavirus entry through the binding to ACE2, however, it less explored that it has also been reported that coronaviruses exploit many other surface molecules according to the cell type in order to make the internalization more efficient [8]. Surface virus-host triggers a subsequent series of biological events including the formation of vesicles, enzymes activation/repression, host molecules recruitment, synthesis of viral components, among others. Integrated multi-omics studies offer an unbiased approach to study the host-virus interactomics that ultimately can result in the detection of therapeutic marks for this novel infection, which has a pivotal role in drug repurposing as well as developing new drugs and vaccines in a precise and efficient manner [9]. Consequently, arousing omics-scale studies on this viral infection are offering a great potential to study the pathobiology of the infection, and ways forward for diagnostic and therapeutic innovation [9].
Since SARS-CoV-2 is highly contagious, this oftentimes restricts the provisions for clinical samples handling in omics research facilities, making it a challenge to implement systems-level molecular studies. Given this limitation, it would be useful for scientists in this field to be aware of trends in omics approaches and computational methods to address COVID-19 related issues. Hence, in this bibliometric analysis and mini-review, we aimed to describe how computational methods or omics approaches were used during the first months of the COVID-19 pandemic. We hope that our work will serve as a roadmap to allow the identification of key knowledge gaps and research priorities. Furthermore, we hope that in future pandemics or infectious outbreaks, this document may be useful in helping future researchers quickly understand how omics data and computational methods can be used to help discern the first unknowns that arise in these scenarios.
2. Materials and methods
2.1. Sources of data and search strategy
Searches were conducted in Scopus and preprint servers (BioRxiv and MedRxiv) on June 19, 2020 (Search formula in supplemental material 1), and it was not constrained regarding language, publication stage and time. Nevertheless, document type was refined to journal articles. Ethical approval was not required in this study, because no human subjects were enrolled.
2.2. Data analysis
We analyzed the bibliographic data through a quantitative analysis approach and a knowledge mapping technique using bibliometric data. For the quantitative analysis the information was sourced from Scopus. Knowledge mapping was performed using VOSviewer (V1.6.14) [10], focalizing on “link strength” of networks based on author keywords, and the text corpus (title and abstract). To identify emerging terms, we manually edited the thesaurus list to exclude expected terms such as COVID-19 and SARS-CoV-2 and to avoid irrelevant terms (i.e., mean and order). We used the full counting method to produce two co-occurrence networks of the most-used terms for author and corpus text keywords, with a minimum of 2 occurrences for each term. Additionally, we extended the analysis by developing a bibliographic coupling graph, where the connection between two publications is determined by the third set of publications (minimum three) that were cited by these two.
2.3. Data collection and mini-review
To identify omics and computational relevant methodologies used in each publication and the purpose for what they were used, two researchers (AR and JM) accessed the abstracts in the reference editor Mendeley (version 1.19.3) where the articles from Scopus were downloaded. Because the search engine in BioRxiv and MedRxiv lacks the option to download the set of retrieved results in any format, two researchers (LV and JM) accessed the abstracts directly on the webpage. To enrich the discussion, we also highlight some of the main findings of the articles selected in the mini-review section.
3. Results
3.1. China and USA leading the publication in indexed and pre-prints articles
By June 19th, 2020 we retrieved 100 articles from Scopus and 176 from MedRxiv and BioRxiv. The distribution of the type of article based on the citation overview tool in Scopus showed that there were 77 original articles, 12 reviews, 6 letters, 2 editorials, 2 notes, and 1 data article. Only 62 of the original articles were eligible for further analysis (Supplemental material 2). As the second consulted database does not have a citation download option, we selected the relevant articles by reading the abstracts and in some cases, we were required to review the methodology session in the publication; 122 fulfilled the inclusion criteria in MedRxiv and BioRxiv.
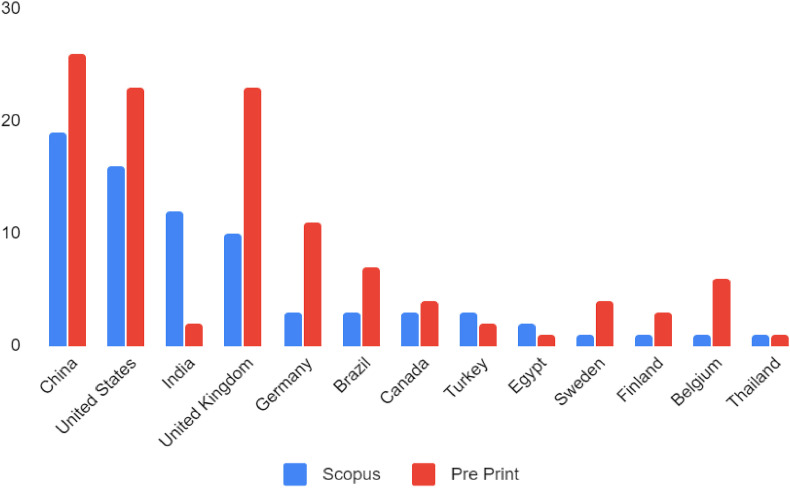
The most active countries publishing about COVID-19 using omics or computational approaches are China, USA, India, and the United Kingdom; Fig. 1 shows the number of published articles per country, differentiating the ones from Scopus and pre-prints articles at MedRxiv and BioRxiv. Table 1 shows bibliometric information of journals that contributed more than 2 articles related to within defined fields research of omics technologies and integrative biology research approaches for combating the COVID-19 pandemic. These journals correspond to 16,88% of the total number of articles found in Scopus. The other 83.12% of the articles are published in 64 different journals, with one article each. These journals are mainly ranked in top 25% of SCImago Journal Rank (SJR), which is referred as Q1. Across the journals, the dominant subject categories are linked to Immunology and Microbiology and Medicine.
Fig. 1.
The countries most active in publications where omic or computational approaches are used to address questions related to COVID-19. The search covered articles published in journals indexed in Scopus or preprints found in MedRxiv and BioRxiv.
Table 1.
Journals that contributed whit more 2 articles.
| Journal | n | SJR | H index | subject categories |
|---|---|---|---|---|
| Viruses | 6 | Q1 | 69 | IM, M |
| Journal Of Medical Virology | 4 | Q2 | 111 | IM, M |
| Nature | 3 | Q1 | 1159 | MU |
| Emerging Microbes And Infections | 3 | Q1 | 38 | IM, M, PTP |
| Cell | 3 | Q1 | 747 | BGM |
| Journal Of Biomolecular Structure And Dynamics | 3 | Q2 | 62 | BGM |
| Computers In Biology And Medicine | 3 | Q2 | 83 | CS, M |
| Pathogens | 2 | Q1 | 29 | BGM, IM, M. |
| Infection Genetics And Evolution | 2 | Q1 | 80 | ABS, BGM, IM, M. |
| Gut | 2 | Q1 | 279 | M |
| Cell Host And Microbe | 2 | Q1 | 163 | IM |
| Elife | 2 | Q1 | 115 | BGM, IM, N. |
| Bioinformatics Oxford England | 2 | N/A | N/A | BGM, CS, MT |
n: number articles; SJR (2019): Q1 corresponds to journal ranking top 25%, Q2 corresponds to journal ranking 25%–50%. H index: expresses the journal's number of articles (h) that have received at least h citations. Subject area: IM: Immunology and Microbiology; M: Medicine; MU: Multidisciplinary; PTP: Pharmacology, Toxicology and Pharmaceutics; BGM: Biochemistry, Genetics and Molecular Biology; CS: Computer Science; ABS: Agricultural and Biological Sciences; N: Neuroscience; MT: Mathematics.
Bibliometric analysis shows that omics and bioinformatics tools are focused on virus animal origin, taxonomy, and receptor entry to the host cell research.
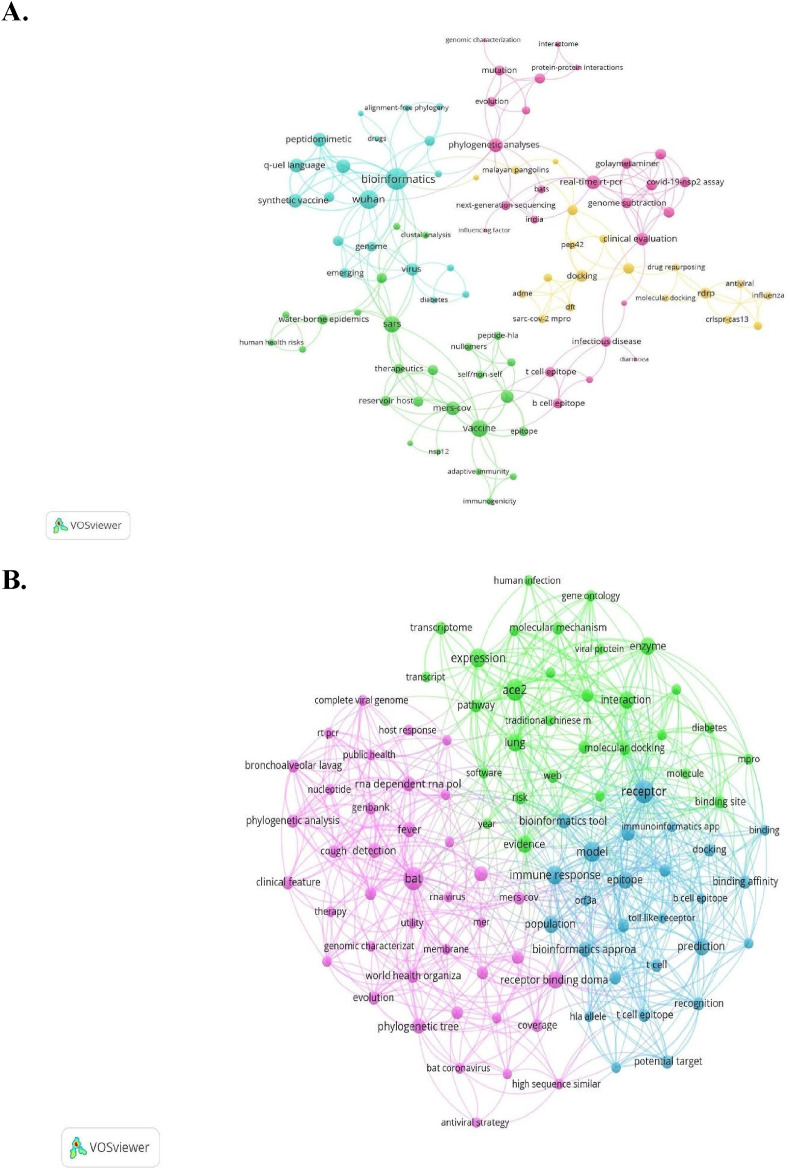
The keyword co-occurrence network was constructed by the VOSviewer software based on author's keywords (Fig. 2 A) and corpus text for Scopus (Fig. 2B), and corpus text for pre-prints articles (Fig. 3 ). The author keywords from Scopus were grouped into four clusters (Fig. 2A). The nodes with the highest frequency were bioinformatics (cluster 1), phylogenetics analysis and RT-PCR (cluster 2), Vaccine (cluster 3) and Docking (cluster 4). As a complement, in the corpus co-occurrence network for Scopus (Fig. 3 B) divided into three clusters, the most frequent terms were bat, receptor binding, and phylogenetic analysis (cluster 1), receptor, model, and prediction (cluster 2) and, ACE2, interaction, and expression (cluster 3). All these frequent keywords allowed us to see that in indexed publications, omics and bioinformatics tools are used to research urgent questions such as the animal origin of the virus, the taxonomy of the new infectious agent, and therapeutics strategies.
Fig. 2.
Keywords co-occurrence network exploring the use of omics and bioinformatic tools to address COVID-19 pandemic, based on 62 articles retrieved from Scopus A. Author's keywords network was built with a total of 133 out of 149 terms (manual edition of the thesaurus list to avoid synonyms, or connectors that had no stand-alone meaning was needed). B. Corpus text network was built with 96 out of 1904 terms.
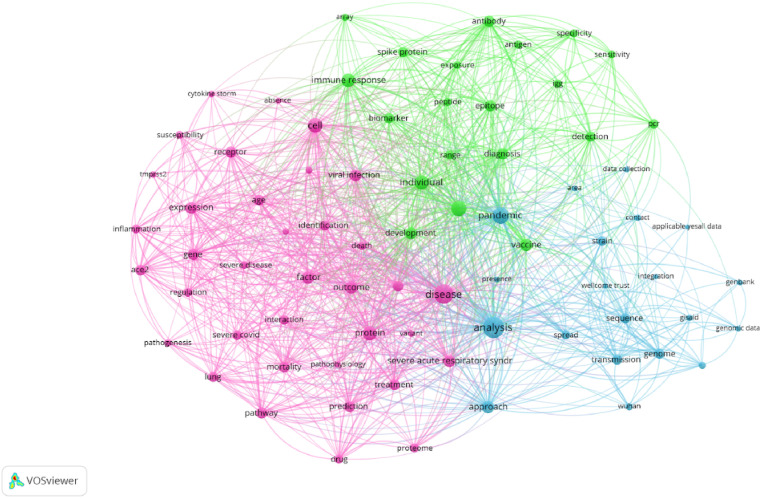
Fig. 3.
Corpus text co-occurrence network exploring the use of omics and bioinformatic tools to address COVID-19 pandemic, based on 122 articles retrieved from MedRxiv and BioRxiv; the network was built with 119 out of 5330 terms.
We also constructed a keyword co-occurrence network, based on corpus text using the pre-print databases MedRxiv and BioRxiv (Fig. 3). The network formed by 76 nodes is divided into 3 clusters; cluster 1 has terms with similar frequencies, such as disease, protein and cell, cluster 2 has 20 terms including immune response, detection, and biomarker and cluster 3, the terms analysis, pandemic and transmission receptor have the highest frequency.
Transcriptomics data and genome sequence analysis facilitated phylogenetic analysis to address the COVID-19 pandemic during the first stages.
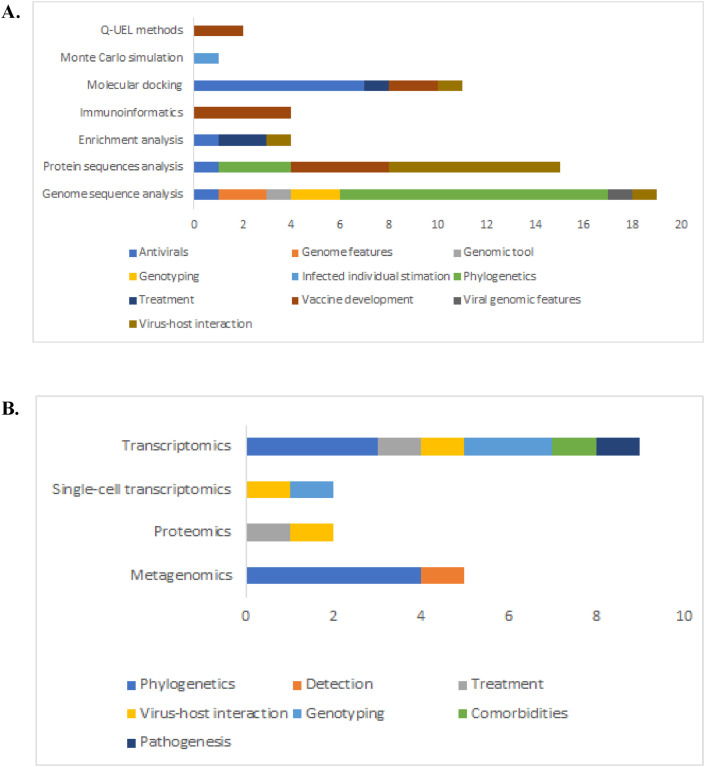
By reading the abstracts and in some cases the methodology session, we identified the omics technology or the bioinformatic approach, and the overall purpose of the research. Genome and protein sequence analysis account for the 43.47% of the Scopus articles, from which 58.33% are focused on establishing SARS-CoV-2 taxonomy and determining the animal origin. (Fig. 4 ). Molecular docking was the third bioinformatic approach most explored by June 19th, 2020, focusing on the identification of potential antiviral drugs (Fig. 4). Transcriptomics and metagenomics approaches were the main omics source data to perform phylogenetic analysis in SARS-CoV-2. Once the taxonomic position was consistently established by different authors, the goal of using the aforementioned approaches was redirected to exploring the virus-host interaction, treatment options, virus detection, and pathogenesis.
Fig. 4.
Omics technologies and bioinformatics approaches, covered by indexed articles in Scopus by June 19th, 2020 to address the COVID-19 pandemic. In colors are shown the main purposes of the publications and in the y-axis A, the bioinformatics approaches and B, omics data used to achieve them. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
China, Italy, and India publications using omics or bioinformatics approaches to study COVID-19 pandemic, between the top 10 most cited articles.
Table 2 shows the top ten cited articles along with the number and the name of the journal, country, number of authors, affiliated institutions, the specific omics or bioinformatic approach used and the purpose of the study. China led the research with 8 of 10 articles, in co-authorship with Australia in 2 articles and 1 with Saudi Arabia and USA. The number of authors varied from 2 to 46, with a mean of 15.7 authors and of 3.4 affiliated institutions per article referent. As it has been pointed out from the above results, Table 2 reaffirms that at the beginning of the pandemic, omics data and bioinformatic tools were mainly used to perform phylogenetic analysis to either taxonomically classify or identify the animal origin of the virus. Wu et al. [11], is the most cited article; published on February 3, 2020, in Nature, by 19 authors from 5 different institutions different from China and one in Australia, followed by Chan et al. [12], whose publication is the continuation of what was presented by Wu et al. [11].
Table 2.
List of the highly cited articles and metrics.
| Title | Citationa | Journal | Country FA | Country CA | # authors | # Affiliated institutions | Methodology | Objective |
|---|---|---|---|---|---|---|---|---|
| A new coronavirus associated with human respiratory disease in China | 404 | Nature | China | China, Australia | 19 | 6 | Metatranscriptomics | Phylogenetics |
| Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan | 187 | Emerging Microbes and Infections | China | China | 7 | 1 | Genome sequence analysis | Phylogenetics |
| Preliminary identification of potential vaccine targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies | 67 | Viruses | China | China | 2 | 1 | In silico protein sequences analysis | Phylogenetics |
| Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms | 54 | Gut | China | China | 46 | 1 | Genome sequence analysis | Phylogenetics |
| Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins | 53 | Nature | China | China, Australia | 29 | 9 | Metagenomics | Phylogenetics |
| RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak | 47 | Emerging Microbes and Infections | China | China | 17 | 4 | Transcriptomics | Phylogenetics |
| Genomic variance of the 2019-nCoV coronavirus | 40 | Journal of Medical Virology | Italy | Italy | 2 | 1 | In silico genome and protein sequence analysis | Antivirals |
| Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV | 31 | Viruses | China | China, Saudi Arabia, USA | 8 | 4 | In silico genome sequence analysis | Phylogenetics |
| Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments | 30 | Veterinary Quarterly | India | India, Iran, Thailand | 8 | 4 | In silico genome sequence analysis | Phylogenetics |
| Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients | 22 | Emerging Microbes and Infections | China | China | 19 | 3 | Transcriptomics | Virus-host interaction |
Citation in Scopus 27 June 2020; FA: first author; CA: Co-authorship.
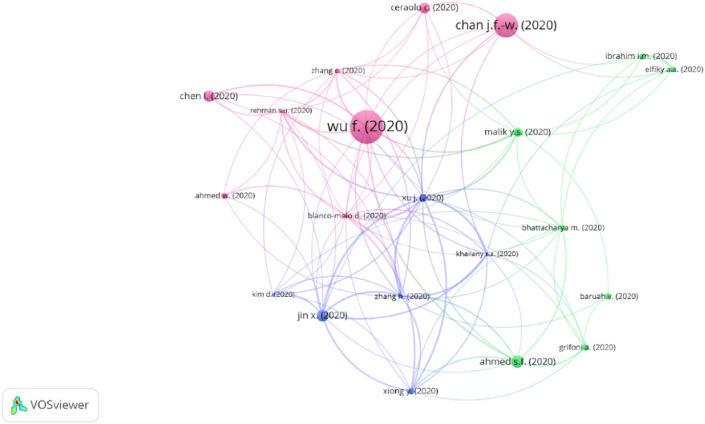
The bibliographic coupling network (Fig. 5 ) shows Wu et al. [11], together with Chan et al. [12], and Chen et al. [13], in the same cluster. These studies gave the first insights about the taxonomy and animal origin of the virus, by working with samples coming from the lung of a patient who worked in the wet-market in Wuhan [11,12], and patients who presumptively were not in contact with the wet-market [13]. In the green cluster, Ahmed et al. [14], and Bhattacharya et al. [15], in independent studies applied immunoinformatics to proposed SARS-CoV-2 epitopes as potential targets for vaccine development. While in the blue cluster, Jin et al. [16], and Xiong et al. [17], in independent studies, gave important contributions to the understanding of the COVID-19 clinical characteristics.
Fig. 5.
Bibliographic coupling network of indexed articles in Scopus using omics technologies and bioinformatics approaches to address the COVID-19 pandemic.
4. Discussion
The outbreak of COVID-19 has caused more than 1.1 million of deaths worldwide, primarily of older-people, according to the World Health Organization as of October 25, 2020 [18], which has caused a rapidly increasing number of publications. Since computational approaches could play a crucial role in different aspects of the pandemic, we aimed to analyze the SARS-CoV-2 literature published by authors that used bioinformatics and omics data to help to make informative and urgent decisions. According to our search by June 19th, 2020, China, the USA, the United Kingdom, and India accounted for the highest proportion of published research. These results are consistent with previous bibliometric analysis regarding the scientific production in bioinformatics around the world that is led by the wealthiest countries [19]. The effort of the Indian government in the development of the bioinformatics infrastructure and human resources is remarkable, which are showing good results during this pandemic. India is not only among the most productive countries but together with China, it is the origin of the most cited articles during this short and active period [20]. In terms of international collaborations, we found only a few of the studies were made with researchers from other countries. This is commensurate with the fact that most of the studies worked with stored sequences. The genome sequence of the SARS-CoV-2 is made available to the public via the U.S National Library of Medicine website (https://www.ncbi.nlm.nih.gov/labs/virus).
Nevertheless, international collaborations have great impact for scientific progress. That is why scholars have used Scientometrics and found that it provides a fast, reliable, and global overview of research [21]. As shown in the bibliometric mapping of keywords in indexed publications and pre-prints articles, omics and bioinformatics tools were used to answer urgent questions such as the animal origin of the virus, its taxonomy, and therapeutics strategies. Wu et al. [11], together with Chan et al. [12], and Chen et al. [13], gave the first insights about the taxonomy and animal origin of the SARS-CoV-2, by working with samples coming from the lung of a patient who worked in the wet-market in Wuhan [11,12] and patients that presumptively were not in contact with this place [13]. Conversely, Ahmed et al. [14], and Bhattacharya et al. [15], in independent studies applied immunoinformatics to proposed SARS-CoV-2 epitopes as potential targets for vaccine development. While Jin et al. [14], and Xiong et al. [15], in independent studies, gave important contributions to the understanding of the COVID-19 clinical characteristics. This bibliometric description and mapping provided a birds-eye view of information on the use of bioinformatics approaches and the use of omics data related research.
4.1. Metagenome assembly suggests pangolins as potential hosts of SARS-CoV-2
Early in the pandemic, it was established that SARS-CoV-2 is a zoonotic virus that jumped from an animal to a human, presumably in the “wet market” in Wuhan China. The closure of this site was part of temporary containment measures, while the identification of the potential intermediate species or natural hosts of the virus, may hold the key to preventing further introductions of the virus into the human population, and it may also provide useful insights to reduce the risk of the future spillover events from animals to humans [22]. Three out of 62 articles are focused on researching the animal origin for SARS-CoV-2 through phylogenetics analysis from metagenomic data of animals viromes and SARS-CoV-2 genome sequences data [[23], [24], [25]]. Lam et al. [25], Wahba et al. [24] and Zhang et al. [23], in independent analysis showed that coronaviruses found in lungs of pangolins bare a similarity between 85.5% and 93% with assembly coverages between 73 and 75%. Manis coronaviruses and Guangdong coronaviruses are closer to SARS-CoV-2 base on genome sequence and the RBD (receptor-binding domain) on the spike protein, respectively. Despite bat coronaviruses (RaTG13) hold a higher genome sequence similarity with SARS-CoV-2, the critical domain in RBD is more conserved between pangolin coronaviruses and SARS-CoV-2.
4.2. Proteomics for host-pathogen interaction studies and researching potential therapeutic targets
Out of the 62 articles related to omics that we found, 13 used proteomics data in primary analysis or secondary analysis combined with bioinformatics tools. Two of the main purposes of the authors who used proteomics were to elucidate the mechanism of host–pathogen interactions, its association with the severity of COVID-19, and to find possible therapeutic targets. Khan and Khan [26], predicted the interaction of the receptor ACE2, which is mainly expressed on the surface of airway epithelium and parenchyma of lung, with the glycoprotein found on the surface of SARS-CoV-2. Also, these authors suggest a possible role of Prevotella in the severity of COVID-19 by increasing severity of pneumonia. Bojkova et al. [27] on the other hand, used primary analysis of proteomics data to identified host cell pathways that are modulated by SARS-CoV-2 infection, which can be useful, as the authors suggests, for developing therapy options for COVID-19 by preventing viral replication in human cells. Robson [28], used protein sequences deposited in GenBank, and simulation tools to predict binding sites used by SARS-CoV-2. The author proposed a sialic acid glycan binding function for the spike protein of SARS-CoV-2, which can be potentially targeted for therapeutic agents to tackle SARS-CoV-2. This would be a different way of interaction between the virus and host cells other than the receptor ACE2.
All studies approaching vaccine development were computational in spirit, and Robson has many publications regarding this matter [[28], [29], [30]]. In one of his studies he found, through a Q-UEL language, that the sequence of amino acids KRSFIEDLLFNKV was particularly well conserved and corresponded to a region of the SARS virus that are believed to be required for virus activation for cell entry [30]. According to the author, this sequence motif and surrounding variations can lay the foundation to outline a specific synthetic vaccine epitope and peptidomimetic agent. Regarding the development of antivirals, they were also mainly approached by in-silico analysis. Studies by Neogi et al. [31], Sepay et al. [32], Kumar et al. [33], among others, aimed to identify drugs or strategies against SARS-CoV-2, such as miRNAs in SARS-CoV-2 [34], the effect of Shengjiang San on SARS-CoV-2 [35], chromone derivatives as inhibitor of SARS-CoV-2 [21], or even the use of chemical compounds from Indian spices as anti SARS-CoV-2 drugs [36].
Phylogenetic analysis, as we mentioned before, were one if the main objectives of the studies included. Lam et al. [25], for instance, focused on identifying SARS-CoV-2 related coronaviruses in Malayan pangolins, finding multiple lineages of pangolin coronavirus similar to SARS-CoV-2, which, according to the authors, suggests that pangolins should be considered as possible reservoirs in the emergence of new coronaviruses and, hence, in order to prevent zoonotic transmission, they should be removed from wet markets. Other authors considered bat_SARS like coronaviruses, as phylogenetically closer to SARS-Cov-2. Therefore, there are some studies comparing structurally the genome and proteome of SARS-Cov-2 isolated from patients, against coronavirus sequences with bat origins [[37], [38], [39]]. Rehman et al. [37], found that mutations in different genomic regions, but especially over the Spike gene, of this new virus, influence its reproductive adaptability, making it better in quickly adapting to the changing environments. Malik et al. [38], also support Rheman findings, by showing that in their genomic analysis, the S gene of SARS-Cov-2 was found to exhibit lower sequence identity with other Beta coronaviruses, which might me explaining their success to jump to the human host. Additionally, since the Spike protein interacts with the human receptor ACE2, these divergent features might be potentially altering functional effects on the interactions with the ACE2 receptor. Likewise, Srinivasan et al. [39] analysis converged to similar conclusions as to the above, adding an extra potential effect of the mutations in the Spike protein, relating to antibody binding sites that were used to inhibit infections with other coronaviruses.
5. Conclusions
Bibliometric analysis and visualized mapping can help monitor research methodologies focused in omics or computational sciences regarding the SARS-CoV-2 recent outbreak. During the first 7 months of the pandemic, the researchers took advantage of stored sequence data to identify and taxonomically and molecularly characterize the etiological agent of the COVID-19 pandemic. This information was essential for the policy makers across the globe to take public measures to track and control the transmission of the virus. The trends in omics and bioinformatics approaches are now moving towards the study of host-virus interaction and characterization of pathogenesis from the perspective of system biology.
Funding sources
None.
Declaration of competing interest
None declared.
Acknowledgement
JM was supported by the Colombian National “Departamento Administrativo de Ciencia, Tecnología e Innovación” (COLCIENCIAS), Ph.D program, number 756-2016. MU was supported by the Centre for Infectious Animal Diseases Initiative of CZU Prague, Ph.D program in Agricultural Specialization. The content is solely the responsibility of the authors and does not necessarily represent the official views of funders.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2020.104162.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhang Y., Geng X., Tan Y., Li Q., Xu C., Xu J. New Understanding of the Damage of SARS-CoV-2 Infection outside the Respiratory System. Biomed. Pharmacother. 2020;127:110195. doi: 10.1016/j.biopha.2020.110195. Elsevier Masson SAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020 Mar;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020 Feb;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qi F., Qian S., Zhang S., Zhang Z., Matsuyama S., Taguchi F. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020 Mar;83(21):11133–11141. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020 May;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venkatakrishnan A.J., Puranik A., Anand A., Zemmour D., Yao X., Wu X. Knowledge synthesis of 100 million biomedical documents augments the deep expression profiling of coronavirus receptors. Elife. 2020 May;9 doi: 10.7554/eLife.58040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fanelli V., Fiorentino M., Cantaluppi V., Gesualdo L., Stallone G., Ronco C. Acute kidney injury in SARS-CoV-2 infected patients. Critical Care. 2020;24 doi: 10.1186/s13054-020-02872-z. BioMed Central Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012 Jun;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ray S., Srivastava S. COVID-19 pandemic: hopes from proteomics and multiomics research. OMICS A J. Integr. Biol. 2020 May;24:457–459. doi: 10.1089/omi.2020.0073. omi.2020.0073. [DOI] [PubMed] [Google Scholar]
- 10.van Eck N.J., Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. 2010 Aug;84(2):523–538. doi: 10.1007/s11192-009-0146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microb. Infect. 2020;9(1):313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3):254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J. Med. Virol. 2020;92(6):618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin X., Lian J.-S., Hu J.-H., Gao J., Zheng L., Zhang Y.-M. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69(6):1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microb. Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization . World Health Organization; 2020. Weekly Epidemiological Update - 27 October 2020.https://www.who.int/publications/m/item/weekly-epidemiological-update---27-october-2020 Available from: [Google Scholar]
- 19.Chasapi A., Promponas V.J., Ouzounis C.A. The bioinformatics wealth of nations. Bioinformatics. 2020 Mar 4;36(9):2963–2965. doi: 10.1093/bioinformatics/btaa132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Som A., Kumari P., Ghosh A. Advancing India's bioinformatics education and research: an assessment and outlook. J. Protein Proteonomics. 2019;10(3):257–267. [Google Scholar]
- 21.Wang M., Liu P., Zhang R., Li Z., Li X. A scientometric analysis of global Health research. Int. J. Environ. Res. Publ. Health. 2020 Apr;17(8):2963. doi: 10.3390/ijerph17082963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shay J.W., Homma N., Zhou R., Naseer M.I., Chaudhary A.G., Al-Qahtani M. Abstracts from the 3rd international genomic medicine conference (3rd IGMC 2015) : Jeddah, Kingdom of Saudi Arabia. BMC Genom. 2016 Jul;17(Suppl 6):487. doi: 10.1186/s12864-016-2858-0. 30 November - 3 December 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C., Zheng W., Huang X., Bell E.W., Zhou X., Zhang Y. Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J. Proteome Res. 2020;19(4):1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahba L., Jain N., Fire A.Z., Shoura M.J., Artiles K.L., McCoy M.J. An extensive meta-metagenomic search identifies SARS-CoV-2-homologous sequences in pangolin lung viromes. mSphere. 2020;5(3) doi: 10.1128/mSphere.00160-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam T.T.-Y., Shum M.H.-H., Zhu H.-C., Tong Y.-G., Ni X.-B., Liao Y.-S. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020;1–4 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 26.Khan A.A., Khan Z. COVID-2019 associated overexpressed Prevotella proteins mediated host-pathogen interactions and their role in coronavirus outbreak. Bioinformatics. 2020;36:4065–4069. doi: 10.1093/bioinformatics/btaa285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583(7816):469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robson B. Bioinformatics studies on a function of the SARS-CoV-2 spike glycoprotein as the binding of host sialic acid glycans. Comput. Biol. Med. 2020;122 doi: 10.1016/j.compbiomed.2020.103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robson B. COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles' heel conserved region to minimize probability of escape mutations and drug resistance. Comput. Biol. Med. 2020:121. doi: 10.1016/j.compbiomed.2020.103749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robson B. Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput. Biol. Med. 2020:119. doi: 10.1016/j.compbiomed.2020.103670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neogi U., Hill K.J., Ambikan A.T., Heng X., Quinn T.P., Byrareddy S.N. Feasibility of known rna polymerase inhibitors as anti-sars-cov-2 drugs. Pathogens. 2020;9(5):320. doi: 10.3390/pathogens9050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sepay N., Sepay N., Al Hoque A., Mondal R., Halder U.C., Muddassir M. In silico fight against novel coronavirus by finding chromone derivatives as inhibitor of coronavirus main proteases enzyme. Struct. Chem. 2020;1 doi: 10.1007/s11224-020-01537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar V., Dhanjal J.K., Kaul S.C., Wadhwa R., Sundar D. With an one and caffeic acid phenethyl ester are predicted to interact with main protease (Mpro) of SARS-CoV-2 and inhibit its activity. J. Biomol. Struct. Dyn. 2020:1–17. doi: 10.1080/07391102.2020.1772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arisan E.D., Dart A., Grant G.H., Arisan S., Cuhadaroglu S., Lange S. The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12(6):614. doi: 10.3390/v12060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X.-L., Yuan Y.-L., Zhang J., Wang R.-F., Ni L.-Q. Exploring potential effect of Shengjiang San on SARS-CoV-2 based on network pharmacology and molecular docking. Chin. Tradit. Herb. Drugs. 2020;51(7):1795–1803. [Google Scholar]
- 36.Umesh U., Kundu D., Selvaraj C., Singh S.K., Dubey V.K. Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1763202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehman S.U., Shafique L., Ihsan A., Liu Q. Evolutionary trajectory for the emergence of novel coronavirus SARS-CoV-2. Pathogens. 2020;9(3):240. doi: 10.3390/pathogens9030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malik Y.S., Sircar S., Bhat S., Sharun K., Dhama K., Dadar M. Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Q. 2020;40(1):68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W. Structural genomics of SARS-COV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12(4):360. doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.