Abstract
To expand the repertoire of chemogenetic tools tailored for molecular and cellular engineering, we describe herein the design of cpRAPID as a circularly permuted rapamycin-inducible dimerization system composed of the canonical FK506-binding protein (FKBP) and circular permutants of FKBP12–rapamycin binding domain (cpFRB). By permuting the topology of the four helices within FRB, we have created cpFRB–FKBP pairs that respond to ligand with varying activation kinetics and dynamics. The cpRAPID system enables chemical-controllable subcellular redistribution of proteins, as well as inducible transcriptional activation when coupled with the CRISPR activation (CRISPRa) technology to induce a GFP reporter and endogenous gene expression. We have further demonstrated the use of cpRAPID to generate chemically switchable split nanobody (designated Chessbody) for ligand-gated antigen recognition in living cells. Collectively, the circular permutation approach offers a powerful means for diversifying the chemogenetics toolbox to benefit the burgeoning synthetic biology field.
Keywords: synthetic biology, chemogenetics, CRISPR, nanobody, antibody engineering
Introduction
Cell signaling involves a dynamic, sophisticated but highly coordinated protein–protein interaction network [1,2]. These subcellular network activities are tightly regulated, with each interaction bearing a unique spatiotemporal pattern to execute proper cellular functions [3]. Several genetically encoded chemical- or light-induced dimerization modules have been developed as synthetic biology tools to remotely control biological processes [4–7]. These molecular tools make it possible to conveniently manipulate protein–protein interaction and rewire cell signaling to decipher the organizational principles of living organism [8,9]. Among them, rapamycin (or its analogs) induced heterodimerization between the FK506-binding protein (FKBP) and the FKBP–rapamycin binding (FRB) domain constitutes one of the most widely used chemically inducible dimerization (CID) systems [10]. Upon addition of rapamycin, proteins of interest fused to FRB or FKBP can be rapidly brought into close proximity to regulate a myriad of cellular activities, such as gene expression, epigenetic modifications, phospholipid metabolism, Ca2+ signaling, and inter-organellar communication [11–16]. The FKBP–FRB-based CID system has also been engineered into chimeric antigen receptors (CARs) to generate small molecule-gated therapeutic T cells for immunotherapy [17]. Compared with optogenetic techniques that are often fraught with limited penetration of light in biological tissues and potential phototoxicity [18–20], chemogenetic approaches are more suited to long-term animal studies. For instance, the Food and Drug Administration-approved rapamycin is used in clinical settings as an immunosuppressant to prevent undesired rejection in organ transplantation [21]. The safe use of this small molecule or its analogs in human body and its ability to rapidly heterodimerize FRB–FKBP make it an ideal ligand to remotely control physiological processes in living organisms.
Circular permutation (CP), an engineering approach to evolve proteins by reorganizing protein domains, can potentially endow improved or new functionality to the scaffold protein [22,23]. CP has been successfully employed to evolve genetically encoded biosensors and actuators, thereby enabling real-time monitoring and interrogation of a myriad of physiological processes both in vitro and in vivo [24–27]. In order to diversity the chemogenetic toolbox for controlling cell signaling, we apply the CP strategy to generate new chemical dimerizers by using the most well-established FKBP–FRB heterodimerization system as a test case. Here, we introduce a circularly permuted rapamycin-inducible dimerization (cpRAPID) system by using novel FRB circular permutant (cpFRB)–FKBP pairs that respond to rapamycin with kinetics and dynamics different from the parental CID system. The cpRAPID system has been applied to precisely control the subcellular localization of proteins and to induce both exogenous and endogenous gene expression when paired with a modified CRISPRa technique [28–30]. Moreover, the cpRAPID system can be engineered into a single-domain VHH antibody [31] to yield chemically switchable split nanobody (designated Chessbody), thereby enabling ligand-gated antibody–antigen interaction in living cells. We anticipate that the similar approach can be extended to other CID systems to greatly diversify genetically encoded molecular tools tailored for precise control of protein actions and biological processes.
Results and Discussions
Design and characterization of cpRAPID
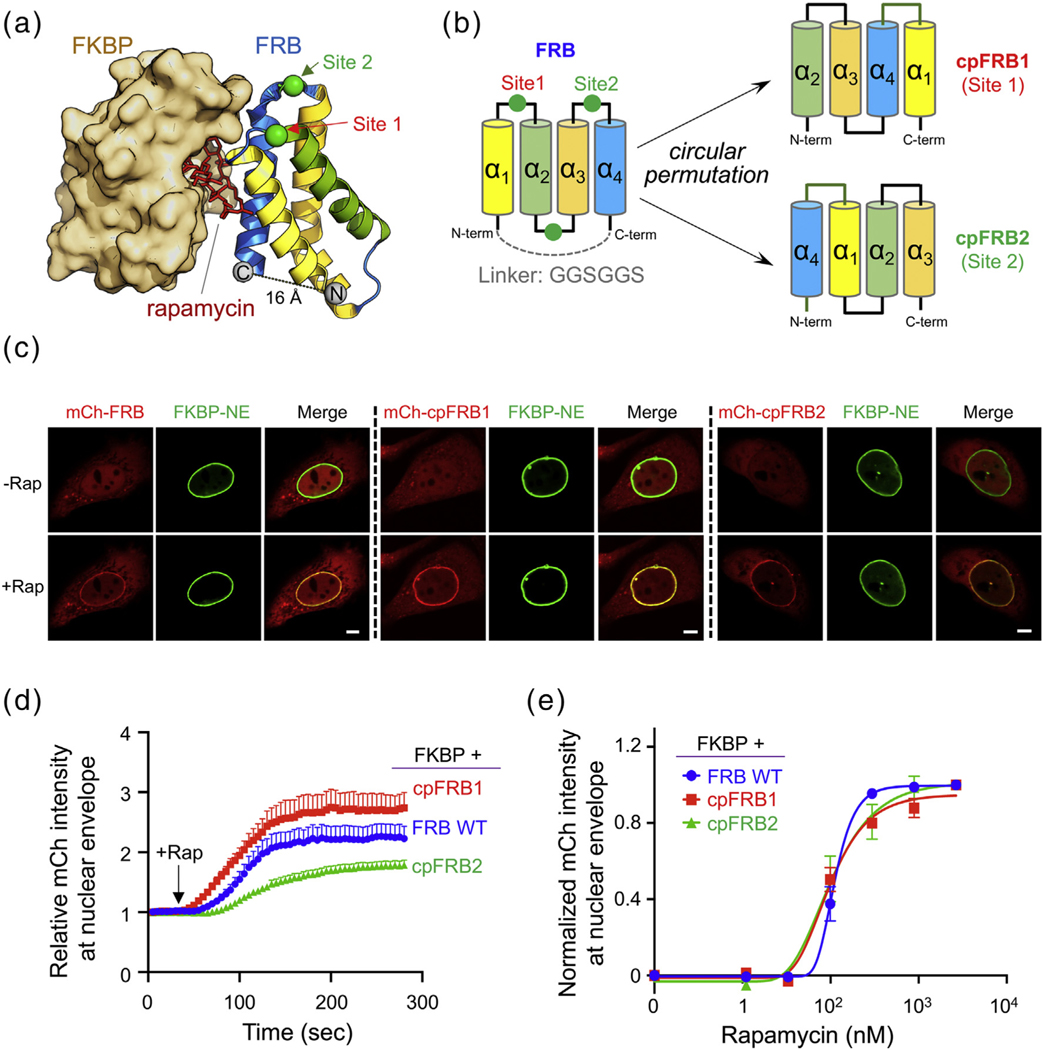
FRB was chosen as the CP target for two reasons: First, FRB has a relatively simple topology with four helices (α1–α4) [32–34]. Second, the N and C termini of FRB were closely positioned in space with an estimated distance of approximately 16 Å, which can be easily bridged by a flexible linker (Figure 1(a) and (b)). To avoid perturbation to the overall structural integrity, we decided to connect the original N and C termini with a GGSGGS linker while creating new termini at three surface-exposed loops that situate between α1 and α2 (site 1 between R30 and N31), between α2 and α3 (between P49 and Q50), or between α3 and α4 (site 2 between S78 and G79) for circular permutation (Figure 1(b)). We anticipate that cpFRBs can maintain similar functions as FRB but exhibit different kinetic and dynamic features compared to FRB. Next, we assessed the rapamycin-inducible heterodimerization between FKBP and the designed cpFRBs. To aid easy visualization of protein–protein interaction at real time in living cells, we anchored the FKBP module to the nuclear envelope (NE) via fusion with mEmerald–Lamin A, and subsequently monitored the subcellular localization of mCherry (mCh)-tagged FRB or cpFRBs before and after rapamycin treatment in mammalian cells (Figure 1(c)). Two newly created circular permutants, cpFRB1 (α2–α3–α4–α1) and cpFRB2 (α4–α1–α2–α3), showed rapamycin-inducible translocation from the nucleoplasm to the NE within 5 min (Figure 1(c), and Supplementary Video 1). By contrast, creating new termini in the loop region between α2 and α3 (in the topology of α3–α 4–α1–α 2) abolished cpFRB–FKBP interaction, likely owing to the disruption of proper folding of engineered FRB. The activation half-lives of the cpRAPID system based on cpFRB1 or cpFRB2 were determined to be 32.6 ± 5.2 or 47.0 ± 3.8 s, respectively (Figure 1(d)), which was comparable to or greater than the half-life of the parental FRB–FKBP pair (t1/2,on = 26.1 ± 3.3 s) during the formation of a ternary complex. The dynamic range of ligand-induced response was found to be slightly higher (cpFRB1) or lower (for cpFRB2) than wild-type (WT) FRB (Figure 1(d)). In parallel, we performed dose–response studies by using the same fluorescence-based cellular assay and titrating increasing amounts of ligand into cpRAPID-expressing cells (Figure 1(e)). The EC50 values for rapamycin in our assay were determined to be 112.1 ± 0.8 nM (WT), 99.4 ± 6.8 nM (cpFRB1), and 100.7 ± 4.8 nM (cpFRB2), suggesting that circular permutation did not compromise, but rather slightly improved the activation profiles of the engineered CID systems.
Figure 1.
Design and characterization of the cpRAPID system composed of FKBP and FRB circular permutants. (a) The 3D structure of the ternary FKBP–rapamycin–FRB complex (PDB entry: 3FAP). The locations of newly created N termini (green spheres; site 1 between r29 and n30 and site 2 between S78 and G79) in two functional circular permutants were indicated on the structure of FRB. FRB was presented as cartoon with the four helices (α1–α4) shown in different colors; FKBP was shown as surface rendering in wheat color; and rapamycin was presented as sticks in red. (b) The domain architecture of FRB and topological views of two functional FRB circular permutants (cpFRB1 and cpFRB2). Two permutation sites (green sphere) were selected to create new N and C termini. The original N and C termini of FRB (separated by ~ 16 Å) was connected by a flexible linker GGSGGS. The cpRAPID system, consisting of cpFRB and FKBP, is designed to enable rapamycin-inducible heterodimerization with kinetic and dynamic features different from the parental FKBP–FRB CID system. (c) Visualization of rapamycin (Rap)-inducible translocation of nucleoplasmic mCherry (mCh)-FRB or mCh-cpFRB (red) toward the NE-anchored FKBP (FKBP–mEmerald–Lamin A; abbreviated as FKBP-NE; green). Shown were representative confocal images of HeLa cells co-expressing the indicated constructs before and after rapamycin treatment (600 nM) for 5 min. The scale bar represents 5 μm. Also see Supplementary Video 1. (d) Representative time courses of Rap-induced NE translocation for wild-type (WT) FRB and cpFRB variants. The mCh signals at NE (as illustrated in (c)) was monitored every 5 s. Black arrow indicated the time point when rapamycin was added. The corresponding half-lives (t1/2) were as follows: 26.1 ± 3.3 s (FRB), 32.6 ± 5.2 s (cpFRB1), and 47.0 ± 3.8 s (cpFRB2). n = 16–41 cells from three independent assays (mean ± s.e.m.). (e) Dose–response curves of for the indicated FRB/cpFRB–FKBP pairs following rapamycin treatment. The normalized NE-resident mCh signals (as seen in (c)) were plotted against increasing doses of the ligand. EC50 values were determined as follows: 112.1 ± 0.8 nM (WT), 99.4 ± 6.8 nM (cpFRB1), and 100.7 ± 4.8 nM (cpFRB2). An asymmetric logistic equation was used to fit the curves. n = 3 independent titration assays. Data were shown as mean ± s.d..
The apparent EC50 values obtained from our cellular assay seemed to be 8- to 9-fold higher than the binding affinity of FRB toward the FKBP–rapamycin complex, with the dissociation constant (Kd = 12 nM) determined in vitro by using purified recombinant proteins [35]. This discrepancy can be explained by two possible reasons. First, not all the rapamycin dissolved in the solvent will permeate through the plasma membrane (PM) of human cells. However, almost all rapamycin may take effect when mixed with recombinant proteins in a test tube. Second, because our cellular assay used nucleoplasmic signals as readout, the effective amounts of rapamycin within the nuclei might be further reduced after traveling across two layers of biological membranes (PM and NE). Regardless, we believe that the relative binding strengths of the three FKBP–FRB/cpFRB pairs tested in our in cellulo assay will likely remain the same as in vitro. Furthermore, because most chemogenetic studies involve the use of mammalian cells, EC50 values calculated from in cellulo assays are expected to provide more practical guidance for future applications. Collectively, we have established a novel CID system, designated cpRAPID, by utilizing FRB circular permutants-FKBP pairs that exhibit varying kinetics and EC50 values toward rapamycin in living human cells.
Ligand-inducible transcriptional activation by combining cpRAPID with CRISPRa
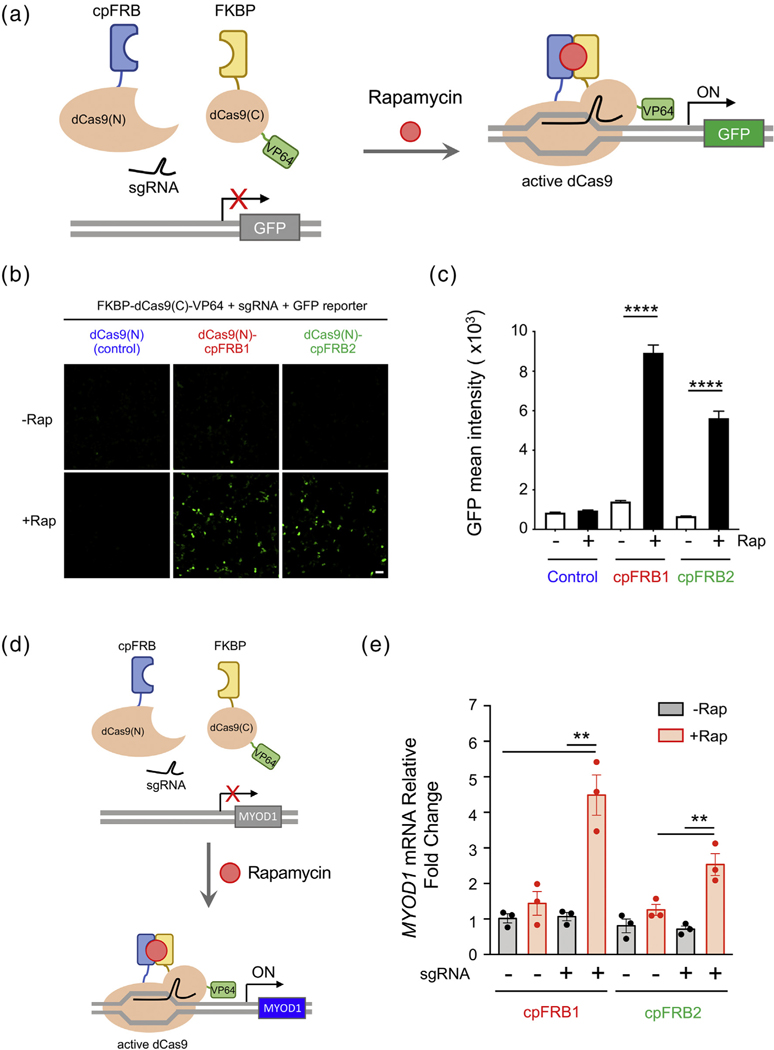
Having validated the in cellulo performance of the cpRAPID system, we set out to explore their potential for controlling gene expression. To test this, we decided to fuse cpFRB and FKBP derived from the cpRAPID system with the N- and C-terminal fragments of a split catalytically dead spCas9 from Streptococcus pyogenes (dCas9) [7,36–38], respectively. We envision that rapamycin-induced heterodimerization between cpFRB and FKBP will lead to the reassembly of a functional dCas9, thereby restoring its sgRNA-guided DNA binding capability (Figure 2(a)). To enable targeted transcriptional activation, the C-terminal fragment of split Cas9 was further fused to VP64, a synthetic protein that contains four copies of herpes simplex viral protein 16 to act as a potent transactivator during gene transcription [39–41]. By inserting sgRNA targeting sequences and a minimal promoter upstream of the reporter gene GFP, we were able to quantitatively compare the levels of gene expression before and after rapamycin treatment (Figure 2(a) and (b)). Upon addition of rapamycin, we found that both cpFRB1- and cpFRB2-based split dCas9–VP64 proteins were able to efficiently turn on gene expression, with the GFP intensities enhanced by 6.4- and 5.6-fold, respectively (Figure 2(c)). As a stringent control, the cells expressing the N-terminal fragment of dCas9 and other necessary components (FKBP-dCas9(C)-VP64 + sgRNA + GFP reporter) did not show significant increase in GFP signals (Figure 2(c)), further attesting to the necessity of ligand-inducible reassembly of dCas9–VP64 for transcriptional activation. We noticed that the cpFRB1 group displayed higher efficiency for gene transcription, likely due to the fact that the cpFRB1-based system has a stronger affinity and faster response to rapamycin when compared with cpFRB2 (Figure 1(d) and (e)). We further moved on to explore whether the cpRAPID–CRISPRa system could be employed to conditionally switch on endogenous gene expression. To test this, we used an sgRNA designed to target the promoter region of the myogenic differentiation 1 (MYOD1) gene and examined the mRNA expression level before and after rapamycin treatment in HEK293T cells. We envisioned the heterodimerization between cpFRB and FKBP would restore the DNA binding capability of split dCas9–VP64 and lead to transcriptional activation of endogenous MYOD1 (Figure 2(d)). Upon addition of rapamycin, we found that both cpFRB1- and cpFRB2-based split dCas9–VP64 were able to enhance the endogenous MyoD1 gene expression by 4.5-fold and 2.5-fold, respectively (Figure 2(e)). Together, we have demonstrated that the cpRAPID system can be combined with CRISPRa to precisely drive the expression of both exogenous and endogenous genes in a ligand-gated manner.
Figure 2.
cpRAPID used for inducible transcriptional activation using CRISPRa. (a) Schematic design of split dCas9–VP64 to conditionally drive GFP reporter gene expression. Upon rapamycin addition, two split dCas9 fragments, comprising dCas9(N)-cpFRBs and FKBP-dCas9(C)-VP64, restore the functional assembly of dCas9–VP64 to enable precise CRISPR activation, thereby promoting GFP expression in the presence of sgRNAs targeted to the promoter region of the reporter gene. (b) Representative confocal images showing the GFP reporter expression before and after rapamycin treatment (200 nM). HEK293T cells were co-transfected with FKBP-dCas9(C)-VP64, sgRNA, GFP-reporter, and the indicated dCas9(N)-cpFRB constructs. Fluorescent images were acquired 48 h after transfection. The scale bar represents 50 μm. (c) Quantification of GFP reporter expression under the indicated conditions shown in (b). n = 693–1790 cells from three independent experiments (mean ± s.e.m.). ****P < .0001 (compared to the non-Rap treated group; two tailed Student’s t-test). (d) Design of split dCas9–VP64 to conditionally drive endogenous gene expression. Upon rapamycin addition, two split dCas9 fragments, comprising dCas9(N)-cpFRBs and FKBP-dCas9(C)-VP64, restore the functional assembly of dCas9–VP64 to activate endogenous MYOD1 transcription in the presence of an sgRNA targeted to the promoter region of the MYOD1 gene. (e) Quantification of rapamycin-induced expression of endogenous MYOD1 in HEK293T cells by real-time qPCR. Cells were co-transfected with FKBP-dCas9(C)-VP64, the indicated dCas9(N)-cpFRB constructs with or without sgRNA. Rapamycin treatment (200 nM) was performed 24 h after transfection, and mRNA was extracted 24 h after treatment for the assay. n = 3; **P < .01 (two tailed Student’s t-test).
Using cpRAPID to create Chessbody
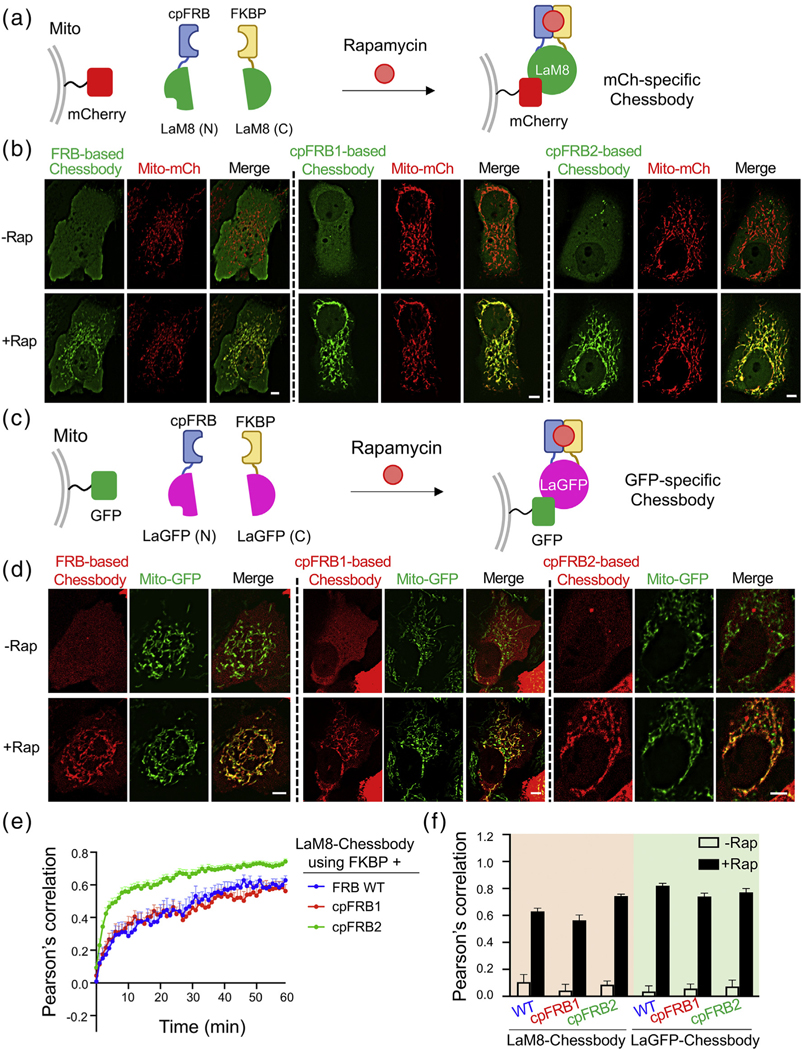
We moved on to further validate the cpRAPID system in a different biological context, i.e., the binary antibody–antigen interaction. Given that the split single-variable domain on a heavy-chain antibody (VHH or more commonly known as nanobody) can be functionally reassembled to restore antigen binding by using genetically encoded dimerizers [42], we reasoned that the chemical-based cpRAPID system can likewise be applied to fulfill the same purpose. Hence, we fused cpFRB or FKBP to the N- and C-terminal fragments of a split nanobody and named the engineered antibody as Chessbody for chemically switchable split nanobody (Figure 3). We used LaM8 and LaGFP, two split nanobodies that specifically recognize mCherry and GFP [43], respectively, as our test cases because of easy visualization of chemical-inducible association between the designed Chessbodies and antigens in living cells (Figure 3(a)–(d)). To ensure a nearly 1:1 expression of the two polypeptides in Chessbody with minimized waste of the two complementary components, we used a bi-cistronic expression vector bearing the T2A self-cleaving peptide sequence [44]. Specifically, a single plasmid containing the GFP-LaM(N)-cpFRB-T2A-FKBP-LaM(C) cassette was used to drive the expression of GFP-tagged, mCherry-specific Chessbody (GFP-LaM(N)-cpFRB + FKBP-LaM(C)), which would permit real-time monitoring of ligand-induced association between Chessbody and the mitochondria-anchored antigen, mCherry (Figure 3(a) and (b)). A similar design was applied to the GFP-recognizing Chessbody made of mCh-LaGFP(N)-cpFRB and FKBP-LaGFP(C) (Figure 3(c) and (d)). In both cases, we observed rapamycin-inducible translocation of the cytosolic Chessbody component toward the mitochondria-anchored target (Figure 3(b)–(d), and Supplementary Video 2), with the degree of colocalization at mitochondria quantified by Pearson’s correlation (Figure 3(e) and (f)). Compared to the parental FKBP–FRB CID system (t1/2, on = 20.0 ± 8.2 min), cpRAPID enabled at least comparable or much faster restoration of split nanobody-antigen binding, with the half-lives ranging from 7.8 ± 1.8 min (cpFRB2-based Chessbody) to 18.0 ± 4.0 min (cpFRB1-based Chessbody) (Figure 3(e)). Worthy to note, the time spent to fully restore a functional split nanobody seemed to be considerably slower than the time required for the ternary complex formation (t1/2 < 1 min; Figure 1(d)). It is likely that the reassembling process might involve the repositioning and/or refolding of the two essential split components, hence requiring more time and energy to counteract entropy increase when the complex becomes more ordered. Furthermore, we noted that, although the cpFRB2–FKBP pair had a relatively weaker EC50 and slower response during the cpFRB2–rapamycin–FKBP ternary complex formation (Figure 1(d)), cpFRB2-based Chessbody was able to more rapidly restore its antigen-recognition ability compared to the FRB- or cpFRB1-based versions. We speculated that the cpFRB2-LaM(N) and FKBP-LaM(C) combinations might render the split nanobody in a more favorable conformation to save the entropy cost during functional reassembly of an intact single-domain antibody. Indeed, if we use a two-phase decay fitting, the half-lives for cpFRB2 were as follows: t1/2, fast = 1.5 ± 0.1 min and t1/2, slow = 14.1 ± 1.5 min, respectively. The fast kinetics was more consistent with the time course of the cpFRB2–rapamycin–FKBP ternary complex formation (t1/2: ~ 50 s; Figure 1(d)). It is likely that the cpFRB2–FKBP combination restores the antigen recognition ability upon the formation of a ternary complex, whereas the other two combinations may take longer time for proper folding of the engineered Chessbody. These interesting points warrant further in-depth investigation in the follow-on studies. Congruently, we have fully established the feasibility of utilizing cpRAPID to generate ligand-switchable antibodies for future biotechnological and biomedical applications, such as the design of chemically controllable CAR T-cells and therapeutic antibodies for precision medicine.
Figure 3.
Use of cpRAPID to design Chessbody for ligand-gated antigen recognition in living cells. (a) Design of a Chessbody that recognizes mCherry upon addition of rapamycin. The N- and C-terminal fragments of a split nanobody (LaM8; split between residues S64 and V65) was fused to cpFRB and FKBP, respectively. The chemical induced dimerization of the cpRAPID system facilitates the reassembly of a functional Chessbody, which restores the antibody–antigen binary interaction between mitochondria (mito)-anchored mCherry (antigen) and the LaM8-specific Chessbody composed of GFP-cpFRB-LaM8(N) and FKBP-LaM8(C). (b) Representative confocal images of HeLa cells co-expressing the indicated Chessbody components (green) and the antigen (red; AKAP1-mCherry or Mito-mCh) before and after rapamycin (600 nM) treatment for 30 min. The scale bar represents 5 μm. (c) Design of a GFP-specific Chessbody made of mCh-cpFRB-LaGFP(N) and FKBP-LaGFP(C), which exhibits rapamycin-inducible binding toward the mitochondria-tethered antigen (NTOM20-GFP). (d) Representative confocal images of HeLa cells co-expressing a GFP-recognizing Chessbody (red; mCh-cpFRB-LaGFP(N) + FKBP-LaGFP(C)) and mitochondria-tethered GFP (green; NTOM20-GFP or Mito-GFP) before and after rapamycin treatment as did in (b). The scale bar represents 5 μm. (e) Real-time monitoring of rapamycin (600 nM) induced restoration of Chessbody–antigen interaction using the indicated FKBP–FRB/cpFRB pairs. The Pearson’s correlation between GFP and mCherry signals at mitochondria was used to quantitate the degrees of colocalization. The corresponding half-lives (t1/2) using a single-phase decay fitting were as follows: 20.0 ± 8.2 min (FRB), 18.0 ± 4.0 min (cpFRB1), and 7.8 ± 1.8 min (cpFRB2). For cpFRB2, if using a two-phase decay fitting, the half-lives were as follows: t1/2, fast = 1.5 ± 0.1 min and t1/2, slow = 14.1 ± 1.5 min. n = 10–23 cells from three independent assays (mean ± s.e.m.). (f) Quantification of Pearson's correlation for the indicated Chessbodies before and after rapamycin treatment (as shown in (b) and (d)). n = 10–23 cells from three independent assays (mean ± s.e.m.).
Conclusions
We report herein the creation of cpRAPID to complement the existing chemogenetic tools by taking a simple circular permutation approach. We have developed two cpFRB–FKBP pairs with diversified activation kinetics and dynamic ranges of response for the rapamycin-based CID system. We have successfully demonstrated the use of cpRAPID to effectively program the transcriptional outputs of both exogenous and endogenous genes, and restore antibody–antigen interaction in a ligand-gated manner within living cells. We anticipate that a similar engineering approach can be extended to other CID systems to further expand the repertoire of synthetic biology toolkit, particularly chemical-controlled protein switches and genetically encoded chemical dimerizers.
Materials and Methods
Molecular cloning and plasmid construction
Plasmids were all created by the standard restriction enzyme digestion and ligation method. KOD Hot Start DNA polymerase (EMD Milipore, MA, USA) was used for PCR amplifications. All the restriction enzymes and the T4 ligase kit were purchased from New England Biolabs (Ipwich, MA, USA).
The circularly permuted FRB gene fragments with two new N termini (N30 and G79) were first synthesized as double-stranded DNA by Genewiz (South Plainfield, NJ, USA). The cDNAs were individually inserted into the mCherry2-C1 vector between the HindIII and EcoRI restriction sites to generate mCh-cpFRB1 and mCh-cpFRB2. A 6-mer linker GGSGGS was used to connect the original N and C termini of FRB. The PCR-amplified FRB fragment was cloned into the same mCh-C1 vector between HindIII and EcoRI sites and used as control. To make NE-tethered FKBP-mEmerald-LaminA, the NE-targeting LaminA cDNA was inserted into a customized mEmerald-C1 vector between the BglII and BamHI sites, followed by FKBP insertion (NheI–EcoRI). To generate mitochondria-targeting mCh or GFP, the cDNA sequences encoding human AKAP11–30 flanked by NheI and BamHI restriction sites and NTOM20 flanked by HindIII and EcoRI were inserted into mCherry-N1 and GFP-N1, respectively. For split dCas9 constructs, FRB was replaced by cpFRB1 and cpFRB2 in the vector dCas9 (N)-FRB-2xNES (Addgene no. 62887) to make dCas9 (N)-cpFRB. The FKBP-dCas9(C)-NLS-VP64, GFP reporter, and sgRNA of the split dCas9 reporter system were purchased from Addgene (nos. 62888, 60718, and 60719). cDNAs encoding the GFP nanobody (LaGFP) was synthesized by Genewiz and inserted into a customized pTriEx-mCh plasmid between the HindIII and XhoI sites to yield pTriEx-mCh-LaGFP. The pTriEx-GFP-LaM8 was made by a similar strategy. Next, codon-optimized FRB/cpFRB1/cpFRB2-T2A-FKBP cDNA fragments with flanking glycine-serine linkers were inserted into pTriEx-mCh-LaGFP or pTriEx-GFP-LaM8 by the NEBuilder HiFi DNA Assembly kit to generate split nanobodies. The LaGFP was split between residues S65 and V66, while the LaM8 was split between residues S64 and V65.
Cell culture and transfection
HeLa and HEK293T cell lines were purchased from ATCC and cultured at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. For fluorescence imaging experiments, cells were seeded in four-well 35-mm glass-bottom dishes (Cellvis, Mountain View, USA) 1 day before transfection. The plasmids were transfected by using the Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. Cells were imaged or extracted for mRNA 18–24 h after transfection.
Time-lapsed confocal imaging and data analysis
Fluorescence imaging was performed on a Nikon Ti2 inverted epifluorescence microscope with a Yokogawa W1 spinning disk scan-head equipped with a LUNV laser and a live-cell culture cage to maintain the temperature at 37 °C with 5% CO2. A 60 × oil lens was used for high-resolution imaging. All data were analyzed with the Nikon NIS-Elements imaging software, with the analyzed results plotted by using the Prism 8.3.0 (GraphPad) software.
To monitor rapamycin-induced cpFRB–FKBP heterodimerization, HeLa cells seeded on glass-bottom dishes were co-transfected with mCh-FRB/cpFRB1/cpFRB2 and FKBP-mEmerald-LaminA. Confocal images were acquired 24 h after transfection. Time-lapsed imaging was performed every 5 s for up to ~ 5 min. Rapamycin (600 nM) was added to induce the interaction in cellulo. To quantify FRB/cpFRB–FKBP binding, we used the region-of-interest toolbox to define the NE regions manually. The “Time Measurement” tool in the Nikon NIS-Elements software was used to determine mCherry intensity. The fluorescence intensity ratio (F/F0) was calculated to examine the kinetics of cpFRB variants. To obtain EC50 values of the cpRAPID system, time-lapsed imaging was carried out by adding increasing concentrations (11–8000 nM) of rapamycin into transfected HeLa cells under the imaging field. The mCherry intensity ratio (NE/nuclear signals) was calculated for each condition.
GFP reporter assay
HEK293T cells seeded on glass-bottom dishes were co-transfected with 100 ng dCas9(N)-cpFRB (dCas9(N) without cpFRB was used as control), 100 ng FKBP-dCas9(C)-NLS-VP64, 100 ng pGL3-Basic-8x-gRNA-eGFP (sgRNA targeting the upstream promoter of EGFP reporter), and 200 ng gRNA-eGFP-Reporter for each well. Eight hours after transfection, cells were added with rapamycin (200 nM) and cultured for an additional 40 h. Confocal images were acquired with 20 × air lens, and mean GFP intensity of cells was measured by the Nikon imaging software.
CRISPRa assay
HEK293 cells seeded on six-well plates (Corning Inc., USA) were co-transfected with 333 ng dCas9(N)-cpFRB1 or 333 ng of FKBP-dCas9(C)-NLS-VP64, 333 ng of sgRNA4_MYOD1 (Addgene no. 64139) for each well. Twenty-four hours after transfection, cells were added with or without rapamycin (200 nM) and cultured for an additional 24 h. Then mRNA was extracted using the Qiagen RNeasy spin prep columns, and reverse transcription PCR was performed using the amfiRivert cDNA Synthesis Platinum Master Mix (genDEPOT). Relative levels of cDNA were detected using Applied Biosystems PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, USA) and Mastercycle Real-Time PCR (Eppendorf, USA). RT-PCR primers targeting MYOD1 include the following: ccgcctgagcaaagtaaatgagg (forward primer) and gcagcagagcctgcaggccctcg (reverse primer). The data were normalized to 18S rRNA.
Real-time monitoring of Chessbody–antigen interactions
To monitor rapamycin-induced nanobdy–antigen interactions, HeLa cells were co-transfected with NTOM20-GFP and pTriEx-mCh-LaGFP(N)-FRB/cpFRB-T2A-FKBP-LaGFP(C) or AKAP11–30-mCh and pTriEx-GFP-LaM8(N)-FRB/cpFRB-T2A-FKBP-LaM8(C). Cells were treated with 600 nM rapamycin, and images were acquired every 1 min for up to 1 h for both the green and red channels. The degrees of colocalization of engineered nanobodies with mitochondria-anchored mCh or GFP were quantified by calculating the Pearson’s correlation coefficients of selected regions-of-interest with the Nikon NIS-Elements software package.
Statistical analysis
Quantitative data are showed as mean ± s.e.m. unless otherwise noted. The analyzed number (n) of samples were listed for each experiment. All the data were plotted using the GraphPad Prism 8.3.0 software. The half-lives were calculated by using the “One phase decay” function in the software package. Statistical analysis was performed by using two-tailed Student’s t-test: *P < .05, **P < .01, ***P < .001, and ****P < .0001.
Supplementary Material
Acknowledgments
This work was supported by the Welch Foundation (BE-1913-20190330 to Y.Z.), the American Cancer Society (RSG-16-215-01-TBE to Y.Z. and RSG-18-043-01-LIB to Y.H.), the National Institutes of Health (R01GM112003 and R01CA232017 to Y.Z.), and the John S. Dunn Foundation (to Y.Z.).
Abbreviations used:
- cpRAPID
circularly permuted rapamycin-inducible dimerization system
- Chessbody
chemically switchable split nanobody
- FKBP
the FK506-binding protein
- FRB
the FKBP–rapamycin binding domain of the mammalian target of rapamycin kinase (mTOR)
- CRISPR
clustered regularly interspaced short palindromic repeats
- GFP
green fluorescent protein
- CID
chemically inducible dimerization
- CAR
chimeric antigen receptor
- VHH
single-variable domain on a heavy-chain antibody
- AKAP
A-kinase anchor protein
Footnotes
Declaration of Competing Interest
None.
Two supplementary videos related to this article can be found online https://doi.org/10.1016/j.jmb.2020.03.033.
References
- [1].Hynes NE, Ingham PW, Lim WA, Marshall CJ, Massague J, Pawson T, Signalling change: signal transduction through the decades, Nat. Rev. Mol. Cell Biol. 14 (2013) 393–398. [DOI] [PubMed] [Google Scholar]
- [2].Silverbush D, Sharan R, A systematic approach to orient the human protein-protein interaction network, Nat. Commun. 10 (2019) 3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gordley RM, Bugaj LJ, Lim WA, Modular engineering of cellular signaling proteins and networks, Curr. Opin. Struct. Biol. 39 (2016) 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Miesenbock G, Optogenetic control of cells and circuits, Annu. Rev. Cell Dev. Biol. 27 (2011) 731–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Atasoy D, Sternson SM, Chemogenetic tools for causal cellular and neuronal biology, Physiol. Rev. 98 (2018) 391–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, et al. , Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins, Proc. Natl. Acad. Sci. U. S. A. 112 (2015) 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zetsche B, Volz SE, Zhang F, A split-Cas9 architecture for inducible genome editing and transcription modulation, Nat. Biotechnol. 33 (2015) 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA, Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems, Annu. Rev. Biophys. 39 (2010) 515–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stanton BZ, Chory EJ, Crabtree GR, Chemically induced proximity in biology and medicine, Science. 359 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fegan A, White B, Carlson JC, Wagner CR, Chemically controlled protein assembly: techniques and applications, Chem. Rev. 110 (2010) 3315–3336. [DOI] [PubMed] [Google Scholar]
- [11].DeRose R, Miyamoto T, Inoue T, Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology, Pflugers Arch. 465 (2013) 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Voss S, Klewer L, Wu YW, Chemically induced dimerization: reversible and spatiotemporal control of protein function in cells, Curr. Opin. Chem. Biol. 28 (2015) 194–201. [DOI] [PubMed] [Google Scholar]
- [13].Rivera VM, Clackson T, Natesan S, Pollock R, Amara JF, Keenan T, et al. , A humanized system for pharmacologic control of gene expression, Nat. Med. 2 (1996) 1028–1032. [DOI] [PubMed] [Google Scholar]
- [14].Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS, Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation, Nature 454 (2008) 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Csordas G, Varnai P, Golenar T, Roy S, Purkins G, Schneider TG, et al. , Imaging interorganelle contacts and local calcium dynamics at the ER–mitochondrial interface, Mol. Cell 39 (2010) 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jing J, Liu G, Huang Y, Zhou Y, A molecular toolbox for interrogation of membrane contact sites, J. Physiol. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wu CY, Roybal KT, Puchner EM, Onuffer J, Lim WA, Remote control of therapeutic T cells through a small molecule-gated chimeric receptor, Science 350 (2015), aab4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tye KM, Deisseroth K, Optogenetic investigation of neural circuits underlying brain disease in animal models, Nat. Rev. Neurosci. 13 (2012) 251–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tan P, He L, Han G, Zhou Y, Optogenetic immunomodulation: shedding light on antitumor immunity, Trends Biotechnol. 35 (2017) 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tyssowski KM, Gray JM, Blue light increases neuronal activity-regulated gene expression in the absence of optogenetic proteins, eNeuro 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Saunders RN, Metcalfe MS, Nicholson ML, Rapamycin in transplantation: a review of the evidence, Kidney Int. 59 (2001) 3–16. [DOI] [PubMed] [Google Scholar]
- [22].Cunningham BA, Hemperly JJ, Hopp TP, Edelman GM, Favin versus concanavalin a: circularly permuted amino acid sequences, Proc. Natl. Acad. Sci. U. S. A. 76 (1979) 3218–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ostermeier M, Designing switchable enzymes, Curr. Opin. Struct. Biol. 19 (2009) 442–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Baird GS, Zacharias DA, Tsien RY, Circular permutation and receptor insertion within green fluorescent proteins, Proc. Natl. Acad. Sci. U. S. A. 96 (1999) 11241–11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kostyuk AI, Demidovich AD, Kotova DA, Belousov VV, Bilan DS, Circularly permuted fluorescent protein-based indicators: history, principles, and classification, Int. J. Mol. Sci. 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Banerjee S, Mitra D, Structural basis of design and engineering for advanced plant optogenetics, Trends Plant Sci. 25 (2020) 35–65. [DOI] [PubMed] [Google Scholar]
- [27].Tischer D, Weiner OD, Illuminating cell signalling with optogenetic tools, Nat. Rev. Mol. Cell Biol. 15 (2014) 551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. , CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes, Cell 154 (2013) 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nguyen NT, He L, Martinez-Moczygemba M, Huang Y, Zhou Y, Rewiring calcium signaling for precise transcriptional reprogramming, ACS Synth. Biol. 7 (2018) 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M, CRISPR–Cas9-based photoactivatable transcription system, Chem. Biol. 22 (2015) 169–174. [DOI] [PubMed] [Google Scholar]
- [31].Muyldermans S, Nanobodies: natural single-domain antibodies, Annu. Rev. Biochem. 82 (2013) 775–797. [DOI] [PubMed] [Google Scholar]
- [32].Leone M, Crowell KJ, Chen J, Jung D, Chiang GG, Sareth S, et al. , The FRB domain of mTOR: NMR solution structure and inhibitor design, Biochemistry 45 (2006) 10294–10302. [DOI] [PubMed] [Google Scholar]
- [33].Choi J, Chen J, Schreiber SL, Clardy J, Structure of the FKBP12–rapamycin complex interacting with the binding domain of human FRAP, Science 273 (1996) 239–242. [DOI] [PubMed] [Google Scholar]
- [34].Liang J, Choi J, Clardy J, Refined structure of the FKBP12–rapamycin–FRB ternary complex at 2.2 Å resolution, Acta Crystallogr. D Biol. Crystallogr. 55 (1999) 736–744. [DOI] [PubMed] [Google Scholar]
- [35].Banaszynski LA, Liu CW, Wandless TJ, Characterization of the FKBP.rapamycin.FRB ternary complex, J. Am. Chem. Soc. 127 (2005) 4715–4721. [DOI] [PubMed] [Google Scholar]
- [36].Kaya H, Ishibashi K, Toki S, A split Staphylococcus aureus Cas9 as a compact genome-editing tool in plants, Plant Cell Physiol. 58 (2017) 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nihongaki Y, Otabe T, Ueda Y, Sato M, A split CRISPR–Cpf1 platform for inducible genome editing and gene activation, Nat. Chem. Biol. 15 (2019) 882–888. [DOI] [PubMed] [Google Scholar]
- [38].Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, et al. , Rational design of a split-Cas9 enzyme complex, Proc. Natl. Acad. Sci. U. S. A. 112 (2015) 2984–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Beerli RR, Segal DJ, Dreier B, Barbas CF 3rd., Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks, Proc. Natl. Acad. Sci. U. S. A. 95 (1998) 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sadowski I, Ma J, Triezenberg S, Ptashne M, GAL4-VP16 is an unusually potent transcriptional activator, Nature 335 (1988) 563–564. [DOI] [PubMed] [Google Scholar]
- [41].Seipel K, Georgiev O, Schaffner W, Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions, EMBO J. 11 (1992) 4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yu D, Lee H, Hong J, Jung H, Jo Y, Oh BH, et al. , Optogenetic activation of intracellular antibodies for direct modulation of endogenous proteins, Nat. Methods 16 (2019) 1095–1100. [DOI] [PubMed] [Google Scholar]
- [43].Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF, et al. , A robust pipeline for rapid production of versatile nanobody repertoires, Nat. Methods 11 (2014) 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, et al. , Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector, Nat. Biotechnol. 22 (2004) 589–594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.