Graphical abstract
Keywords: CRISPR, Cas9, HLA, Disease models, CAR T
Abstract
CRISPR/Cas systems are popular genome editing tools that belong to a class of programmable nucleases and have enabled tremendous progress in the field of regenerative medicine. We here outline the structural and molecular frameworks of the well-characterized type II CRISPR system and several computational tools intended to facilitate experimental designs. The use of CRISPR tools to generate disease models has advanced research into the molecular aspects of disease conditions, including unraveling the molecular basis of immune rejection. Advances in regenerative medicine have been hindered by major histocompatibility complex-human leukocyte antigen (HLA) genes, which pose a major barrier to cell- or tissue-based transplantation. Based on progress in CRISPR, including in recent clinical trials, we hypothesize that the generation of universal donor immune-engineered stem cells is now a realistic approach to tackling a multitude of disease conditions.
1. Introduction
Currently trending genome-editing tools support the study of gene functions and disease modeling by altering genes with programmable nucleases to mimic uncharacterized genotypes. The ability to genetically manipulate biological systems holds enormous potential across biotechnology, basic science, and medicine. The CRISPR/Cas9 system belongs to a class of programmable nucleases and permits precise alteration of eukaryotic genomes. These nucleases act by inducing targeted double-stranded DNA breaks (DSBs) on chromosomes, followed by activation of the homology-directed repair (HDR) pathway. The HDR pathway requires exogenous DNA with short sequences homologous to the acceptor sites to act as templates for repairing the underlying sequence-specific DSBs. Activation of the endogenous HDR pathway can precisely alter the genome at predefined sequences [1], [2]. However, despite its enormous potential, use of HDR-gene editing approaches is limited by the predominance of the non-homologous end joining (NHEJ) pathway in repair of DSBs. The NHEJ pathway has been used to study gene functions via disruption of expression due to formation of indels (insertions or deletions). However, CRISPR systems have been reported to utilize this pathway to restore the functions of genes disrupted by disease mutations. For example, CRISPR systems have been used to repair CYBB gene mutations causing X-linked chronic granulomatous disease (XCGD) [3] and FANCA mutations causing Fanconi anemia (FA) [4]. The HDR pathway is restricted mainly to the G2-S phases of the cell cycle and depends on availability of sister chromatids, whereas the NHEJ pathway involves end-to-end ligation of DSBs and takes place throughout the cell cycle [5], [6]. Reversible cell synchronization in the G2/M phase has been shown to improve donor integration efficiency during gene editing [7]. Inhibition of the ligase IV-dependent NHEJ DNA repair pathway by small molecule inhibitors like SCR7 has demonstrated an increase in homologous recombination (HR) in mammalian cell lines and mouse models [8], [9], [10]. Therefore, control of delivery time of the CRISPR/Cas9 system along with inhibition of the NHEJ repair mechanism can greatly improve genome editing efficiency. Various small molecules that can enhance HDR efficiency are listed in Table 1.
Table 1.
Summary of HDR-enhancing small molecules.
| Small Molecule | Target | Mechanism | Effect on HDR | Reference |
|---|---|---|---|---|
| SCR7 | Inhibition of NHEJ | Inhibition of ligase IV | Decrease NHEJ repair and increase HDR in several cell lines and mice and rats with ds and ss donors | [9], [10], [11], [12], [13], [14] |
| i53 | Inhibition of NHEJ | Inhibition of 53BP1, an important regulator of the DSB repair pathway | Increased HDR by 5–6 fold with ss and ds donors | [15] |
| STL127705 | Inhibition of NHEJ | Inhibitor of the DNA repair protein Ku70/80 | 2.8 to 7.2-fold and 2.3 to 4-fold increase in genome editing with Cas9n and Cpf1, respectively | [16], [17] |
| KU0060648 | Inhibition of NHEJ | Inhibition of DNA-protein kinase catalytic subunits | Increased HDR in HEK293T cells with reduced NHEJ frequency | [18] |
| NU7026 | Inhibition of NHEJ | Inhibition of DNA-protein kinase catalytic subunits | Reduced NHEJ frequency with increased KI efficiency in hiPSCs using Cas9 or Cpf1 | [19] |
| NU7441 | Inhibition of NHEJ | Inhibition of DNA-protein kinase catalytic subunits | Increased HDR in HEK293T and hiPSCs by 13.4 fold in zebrafish embryos | [18], [20], [21] |
| VE-822 | Inhibition of NHEJ | Inhibition of ataxia telangiectasia mutated and Rad3-related kinase (ATR) | 5.9-fold increased HDR in hiPSCs with ss and ds donors after DSB induced by Cpf1 | [22] |
| AZD7762 | Inhibition of NHEJ | Inhibitor of checkpoint kinase CHEK1 | 2.7-fold increased HDR in hiPSCs with ss and ds donors after DSB induced by Cpf1 | [22] |
| M3814 | Inhibition of NHEJ | Inhibition of DNA-protein kinase catalytic subunits | Increased KI in hiPSC and K562 cells using Cas9 or Cpf1 | [23] |
| RS-1 | Direct increase of HDR | Enhance binding of RAD51 with ssDNA | Increased HDR in HEK293T cells, HeLa cells, and rabbit and bovine embryos | [24], [25], [26], [27] |
| Mimosin, thymidine, hydroxy urea, lovastatin | Regulation of cell cycle | G1/S blocker | Increased HDR in neonatal fibroblasts | [6] |
| Nocodazole | Regulation of cell cycle | G2/M blocker | Increased HDR in HEK293T and hPSC using ss or ds donors | [6], [20] |
| XL413 | Regulation of cell cycle | G1/S blocker | Increased HDR in HSPCs, K562 cells, and T cells with ss or ds donors | [28] |
| Aphidicolin | Regulation of cell cycle | G1/S blocker | Increased HDR in HEK293T and neonatal fibroblasts using ss donor | [6] |
| ABT263 | Regulation of cell survival | BCL inhibitor | 70% increase in HDR in hiPSC | [29] |
| ABT751 | Regulation of cell cycle | G2/M blocker | Increased HDR in hPSCs with ds donor | [7] |
| Valproic acid (VPA) | Undetermined pathway | Histone deacetylase inhibitor | Increased HDR in hESCs with ds donor | [30] |
| Resveratrol | Suppression of NHEJ | Downregulation of LIG4, PRKDC, KU70, and KU80 | Increased HDR in porcine fetal fibroblast using ds donor | [14] |
| Brefeldin A | Undetermined pathway | Inhibition of intracellular transport from ER to golgi | Increased HDR in mouse ESCs using ds donor | [31] |
| L755507 | Undermined pathway | Agonist for β3-adrenergic receptor | Increased HDR in mouse ESCs using ds donor | [14], [31] |
**NHEJ – non-homologous end joining; HDR – homology-directed repair; ss – single stranded; ds – double stranded; hiPSCs – human-induced pluripotent stem cells; hPSCs – human pluripotent stem cells; ESCs – embryonic stem cell; KI – knock-in; ER – endoplasmic reticulum.
In this review, we outline the structural and molecular frameworks of the well-characterized type II CRISPR system and several computational tools intended to facilitate experimental design. The gene-editing approaches have been expanded to progenitor cells, including induced pluripotent stem cells, and in vivo in a wide array of higher mammals developed as disease models, which we discuss in this review.
2. Programmable nucleases
DSB-inducing programmable nucleases facilitate precise editing at endogenous genomic loci to enable methodical interrogation of genes and gene variations in a broad range of species [32]. Programmable nucleases include the first truly targetable zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the popular RNA-guided engineered nucleases (RGENs), the most famous of which is the bacterial and archaean clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated system (Cas) [1], [33], [34], [35], [36]. Though these nucleases have unique characteristics, they share a common mode of action: cleaving chromosomal DNA at specific sites to trigger endogenous DNA repair mechanisms and produce targeted genome modifications. In this section, we review the development of and advances in the popular CRISPR/Cas endonucleases.
2.1. CRISPR system: A new generation of programmable nucleases
CRISPR/Cas systems have emerged as an efficient, potentially facile alternative to ZFNs and TALENs for inducing DSB-mediated gene alterations [37]. The CRISPR system is a part of the bacterial and archaean acquired immune system that identifies and destroys foreign DNA via RNA-guided DNA cleavage [38]. These systems have been classified into three types (I-III) across various bacterial and archaeal hosts. CRISPR systems comprise Cas genes; non-coding RNAs; and distinctive, repetitive elements (direct repeats) interspersed with short variable sequences called protospacers that together constitute the CRISPR RNA (crRNA) [38], [39]. Each protospacer is associated with a protospacer adjacent motif (PAM) that is responsible for sequence identification and varies depending on CRISPR system [40], [41]. CRISPR/Cas systems assemble mature crRNA with Cas proteins to form crRNA-effector complexes that interrogate target DNA and destroy matching sequences in foreign genomic material [42]. By manipulating the guide RNA (gRNA) component of the crRNA, a CRISPR/Cas system can be directed to target virtually any DNA sequence, and it has been shown to be applicable to human cells [43], [44].
The best characterized CRISPR systems are the type II systems, which include the Cas9 nucleases, gRNA encoding the crRNA array, and trans-activating crRNA (tracrRNA). TracrRNA is required to process the crRNA assembly into discrete units containing the 20 bp guide sequences required for Cas9 specificity and partial direct repeats [36], [45]. Streptococcus pyogenes-derived CRISPR/Cas (spCas/SpyCas9) targets DNA that contains 5′-NGG-3′ as its PAM. On the other hand, CRISPR/Cas derived from S. thermophilus targets 5′-NNAGAA-3′ for CRISPR1 (st1Cas9) and 5′-NGGNG-3′ (st3Cas9) for CRISPR3; Staphylococcus aureus (SaCas9) targets 5′-NNGRT-3′; Acidaminococcus sp. (Cas12a) and Lachnospiraceae bacterium (Cpf1) recognize 5′-TTTN-3′, and Neisseria meningitidis recognizes 5′-NNNNGATT-3′ [47], [48], [49], [50], [51], [52], [53].
2.1.1. Molecular structure of the CRISPR/Cas9 system
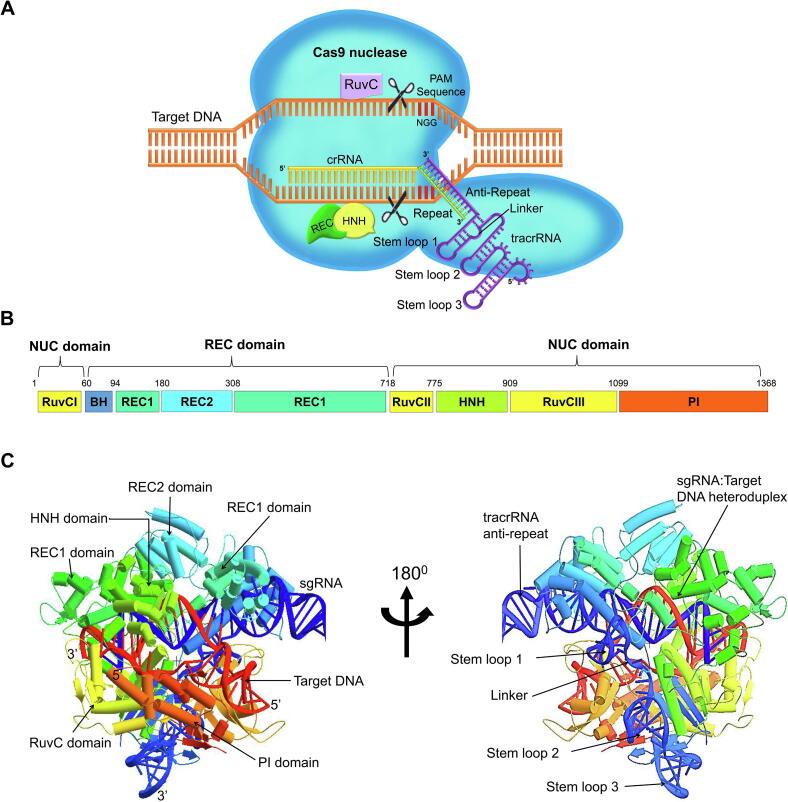
The popularity of the CRISPR/spCas9 system has been attributed to its simplicity in designing single guide RNAs (sgRNAs) via synthetic fusion of tracrRNA and crRNA, which can be modified to cleave virtually any sequence preceding the 5′-NGG-3′ PAM sequence [50], [51]. These RNA-guided nuclease functions can be performed in mammalian cells via heterologous expression of the requisite human codon-optimized RNA components and optimized Cas9 [54]. Analyses of the Cas9 crystal structure have revealed two lobes, a recognition (REC) lobe and a nuclease (NUC) lobe, with adjacent active sites. The surface area between the two structural lobes was reported to be 1034 Å [55]. A schematic representation of the CRISPR/Cas9 system is shown in Fig. 1A.
Fig. 1.
Structure of Streptococcus pyogenes Cas9-sgRNA complex. A. Schematic representation of the CRISPR/Cas9 system. B. Domain organization of the spCas system. The bilobed architecture of Cas9 consists of a recognition (REC) domain and a nuclease (NUC) lobe. C. Crystal structure of spCRISPR/Cas9 endonuclease in complex with sgRNA and double-stranded target DNA primed for specific DNA cleavage (PDB id: 5F9R). The REC domain consists of three regions, a long α helix called bridge helix (BH) and the REC1 and REC2 domains, while the NUC domain consists of RuvC, HNH, and PAM-interacting (PI) domains. The REC1 and REC2 domains interact with repeat-anti-repeat duplex and are essential for recognition of sgRNA and target DNA. The PI domain recognizes a PAM sequence on a non-complementary DNA strand. The RuvC domain is assembled into three split motifs containing an RNase H fold and interfaces with the PI domain to form a positively charged surface that interacts with the 3′ tail of sgRNA. The HNH and RuvC nuclease domains are essential for cleavage specificity of complementary and non-complementary strands of the target DNA, respectively [56].
The REC lobe consists of a long bridge helix (residues 60–93), REC1 domain (residues 94–179 and 308–718), and REC2 domain (residues 180–307). The REC1 protein consists of 25 α helices (α2-α5 and α12-α32) and two β sheets (β6, β10 and β7-β9) and adopts an elongated α-helical structure, whereas REC2 adopts a six-helix bundle (α6-α11) structure. On the other hand, the NUC lobe contains a structural core that forms the RuvC domain (residues 1–59, 718–769, and 909–1098), HNH domain (residues 775–908), and the PAM-interacting (PI) domain (residues 1099–1368) (Fig. 1B) [56]. The RuvC domain consists of three RuvC motifs (RuvC I-III) made up of six mixed β sheets flanked by α helices; it has a positive charge due to its interface with the PI domain, and it interacts with the sgRNA tail. A structural comparison analysis identified four catalytic residues (aspartic acid 10 (Asp10), glutamic acid 762 (Glu762), histidine 983 (His983), and Asp986) in the Cas9 RuvC domain that are structurally similar to an RNase H fold found in members of the retroviral integrase superfamily, suggesting a two-metal-ion catalytic cleavage mechanism for non-target DNA cleavage [55], [56]. Asp10 is reported to be critical for non-complementary DNA cleavage, and Cas9 cleavage activity depends on Mg2+ ions [56], [57], [58]. The HNH domain has only a few contact points with the rest of the protein, lies between the RuvC II and III motifs, and comprises four α helices that surround dual-stranded antiparallel β sheets [56]. The HNH nuclease active site is made up of three catalytic residues (eg-Asp40, His41, and Asn62) and is said to cleave DNA via a single metal mechanism [59]. The crystal structure of Streptococcus pyogenes CRISPR/Cas9 endonuclease in complex with sgRNA is shown in Fig. 1C. The REC and NUC lobes are connected by two link segments, one of which forms an arginine-rich bridge. Because sgRNA and its target DNA heteroduplex are negatively charged, they interact in the positively charged groove between the REC and NUC lobes. Whereas the REC-recognition lobe is essential for binding sgRNA and DNA, the NUC-nuclease lobe contains the HNH and RuvC nucleases that cleave the complementary and non-complimentary strands of the target DNA, respectively. Alignment of the bound complementary DNA and the RuvC domain active site suggests that the PI domain recognizes PAM sequences in non-complementary DNA (Fig. 1A and 1C) [56].
Cas9 has also been reported to be mutable at either HNH (H840A) or RuvC (D10A) and transforms it into a nickase capable of single-strand nicks. However, mutating both nuclease domains yields dead Cas9 (dCas9) that retains its RNA-guided DNA binding ability but has no cleavage activity [60].
2.1.2. Mechanism of action of CRISPR/Cas9
Cas9 is a powerful, highly specific, and efficient genome editing tool that has been studied extensively. Structural and mechanistic studies have provided a fundamental understanding of the mechanism that regulates the CRISPR/Cas9 system, along with a framework for structure-based rational design to improve its efficiency and minimize its off-target results.
Formation of the Cas9-sgRNA complex is important for the transition from inactive to active conformation of Cas9 upon PAM recognition [36], [50]. Accurate sgRNA-target DNA complementarity is necessary for Cas9-mediated targeting and cleavage of DNA, though imperfect base pairing at non-seed regions is tolerated within target binding specificity [61]. Cas9 forms a Cas9-sgRNA binary complex by recognizing the PAM-proximal guide region and repeat: anti-repeat sgRNA duplex. This Cas9-sgRNA complex searches DNA sites for complementary target sequences and requires complementary base pairing between the 20-bp spacer sequence and the protospacer in the target DNA and the presence of a PAM sequence adjacent to the target site to form the final Cas9-sgRNA-target DNA ternary complex [62]. Interaction between the Cas9 complex and the target DNA occurs through a three-dimensional collision that allows rapid dissociation of Cas9 from DNA that does not contain the right PAM sequences. This dwell time depends on gRNA complementarity with the adjacent DNA when the correct PAM is present [63], [64], [65].
The PI domain recognizes PAM sequences on non-complementary DNA to form the 20-bp guide-target DNA heteroduplex formation recognized by Cas9 in a sequence-independent manner and triggers local DNA melting at PAM-adjacent nucleation sites prior to formation of the ternary complex. Next, RNA strand invasion forms an RNA-DNA hybrid and displaces a DNA R-loop strand from the PAM-proximal end to the PAM-distal end [65], [66]. The REC1 (Asn497, Trp659, Arg661, and Gln695), RuvC (Gln926), and PI (Glu1108) domains interact with the phosphate backbone groups of the target DNA (nucleotides 1′, 9′-11′, 13′, and 20′). The REC1 (Leu169, Tyr450, Met495, Met694, and His698) and RuvC (Ala728) domains interact via Van der Waals interactions with the C2′ atoms of the target DNA at nucleotides 5′, 7′, 8′, 11′, 19′, and 20′. Assembly of the ternary complex follows cleavage of the complementary strand by the HNH domain and cleavage of the non-complementary strand by the RuvC domain in the sgRNA-target heteroduplex complex [56], [61], [67]. Each domain cleaves a strand of the DNA three base pairs from the NGG PAM, producing a blunt-ended DSB [65].
Post cleavage, Cas9 remains bound to the target DNA until it is displaced by other cellular processes for recycling [65], [68]. Cas9 nickase produces single-strand breaks by targeting only a single strand of the DNA duplex. Staggered-end DNA can be generated by pairing sense and anti-sense targeting sgRNAs with Cas9 nickases. DSBs generated by such double nicks can enhance the specificity of genome editing [68]. DNA cleavage can be repaired by either the NHEJ pathway or the HDR pathway.
2.1.3. Tools to design sgRNAs
sgRNA design is critical to CRISPR-based screening techniques. Each sgRNA contains a target DNA complementary spacer sequence that guides the Cas9 protein to the target sequences. The cleavage efficiency and binding of the CRISPR system require different complementarity status within the 20 bp region. sgRNA design aims to identify specific target sites in the genome by simply scanning PAM sequences, such as the 5′-NGG-3′ of spCas9, but that process faces several challenges. Currently, many tools have been developed for sgRNA design, and they vary in design specifications, genomes, parameters, and so on [69], [70]. Complementarity between the 5′-end 20-nt sgRNA sequence and the target DNA should theoretically be sufficient for sgRNA-Cas9 complex formation and cleavage, but previous studies suggest that sgRNAs have variable cleavage efficiencies. Researchers need to select the best sgRNAs as input sequences by identifying potential off targets and their relative cleavage rates, and that process can be facilitated by the many computational tools summarized in Table 2. Highly stable binding requires seven to nine matched bases proximal to the PAM region; even a few mismatches in this region can hinder cleavage. As few as four mismatches of up to eleven bps in length in the PAM-proximal region have been shown to hinder cleavage but not binding by allowing formation of stable complexes. This suggests an extremely slow rejection that could sequester Cas9-RNA [71], [72]. The off-target effects of sgRNA have been widely investigated using in vitro genome-wide assays that align spacer sequences to the genome [60] with LAM-HTGTS [73], Digenome-seq [74], SITE-Seq [75], CIRCLE-seq [76], DISCOVER-seq [77], dCas9 ChIP-seq [61], and GUIDE-seq [78]. Although the studies reached different conclusions based on analysis method, they all agreed on the importance of PAM, the 7–10-nucleotide seed sequence proximal to PAM, and the concentrations of Cas9 and gRNA for efficient cleavage. Tools designed to aid sgRNA designing by analyzing their efficiencies are summarized in Table 2.
Table 2.
Available tools to design sgRNA and to evaluate on-target efficiency/off-target effect.
| Tool name | Description | On-target prediction | Off-target prediction | Source | Reference |
|---|---|---|---|---|---|
| AttnToCrispr | To determine CRISPR Cas9 and Cas12a specificity and efficiency | Yes | Yes | https://github.com/qiaoliuhub/AttnToCrispr | [79] |
| Breaking-Cas | To design sgRNA for SpCas9, SaCas9, and Cpf1 and to evaluate probable off target sites | Yes | Yes | https://bioinfogp.cnb.csic.es/ | [80] |
| CHOPCHOP | Selection of the optimal CRISPR/cas9 sequence gene; identifying sgRNA targeting Cas9 and its variants and Cpf1. | Yes | Yes | https://chopchop.cbu.uib.no | [81], [82], [83] |
| CRISPR-RGEN Tools | Potential off-target sites in input sequence or given genome | No | Yes | http://www.rgenome.net/ | [84], [85], [86] |
| CRISPOR | To identify sgRNA sequence in input, rank them according to efficiency score, and predict off-target and on-target activity | Yes | Yes | http://crispor.tefor.net/ | [87], [88] |
| CCTop | Identifying sgRNA targeting Cas9 and its variants and Cpf1. | Yes | Yes | https://crispr.cos.uni-heidelberg.de/ | [89] |
| CRISPR-ERA | To design sgRNAs for genome editing, repression, and activation | Yes | No | http://crispr-era.stanford.edu/ | [90] |
| CRISPRdirect | To design sgRNAs | No | Yes | https://crispr.dbcls.jp/ | [91] |
| CRISPRscan | To design sgRNAs for Cas9 and Cpf1 | Yes | Yes | https://www.crisprscan.org/ | [92] |
| CRISPETa | Flexible and scalable paired sgRNA design based on an empirical scoring model | Yes | Yes | http://crispeta.crg.eu/ | [93] |
| CRISPRseek | To design sgRNAs for target sequences identified in wide variety of genome-wide analyses | Yes | Yes | http://bioconductor.org/ | [94] |
| CRISPR-GE | Experimental design and mutation analysis for genome editing in the Cpf1/CRISPR/Cas9 system | Yes | Yes | http://skl.scau.edu.cn/ | [95] |
| Cas-OFFFinder | To identify potential off-target sites in a given genome or user-defined sequences | No | Yes | http://www.rgenome.net/cas-offinder/ | [86] |
| CLD | To predict a large fraction of functional sgRNAs | Yes | Yes | https://github.com/boutroslab/cld | [96] |
| CFD | Off-target evaluation | No | Yes | https://portals.broadinstitute.org/gpp/public/software/index | [97] |
| CRISTA | To design sgRNAs for targeting sequences identified in a wide variety of genome-wide analyses | Yes | Yes | http://crista.tau.ac.il/ | [98] |
| CRISPR-OFF | Off-target evaluation | No | Yes | https://rth.dk/resources/crispr/ | [99] |
| CRISPR optimal target finder | Off-target evaluation | No | Yes | http://targetfinder.flycrispr.neuro.brown.edu/ | [100] |
| COSMID | To identify potential off-target sites with the specified number of mismatched bases and insertions or deletions compared with the guide strand | Yes | Yes | https://crispr.bme.gatech.edu/ | [101] |
| CRISPR.mit | To design sgRNAs and evaluate off-target effects | Yes | Yes | https://zlab.bio/guide-design-resources | [60] |
| CROP-IT | To design sgRNAs and evaluate off-target effects | Yes | Yes | http://cheetah.bioch.virginia.edu/AdliLab/CROP-IT/homepage.html | [102] |
| CRISPR MultiTargeter | To identify unique isoform-specific sgRNA sites and common sgRNA sites | Yes | Yes | http://www.multicrispr.net/ | [103] |
| CASPER | To accommodate use of the CRISPR/Cas system for genome editing, multitargeting analysis, and multispecies population analysis. | Yes | Yes | https://github.com/TrinhLab/CASPER | [104] |
| CRISPy-web | To design sgRNAs for a user-provided microbial genome (non-model organisms) | Yes | Yes | https://crispy.secondarymetabolites.org/ | [105] |
| CRISPR-PLANT | To evaluate off-target sites for seven genomes of model and crop plants | Yes | Yes | https://www.genome.arizona.edu/crispr2/ | [106], [107] |
| DeepCRISPR | To design sgRNAs with on-target and off-target site prediction with deep learning | Yes | Yes | http://www.deepcrispr.net/ | [108] |
| E-CRISP | To design sgRNAs | Yes | Yes | http://www.e-crisp.org/E-CRISP/ | [109] |
| EuPaGDT | To design gRNAs for eukaryotic pathogens | Yes | Yes | http://grna.ctegd.uga.edu | [110] |
| Elevation | Cloud-based machine learning platform for off-target and on-target prediction | Yes | Yes | https://crispr.ml/ | [111] |
| FlashFry | To design sgRNAs and predict on-target and off-target sites with unconstrained number of mismatches | Yes | Yes | http://mckennalab.org/FlashFry/ | [112] |
| GuideScan | To design high-density sets of sgRNAs for single and paired sgRNA genome-wide screens and correctly determine off-target sites | Yes | Yes | http://www.guidescan.com/ | [113] |
| GT-Scan | To identify unique genome targets | https://gt-scan.csiro.au/ | [114] | ||
| PROTOSPACER | Offline software for flexible design of Cas9 sgRNAs with a graphical user interface with a file-based database and third-party sequence mapping tools to maximize flexibility and information retrieval when designing sgRNAs | Yes | Yes | http://www.protospacer.com/ | [115] |
| SgRNA designer | To design sgRNAs for SaCas9 and SpCas9 | Yes | Yes | https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design | [97], [116] |
| sgRNAcas9 | Design of gRNAs for Cas9s from S. aureus and S. thermophilus 3 and Cpf1 | Yes | Yes | http://www.biootools.com/software.html | [117] |
| sgRNA Scorer 2.0 | Design of gRNAs for Cas9s from S. aureus and S. thermophilus 3 and Cpf1 | Yes | No | http://crispr.med.harvard.edu/sgRNAScorerV2 | [118] |
| WU-CRISPR | To design high-density sets of sgRNAs for single and paired sgRNA genome-wide screens and correctly determine off-target sites | Yes | Yes | http://crispr.wustl.edu | [97] |
To further improve DNA targeting efficiency by processing only correct targets, systematic investigations into the binding kinetics of Cas9-RNA need to be conducted. How dissociation kinetics vary according to mismatches also needs to be established to determine the rejection rates of mismatched/partially matched sequences [71].
One target efficiency detection model was proposed by Doench et al., who created a pool of sgRNAs and screened all possible target sites across six endogenous mouse and three endogenous human genes. They used antibody staining and flow cytometry to quantitatively assess 1841 sgRNAs for ability to produce null target gene alleles. Using those data, they generated a predictive model of sgRNA activity to improve sgRNA design for gene screening and editing. That model was made into an online tool that rates active sgRNAs targeting any gene of interest with score between 0 and 1, with higher values indicating higher efficiency [119]. Several tools (CHOPCHOP, E-CRISP, WU-CRISPR, CRISPR library designer, CRISPRpred, CRISPETa, and CRISPOR [81], [83], [88], [93], [96], [109], [120], [121]) use that model, which is driven by the hypothesis that sgRNA knockout efficiency can be empirically scored considering the effects of genome context factors. Another sgRNA design method predicts sgRNA efficiencies considering features from a training model. Available tools using that method include SSC, CRISPRscan, sgRNA Designer, and Deep CRISPR [92], [97], [108], [118], [122], [123]. These comprehensive computational platforms unify sgRNA on- and off-target predictive scores into one framework with deep learning and surpass performance of state-of-the-art in silico tools.
The advances in CRISPR techniques and volume of genome-editing data being accumulated are presenting new computational challenges. The in silico design of sgRNAs has become a key issue for successful gene-editing using CRISPR systems, and continuous efforts are being made to refine it [124]. sgRNA efficiency is significantly influenced by the nucleotide composition of DNA proximal to or downstream of PAM. Nucleotides upstream of PAM do not significantly affect sgRNA efficiency. For efficient sgRNA design associated with Cas9, human and mouse cell lines prefer guanines at the −1 and −2 positions proximal to PAM. On the other hand, thymidine at the + 4/-4 positions is disfavored close to PAM [122], [125], [126]. Efficient in silico models that integrate heterogeneous genome editing data can be built to derive unbiased sgRNA design rules and improve sgRNA design [119]. A recent development in sgRNA design involves an in vivo library-on-library method that can simultaneously assess sgRNA activity across about 1400 genomic loci to unravel underlying epigenetic parameters and nucleotide sequences related to sgRNA knockout (KO). Such research has found that sequence composition and locus accessibility are important factors of sgRNA activity [118].
Given the number of features involved, data modeling most commonly uses machine learning methods. The construction of such models requires that many sgRNAs be tested experimentally to build a robust dataset that can enable prediction of efficiency. Therefore, researchers have typically adopted biological enrichment schemes that monitor events affecting observable biological phenotypes such as cell survival [127]. Other factors that influence sgRNA activity can be discovered by incorporating additional datasets, modeling approaches, and activity readouts. All those features have been shown to correlate with the on-target activity of Cas9-sgRNA complexes, so they can help enable effective use of CRISPR technology to probe gene functions and edit genomes.
2.2. Recent development in CRISPR/Cas system for improved genome editing
Based on recent advances in crystal structure analysis of the CRISPR/Cas system, many Cas variants have been developed for improved editing efficiency and performance [128]. After successful application of SpCas9 as a tool for genome editing in mammalian cells, other Cas9 proteins have been developed. For example, SaCas9 (Staphylococcus aureus) [129], Nme2Cas9 (Neisseria meningitides) [130], AsCpf1 (Acidaminococcus sp), LbCpf1 (Lachnospiraceae sp) [49], [131], and recently identified AacCas12b (Alicyclobacillus acidoterrestris) [132] have shown comparable or higher genome editing efficiency to that of SpCas9 [128]. Furthermore, a recently reported type VI CRISPR-associated RNA targeting Cas13a and Cas13b system can specifically cleave strands of RNA [133], [134], [135].
Recent technological advances have enabled development of nuclease-deficient, catalytically-inactive dead Cas9 (dCas9) or paired nickase (Cas9N) [136] that can be fused with various transcription regulatory domains creating either CRISPR activator (CRISPRa) or CRISPR inhibitors (CRISPRi) that can specifically activate or silence the target gene [137]. In addition, the recently reported D10A cas9 nickase-based cytidine base editors (CBEs) and adenine base editors (ABEs) can generate A to T and C to G transitions, respectively, causing many double-stranded breaks in the targeted genomic region [138]. In addition, the Liu group developed high-efficiency H840A Cas9 nickase-based prime editors (PEs) designed to restore both strands to the desired sequence instead of the original without DSB or donor DNA template [139]. Thus, application of the CRISPR/Cas system, which can modulate target gene expression or transcript levels in a PAM-independent and transient manner, could provide a controllable approach for disease modelling and treatment.
3. Development of models for CRISPR-based correction of defective genes
The CRISPR/Cas9 system has been widely used to create mutations in target genes and generate sequence-specific genomic modification through HDR. CRISPR has been used for other purposes such as regulation of endogenous gene expression, live-cell imaging of chromosomal loci, epigenome engineering, and high-throughput screening and editing of RNA [137]. Given its precision, simplicity, and robustness, CRISPR has emerged as a tool for generating novel in vitro and in vivo disease models and has the potential to connect biological discoveries with clinical therapeutic approaches. In this section, we review the use of CRISPR/Cas9 systems to develop various disease models.
3.1. Pluripotent stem cell technology: Toward gene correction in humans
The technique of culturing cells in a dish has been the backbone of basic biomedical research for decades because it can provide insights into normal and pathological cellular processes. Regenerative medicine and cell therapy aim to replace unhealthy or diseased cells with healthy new cells. Primary cells also can be genetically manipulated and re-implanted into patients to cure or treat genetic diseases [140]. However, the limited lifespan of primary human cells in culture is a drawback that limits inquiries into regulation of tissue formation, regeneration, and repair [141]. Most human cell lines used today have been derived from tumors or transformed derivatives of native tissues that carry mutations; the inability to faithfully adapt these human cells for in vitro growth limits research [141], [142]. However, advances in reprogramming human somatic cells into induced pluripotent stem cells (iPSCs) have begun to address correction of defective genes in mammals.
Pluripotent stem cells (PSCs) or typical embryonic stem cells (ESCs) derived from the inner cell mass of blastocysts can differentiate into specialized cells to recreate specific tissues such as lungs or brain [143]. Thus, PSCs are an immortal population that can differentiate into virtually any cell type within the human body and enable establishment of new models of mammalian development and a new source of cells for regenerative medicine [144], [145]. The immortality of PSCs could help to generate clinically relevant cell populations for aid in tissue repair and regenerative medicine, although they show difficulty in efficient genome editing [143]. On the contrary, iPSCs are easier to manipulate, but their applications in regenerative medicine approaches should be considered prior to human studies [146]. Issues such as immune rejections, carcinogenicity, genomic instability, lack of in situ integration and lack of quality controls have hindered application of iPSCs to routine clinical use [146]. However, a recent study published in a reputed journal has used autologous patient-derived iPSCs to dopaminergic progenitor cells to treat Parkinson’s disease. Although this study is still in its infancy, it opens up numerous avenues toward application of iPSCs to treat human diseases [147].
iPSCs have made progress toward modeling of numerous human genetic disorders. The scope of CRISPR/Cas9 gene editing systems has been expanded to disease-focused research through production and characterization of patient-derived iPSCs that carry specific genetic diseases. CRISPR/Cas9 editing systems thus allow either repair of genetic mutations in patient-derived iPSC models or generation of disease mutations in healthy iPSCs to generate disease models. An example of the application of iPSCs toward modeling of human diseases was described for the multifactorial neurodegenerative Parkinson’s disease (PD) [148]. Some PD cases are due to autosomal dominant mutations in SNCA genes that encode α-synuclein responsible for synaptic transmission and vesicle transport. Arias-Fuenzalida et al. generated isogenic-biallelic mutations in the SNCA gene using healthy iPSCs and FACS-assisted CRISPR/Cas9 editing to develop a PD-specific model [148]. CRISPR/Cas9 also has been used to derive isogenic CSB/ERCC6 gene corrections in iPSCs derived from a rare autosomal recessive disorder cockayne syndrome (CS) patient [149]. Another recent report regarding the use of CRISPR/Cas9 to correct gene defects was demonstrated in amyotrophic lateral sclerosis (ALS), which is commonly caused by C9orf72 mutations. CRISPR/Cas9-mediated excision of C9orf72 repeat expansion reversed the pathophysiological effects of astrocytes in human iPSCs harboring these gene mutations [150].
iPSCs have the capability to differentiate into certain cell types relevant to disease phenotypes [151], [152]. This differentiation capability make iPSCs an important asset to model numerous diseases, as described in the case of type 1 diabetes (T1D) by Leite et al. [153]. Autoimmune T1D results in destruction of pancreatic β cells that secrete insulin. Leite et al. used patient-derived endocrine cells and autologous immune cells to model T1D using an in vitro platform. They demonstrated a cell-type specific immune response against iPSC-derived β cells and a reduced effect against α cells [153]. Leite et al. demonstrated use of CRISPR/Cas9 systems to ablate HLA class I expression in iPSC-β cells, which resulted in reduced T cell activation, the significance of which has been described in the following sections (Section 4). iPSC-derived neurons also have been recently used to generate a CRISPR interference-based platform to study essential/non-essential gene functions [155]. Disease-in-a-dish in vitro iPSC models have been developed for numerous diseases including Shwachman-Bodian-Diamond syndrome [141], adenosine deaminase deficiency-related severe combined immunodeficiency [141], Duchenne and Becker muscular dystrophies [141], [154], Gaucher disease type III [141], Parkinson’s disease [141], [156], [157], Huntington’s disease [141], [158], Alzheimer’s disease [159], rheumatoid arthritis and osteoarthritis [160], types 1 and 2 diabetes [141], [153], [161], the carrier state of Lesch-Nyhan syndrome [141], and Down syndrome/trisomy [141].
These disease models could also be used for high-throughput screening of therapeutic drugs. Advances in genotyping and next-generation sequencing techniques have facilitated identification of genetic mutations responsible for disease pathology and phenotypes [162]. Drugs elicit different responses in different patient-derived iPSCs, probably due to the different genetic backgrounds of individual donors [163]. Derivation of diseases models by inducing specific mutations in healthy iPSC cell lines (isogenic controls) allows identification of causative lesions that define cellular phenotype, resulting in disease. Such comparisons are not possible by comparing iPSCs-derived from patients and healthy individuals due to the inheritance patterns of single-nucleotide polymorphisms (SNPs) among individuals [162], [163]. In addition to establishment of disease models without needing patient samples to screen for drug recovery efficacy, this strategy of disease modeling might be used to model various genetic conditions for which obtaining patient samples is problematic. The application of CRISPR-mediated gene correction strategies in pluripotent cells could confirm validity and safety before clinical applications while eliminating the need for murine models for validation. Disease-specific pluripotent cells capable of differentiating into the various tissues affected by each condition would provide insights into the pathophysiology of disease in a controlled, in vitro human system and allow screening of potential drugs in that system.
Nonetheless, certain challenges in the use of iPSC disease models need to be addressed. For example, the rarity of certain genetic diseases limits the ability to build patient-derived cell models, and genetic variations among populations could result in inappropriate interpretations of disease phenotypes in vitro [141], [164], [165]. The number and use of control cell lines during iPSC development, nuclease design, disease modeling, and drug screening also are controversial. Previously, the use of control cells from healthy family members was considered adequate, but isogenicity and genome sharing among family members could influence cellular disease phenotypes [166]. However, this problem can be tackled by the use of gene editing techniques to generate disease models in iPSCs derived from healthy individuals and eliminates the genetic modifiers that differ among patient-derived iPSCs and controls [162]. Another issue regarding use of iPSCs as disease models is the somatically acquired mutations in disease-affected cells, contributing to disease pathologies. Such mutations may not be present in iPSCs or iPSC-derived cells, though unrelated somatic mutations that have accumulated in cells used for reprogramming also would be propagated to iPSCs [162], [167]. Advances in single-cell and deep-genome sequencing techniques can be utilized to study the effects of such mutations in diseases. Another issue regarding use of iPSCs arises due to the high clonal diversity of human iPSCs. Patient-derived iPSCs may not always be easy to derive, handle, or transfect; thus, protocols have to be adjusted constantly for individual cell lines. This may not be required when using universal human iPSC cell lines [166].
3.2. Transgenic animal models for in vivo gene editing
The CRISPR system has evolved at a rapid pace; during the past few years, its applications have expanded far beyond DNA mutagenesis [168]. Heritable gene editing has been achieved using the CRISPR/Cas9 system and generated a variety of transgenic animals. Those studies delivered the CRISPR/Cas9 system directly into fertilized zygotes using methods such as microinjection and electroporation incorporating DNA [169], [170]. However, application of CRISPR to somatic tissues in postnatal animals has been challenging due to larger transgene size and existing delivery strategies for Cas9 in vivo [129]. To address that issue, various vectors, such as adeno-associated virus, lentivirus, and particle-mediated delivery, have been developed to offer low immunogenic potential, broad serotype specificity, and reduced oncogenic potential and host-genome interactions with the delivery of Cas9 [171], [172]. Direct injection of nuclease-encoding messenger RNA into early mammalian embryos has proven quite effective in generating germline modifications, adding another tool for mammalian genetics [173].
CRISPR/Cas9 genome engineering has been successfully applied in many model organisms, including mice, cynomolgus monkeys, rats, pigs, Caenorhabditis elegans, Xenopus tropicalis, and Drosophila [169], [174]. Among them, the mouse model is the most popular and has been used in developing many nuclease-directed systems, including CRISPR/Cas9, and brings a new level of flexibility to genomic manipulation [168]. The collective research using CRISPR/Cas9 for in vivo gene therapeutics in animal models of various human genetic diseases has demonstrated the potential applicability of the system for human use. These research advances suggest that CRISPR/Cas9 could be used to correct disease-driving mutations in vivo [164], [169].
CRISPR/Cas9 systems have been routinely used to generate single-nucleotide polymorphism (SNP) animal models of human disease in rodents [169], [175], [176]. Such models provide a functional insight into human genetics that allow development of potential new therapies. An example of a pathological SNP modelled in mice was identified by human genome-wide association studies (GWAS) in the STXBP5 gene (rs1039084 A > G). STXBP5 acts as a regulator of platelet secretion in humans. The SNP-rs1039084 A > G has been reproduced by CRISPR in the mouse to display nearly the same thrombosis phenotype, confirming the causality of this SNP in humans [169], [177].
The most popular application of CRISPR/Cas9 to the study of gene functions and drug discovery has been generation of specific gene knockouts. Conditional transgenesis in adult mice has been attempted to enable tissue-specific and inducible Cas9-mediated mutagenesis. Cre-mediated systems have been modified by including CRISPR targeting systems to enable them to express Cas9 under control of strong promoters such as CAGs [178]. Doxycycline-inducible systems have also been attempted, as demonstrated recently by Dow et al., to provide Cas9 and sgRNA to the germlines of animals. These models can target individual and multiple tissues and have not been restricted by the exogenous sgRNA delivery efficiency required to extinguish the expression of Cas9 after gene editing [179]. CRISPR systems also have been successfully utilized to insert LoxP sites surrounding various exons in mice. This strategy was described as Easi-CRISPR (Efficient additions with ssDNA inserts-CRISPR) that efficiently allows creation of conditional knock-in as well as knock-out models [180].
CRISPR also holds great promise for use in larger vertebrate model systems that reflect human disease progression better than mouse models. For example, pigs closely resemble human physiology and can be a highly relevant model for human diseases, as demonstrated by research into familial adenomatous polyposis and cystic fibrosis (CF). The existing mouse models of these monogenic diseases did not accurately resemble disease progression or treatment, and the pig models were more accurate. CRISPR can be applied to create monoallelic and biallelic mutants in higher animal models such as pigs, providing a gateway for improved study of cardiovascular diseases, metabolic treatments, and wound-healing. With implementation of new tools and technologies, these models could improve future drug discovery and treatment research [181], [182]. Homozygous Pink1/Parkin knockout pigs have been developed as models for PD using CRISPR/Cas9 systems and somatic cell nuclear transfer (SCNTs) producing gene-edited animals with single identical mutations [183]. CRISPR/Cas9-based genome editing strategies have heightened the capacity to genetically modify pigs to engineer organs for xenotransplantation [184]. Toward such goals, CRISPR/Cas9 has been utilized to inactivate copies of the porcine endogenous retrovirus (PERVs) in pigs that could potentially be passed to humans [185]. Wu et al. demonstrated disruption of pancreatogenesis in pig embryos utilizing the CRISPR/Cas9 system to target the PDX1 gene. These models could serve as a platform for xeno-generation of human organs in pigs when combined with chimeric-competent human iPSC technologies [186].
Another advantage offered by CRISPR/Cas9 is its ability to target multiple genes simultaneously, offering a way to analyze multiallelic mutants in a single step and removing the need to breed independent strains [168]. An example of this strategy was demonstrated by Li et al., who generated GGTA1/iGb3S and GGTA1/CMAH double knockout and GGTA1/iGb3S/CMAH triple knockout pigs that are naturally deleted in humans using the CRISPR/Cas9 system, indicating potential for xenotransplantation [187]. Multiple double-stranded breaks in DNA can also lead to genomic structural variations (SVs). SVs are large structural differences in genomic DNA that lead to chromosomal duplications, deletions, inversions, insertions, translocations, or combinations of these. Numerous human disorders are associated with such SVs involving several genes, and CRISPR/Cas9 systems have facilitated a way of modeling these diseases in mouse models [188], [189], [190], [191]. Application of CRISPR/Cas9 to model human diseases containing SVs has been demonstrated for Downs syndrome using a CRISpr MEdiated REarrangement (CRISMERE) strategy [188]. However, a drawback of targeting multiple sites using CRISPR is the sequence variations caused at breakpoints due to the randomness of NHEJ and the accessibility of CRISPR target sites that affect sgRNA efficiency [190]. CRISPR/Cas9 has been adapted for direct in vivo delivery systems that enable specific, multiplexed gene modifications in select mouse tissues. Proof-of-concept therapeutic models of CRISPR have been delivered via mosaic delivery systems that disrupt or modify target genes in subsets of cells that surround normal tissues [192]. CRISPR/Cas9 offers surgical-precision editing that leaves behind little or no trace of the entire process [192]. The future of genome editing could lie in successful, direct application of CRISPR in vivo, which could have multiple applications in precision medicine.
3.3. Organoid-based models offer futuristic approaches to gene correction
Organoids are 3D arrangements of specific cell types derived from stem cells that self-organize spatially and have restricted lineage commitments to represent the key structural and functional properties of organs [193]. Organoids that represent multiple niche interactions have been successfully established from adult stem cells and PSCs derived from mice and humans for multiple organs, such as the esophagus, salivary gland, prostate, fallopian tube, brain, liver, kidney, stomach, intestine, ovary, lung, and pancreas [194], [195]. Technologies such as adeno-associated viral vectors and other lentiviral expression systems allow for genetic manipulation of organoids to successfully develop gene therapy strategies and disease models [196].
Because the genetic signatures of tissues can be retained in organoids, several genetic disorders, such as the autosomally recessive CF disorder caused by mutations in the CF transmembrane conductance regulator (CFTR) gene, have been successfully modeled using organoids [195], [197], [198]. Schwank et al. demonstrated the successful application of CRISPR/Cas9-mediated correction of mutations in the CFTR regions of organoids derived from the intestines of two CF patients via homologous recombination [198]. The genome-edited organoids showed corrected CFTR gene expression with fully functional protein expression, indicating that CRISPR/Cas9 is a potential gene therapy strategy [198]. CFTR gene correction using CRISPR/Cas9 has been demonstrated in patient-derived iPSCs [199]. Organoids derived from CF patients have been deposited into biobanks and could have therapeutic applications in the near future [194].
Differentiated kidney organoids have been used to model congenital kidney disorders following introduction of disease mutations in human PSCs (hPSCs). This strategy was demonstrated by Freedman et al., who generated an in vitro human model of polycystic kidney disease (PKD) [201]. They used CRISPR to generate biallelic mutants of the PKD1 and PKD2 genes, which encode polycystin-1 (PC1) and polycystin-2 (PC2), respectively. These knockout cell-derived kidney organoids formed cysts that are characteristic of the disease in kidney tubules in vitro [201]. Genetically edited, podocalyxin gene-mutated hPSC cell lines have been developed using this strategy. Those gene-defective kidney organoids displayed junctional organization defects in podocyte-like cells, recapitulating glomerulopathy-like phenotypes in vitro [201].
Other kidney diseases have been studied in H9 human embryonic stem cells (hESCs) that were edited using CRISPR/Cas9 to target the mutations in the PKD1/PKD2 genes that cause PKD and autosomally dominant PKD [201]. The kidney organoids derived from PKD1 or PKD2 KO cells displayed about 6% cyst formation compared with wildtype kidney organoids, which resembles the clinical symptoms of PKD [202], [203]. The application of CRISPR/Cas9-modified organoids has been extended to model diseases such as dyskeratosis congenital disease [204], monogenic diabetes [205], microcephaly [206], autism spectrum disorders [207], multiple intestinal atresia [208], and microvillus inclusion disease [209].
Therefore, organoids have great potential in cancer research, regenerative medicine, disease modeling, and drug screening, and they are a potential source of cells or whole organs for transplantation [210]. Combining iPSC generation strategies with CRISPR/Cas9-HDR gene correction and organoid generation would offer a personalized approach to correcting even rare genetic disorders and organ transplantation. Though use of iPSCs in regenerative medicine is currently hindered by various drawbacks, they hold great promise in this field. Numerous strategies are being investigated to aid generation of “safe” iPSCs that could have a positive influence on regenerative medicine [211], [212].
4. Stem cell immune engineering to overcome HLA barriers in regenerative medicine
Despite the progress in regenerative therapies to replace or restore dysfunctional cells or tissues, immuno-incompatibility is a major barrier to clinical application. A host immune response called graft-versus-host disease (GVHD) can be triggered by the highly polymorphic human leukocyte antigen (HLA) system upon transplantation of cells or tissues into immune-competent individuals [212]. GVHD is triggered by alloreactive T-cells, which are the main cellular mediators that recognize non-self HLA molecules on the surface of allogeneic cells by a process called allorecognition [213]. Allogeneic donor cells can trigger the adaptive immune system via B lymphocytes, T lymphocytes, or natural killer (NK) cells [214], [215]. CRISPR/Cas9 systems used with iPSC technology could establish universal donor iPSC cell banks to match the diversity of HLA phenotypes. The differentiation of such gene-engineered-iPSCs into desired cell types could be used as a strategy for regenerative medicine [216].
The HLA genes belong to the major histocompatibility complex (MHC) group of membrane-bound glycoproteins that comprises three classes, though MHC class III molecules are not involved in immunogenicity [215]. MHC class I genes are expressed on the surface of all nucleated cells and platelets and comprise three major genes, HLA-A, HLA-B, and HLA-C [217]. The polymorphism of MHC class I genes has been attributed to an alpha heavy chain, and its surface expression requires β2-microglobulin encoded by the B2M gene. These class I genes enable elimination of cells expressing foreign or viral antigens by presenting intracellularly processed peptides to CD8+ cytotoxic T cells [218], [219]. MHC class II genes include HLA-DR, HLA-DP, and HLA-DQ and are expressed only on specialized antigen-presenting cells such as macrophages, dendritic cells, and B cells. Non-self-antigens presented by MHC class II molecules trigger immune responses via CD4+ T helper cells. The class II proteins contain polypeptide α and β chains and require the CIITA transcription factor to activate HLA II gene expression [218], [219]. HLA homozygosity at HLA-A, -B, -C and -DRB1 plays an important role during transplantations at allelic levels. HLA homozygous haplotypes have been defined as having only one allele at each of these four loci, whereas HLA heterozygosity is defined as two alleles at any of these loci [220].
Many groups have attempted to achieve a universal donor using different strategies. Early strategies to achieve successful engraftment included identification of HLA homozygous iPSC cell lines at HLA-A, HLA-B, and HLA-DR loci [221], [222], [223]. Those homozygous cell lines, however, need to be matched at the HLA C1/C2 ligands to prevent attack by natural killer (NK) cells [224]. NK cells trigger an allogenic response when they do not detect HLA class I expression on cell surfaces through two inhibitory receptors, the killer cell immunoglobulin-like receptor (KIR) and NKG2A receptor [224]. Precise CRISPR-based gene editing strategies have been used to efficiently target the exons of the MHC class I HLA-A, HLA-B, and HLA-C genes [225]. Various strategies have reduced HLA class I gene expression through knockout of B2M [226], [227], [228], [229], [230]. However, those cells could be targeted by host NK cells due to their deficiency of class I genes. Immune responses triggered via NKG2A receptors can be suppressed by HLA-E surface expression fused to the B2M promoter [228]. However, HLA-E overexpression does not suppress KIRD2+ NK cells. The KIR2DL receptor requires HLA-C or HLA-G expression, as demonstrated by Han et al., who ablated the expression of HLA-A, HLA-B, HLA-C, and HLA class II genes in human iPSCs [224], [228], [231]. The HLA-deficient cells were modified to express HLA-G and the immunomodulatory factors PD-L1 and CD47, which resulted in a blunted T cell response [231]. A recent study has demonstrated generation of hypoimmunogenic human PSCs (hPSCs) by knocking out B2M to ablate HLA-class 1 and replace it with HLA-G1 within the frame of B2M. These engineered hPSCs are reported to be compatible with CD8 + T cells and NK cell-mediated immunotoxicity [232]. HLA-A and HLA-B carry Bw4 motifs that can be recognized by the KIR3DL1 receptors of NK cells [224]. HLA-C(C1) can suppress KIR2DL2/3 receptors, and HLA-C(C2) suppresses KIR2DL1 receptors [233]. On this basis, Xu et al. demonstrated that sequential deletion of HLA-A and -B in iPSCs using CRISPR/Cas9 while retaining expression of HLA-C allowed cells to evade CD8+ T cells and NK cells more efficiently than B2M KO [234]. However, minor histocompatibility antigens on those MHC-edited cells could trigger snap responses in patients, diminishing the T cell repertoire [235].
HLA class II proteins present peptides to CD4+ T helper cells and have been manipulated in ways similar to those described for class I genes. The CIITA transcription factor is essential for class II gene expression, and several studies have attempted CIITA KO using CRISPR/Cas9 [228], [231], [234]. Deuse et al. generated HLA-I and HLA-II KO mouse and human iPSCs using CRISPR/Cas9, generating hypoimmunogenic iPSCs that could evade immune rejection [228]. However, depending on the cell fate of those modified stem cells, deletion of important HLA genes could affect antigen presentation. For example, dendritic cells derived from CIITA-deleted iPSCs could have ablated antigen-presenting functions. Various strategies aimed toward generation of universal donor stem cells are highlighted in Fig. 2.
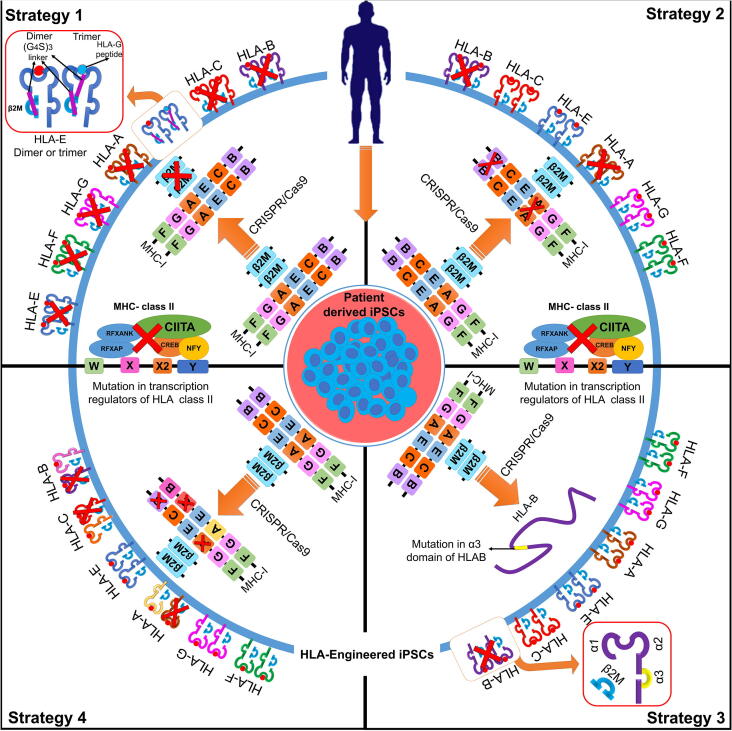
Fig. 2.
CRISPR-based HLA immunoengineering toward development of immunocompatible-induced pluripotent stem cells. The figure represents various strategies aimed towards generation of HLA-immunocompatible iPS cell lines by selectively targeting HLA class I and/or II using the CRISPR/Cas9 system. Strategy 1: Selective knockout of transactivators for HLA class I-B2M gene and HLA class II-CIITA using a CRISPR/Cas9 system prevents immune response from cytotoxic CD8+ and CD4+ T cells [229]. However, lack of HLA surface expression can activate NK cells, risking proliferation of infected cells and increasing the risk of cancer. Exogenous surface expression of HLA E single-chain dimers fused to B2M promoter or as trimers containing HLA G signal peptides in cells lacking surface expression of HLA-A, B or C escapes allogenic recognition by CD8+ T cells while conferring resistance to NK cell mediated lysis [236]. Strategy 2: HLA-C plays a pivotal role in suppressing activation of NK cells. CRISPR/Cas9-mediated disruption of HLA-A and HLA-B biallelically, while retaining HLA-C along with other non-canonical HLA class I in iPSCs expands donor compatibility while suppressing T cell and NK cell activation. The immunocompatibility of HLA-C-retained iPSCs can be improved by targeted knockout of class II transactivator-CIITA, suppressing activation of CD4+ T cells [234]. Strategy 3: The CRISPR/Cas9 system has been demonstrated to increase immunocompatibility by knocking out heterozygous HLA-B in homozygous HLA-A iPSCs with an HLA haplotype HLA-A (24:02, 24:02) and HLA-B (40:01, 54:01). HLA-B-engineered iPSCs were generated by specifically mutating the alpha 2–3 domains that form the B2M binding groove. The HLA-KO homozygous iPSCs exhibit less immunogenicity while maintaining pluripotency [237]. Strategy 4: Generation of pseudo-homozygous iPSCs using the CRISPR/Cas9 system. HLA homozygous cell line stocks have been proposed to match a major percentage of the population [226]. CRISPR/Cas9 gene editing can be used to generate class I HLA haploid iPSCs by targeted allele-specific HLA disruption in HLA heterozygous iPSCs to generate “pseudo-homozygous” iPSCs with similar donor potential to HLA homozygous iPSCs [234].
The use of genome editing tools such as the CRISPR/Cas9 system with gene knock-in strategies opens avenues that could theoretically change the serotype of any HLA genes to better match donors to recipients. This strategy could potentially generate fully functional universal donor cell lines that would enable transplantation into any patient.
5. Transition of a CRISPR/Cas9-based genome editing system to the clinical system
CRISPR/Cas systems have applicability far beyond the disease modeling and universal cell line generation strategies reviewed in this article. Advanced clinically approved gene editing strategies rely on ex vivo gene manipulation that can provide therapeutic advantage following transplantation of modified cells into recipients. Currently, clinical protocols are in progress to study application of CRISPR/Cas9 technology in lung, renal, bladder, and esophageal cancer [128]. In particular, a gene editing approach using engineered T cells overexpressing chimeric antigen receptors (CARs) has been successfully used in T cell-based immunotherapies [238], [239]. Researchers have successfully used multiplex genome editing to generate CAR-T cells resistant to inhibitory programmed cell death protein 1 (PD-1) that prevents recognition of tumor cells by T-cells [239], [240], [241]. In fact, knock out of PD-1 in autologous T-cells and infusion back into cancer patients was the first reported use of CRISPR/Cas9 system in a human clinical trial in Europe (NCT03399448) and China (NCT03081715) [242]. Several clinical trials registered at www.clinicaltrials.gov (NCT04438083, NCT03545815, NCT04037566, NCT04035434) are studying the safety and efficacy of CRISPR/Cas9-engineered T cells to treat cancer [243]. The first reported CRISPR treatment for a rare condition called Leber congenital amaurosis 10 (LCA10) used ex vivo correction of patient-derived retinal cells. The CEP290 gene mutation responsible for LCA10 was deleted using CRISPR/Cas9 in retinal cells that were then re-implanted into the eye [244], [245]. Human trials based on the CRISPR/Cas system have been undertaken for treatment of various genetic diseases like β-thalassemia (NCT03655678) (NCT0372832) and Kabuki syndrome 1 (NCT03855631) [246] (https://clinicaltrials.gov/). Table 3 summarizes the application and advances of CRISPR/Cas system in clinical trials. Collectively, the success of these clinical trials suggests the potential of development and establishment of protocols for treatment of various diseases and cancer using a CRSISR/Cas9-based genome editing system.
Table 3.
The CRISPR/Cas-based cell therapy in clinical trial. Data
| Clinical Trial Identifier | Trial name | Disease | Target gene | Interventions | Phase of study | Group | Status |
|---|---|---|---|---|---|---|---|
| NCT03745287 | A safety and efficacy study evaluating CTX001 in subjects with severe sickle cell disease | Sickle cell disease | BCL11A | Autologous CD34+ HSPCs | Phase I/II | Vertex Pharmaceuticals Incorporated, USA | Recruiting |
| NCT03655678 | A safety and efficacy study evaluating CTX001 in subjects with transfusion‐dependent β‐thalassemia | β‐thalassemia | BCL11A | Autologous CD34+ HSPCs | Phase I/II | Vertex Pharmaceuticals Incorporated, USA | Recruiting |
| NCT03728322 | iHSCs with gene correction of HBB intervention subjects with β-thalassemia mutations | β‐thalassemia | HBB | CRISPR–Cas9-treated induced hematopoietic stem cells | Early phase | Allife Medical Science and Technology Co., Ltd, China | Not yet recruiting |
| NCT03872479 | Single ascending dose study in participants with LCA10 | Leber congenital amaurosis type 10 | CEP290 | Subretinal injection of Drug AGN-151587 | Phase I/II | Allergan, USA | Recruiting |
| NCT03166878 | A study evaluating UCART019 in patients with relapsed or refractory CD19 + leukemia and lymphoma | B cell leukemia, B cell lymphoma | TCR and B2M | CRISPR–Cas9 treated CD19-directed chimeric antigen receptor T (CAR T) cells (UCART019) | Phase I/II | Chinese PLA General Hospital, China | Recruiting |
| NCT03081715 | PD‐1 knockout engineered T cells for advanced esophageal cancer | recurrent or metastatic esophageal cancer | PD‐1 | Autologous T‐cells | Phase II | Hangzhou Cancer Hospital, China | Completed |
| NCT02793856 | PD‐1 knockout engineered T cells for metastatic non‐small cell lung cancer | Stage IV non‐small cell lung cancer | PD‐1 | Autologous T‐cells | Phase I | Sichuan University, China | Active, not recruiting |
| NCT03545815 | Study of CRISPR-Cas9 mediated PD-1 and TCR gene-knocked out mesothelin-directed CAR-T cells in patients with mesothelin-positive multiple solid tumors. | Solid tumor, adult | PD1 and TCR | CRISPR–Cas9-treated CAR T cell infusions (ex vivo) | Phase I | Chinese PLA General Hospital, China | Recruiting |
| NCT03399448 | NY-ESO-1-redirected CRISPR (TCRendo and PD1) edited T cells (NYCE T Cells) | Multiple myeloma, melanoma, synovial sarcoma | PD-1 and TCR | CRISPR–Cas9-treated autologous T cells (ex vivo), combined with chemotherapy agents | Phase I | University of Pennsylvania, USA | Terminated |
| NCT04037566 | CRISPR (HPK1)-edited CD19-specific CAR-T cells (XYF19 CAR-T cells) for CD19 + leukemia or lymphoma | CD19-positive leukemia, lymphoma | HPK1 | CRISPR–Cas9-treated autologous CD19-directed T cells | Phase I | Xijing Hospital, China | Recruiting |
| NCT03164135 | Safety of transplantation of CRISPR CCR5-modified CD34 + cells in HIV subjects with hematological malignancy | HIV-1-infection | CCR5 | Transplantation of CRISPR–Cas9-treated CD34 + HSPCs (ex vivo) | N/A | Academy of Military Medical Sciences, China | Recruiting |
| NCT03057912 | Safety and efficacy study of TALEN and CRISPR/Cas9 in treatment of HPV-related cervical intraepithelial neoplasiaⅠ | HPV-related malignant neoplasm | HPV E6/E7, 16 and 18 | CRISPR/Cas9-HPV18 E6/ E7T2 | Phase I | Sun Yat-Sen University | Not yet recruiting |
| NCT03855631 | Exploiting epigenome editing in Kabuki Syndrome: a new route toward gene therapy for rare genetic disorders (Epi-KAB) | Kabuki syndrome 1 | KMT2D | Unknown (ex vivo) | N/A | University Hospital, Montpellier, France | Active, not recruiting |
available at https://clinicaltrials.gov/
6. Concluding remarks
CRISPR/Cas systems have been efficiently used to generate disease models, and great strides have been made toward clinical applications. Research evidence demonstrates that genome editing systems such as CRISPR/Cas9 can make significant contributions to therapeutic strategies for various human diseases by directly interfering with target genes or deriving multifunctional tools. The news of the first CRISPR-gene-edited babies in China back in 2018 shocked the scientific world. That work, conducted by He Jiankui et al., was a direct violation of the international scientific consensus that gene editing using CRISPR/Cas9 is still in its infancy and is not ready to use in germline modifications of humans, which could be passed through generations [247]. However, CRISPR technologies have progressed ethically toward clinical trials. Development of CRISPR/Cas9 genome editing technology has enabled therapeutic application of CAR-T therapy through targeted intervention of endogenous genes [248]. Engineered CAR-T cells called iCARTs can be generated from patient-derived iPSCs with HLA-independent customizable antigen recognition, and they have been shown to kill cancer cells [248], [249], [250]. Together, these studies show that patient-derived iPSCs or hESCs used to generate NK cells in combination with CRISPR/Cas9-mediated gene modification could be an efficient cancer immunotherapy. Such studies provide hope that CRISPR will advance as an off-the-shelf cell therapeutic approach.
Although hindered by drawbacks such as potential off-target effects, Cas9 immunity, and HDR efficiency for gene correction, this genome editing technology is progressing toward clinical application. Much research is being conducted to tackle such drawbacks; for example, the specificity of CRISPR can be improved by modifying Cas9 construction [251], optimizing sgRNA designs [97], [252], or using Cas9 mRNA for transient Cas9 expression [44]. The efficiency of HDR has been improved utilizing single-stranded donors [253], [254] and small molecules [14], [16], varying the lengths of homologous arms [255], or using modified double-stranded donors [256]. The immunogenicity of CRISPR/Cas9 can be addressed utilizing non-viral delivery systems such as nanoparticles or lipid-based vectors [257], [258]. Deeper exploration of this genome editing technology has the potential to uncover fundamental biological mechanisms that drive many diseases and provide permanent cures for previously untreatable diseases. The compatibility of CRISPR systems with various biological systems, including stem cells, makes them an incredible tool. However, use of stem cells is surrounded by controversies including the ethical issues surrounding embryonic stem cells (ESCs) and somatic cell nuclear transfer (SCNTs), carcinogenicity, and genetic instability factors surrounding iPSCs [146], [259], [260]. iPSCs are not hindered by ethical issues, and progress is being made toward derivation of clinically relevant “safe” iPSCs. Efforts are being made to replace the oncogenic genes in the reprogramming cocktail that typically include Oct4, Sox2, Nanog, and c-Myc with non-oncogenic genes such as Nkx3.1 or by eliminating them completely [261], [262], [263], [264], [265]. Gene-corrected iPSCs could be differentiated into specific cell types, and such “safe” stem cell lines could be developed as universal donor stem cells that can be transplanted into any patient and can be realistically achieved using CRISPR systems to revolutionize the future of regenerative medicin.
CRediT authorship contribution statement
Ainsley Mike Antao: Conceptualization, Software, Writing - original draft. Janardhan Keshav Karapurkar: Conceptualization, Software, Writing - original draft. Dong Ryul Lee: Writing - review & editing. Kye-Seong Kim: Supervision, Writing - review & editing. Suresh Ramakrishna: Conceptualization, Funding acquisition, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank all the members of Suri’s laboratory and Kye-Seong Kim’s laboratory for their helpful insights. This work was supported by a grant from the National Research Foundation of Korea (NRF) (2017M3A9C6061361).
Contributor Information
Kye-Seong Kim, Email: ks66kim@hanyang.ac.kr.
Suresh Ramakrishna, Email: suri28@hanyang.ac.kr.
References
- 1.Kim H., Kim J.S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 2.Maggio I., Gonçalves M.A. Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol. 2015;33:280–291. doi: 10.1016/j.tibtech.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Sürün D., Schwäble J., Tomasovic A., Ehling R., Stein S., Kurrle N. High efficiency gene correction in hematopoietic cells by donor-template-free CRISPR/Cas9 genome editing, molecular therapy. Nucleic Acids. 2018;10:1–8. doi: 10.1016/j.omtn.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Román-Rodríguez F.J., Ugalde L., Álvarez L., Díez B., Ramírez M.J., Risueño C. NHEJ-mediated repair of CRISPR-Cas9-induced DNA breaks efficiently corrects mutations in HSPCs from patients with fanconi anemia. Cell Stem Cell. 2019;25:607–621.e607. doi: 10.1016/j.stem.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang D., Scavuzzo M.A., Chmielowiec J., Sharp R., Bajic A., Borowiak M. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci Rep. 2016;6:21264. doi: 10.1038/srep21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh P., Schimenti J.C., Bolcun-Filas E. A mouse geneticist's practical guide to CRISPR applications. Genetics. 2015;199:1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 10.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava M., Nambiar M., Sharma S., Karki S.S., Goldsmith G., Hegde M. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 12.Hu Z., Shi Z., Guo X., Jiang B., Wang G., Luo D. Ligase IV inhibitor SCR7 enhances gene editing directed by CRISPR-Cas9 and ssODN in human cancer cells. Cell Biosci. 2018;8:12. doi: 10.1186/s13578-018-0200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y., Chen W., Zhang X., Yu L., Dong W., Pan S. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing in rats by inhibiting NHEJ and using Cas9 protein. RNA Biol. 2016;13:605–612. doi: 10.1080/15476286.2016.1185591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G., Zhang X., Zhong C., Mo J., Quan R., Yang J. Small molecules enhance CRISPR/Cas9-mediated homology-directed genome editing in primary cells. Sci Rep. 2017;7:8943. doi: 10.1038/s41598-017-09306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canny M.D., Moatti N., Wan L.C.K., Fradet-Turcotte A., Krasner D., Mateos-Gomez P.A. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat Biotechnol. 2018;36:95–102. doi: 10.1038/nbt.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riesenberg S., Maricic T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat Commun. 2018;9:2164. doi: 10.1038/s41467-018-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weterings E., Gallegos A.C., Dominick L.N., Cooke L.S., Bartels T.N., Vagner J., Matsunaga T.O., Mahadevan D. A novel small molecule inhibitor of the DNA repair protein Ku70/80. DNA repair. 2016;43:98–106. doi: 10.1016/j.dnarep.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Robert F., Barbeau M., Éthier S., Dostie J., Pelletier J. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med. 2015;7:93. doi: 10.1186/s13073-015-0215-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shahar O.D., Kalousi A., Eini L., Fisher B., Weiss A., Darr J. A high-throughput chemical screen with FDA approved drugs reveals that the antihypertensive drug Spironolactone impairs cancer cell survival by inhibiting homology directed repair. Nucleic Acids Res. 2014;42:5689–5701. doi: 10.1093/nar/gku217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J.P., Li X.L., Li G.H., Chen W., Arakaki C., Botimer G.D. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017;18:35. doi: 10.1186/s13059-017-1164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aksoy Y.A., Nguyen D.T., Chow S., Chung R.S., Guillemin G.J., Cole N.J. Chemical reprogramming enhances homology-directed genome editing in zebrafish embryos. Commun Biol. 2019;2:198. doi: 10.1038/s42003-019-0444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X., Chen X., Jin Y., Ge W., Wang W., Kong L. Small molecules promote CRISPR-Cpf1-mediated genome editing in human pluripotent stem cells. Nat Commun. 2018;9:1303. doi: 10.1038/s41467-018-03760-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riesenberg S., Chintalapati M., Macak D., Kanis P., Maricic T., Pääbo S. Simultaneous precise editing of multiple genes in human cells. Nucleic Acids Res. 2019;47:e116. doi: 10.1093/nar/gkz669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J., Yang D., Xu J., Zhu T., Chen Y.E., Zhang J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun. 2016;7:10548. doi: 10.1038/ncomms10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinder J., Salsman J., Dellaire G. Nuclear domain 'knock-in' screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015;43:9379–9392. doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamas-Toranzo I., Martínez-Moro A., E O.C., Millán-Blanca G., Sánchez J.M., Lonergan P., Bermejo-Álvarez P. RS-1 enhances CRISPR-mediated targeted knock-in in bovine embryos. Mol Reprod Dev. 2020;87:542–549. doi: 10.1002/mrd.23341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeon I.S., Shin J.C., Kim S.R., Park K.S., Yoo H.J., Lee K.Y. Role of RS-1 derivatives in homology-directed repair at the human genome ATG5 locus. Arch Pharmacal Res. 2020;43:639–645. doi: 10.1007/s12272-020-01226-1. [DOI] [PubMed] [Google Scholar]
- 28.Wienert B., Nguyen D.N., Guenther A., Feng S.J., Locke M.N., Wyman S.K. Timed inhibition of CDC7 increases CRISPR-Cas9 mediated templated repair. Nat Commun. 2020;11:2109. doi: 10.1038/s41467-020-15845-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X.L., Li G.H., Fu J., Fu Y.W., Zhang L., Chen W. Highly efficient genome editing via CRISPR-Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res. 2018;46:10195–10215. doi: 10.1093/nar/gky804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takayama K., Igai K., Hagihara Y., Hashimoto R., Hanawa M., Sakuma T. Highly efficient biallelic genome editing of human ES/iPS cells using a CRISPR/Cas9 or TALEN system. Nucleic Acids Res. 2017;45:5198–5207. doi: 10.1093/nar/gkx130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu C., Liu Y., Ma T., Liu K., Xu S., Zhang Y. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J., McLachlan A., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y.-G., Cha J., Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christian M., Cermak T., Doyle E.L., Schmidt C., Zhang F., Hummel A. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaj T., Gersbach C.A., Barbas C.F. 3rd, ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiedenheft B., Sternberg S.H., Doudna J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 39.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science (New York, NY) 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 41.Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 42.Jiang F., Doudna J.A. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol. 2015;30:100–111. doi: 10.1016/j.sbi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E. RNA-guided human genome engineering via Cas9. Science (New York, N.Y.) 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garneau J.E., Dupuis M., Villion M., Romero D.A., Barrangou R., Boyaval P. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 47.Xie H., Tang L., He X., Liu X., Zhou C., Liu J. SaCas9 requires 5'-NNGRRT-3' PAM for sufficient cleavage and possesses higher cleavage activity than SpCas9 or FnCpf1 in human cells. Biotechnol J. 2018;13:e1700561. doi: 10.1002/biot.201700561. [DOI] [PubMed] [Google Scholar]
- 48.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao L., Cox D.B.T., Yan W.X., Manteiga J.C., Schneider M.W., Yamano T. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017;35:789–792. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Magadán A.H., Dupuis M., Villion M., Moineau S. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS ONE. 2012;7:e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y., Heidrich N., Ampattu B.J., Gunderson C.W., Seifert H.S., Schoen C. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 55.Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S., Kaplan M., Iavarone A.T., Charpentier E., Nogales E., Doudna J.A. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science (New York, N.Y.) 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biertümpfel C., Yang W., Suck D. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature. 2007;449:616–620. doi: 10.1038/nature06152. [DOI] [PubMed] [Google Scholar]
- 58.Li C.L., Hor L.I., Chang Z.F., Tsai L.C., Yang W.Z., Yuan H.S. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. The EMBO journal. 2003;22:4014–4025. doi: 10.1093/emboj/cdg377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao L.N., Mondal D., Warshel A. Exploring alternative catalytic mechanisms of the Cas9 HNH domain. Proteins. 2020;88:260–264. doi: 10.1002/prot.25796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu X., Scott D.A., Kriz A.J., Chiu A.C., Hsu P.D., Dadon D.B. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang F., Zhou K., Ma L., Gressel S., Doudna J.A. A Cas9–guide RNA complex preorganized for target DNA recognition. Science (New York, N.Y.) 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- 63.Knight S.C., Xie L., Deng W., Guglielmi B., Witkowsky L.B., Bosanac L., Zhang E.T., El Beheiry M., Masson J.B., Dahan M., Liu Z., Doudna J.A., Tjian R. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science (New York, N.Y.) 2015;350:823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- 64.Ma H., Tu L.C., Naseri A., Huisman M., Zhang S., Grunwald D. CRISPR-Cas9 nuclear dynamics and target recognition in living cells. J Cell Biol. 2016;214:529–537. doi: 10.1083/jcb.201604115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szczelkun M.D., Tikhomirova M.S., Sinkunas T., Gasiunas G., Karvelis T., Pschera P., Siksnys V., Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang F., Doudna J.A. CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 68.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu L.J. Overview of guide RNA design tools for CRISPR-Cas9 genome editing technology. Front. Biol. 2015;10:289–296. [Google Scholar]
- 71.Singh D., Sternberg S.H., Fei J., Doudna J.A., Ha T. Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat Commun. 2016;7:12778. doi: 10.1038/ncomms12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., Alt F.W. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. 231 p following 243. [DOI] [PubMed] [Google Scholar]
- 75.Cameron P., Fuller C.K., Donohoue P.D., Jones B.N., Thompson M.S., Carter M.M. Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat Methods. 2017;14:600–606. doi: 10.1038/nmeth.4284. [DOI] [PubMed] [Google Scholar]
- 76.Tsai S.Q., Nguyen N.T., Malagon-Lopez J., Topkar V.V., Aryee M.J., Joung J.K. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 2017;14:607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wienert B., Wyman S.K., Richardson C.D., Yeh C.D., Akcakaya P., Porritt M.J. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science (New York, N.Y.) 2019;364:286–289. doi: 10.1126/science.aav9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Q., He D., Xie L. Prediction of off-target specificity and cell-specific fitness of CRISPR-Cas System using attention boosted deep learning and network-based gene feature. PLoS Comput Biol. 2019;15:e1007480. doi: 10.1371/journal.pcbi.1007480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oliveros J.C., Franch M., Tabas-Madrid D., San-León D., Montoliu L., Cubas P. Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res. 2016;44:W267–W271. doi: 10.1093/nar/gkw407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hwang G.-H., Park J., Lim K., Kim S., Yu J., Yu E. Web-based design and analysis tools for CRISPR base editing. BMC Bioinf. 2018;19:542. doi: 10.1186/s12859-018-2585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park J., Bae S., Kim J.-S. Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics. 2015;31:4014–4016. doi: 10.1093/bioinformatics/btv537. [DOI] [PubMed] [Google Scholar]
- 86.Bae S., Park J., Kim J.-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Concordet J.-P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.-B. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stemmer M., Thumberger T., del Sol Keyer M., Wittbrodt J., Mateo J.L. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu H., Wei Z., Dominguez A., Li Y., Wang X., Qi L.S. CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics (Oxford, England) 2015;31:3676–3678. doi: 10.1093/bioinformatics/btv423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Naito Y., Hino K., Bono H., Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2014;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pulido-Quetglas C., Aparicio-Prat E., Arnan C., Polidori T., Hermoso T., Palumbo E. Scalable design of paired CRISPR guide RNAs for genomic deletion. PLoS Comput Biol. 2017;13:e1005341. doi: 10.1371/journal.pcbi.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu L.J., Holmes B.R., Aronin N., Brodsky M.H. CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PloS one. 2014;9:e108424. doi: 10.1371/journal.pone.0108424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xie X., Ma X., Zhu Q., Zeng D., Li G., Liu Y.-G. CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Mol Plant. 2017;10:1246–1249. doi: 10.1016/j.molp.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 96.Heigwer F., Zhan T., Breinig M., Winter J., Brügemann D., Leible S. CRISPR library designer (CLD): software for multispecies design of single guide RNA libraries. Genome Biol. 2016;17:55. doi: 10.1186/s13059-016-0915-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abadi S., Yan W.X., Amar D., Mayrose I. A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput Biol. 2017;13:e1005807. doi: 10.1371/journal.pcbi.1005807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alkan F., Wenzel A., Anthon C., Havgaard J.H., Gorodkin J. CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol. 2018;19:177. doi: 10.1186/s13059-018-1534-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xiao A., Cheng Z., Kong L., Zhu Z., Lin S., Gao G. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics. 2014;30:1180–1182. doi: 10.1093/bioinformatics/btt764. [DOI] [PubMed] [Google Scholar]
- 101.Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G. COSMID: a web-based tool for identifying and validating CRISPR/Cas off-target sites, molecular therapy. Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singh R., Kuscu C., Quinlan A., Qi Y., Adli M. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 2015;43:e118. doi: 10.1093/nar/gkv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Prykhozhij S.V., Rajan V., Gaston D., Berman J.N. Correction: CRISPR multitargeter: a web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PLoS ONE. 2015;10:e0138634. doi: 10.1371/journal.pone.0119372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mendoza B.J., Trinh C.T. Enhanced guide-RNA design and targeting analysis for precise CRISPR genome editing of single and consortia of industrially relevant and non-model organisms. Bioinformatics (Oxford, England) 2018;34:16–23. doi: 10.1093/bioinformatics/btx564. [DOI] [PubMed] [Google Scholar]
- 105.Blin K., Pedersen L.E., Weber T., Lee S.Y. CRISPy-web: an online resource to design sgRNAs for CRISPR applications. Synth Syst Biotechnol. 2016;1:118–121. doi: 10.1016/j.synbio.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Minkenberg B., Zhang J., Xie K., Yang Y. CRISPR-PLANT v2: an online resource for highly specific guide RNA spacers based on improved off-target analysis. Plant Biotechnol J. 2019;17:5. doi: 10.1111/pbi.13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie K., Zhang J., Yang Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR–Cas9-mediated genome editing in model plants and major crops. Mol Plant. 2014;7:923–926. doi: 10.1093/mp/ssu009. [DOI] [PubMed] [Google Scholar]
- 108.Chuai G., Ma H., Yan J., Chen M., Hong N., Xue D. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018;19:80. doi: 10.1186/s13059-018-1459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Heigwer F., Kerr G., Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 110.Peng D., Tarleton R. EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb Genomics. 2015;1 doi: 10.1099/mgen.0.000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Listgarten J., Weinstein M., Kleinstiver B.P., Sousa A.A., Joung J.K., Crawford J. Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat Biomed Eng. 2018;2:38–47. doi: 10.1038/s41551-017-0178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McKenna A., Shendure J. FlashFry: a fast and flexible tool for large-scale CRISPR target design. BMC Biol. 2018;16:74. doi: 10.1186/s12915-018-0545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Perez A.R., Pritykin Y., Vidigal J.A., Chhangawala S., Zamparo L., Leslie C.S. GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol. 2017;35:347–349. doi: 10.1038/nbt.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.O'Brien A., Bailey T.L. GT-Scan: identifying unique genomic targets. Bioinformatics (Oxford, England) 2014;30:2673–2675. doi: 10.1093/bioinformatics/btu354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.MacPherson C.R., Scherf A. Flexible guide-RNA design for CRISPR applications using Protospacer Workbench. Nat Biotechnol. 2015;33:805–806. doi: 10.1038/nbt.3291. [DOI] [PubMed] [Google Scholar]
- 116.Sanson K.R., Hanna R.E., Hegde M., Donovan K.F., Strand C., Sullender M.E. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun. 2018;9:5416. doi: 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Duan J., Lu G., Xie Z., Lou M., Luo J., Guo L. Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Res. 2014;24:1009–1012. doi: 10.1038/cr.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chari R., Mali P., Moosburner M., Church G.M. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods. 2015;12:823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rahman M.K., Rahman M.S. CRISPRpred: a flexible and efficient tool for sgRNAs on-target activity prediction in CRISPR/Cas9 systems. PLoS ONE. 2017;12:e0181943. doi: 10.1371/journal.pone.0181943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wong N., Liu W., Wang X. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015;16:218. doi: 10.1186/s13059-015-0784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xu H., Xiao T., Chen C.H., Li W., Meyer C.A., Wu Q. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chari R., Yeo N.C., Chavez A., Church G.M. sgRNA scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity. ACS Synth Biol. 2017;6:902–904. doi: 10.1021/acssynbio.6b00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chuai G.H., Wang Q.L., Liu Q. In silico meets in vivo: towards computational CRISPR-based sgRNA design. Trends Biotechnol. 2017;35:12–21. doi: 10.1016/j.tibtech.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 125.Bae S., Kweon J., Kim H.S., Kim J.S. Microhomology-based choice of Cas9 nuclease target sites. Nat Methods. 2014;11:705–706. doi: 10.1038/nmeth.3015. [DOI] [PubMed] [Google Scholar]
- 126.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science (New York, N.Y.) 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hiranniramol K., Chen Y., Liu W., Wang X. Generalizable sgRNA design for improved CRISPR/Cas9 editing efficiency. Bioinformatics. 2020;36:2684–2689. doi: 10.1093/bioinformatics/btaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pickar-Oliver A., Gersbach C.A. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Edraki A., Mir A., Ibraheim R., Gainetdinov I., Yoon Y., Song C.Q. High-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol Cell. 2019;73:714–726.e714. doi: 10.1016/j.molcel.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Strecker J., Jones S., Koopal B., Schmid-Burgk J., Zetsche B., Gao L. Engineering of CRISPR-Cas12b for human genome editing. Nat Commun. 2019;10:212. doi: 10.1038/s41467-018-08224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Smargon A.A., Cox D.B.T., Pyzocha N.K., Zheng K., Slaymaker I.M., Gootenberg J.S. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65:618–630.e617. doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Abudayyeh O.O., Gootenberg J.S., Franklin B., Koob J., Kellner M.J., Ladha A., Joung J., Kirchgatterer P., Cox D.B.T., Zhang F. A cytosine deaminase for programmable single-base RNA editing. Science (New York, N.Y.) 2019;365:382–386. doi: 10.1126/science.aax7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J. RNA editing with CRISPR-Cas13. Science (New York, N.Y.) 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Grünewald J., Zhou R., Iyer S., Lareau C.A., Garcia S.P., Aryee M.J. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat Biotechnol. 2019;37:1041–1048. doi: 10.1038/s41587-019-0236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mironov V., Visconti R.P., Markwald R.R. What is regenerative medicine? Emergence of applied stem cell and developmental biology. Expert Opin Biol Therapy. 2004;4:773–781. doi: 10.1517/14712598.4.6.773. [DOI] [PubMed] [Google Scholar]
- 141.Park I.H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Grimm S. The art and design of genetic screens: mammalian culture cells. Nat Rev Genet. 2004;5:179–189. doi: 10.1038/nrg1291. [DOI] [PubMed] [Google Scholar]
- 143.Yang G., Huang X. Methods and applications of CRISPR/Cas system for genome editing in stem cells. Cell Regeneration (London, England) 2019;8:33–41. doi: 10.1016/j.cr.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lowry W.E., Richter L., Yachechko R., Pyle A.D., Tchieu J., Sridharan R., Clark A.T., Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Murry C.E., Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 146.Deinsberger J., Reisinger D., Weber B. Global trends in clinical trials involving pluripotent stem cells: a systematic multi-database analysis. NPJ Regener Med. 2020;5:15. doi: 10.1038/s41536-020-00100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schweitzer J.S., Song B., Herrington T.M., Park T.-Y., Lee N., Ko S. Personalized iPSC-derived dopamine progenitor cells for Parkinson’s disease. N Engl J Med. 2020;382:1926–1932. doi: 10.1056/NEJMoa1915872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Arias-Fuenzalida J., Jarazo J., Qing X., Walter J., Gomez-Giro G., Nickels S.L. FACS-assisted CRISPR-Cas9 genome editing facilitates parkinson's disease modeling. Stem Cell Rep. 2017;9:1423–1431. doi: 10.1016/j.stemcr.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang S., Min Z., Ji Q., Geng L., Su Y., Liu Z. Rescue of premature aging defects in Cockayne syndrome stem cells by CRISPR/Cas9-mediated gene correction. Protein Cell. 2020;11:1–22. doi: 10.1007/s13238-019-0623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhao C., Devlin A.C., Chouhan A.K., Selvaraj B.T., Stavrou M., Burr K. Mutant C9orf72 human iPSC-derived astrocytes cause non-cell autonomous motor neuron pathophysiology. Glia. 2020;68:1046–1064. doi: 10.1002/glia.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bragança J., Lopes J.A., Mendes-Silva L., Almeida Santos J.M. Induced pluripotent stem cells, a giant leap for mankind therapeutic applications. World J Stem Cells. 2019;11:421–430. doi: 10.4252/wjsc.v11.i7.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 153.Leite N.C., Sintov E., Meissner T.B., Brehm M.A., Greiner D.L., Harlan D.M. Modeling type 1 diabetes in vitro using human pluripotent stem cells. Cell Rep. 2020;32:107894. doi: 10.1016/j.celrep.2020.107894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ebert A.D., Yu J., Rose F.F., Jr., Mattis V.B., Lorson C.L., Thomson J.A. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tian R., Gachechiladze M.A., Ludwig C.H., Laurie M.T., Hong J.Y., Nathaniel D. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron. 2019;104:239–255.e212. doi: 10.1016/j.neuron.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Soldner F., Hockemeyer D., Beard C., Gao Q., Bell G.W., Cook E.G. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ke M., Chong C.-M., Su H. Using induced pluripotent stem cells for modeling Parkinson's disease. World J Stem Cells. 2019;11:634–649. doi: 10.4252/wjsc.v11.i9.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lim R.G., Quan C., Reyes-Ortiz A.M., Lutz S.E., Kedaigle A.J., Gipson T.A. Huntington's disease iPSC-derived brain microvascular endothelial cells reveal WNT-mediated angiogenic and blood-brain barrier deficits. Cell Rep. 2017;19:1365–1377. doi: 10.1016/j.celrep.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Freude K., Pires C., Hyttel P., Hall V.J. Induced pluripotent stem cells derived from Alzheimer's disease patients: the promise, the hope and the path ahead. J Clin Med. 2014;3:1402–1436. doi: 10.3390/jcm3041402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee J., Kim Y., Yi H., Diecke S., Kim J., Jung H. Generation of disease-specific induced pluripotent stem cells from patients with rheumatoid arthritis and osteoarthritis. Arthritis Res Therapy. 2014;16:R41. doi: 10.1186/ar4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kudva Y.C., Ohmine S., Greder L.V., Dutton J.R., Armstrong A., De Lamo J.G. Transgene-free disease-specific induced pluripotent stem cells from patients with type 1 and type 2 diabetes. Stem Cells Transl Med. 2012;1:451–461. doi: 10.5966/sctm.2011-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sterneckert J.L., Reinhardt P., Schöler H.R. Investigating human disease using stem cell models. Nat Rev Genet. 2014;15:625–639. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- 163.Bassett A.R. Editing the genome of hiPSC with CRISPR/Cas9: disease models. Mamm Genome. 2017;28:348–364. doi: 10.1007/s00335-017-9684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cai L., Fisher A.L., Huang H., Xie Z. CRISPR-mediated genome editing and human diseases. Genes Dis. 2016;3:244–251. doi: 10.1016/j.gendis.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kajiwara M., Aoi T., Okita K., Takahashi R., Inoue H., Takayama N., Endo H., Eto K., Toguchida J., Uemoto S., Yamanaka S. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12538–12543. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Merkert S., Martin U. Site-specific genome engineering in human pluripotent stem cells. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17071000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Poduri A., Evrony G.D., Cai X., Walsh C.A. Somatic mutation, genomic variation, and neurological disease. Science (New York, N.Y.) 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dow L.E. Modeling disease in vivo with CRISPR/Cas9. Trends Mol Med. 2015;21:609–621. doi: 10.1016/j.molmed.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Birling M.C., Herault Y., Pavlovic G. Modeling human disease in rodents by CRISPR/Cas9 genome editing. Mamm Genome. 2017;28:291–301. doi: 10.1007/s00335-017-9703-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pritchard C.E.J., Kroese L.J., Huijbers I.J. Direct generation of conditional alleles using CRISPR/Cas9 in mouse zygotes. Methods Mol Biol. 2017;1642:21–35. doi: 10.1007/978-1-4939-7169-5_2. [DOI] [PubMed] [Google Scholar]
- 171.Mingozzi F., High K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 172.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Carroll D. A CRISPR approach to gene targeting. Mol Ther. 2012;20:1658–1660. doi: 10.1038/mt.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yang X. Applications of CRISPR-Cas9 mediated genome engineering. Mil Med Res. 2015;2:11. doi: 10.1186/s40779-015-0038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Akkaya M., Bansal A., Sheehan P.W., Pena M., Cimperman C.K., Qi C.F. Testing the impact of a single nucleotide polymorphism in a Plasmodium berghei ApiAP2 transcription factor on experimental cerebral malaria in mice. Sci Rep. 2020;10:13630. doi: 10.1038/s41598-020-70617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lee H.K., Willi M., Smith H.E., Miller S.M., Liu D.R., Liu C. Simultaneous targeting of linked loci in mouse embryos using base editing. Sci Rep. 2019;9:1662. doi: 10.1038/s41598-018-33533-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhu Q.M., Ko K.A., Ture S., Mastrangelo M.A., Chen M.H., Johnson A.D. Novel thrombotic function of a human SNP in STXBP5 revealed by CRISPR/Cas9 gene editing in mice. Arterioscler Thromb Vasc Biol. 2017;37:264–270. doi: 10.1161/ATVBAHA.116.308614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ahmad G., Amiji M. Use of CRISPR/Cas9 gene-editing tools for developing models in drug discovery. Drug Discovery Today. 2018;23:519–533. doi: 10.1016/j.drudis.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 179.Dow L.E., Fisher J., O'Rourke K.P., Muley A., Kastenhuber E.R., Livshits G. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015;33:390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Quadros R.M., Miura H., Harms D.W., Akatsuka H., Sato T., Aida T. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 2017;18:92. doi: 10.1186/s13059-017-1220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Flisikowska T., Merkl C., Landmann M., Eser S., Rezaei N., Cui X., Kurome M., Zakhartchenko V., Kessler B., Wieland H. A porcine model of familial adenomatous polyposis. Gastroenterology. 2012;143:1173–1175. doi: 10.1053/j.gastro.2012.07.110. e1177. [DOI] [PubMed] [Google Scholar]
- 182.Rogers C.S., Stoltz D.A., Meyerholz D.K., Ostedgaard L.S., Rokhlina T., Taft P.J., Rogan M.P., Pezzulo A.A., Karp P.H., Itani O.A. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science (New York, N.Y.) 2008;321:1837–1841. doi: 10.1126/science.1163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhou X., Xin J., Fan N., Zou Q., Huang J., Ouyang Z. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci. 2015;72:1175–1184. doi: 10.1007/s00018-014-1744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sykes M., Sachs D.H. Transplanting organs from pigs to humans. Science Immunology. 2019;4:eaau6298. doi: 10.1126/sciimmunol.aau6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Niu D., Wei H.-J., Lin L., George H., Wang T., Lee I.-H. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science (New York, N.Y.) 2017;357:1303–1307. doi: 10.1126/science.aan4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wu J., Vilarino M., Suzuki K., Okamura D., Bogliotti Y.S., Park I. CRISPR-Cas9 mediated one-step disabling of pancreatogenesis in pigs. Sci Rep. 2017;7:10487. doi: 10.1038/s41598-017-08596-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Li P., Estrada J.L., Burlak C., Montgomery J., Butler J.R., Santos R.M. Efficient generation of genetically distinct pigs in a single pregnancy using multiplexed single-guide RNA and carbohydrate selection. Xenotransplantation. 2015;22:20–31. doi: 10.1111/xen.12131. [DOI] [PubMed] [Google Scholar]
- 188.Birling M.C., Schaeffer L., André P., Lindner L., Maréchal D., Ayadi A. Efficient and rapid generation of large genomic variants in rats and mice using CRISMERE. Sci Rep. 2017;7:43331. doi: 10.1038/srep43331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Boroviak K., Doe B., Banerjee R., Yang F., Bradley A. Chromosome engineering in zygotes with CRISPR/Cas9. Genesis. 2016;54:78–85. doi: 10.1002/dvg.22915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kraft K., Geuer S., Will Anja J., Chan Wing L., Paliou C., Borschiwer M. Engineering of Structural Variants using CRISPR/Cas in Mice. Cell Rep. 2015;10:833–839. doi: 10.1016/j.celrep.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 191.Zhang L., Jia R., Palange N.J., Satheka A.C., Togo J., An Y., Humphrey M., Ban L., Ji Y., Jin H., Feng X., Zheng Y. Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PloS one. 2015;10:e0120396. doi: 10.1371/journal.pone.0120396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sánchez-Rivera F.J., Papagiannakopoulos T., Romero R., Tammela T., Bauer M.R., Bhutkar A. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science (New York, N.Y.) 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 194.Dutta D., Heo I., Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 195.Driehuis E., Clevers H. CRISPR/Cas 9 genome editing and its applications in organoids. Am J Physiol-Gastrointestinal Liver Physiol. 2017;312:G257–G265. doi: 10.1152/ajpgi.00410.2016. [DOI] [PubMed] [Google Scholar]
- 196.Huang C.-H., Lee K.-C., Doudna J.A. Applications of CRISPR-Cas enzymes in cancer therapeutics and detection. Trends Cancer. 2018;4:499–512. doi: 10.1016/j.trecan.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lokody I. Genetic therapies: correcting genetic defects with CRISPR–Cas9. Nat Rev Genet. 2013;15:63. doi: 10.1038/nrg3656. [DOI] [PubMed] [Google Scholar]
- 198.Schwank G., Koo B.-K., Sasselli V., Dekkers J.F., Heo I., Demircan T. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 199.Firth A.L., Menon T., Parker G.S., Qualls S.J., Lewis B.M., Ke E. Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Rep. 2015;12:1385–1390. doi: 10.1016/j.celrep.2015.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Freedman B.S., Brooks C.R., Lam A.Q., Fu H., Morizane R., Agrawal V. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun. 2015;6:8715. doi: 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.van Adelsberg J.S., Frank D. The PKD1 gene produces a developmentally regulated protein in mesenchyme and vasculature. Nat Med. 1995;1:359–364. doi: 10.1038/nm0495-359. [DOI] [PubMed] [Google Scholar]
- 203.Morizane R., Bonventre J.V. Kidney organoids: a translational journey. Trends Mol Med. 2017;23:246–263. doi: 10.1016/j.molmed.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.García M.S.F., Teruya-Feldstein J. The diagnosis and treatment of dyskeratosis congenita: a review. J Blood Med. 2014;5:157. doi: 10.2147/JBM.S47437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Maxwell K.G., Augsornworawat P., Velazco-Cruz L., Kim M.H., Asada R., Hogrebe N.J. Gene-edited human stem cell–derived β cells from a patient with monogenic diabetes reverse preexisting diabetes in mice. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.aax9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Omer Javed A., Li Y., Muffat J., Su K.-C., Cohen M.A., Lungjangwa T. Microcephaly modeling of kinetochore mutation reveals a brain-specific phenotype. Cell Rep. 2018;25:368–382.e365. doi: 10.1016/j.celrep.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wang P., Mokhtari R., Pedrosa E., Kirschenbaum M., Bayrak C., Zheng D. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol Autism. 2017;8:11. doi: 10.1186/s13229-017-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bigorgne A.E., Farin H.F., Lemoine R., Mahlaoui N., Lambert N., Gil M. TTC7A mutations disrupt intestinal epithelial apicobasal polarity. J Clin Investig. 2014;124:328–337. doi: 10.1172/JCI71471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yin X., Mead B.E., Safaee H., Langer R., Karp J.M., Levy O. Engineering stem cell organoids. Cell Stem Cell. 2016;18:25–38. doi: 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 211.Moradi S., Mahdizadeh H., Šarić T., Kim J., Harati J., Shahsavarani H. Research and therapy with induced pluripotent stem cells (iPSCs): social, legal, and ethical considerations. Stem Cell Res Ther. 2019;10:341. doi: 10.1186/s13287-019-1455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Blazar B.R., Murphy W.J., Abedi M. Advances in graft-versus-host disease biology and therapy. Nat Rev Immunol. 2012;12:443–458. doi: 10.1038/nri3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Quach D.H., Becerra-Dominguez L., Rouce R.H., Rooney C.M. A strategy to protect off-the-shelf cell therapy products using virus-specific T-cells engineered to eliminate alloreactive T-cells. J Transl Med. 2019;17:240. doi: 10.1186/s12967-019-1988-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Geller M.A., Miller J.S. Use of allogeneic NK cells for cancer immunotherapy. Immunotherapy. 2011;3:1445–1459. doi: 10.2217/imt.11.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hoornaert C.J., Le Blon D., Quarta A., Daans J., Goossens H., Berneman Z. Concise review: innate and adaptive immune recognition of allogeneic and xenogeneic cell transplants in the central nervous system. Stem Cells Transl Med. 2017;6:1434–1441. doi: 10.1002/sctm.16-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Koga K., Wang B., Kaneko S. Current status and future perspectives of HLA-edited induced pluripotent stem cells. Inflamm Regeneration. 2020;40:23. doi: 10.1186/s41232-020-00132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Allcock R.J. The major histocompatibility complex: a paradigm for studies of the human genome. Methods Mol Biol. 2012;882:1–7. doi: 10.1007/978-1-61779-842-9_1. [DOI] [PubMed] [Google Scholar]
- 218.Wieczorek M., Abualrous E.T., Sticht J., Álvaro-Benito M., Stolzenberg S., Noé F. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front Immunol. 2017;8 doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Shlomchik W.D. Graft-versus-host disease. Nat Rev Immunol. 2007;7:340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- 220.Morishima Y., Morishima S., Murata M., Arima N., Uchida N., Sugio Y. Impact of homozygous conserved extended HLA haplotype on single cord blood transplantation: lessons for induced pluripotent stem cell banking and transplantation in allogeneic settings. Biol Blood Marrow Transpl. 2020;26:132–138. doi: 10.1016/j.bbmt.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 221.Turner M., Leslie S., Martin Nicholas G., Peschanski M., Rao M., Taylor Craig J. Toward the development of a global induced pluripotent stem cell library. Cell Stem Cell. 2013;13:382–384. doi: 10.1016/j.stem.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 222.Taylor Craig J., Peacock S., Chaudhry Afzal N., Bradley J.A., Bolton Eleanor M. Generating an iPSC Bank for HLA-matched tissue transplantation based on known donor and recipient HLA types. Cell Stem Cell. 2012;11:147–152. doi: 10.1016/j.stem.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 223.Nakatsuji N., Nakajima F., Tokunaga K. HLA-haplotype banking and iPS cells. Nat Biotechnol. 2008;26:739–740. doi: 10.1038/nbt0708-739. [DOI] [PubMed] [Google Scholar]
- 224.Ichise H., Nagano S., Maeda T., Miyazaki M., Miyazaki Y., Kojima H. NK cell alloreactivity against KIR-ligand-mismatched HLA-haploidentical tissue derived from HLA haplotype-homozygous iPSCs. Stem Cell Rep. 2017;9:853–867. doi: 10.1016/j.stemcr.2017.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hong C.H., Sohn H.J., Lee H.J., Cho H.I., Kim T.G. Antigen presentation by individually transferred HLA class i genes in HLA-A, HLA-B, HLA-C null human cell line generated using the multiplex CRISPR-Cas9 system. J Immunother. 2017;40:201–210. doi: 10.1097/CJI.0000000000000176. [DOI] [PubMed] [Google Scholar]
- 226.Riolobos L., Hirata R.K., Turtle C.J., Wang P.-R., Gornalusse G.G., Zavajlevski M. HLA engineering of human pluripotent stem cells. Mol Ther. 2013;21:1232–1241. doi: 10.1038/mt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang D., Quan Y., Yan Q., Morales J.E., Wetsel R.A. Targeted disruption of the β2-microglobulin gene minimizes the immunogenicity of human embryonic stem cells. Stem Cells Transl Med. 2015;4:1234–1245. doi: 10.5966/sctm.2015-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Deuse T., Hu X., Gravina A., Wang D., Tediashvili G., De C. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat Biotechnol. 2019;37:252–258. doi: 10.1038/s41587-019-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mattapally S., Pawlik K.M., Fast V.G., Zumaquero E., Lund F.E., Randall T.D. Human leukocyte antigen class I and II knockout human induced pluripotent stem cell-derived cells: universal donor for cell therapy. J Am Heart Assoc. 2018;7:e010239. doi: 10.1161/JAHA.118.010239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Norbnop P., Ingrungruanglert P., Israsena N., Suphapeetiporn K., Shotelersuk V. Generation and characterization of HLA-universal platelets derived from induced pluripotent stem cells. Sci Rep. 2020;10:8472. doi: 10.1038/s41598-020-65577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Han X., Wang M., Duan S., Franco P.J., Kenty J.H.-R., Hedrick P. Generation of hypoimmunogenic human pluripotent stem cells. Proc Natl Acad Sci. 2019;116:10441–10446. doi: 10.1073/pnas.1902566116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Shi L., Li W., Liu Y., Chen Z., Hui Y., Hao P. Generation of hypoimmunogenic human pluripotent stem cells via expression of membrane-bound and secreted β2m-HLA-G fusion proteins. Stem Cells. 2020;38:1423–1437. doi: 10.1002/stem.3269. [DOI] [PubMed] [Google Scholar]
- 233.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 234.Xu H., Wang B., Ono M., Kagita A., Fujii K., Sasakawa N. Targeted disruption of HLA genes via CRISPR-Cas9 generates iPSCs with enhanced immune compatibility. Cell Stem Cell. 2019;24:566–578.e567. doi: 10.1016/j.stem.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 235.Trounson A., Boyd N.R., Boyd R.L. Toward a universal solution: editing compatibility into pluripotent stem cells. Cell Stem Cell. 2019;24:508–510. doi: 10.1016/j.stem.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 236.Gornalusse G.G., Hirata R.K., Funk S.E., Riolobos L., Lopes V.S., Manske G. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat Biotechnol. 2017;35:765–772. doi: 10.1038/nbt.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Jang Y., Choi J., Park N., Kang J., Kim M., Kim Y. Development of immunocompatible pluripotent stem cells via CRISPR-based human leukocyte antigen engineering. Exp Mol Med. 2019;51:1–11. doi: 10.1038/s12276-018-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J., Hamieh M., Cunanan K.M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ren J., Zhang X., Liu X., Fang C., Jiang S., June C.H. A versatile system for rapid multiplex genome-edited CAR T cell generation. Oncotarget. 2017;8:17002–17011. doi: 10.18632/oncotarget.15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Schumann K., Lin S., Boyer E., Simeonov D.R., Subramaniam M., Gate R.E., Haliburton G.E., Ye C.J., Bluestone J.A., Doudna J.A., Marson A. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci USA. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ren J., Liu X., Fang C., Jiang S., June C.H., Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin Cancer Res J Am Assoc Cancer Res. 2017;23:2255–2266. doi: 10.1158/1078-0432.CCR-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Torikai H., Reik A., Soldner F., Warren E.H., Yuen C., Zhou Y. Toward eliminating HLA class I expression to generate universal cells from allogeneic donors. Blood. 2013;122:1341–1349. doi: 10.1182/blood-2013-03-478255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Stadtmauer E.A., Fraietta J.A., Davis M.M., Cohen A.D., Weber K.L., Lancaster E. CRISPR-engineered T cells in patients with refractory cancer. Science (New York, N.Y.) 2020;367 doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ledford H. CRISPR treatment inserted directly into the body for first time. Nature. 2020;579:185. doi: 10.1038/d41586-020-00655-8. [DOI] [PubMed] [Google Scholar]
- 245.Yu W., Wu Z. In vivo applications of CRISPR-based genome editing in the retina. Front Cell Dev Biol. 2018;6:53. doi: 10.3389/fcell.2018.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Maeder M.L., Stefanidakis M., Wilson C.J., Baral R., Barrera L.A., Bounoutas G.S. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 247.Cyranoski D. The CRISPR-baby scandal: what's next for human gene-editing. Nature. 2019;566:440–442. doi: 10.1038/d41586-019-00673-1. [DOI] [PubMed] [Google Scholar]
- 248.Al Abbar A., Ngai S.C., Nograles N., Alhaji S.Y., Abdullah S. Induced pluripotent stem cells: reprogramming platforms and applications in cell replacement therapy. Biores Open Access. 2020;9:121–136. doi: 10.1089/biores.2019.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Themeli M., Kloss C.C., Ciriello G., Fedorov V.D., Perna F., Gonen M. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat Biotechnol. 2013;31:928–933. doi: 10.1038/nbt.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lei F., Zhao B., Haque R., Xiong X., Budgeon L., Christensen N.D. In vivo programming of tumor antigen-specific T lymphocytes from pluripotent stem cells to promote cancer immunosurveillance. Cancer Res. 2011;71:4742–4747. doi: 10.1158/0008-5472.CAN-11-0359. [DOI] [PubMed] [Google Scholar]
- 251.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 253.Okamoto S., Amaishi Y., Maki I., Enoki T., Mineno J. Highly efficient genome editing for single-base substitutions using optimized ssODNs with Cas9-RNPs. Sci Rep. 2019;9:4811. doi: 10.1038/s41598-019-41121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Bai H., Liu L., An K., Lu X., Harrison M., Zhao Y. CRISPR/Cas9-mediated precise genome modification by a long ssDNA template in zebrafish. BMC Genom. 2020;21:67. doi: 10.1186/s12864-020-6493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yao X., Zhang M., Wang X., Ying W., Hu X., Dai P. Tild-CRISPR allows for efficient and precise gene knockin in mouse and human cells. Dev Cell. 2018;45:526–536.e525. doi: 10.1016/j.devcel.2018.04.021. [DOI] [PubMed] [Google Scholar]
- 256.Yu Y., Guo Y., Tian Q., Lan Y., Yeh H., Zhang M. An efficient gene knock-in strategy using 5′-modified double-stranded DNA donors with short homology arms. Nat Chem Biol. 2020;16:387–390. doi: 10.1038/s41589-019-0432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zuris J.A., Thompson D.B., Shu Y., Guilinger J.P., Bessen J.L., Hu J.H. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.de Wert G., Mummery C. Human embryonic stem cells: research, ethics and policy. Hum Reprod. 2003;18:672–682. doi: 10.1093/humrep/deg143. [DOI] [PubMed] [Google Scholar]
- 260.Lewandowski J., Kurpisz M. Techniques of human embryonic stem cell and induced pluripotent stem cell derivation. Archivum Immunologiae et Therapiae Experimentalis. 2016;64:349–370. doi: 10.1007/s00005-016-0385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wernig M., Meissner A., Cassady J.P., Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 262.Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 263.Seki T., Fukuda K. Methods of induced pluripotent stem cells for clinical application. World J Stem Cells. 2015;7:116–125. doi: 10.4252/wjsc.v7.i1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Mai T., Markov G.J., Brady J.J., Palla A., Zeng H., Sebastiano V. NKX3-1 is required for induced pluripotent stem cell reprogramming and can replace OCT4 in mouse and human iPSC induction. Nat Cell Biol. 2018;20:900–908. doi: 10.1038/s41556-018-0136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Menendez S., Camus S., Herreria A., Paramonov I., Morera L.B., Collado M. Increased dosage of tumor suppressors limits the tumorigenicity of iPS cells without affecting their pluripotency. Aging Cell. 2012;11:41–50. doi: 10.1111/j.1474-9726.2011.00754.x. [DOI] [PubMed] [Google Scholar]