Abstract
Indigenous leafy green vegetable crops provide a promising nutritious alternative for East African agriculture under a changing climate; they are better able to cope with biotic and abiotic stresses than cosmopolitan vegetable crops. To verify our hypothesis that the associated microbiome is involved, we studied archaeal and bacterial communities of four locally popular leafy green crops in Uganda (Bidens pilosa, Solanum scabrum, Abelmoschus esculentus, and Gynandropsis gynandra) and of four plant microhabitats (phyllosphere, root endosphere, rhizosphere, and soil) by complementary analyses of amplicon and isolate libraries. All plants shared an unusually large core microbiome, comprising 18 procaryotic families but primarily consisting of Bacillus, Sphingobium, Comamonadaceae, Pseudomonas, and one archaeon from the soil crenarchaeotic group. Microbiome composition did not differ significantly for plant species but differed for microhabitats. The diversity was, in general, higher for bacteria (27,697 ASVs/H = 6.91) than for archaea (2,995 ASVs/H = 4.91); both groups form a robust network of copiotrophic bacteria and oligotrophic archaea. Screening of selected isolates for stress and plant health protecting traits showed that strains of Bacillus and Sphingomonas spp. div. constituted a substantial portion (15–31%) of the prokaryotic plant-associated communities. Across plant species, microbiota were characterized by a high proportion of potential copiotrophic and plant-beneficial species, which was not specific by plant species. The use of identified plant-beneficial isolates could provide the basis for the development of consortia of isolates for both abiotic and biotic stress protection to improve plant and ecosystem health, ensuring food security in East Africa.
Keywords: plant microbiome, archaea, leafy green vegetables, PGPR, amplicon analysis
Introduction
The adoption of sustainable agriculture practices is necessary in order to feed growing populations, especially in Sub-Saharan Africa, where food insecurity and malnutrition indices are the most alarming globally (Sikora et al., 2019). Sustainable agricultural practices are additionally important for biodiversity and ecosystem services, the maintenance of which is a global challenge and which is attracting increasing attention (Mariotte et al., 2018). Eastern African farming systems comprise a tapestry of crops and livestock, which is dominated by smallholders and characterized by low productivity (Vanlauwe et al., 2014). A key factor explaining this low productivity is the high pest and pathogen pressure, and consequent production losses due to bioconstraints (Venkateswarlu et al., 2012; Dean et al., 2012; CIAT; BFS/USAID, 2017). The application of synthetic, chemical pesticides is a common coping strategy by farmers, but often using inappropriate, adulterated, out of date products, or generic compounds against which resistance has long since built up (Coyne et al., 2019). Consequently, in these smallholder systems, excessive misuse of pesticides may prevail, especially on crops which are prone to diseases and pests, such as vegetables (James et al., 2010). The detrimental effects of pesticide misuse are mostly reported in relation to human and animal health, but there are important considerations for ecosystem health too (Rosenstock et al., 1991; al-Saleh, 1994; Lemaire et al., 2006). The challenge therefore, is how to reduce the use of, and reliance on, pesticides and simultaneously improve yields while maintaining, facilitating, or enhancing biodiversity in farming systems.
A range of options is available to reduce dependency on synthetic pesticides, while their suitability depends on prevailing circumstances (Bender et al., 2016; Sikora et al., 2019). With the domestication of crops, their commercial exploitation, and the focus on breeding for ever-higher yielding cultivars, there has been a concomitant loss of resistance to stress factors as well as a decrease in microbial diversity (Mariotte et al., 2018; Cordovez et al., 2019). In contrast, indigenous plants that are less commercially exploited and less highly bred, but locally produced or gathered from natural habitats even, tend to be more robust and resilient (Venkateswarlu et al., 2012). In addition to their nutritional value, such properties have led to an increased interest in indigenous leafy green vegetables in Africa, where there is a need to raise the daily nutritional intake (Cernansky, 2015). Preferred traits include high levels of protein, iron, and other valuable nutrients, as well as their ability to better withstand biotic and abiotic stresses, compared to popular non-native vegetables, e.g., kale and cabbage (Kumar et al., 2010; Bartolome et al., 2013; Onyango et al., 2013; Cernansky, 2015; Ronoh et al., 2019). Further, these plants have a short and adapted life cycle, resulting in a lower vulnerability to irregular rainfall due to climate change. Speculation remains, however, why these plants are so robust and whether this robustness can be transferred between crops.
Plant fitness is a phenotypic expression and genotypically determined but can be modified by external factors, such as their associated microbiota. Plants and their associated microbiota combined represent a functional unity, the plant holobiont (Vandenkoornhuyse et al., 2015). Plant species are host to a high diversity and complexity of microbial communities, which vary depending on various external influences (Yeoh et al., 2017). These microbial communities can influence plant growth, productivity, adaptation, and health (Bulgarelli et al., 2013; Berg et al., 2016). Modes of action include nutrient supply, plant hormone production, and antagonism toward pathogens (Berg, 2009; Lugtenberg and Kamilova, 2009). Plant-microbe interactions have largely focused on bacteria and fungi, although archaea are widespread and stable components of plant microbiomes (Hardoim et al., 2015; Moissl-Eichinger et al., 2018). They have the potential to directly interact with the host plant by supporting nutrient supply and growth promotion via auxin biosynthesis (Taffner et al., 2018, 2019), while antagonistic properties are not yet known (Moissl-Eichinger and Huber, 2011). Our hypothesis is that the microbiome strongly contributes to the fitness and health of the indigenous leafy green vegetables and that the archaeal community is an important component. Identification of key species within these communities may be crucial to develop suitable biologically based options toward increased robustness and health in crops, and consequently toward the sustainable improvement of smallholder crop production systems in rural areas of Africa.
This study was aimed at characterizing the microbial communities of four leafy green crops grown in rural, smallholder conditions in Uganda, including blackjack (Bidens pilosa L.), nightshade (Solanum scabrum Mill.), okra (Abelmoschus esculentus Moench), and spiderwisp [Gynandropsis gynandra (L.) Briq.] and to assess the role of the bacterial and archaeal community on plant health. To achieve this, we combined next-generation sequencing and characterization of bacterial isolates as well as screening for antagonism toward five phytopathogenic fungi, including species of the top 10 economically important crop pathogens worldwide (Botrytis cinerea, Fusarium oxysporum, Fusarium verticillioides, Sclerotium rolfsii, and Verticillium dahliae; Dean et al., 2012).
Materials and Methods
Experimental Design and Sampling Procedure
The leafy green vegetables blackjack, okra, nightshade, and spiderwisp were sampled in Kasangati, Uganda (0° 26' 33''N, 32° 36' 19''E) in April 2017. Four samples, each consisting of a single plant (blackjack, spiderwisp, and okra) or three individual plants (nightshade), were gently removed with the aid of a spade, placed in sealed air-tight plastic bags, stored in a cool box, and transferred to the laboratory; four bulked soil samples were also collected and stored in separate plastic bags. Soil parameters were analyzed by “AGROLAB Agrar und Umwelt GmbH” (Sarstedt, Germany). The soil texture was sandy loam with pH = 5.9, organic matter content of 3.7%, and nutrient contents of K = 413 mg kg−1, P = 86 mg kg−1, and Mg = 214 mg kg−1. In order to homogenize the samples, 3 g of the phyllosphere (plant leaves and stalks), 5–10 g root material with adhering soil, and 5 g of soil per replicate were physically mixed in a BagMixer (Interscience, St. Nom, France) with 15 ml of 0.85% NaCl. Samples of root-adhering soil are further called rhizosphere. To obtain root endosphere samples, root samples were further surface sterilized with a 4% sodium hypochloride solution (NaClO) for 3 min, washed four times with 0.85% NaCl, resuspended in 15 ml NaCl, and then physically crushed with a sterile mortar and pestle. Samples were centrifuged at 16,500 g for 20 min at 4°C, and DNA extracts were then stored at −70°C for further processing.
Isolation and CFU Determination of Bacterial Strains
Bacterial strains were isolated according to the protocol of Bragina et al. (2012). Briefly, 100 μl of the 15 ml 0.85% NaCl suspensions of each microhabitat-sample were plated onto NBII agar (Sifin, Berlin, Germany) plates in dilutions ranging from 10−2 to 10−5, incubated for 5 days at 20°C, and number of colony forming units (CFUs) determined and equated to fresh weight of the samples. A total of 512 randomly selected CFUs were isolated and stored in 20% glycerol at −70°C for further characterization.
Screening of Antagonistic Bacteria and Antifungal VOCs Production
The 512 bacterial isolates were each streaked onto a Waksman Agar (WA)-plate and exposed to a fungal pathogen, following the protocols of Berg et al. (2006). The fungal phytopathogens B. cinerea, F. oxysporum, F. verticillioides, S. rolfsii, and V. dahliae were obtained from the strain collection of the Institute of Environmental Biotechnology (Graz University of Technology, Austria). Screenings were performed in triplicate and evaluated according to their antagonistic activity against pathogens according to Wolfgang et al. (2019). Isolates with strong antagonistic activity were further tested for volatile organic compound (VOC) production, using a two-clamp VOC assay (Cernava et al., 2015).
BOX-PCR Fingerprinting and Sequencing of Antagonistic Bacteria
BOX-PCR was performed to resolve bacterial genetic diversity, according to the protocol of Rademaker and de Bruijn (1997). Shortly, colonies of 20 bacterial isolates with strong antagonism against all tested pathogens were solubilized, transferred into glass-bead filled tubes, ribolyzed, and centrifuged. PCR amplification was conducted using the BOXA1R primer 5'-CTA CGG CAA GGC GAC GCT GAC G-3'. After separation by gel electrophoresis, resulting band pattern was compared with “Gel Compar II” V.5.1 (Applied Maths, Kortrijk, Belgium). Different isolates were further sequenced based on the 16S rRNA gene fragment and taxonomically identified by manual BLAST search.1
Abiotic Stress Assays and Phosphate-Solubilization Tests
Bacterial isolates with antagonistic activity toward the tested phytopathogens were additionally screened for resistance to abiotic stress, including drought, salinity, and reactive oxygen, as well as their potential to solubilize phosphate, as described by Zachow et al. (2013). In reactive oxygen species stress assays, bacterial isolates were cultivated overnight in LB (Lennox) medium (Carl Roth, Karlsruhe, Germany). Overnight cultures (5 μl) were added to 96-well plates filled with 195 μl LB in 10 different concentrations of tellurite (1, 3, 5, 7, 9, 10, 13, 15, 18, and 20 μg/ml), and hydrogen peroxide (from 100 to 1,500 μmol in 200 μmol steps, 1,750–4,000 μmol in 250 μmol steps), respectively. Growth of each isolate was measured after 24 h incubation at 30°C under agitation in four replicates using a plate reader (Infinite 200, Tecan Trading AG, Switzerland) at a wavelength of 600 nm (OD600). For evaluation of the tolerated osmolarity level, bacterial isolates were cultivated in LB media with various NaCl concentrations (from 0 to 15% in steps of 1%). Growth was measured in four replicates after 24, 48, 72, and 144 h using the plate reader at 600 nm. In order to test for drought resistance, 20 μl of an overnight culture were dried under sterile conditions in a 96-well plate and were resuspended in 20 μl 0.9% NaCl after 1, 2, 5, 7, 14, 60, and 80 days. Further, 10 μl of the resuspended cells was dropped onto LB-agar plates in a dilution series, incubated and number of CFUs determined.
Screening for Plant-Growth Promoting Activities
Growth-promoting activities of bacterial isolates were tested on tomato (Solanum lycopersicum L. cv. Moneymaker, Austrosaat AG, Austria) plants according to Zachow et al. (2013). Tomato seeds were primed with bacterial cultures derived from three NBII plates grown overnight at 30°C suspended in 20 ml sterile water and were incubated for 4 h under agitation. Number of CFU per ml and OD600 of the suspensions were determined before seed priming (Table 1). Two germination pouches per strain were prepared with 8–9 seeds each. After 15 days, plants were harvested, leaf and root fresh weight were recorded. Roots were further pestled, and suspensions were plated on NBII plates in a dilution series for CFU determination. Plant growth was analyzed using Mann–Whitney U test.
Table 1.
Abiotic stress confrontation assays.
| Strain-ID | Species | Origin | Drought | H2O2 (μmol/ml) | NaCl (24 h) | NaCl (48 h) | NaCl (72 h) | NaCl (6 days) | OD600 (ONC) | CFU/ml priming suspension | ΔRFW (%) | ΔSFW (%) | ΔGR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Soil-I-11 | Bacillus sp. | Soil | Tolerant | 0 | 0 | 0 | 8% | 6% | 1.8 | 1013 | 67 | 46 | + |
| Soil-I-14 | Bacillus sp. | Soil | Tolerant | 2,000 | 5% | 7% | 7% | 7% | 1.6 | 4.2*1014 | 83 | 6 | + |
| Soil-I-45 | Sphingomonas sp. | Soil | Tolerant | 0 | 0 | 0 | 8% | 11% | 2.3 | 3.2*1014 | −11 | 11 | + |
| ORE-30 | Sphingomonas sp. | Okra root | Tolerant | 100 | 0 | 0 | 8% | 10% | 2.6 | >3*1014 | 0 | −24 | + |
| ORE-44 | Sphingomonas sp. | Okra root | Tolerant | 0 | 0 | 0 | 8% | 11% | 2.8 | >3*1014 | 21 | −10 | 0 |
| NSRE-37 | Bacillus sp. | Nightshade root | Tolerant | 900 | 0 | 0 | 0 | 0 | 1.7 | 5*1012 | 88 | 56 | + |
Growth after desiccation was measured by CFU/ml: tolerant: CFU above 105 after drought for 88 days. Reactive oxygen species test performed with hydrogen peroxide (H2O2): highest concentration of H2O2, the isolate could still tolerate. NaCl(x): maximum concentration (w/v) of NaCl in growth medium resulting in increased OD600 relative to the control after time x. Increase in root fresh weight (ΔRFW), shoot fresh weight (ΔSFW), and germination rate (ΔGR; +, increase; −, decrease; 0, no effect) relative to the corresponding control group.
Isolation of Total-Community DNA and Illumina Sequencing
Community DNA pellets from each microhabitat of soil, rhizosphere, root-endosphere, and phyllosphere of the four leafy green crops were subjected to PCR-based barcoding. First, extraction of DNA pellets was conducted using “FastDNA Spin Kit for soil” (MP Biomedical, Eschwege, Germany). PCR-products were purified with GENECLEAN TurboTM Kit (MP Biomedicals, Eschwege, Germany), following the manufacturer’s instructions for genomic DNA. The bacterial PCR approach was carried out with the Illumina barcode universal bacterial primer set 515f-806r (Caporaso et al., 2011) and PNA Mix (Lundberg et al., 2013) to remove host plastid and mitochondrial DNA. In order to amplify the archaeal 16S rRNA gene, a nested PCR was performed using the archaea-specific primers 344f and 915r in the first PCR. In a second PCR approach, the modified primer pair S-D-Arch-0349-a-S-17/S-D-Arch-0519-a-A-16 (here 349f/519r; Klindworth et al., 2013) with an additional 10 bp primer-pad (TATGGTAATT/AGTCAGCCAG) and linker (GT/GG) was used, according to the protocols of the Earth Microbiome Project (Walters et al., 2016). In a third PCR, Golay barcodes were annealed (Hamady et al., 2008). All PCR reactions were conducted as previously described (Taffner et al., 2019). Bacterial and archaeal PCR reactions were conducted in triplicate, purified with the Wizard SV Gel and PCR clean-up system (Promega, Madison, WI, United States), and pooled to equimolarity. Sequencing was carried out by Eurofins MWG Operon (Eurofins, Ebersberg, Germany) with an Illumina HiSeq 2500 system.
Bioinformatic Processing of 16S rRNA Gene Fragments
The generated 16S rRNA gene libraries were pre-processed using QIIME version 1.9.1 (Caporaso et al., 2010) and QIIME2 (version 2018.2, Bolyen et al., 2019). First, read quality was checked with FastQC,2 reads were joined, and barcodes were extracted in QIIME1. Sequences were demultiplexed using the q2_demux plugin and denoised using q2_dada2 (Callahan et al., 2016) Taxonomy was assigned using a naïve Bayes taxonomy classifier (Bokulich et al., 2018) implemented in QIIME2. For taxonomic assignment, SILVA reference data base version 128 was used for bacteria and Silva 16S Archaeal database (349af–519ar 99, otusversion 128) for archaea with a 97 and 99% similarity cut-off, respectively (Quast et al., 2013). Amplicon sequence variants (ASVs) assigned to mitochondria or chloroplasts were removed using taxonomy-based filtering. ASVs were aligned with q2_mafft (Katoh and Standley, 2013), and a phylogenetic tree was constructed with q2_fasttree2 (Price et al., 2010). For estimating diversity metrics, sequence tables were rarefied to 1,210 ASVs (archaea) and 7,444 ASVs (bacteria). For evaluating alpha diversity, Kruskal-Wallis test (all groups and pairwise), alpha rarefaction, Shannon and Faith’s phylogenetic diversity index (Faith, 1992) were calculated. Beta diversity was analyzed by principal coordinate analysis (PCoA) plots and ANOSIM based on phylogenetic distance metrics of weighted UniFrac distances (Lozupone et al., 2007) and visualized with the emperor plugin (Vázquez-Baeza et al., 2013). The ANOSIM test was performed with 999 permutations. To test for the influence of microhabitat and plant species, these variables were tested using the plugin Adonis (Anderson, 2001) for bacteria and archaea. To test for significant differences in abundances of identified antagonistic taxa, the bacterial dataset was analyzed using the LEfSe algorithm implemented in https://www.microbiomeanalyst.ca (Chong et al., 2020). The dataset was filtered using the default settings (minimum count for reads of 4, minimum prevalence in samples 20%, low variance filtered based on 10% interquantile range, LDA score = 2.0), rarefied to minimum library size and scaled using total sum scaling. Taxa were compared on family level between each plant species (all microhabitats combined) and soil, as well as within a single plant species between microhabitats. Cytoscape 3.3.0 software was used to visualize the bacterial distribution and network of the core genera (Shannon et al., 2003). ASVs, that were found in >75% of the plant samples, were assigned as interspecific core ASVs of the plant species. ASVs were assigned to genus level, and data of all four plants were combined. Taxa represented in ≥50% of samples across the dataset were assigned as intraspecific core genera. Abundant sequences with a low taxonomical resolution were additionally assigned by using the nucleotide BLAST search.3
Nucleotide Sequence Accession Numbers
The 16S rRNA gene fragment amplicon library was submitted to the European Nucleotide Archive (ENA) and can be found under the accession number PRJEB39392.
Results
General Community Structure of Prokaryotes Associated With Leafy Green Vegetables
Sequencing of the 16S rRNA gene fragments originating from the phyllosphere, root-endosphere, rhizosphere, and soil of the leafy greens blackjack, nightshade, okra, and spiderwisp resulted in a total of 10,688,730 high quality bacterial reads and 2,692,299 archaeal reads. After taxonomy-based filtering of mitochondria and chloroplast sequences, the datasets comprised 9,795,981 bacterial reads and 2,663,458 archaeal reads, clustered in a total of 27,697 and 2,995 distinct ASVs, respectively. Unassigned sequences remained in the dataset because we expected a considerable and potentially important part of microbes to be still unknown to science.
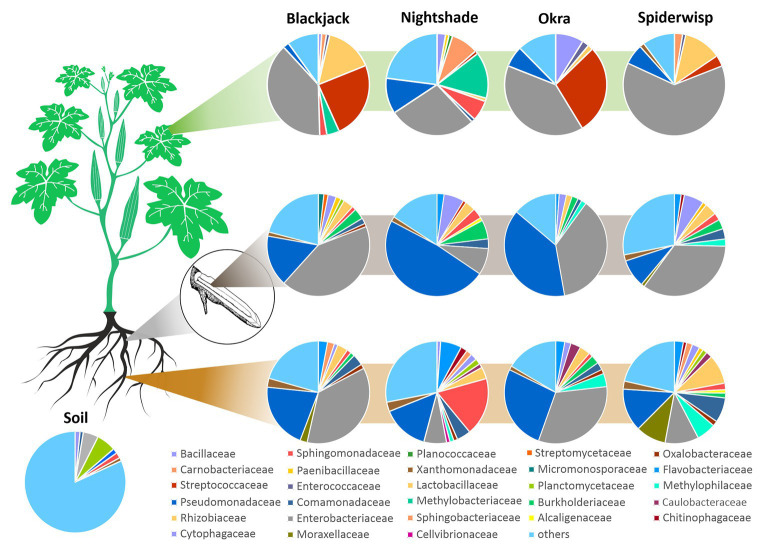
The bacterial core microbiome revealed similarities and differences between the phytobiome composition in respect to the plant genotype and microhabitat (Figure 1). In the phyllosphere, Enterobacteriaceae (42.2%) and Streptococcaceae (14.4%) were dominant in the bacterial community, whereas in the root endosphere and rhizosphere Enterobacteriaceae (30.7 and 21.6%, respectively) and Pseudomonadaceae (28.0 and 19.0%, respectively) were predominant. In general, Sphingomonadaceae (4.2%), Lactobacillaceae (3.3%), Bacillaceae (2.9%), Rhizobiaceae (2.7%), Comamonadaceae (2.5%), Flavobacteriaceae (2.0%), and Xanthomonadaceae (1.5%) were ubiquitous but less abundant. In the phyllosphere of blackjack and Okra, Streptococcaceae were dominant, representing around a quarter of the core microbiome. Blackjack and spiderwisp both harbored Lactobacillaceae with 12.0–15.3% in the phyllosphere. Bacillaceae and Pseudomonadaceae were present in the core microbiome of each crop in each microhabitat (1.09–6.33%), with the exception in the spiderwisp phyllosphere, where no Bacillaceae were found. Throughout all microhabitats and crops, the fraction of families with an abundance lower than 1% (“others”) was relatively high (13.9–21.6%). These bacteria mainly belonged to the families Oxalobacteraceae (0.9%), Caulobacteraceae (0.9%), unidentified Acidobacteria (0.9%), Sphingobacteriaceae (0.8%), Paenibacillaceae (0.8%), Rhizobiales (0.7%), Chitinophagaceae (0.7%), Planctomycetaceae (0.6%), Enterococcaceae (0.6%), and Alcaligenaceae (0.5%).
Figure 1.
Bacterial core microbiome of leafy greens. The composition of the microbiome of blackjack, nightshade, okra and spiderwisp, and their microhabitats, displayed at the family level: phyllosphere (green stripe), root-endosphere (gray stripe), and rhizosphere (brown stripe). Families with abundances below 1% of total microbiome are captured within “others.”
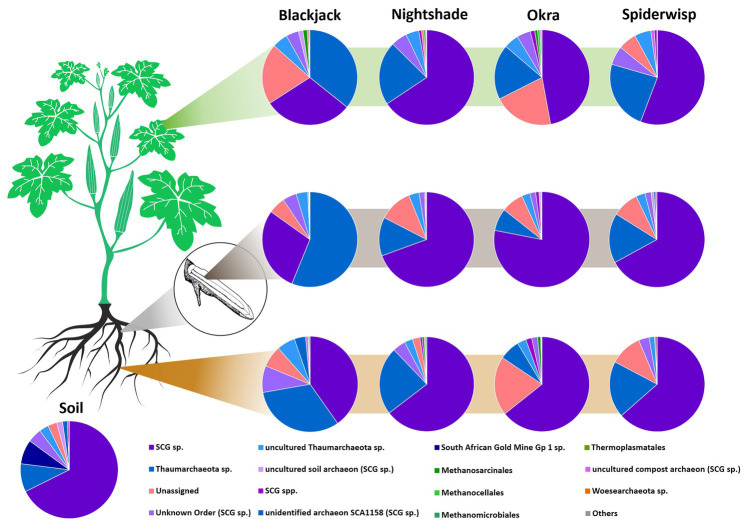
The archaeal communities (Figure 2) were clearly dominated by the phylum Thaumarchaeota (89.0%). In general, a high proportion of unassigned reads of up to 20.7% was detected, which were especially associated with blackjack and okra phyllospheres. In all four leafy green crops, Euryarchaeota were present but in low relative abundances (0.7–1.0%), except in spiderwisp, in which no Euryarchaeota were detected. At the class level, archaea of the soil crenarchaeotic group (SCG) were relatively abundant (56.2%), followed by unassigned Thaumarchaeota (22.9%). Archaea of the SCG were especially abundant in nightshade and spiderwisp. Methanogenic archaea of the class Methanomicrobia were mainly found in phyllosphere and the root-endosphere samples, except in spiderwisp.
Figure 2.
Archaeal community in leafy greens. The composition of the archaeal community of blackjack, nightshade, okra and spiderwisp, and their microhabitats, displayed at the order level: phyllosphere (green stripe), root-endosphere (gray stripe), and rhizosphere (brown stripe).
Bacterial Diversity Associated With Leafy Green Vegetables
Diversity metrics based on phylogeny were calculated with QIIME2 to determine similarities and dissimilarities of the bacterial community of the leafy green crops. Shannon’s diversity in the plant-microhabitats decreased from rhizosphere to phyllosphere, with an exception for nightshade, which showed the least diversity in root endosphere. However, diversity in the rhizosphere of nightshade was the highest comparing all plants [H(nightshade) = 7.81 ± 0.21]. Diversity within the bulk soil samples was higher [H(soil) = 9.41 ± 0.42], however, compared to the plant species [H(mean) = 6.91 ± 0.16; ranging from H = 5.31 to H = 6.24].
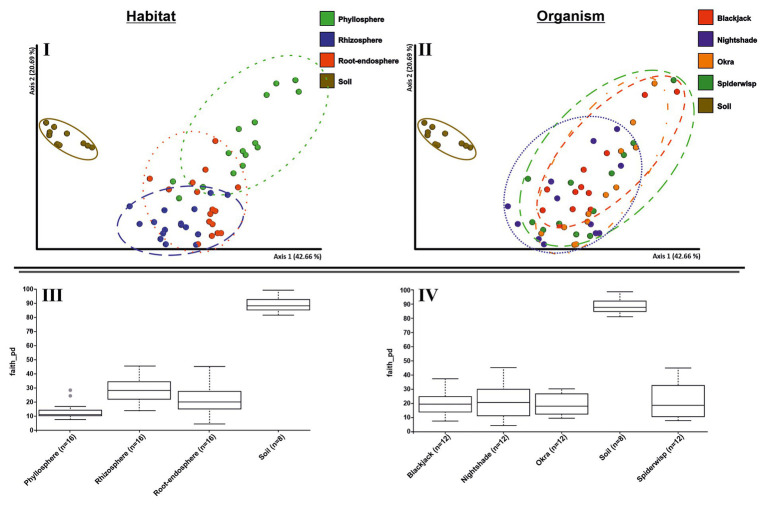
Between microhabitats, a cluster formation (Figure 3I) as well as a trend from rhizosphere to phyllosphere, was observed, whereas the rhizosphere bacterial community overlapped, to some extent, with the root endosphere. However, phyllosphere communities were more distinct, while the soil showed a clear cluster, which was significantly different to the other microhabitats with quantitative measures (ANOSIM: R = 0.504 and p ≤ 0.001). When assigning the same communities to their respective plant species (Figure 3II), no distinct clustering could be detected (ANOSIM: R = 0.048 and p = 0.064). Only nightshade had a slightly different clustering pattern. When investigating relationships within and between plants (within-sample), the alpha diversity index was significantly different between microhabitats (Figure 3III; p = 0.001), but not between the four plant species (Figure 3IV; p = 0.080). The overall group of microhabitats differed in diversity, but with respect to pairwise investigations, this was due to differences in rhizospheres as well as root-endosphere to phyllosphere. Further, group statistics showed that bacterial alpha diversity of the tested leafy green crops was not plant species specific. However, PCoA and Kruskal-Wallis test revealed that microbial diversity was microhabitat-specific. The factor “habitat” explained more variance within the bacterial dataset than the factor “organism” in both Bray-Curtis and weighted UniFrac distances (Supplementary Table 1).
Figure 3.
Bacterial alpha and beta diversity of leafy green vegetables. Principal coordinate analysis (PCoA) plots of the 16S rRNA amplicon datasets of four crops (blackjack, nightshade, okra, and spiderwisp) constructed based on phylogenetic distance metrics (weighted UniFrac). The distance between the data points negatively correlates with the similarity of the communities. (I) Clusters of the communities based on microhabitat (phyllosphere, rhizosphere, root-endosphere, and soil), and (II) based on plant species (blackjack, nightshade, okra, spiderwisp, and soil). Comparison of bacterial alpha diversity based on Faith’s phylogenetic diversity of the microhabitats (III) and plant species (IV).
Archaeal Diversity Associated to Leafy Green Vegetables
Archaeal alpha diversity indices had similar values in all plant species [H(all) = 4.51–4.95], with the highest archaeal diversity in nightshade [H(nightshade) = 4.95 ± 0.21]. Within plant-associated communities, the diversity of the microhabitats differed only slightly, between the root-endosphere [H(endosphere) = 4.42 ± 0.37] and the rhizosphere [H(rhizosphere) = 4.92 ± 0.23; Figure 4III]. Alpha diversity of archaeal communities in bulk soil was higher than in plant-associated communities [H(soil) = 5.26 ± 0.27; Figure 4IV].
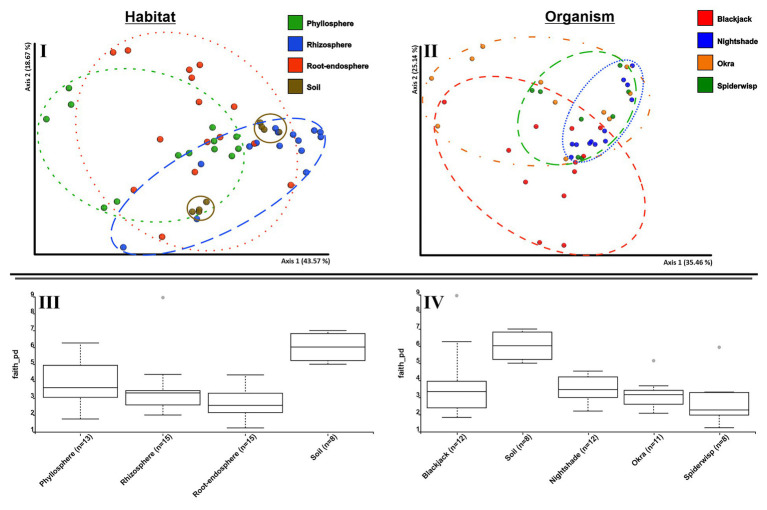
Figure 4.
Alpha and beta diversity analyses of archaeal communities associated to leafy green vegetables. PCoA plots based on weighted UniFrac distance metrics show the archaeal community of phyllosphere, rhizosphere, root-endosphere, and soil microhabitats (I) of the four leafy green crops blackjack, nightshade, okra, and spiderwisp (II); comparison of alpha diversity based on Faith’s phylogenetic diversity of the microhabitats (III) and plant species (IV).
The alpha and beta diversity indices of the archaeal community are presented in Figure 4. In a PCoA-plot (Figure 4I), soil and the rhizosphere communities were clustered, whereas diversities in the root endosphere and phyllosphere were more dispersed. Again, a pattern, from rhizosphere to phyllosphere, was evident, as indicated by the overlapping clusters, with the soil diversity occurring within the rhizosphere diversity. In general, microhabitats showed significant differences in diversity (ANOSIM: R = 0.226; p = 0.001), with soil showing the highest diversity (Figure 4III). Analyzing the beta diversity for plant-type-specific differences, a cluster formation of nightshade and spiderwisp could be seen (Figure 4II). These plant species specific differences were confirmed by ANOSIM-test (R = 0.131; p = 0.002) and were found to be due to nightshade and blackjack (q < 0.05) based on pairwise comparison. However, spiderwisp and okra showed similarities (q > 0.377). Further, alpha diversity analysis with Kruskal-Wallis (all groups and pairwise) confirmed that archaeal diversity differed, depending on the microhabitat (p = 0.001) as well as the plant species (p = 0.01), which is due to the significantly different diversity of nightshade (Figures 4III,IV). However, pairwise comparison did not establish any differences between plant species (q > 0.08). Soil archaeal diversity was significantly different to all plant-associated microhabitats (q < 0.004), as well as the phyllosphere to root-endosphere (q = 0.038). However, the factors microhabitat and plant species explain <35% of the variance within the archaeal dataset (Supplementary Table 2), indicating other important factors determining archaeal community composition.
Analysis of the Core Microbiota of Leafy Green Vegetables
Microbial core communities across blackjack, nightshade, okra, and spiderwisp were cross-linked based on taxonomic analysis at the family level and were visualized as a network (Figure 5). In total, 91 features were identified on genus level, with just one belonging to Archaea. A large core microbiome of 18 families, such as Bacillus, Sphingobium, Comamonadaceae gen., Pseudomonas, and Rhizobiaceae gen. (including the archaeal SCG), mainly assigned to Proteobacteria, were shared between all four crops. An additional 11 families, also mostly Proteobacteria, were common in blackjack, okra, and spiderwisp, thus communities associated with nightshade were more specific. Nightshade and okra shared specific taxa of the genus Carnobacterium, while blackjack and spiderwisp both shared Weissella and Acinetobacter. Each crop was associated with specific bacterial families that were unique in the core microbiome of the respective plant species. The number of such distinctive communities were ranged from five (spiderwisp) to nine (blackjack and nightshade) and 11 (okra).
Figure 5.
Feature network based on taxonomic analysis at the genus level. Each node represents a family of the core microbiome and is colored according to its phylum. If families were only represented by one genus within the core microbiome, the corresponding genus is added in brackets. Cross-linked nodes express families shared between the plants blackjack, nightshade, okra, and spiderwisp. (I) Nightshade. (II) Okra. (III) Blackjack. (IV) Spiderwisp.
Screening and Identification of Bacterial Antagonists Against Biotic and Abiotic Stresses
Of the 512 randomly selected bacterial isolates taken from the four leafy green crops and bulk soil, 108 isolates showed a high antagonistic activity (clear halo between fungi and bacteria ≥ 5 mm) against at least one pathogen (B. cinerea, F. oxysporum, F. verticillioides, S. rolfsii, and V. dahliae), and 23 isolates against four pathogens (Figure 6). Screening test results against V. dahliae needed a separate evaluation category as the culturing of the fungi required a different procedure and was, therefore, not included into the Venn diagram. A total of 44 bacterial isolates were highly active against V. dahliae. Based on these results, a selection of 24 antagonists, mostly antagonistic against all tested pathogens, were chosen for further characterization; 12 of the isolates originated from soil, nine were isolated from root endosphere, and three from the rhizosphere. Genetic characterization of the 24 antagonistic isolates undertaken using BOX-PCR and 16S sequencing identified 16 isolates as Bacillus sp. with suggested species B. siamensis, B. velenzensis, B. amyloliquefaciens, B. methylotrophicus, B. vallismortis, and B. subtilis. A further eight isolates were assigned to Sphingomonas sp. with hits for S. echinoides and S. glacialis. Combining the alignment results with similarity pattern of BOX PCR bands, isolates were clustered into five similarity groups (Supplementary Table 3).
Figure 6.

Number of bacterial isolates with antagonistic effects against fungal pathogens. Fungal pathogens included Fusarium oxysporum, Fusarium verticillioides, Sclerotium rolfsii, and Botrytis cinerea. Only bacterial antagonists showing high antagonistic activity were assigned to their respective fungi. Graph was generated using VENNY 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/).
The resistance to abiotic stresses of antagonistic bacterial strains was further characterized in order to evaluate their potential for application as future biocontrol agents (BCAs). Therefore, abiotic stress tests comprising reactive oxygen species stress tests were conducted (Table 1).
The desiccation assay showed that all tested bacterial isolates were highly resistant to drought with a CFU/ml of above 105 after 88 days. The ability to resist reactive oxygen could not be shown in the tellurite assay, but when using hydrogen peroxide as stressor three isolates could still be cultured. Isolate “Soil-I-14” showed H2O2-tolerance as well as salt tolerance to high levels of NaCl after 24 h. Other isolates needed a longer period to adapt to higher NaCl concentrations and showed tolerance only after an adaption phase of 72 h (Table 1).
Further characterization of the mechanism of antagonism using two clamp VOC assays (TCVAs) showed no antagonistic effects of the bacterial isolates against the pathogens, based on VOCs.
Plant Growth Promotion of Bacterial Antagonists
Priming of tomato seeds with the six bacterial antagonistic strains resulted in a significantly increased fresh biomass of leaves (p = 0.039) and whole seedlings (p = 0.020) relative to the control when using Bacillus strains. Priming with Sphingomonas species showed no significant effect on both root and leaves growth (Table 1). The strongest plant growth-promoting effect was observed when using a Bacillus strain derived from roots of nightshade (strain NSRE-37). None of the bacterial isolates showed signs of phosphate solubilization.
Localization of Antagonists Within the Microbial Network of Leafy Green Vegetable Crops
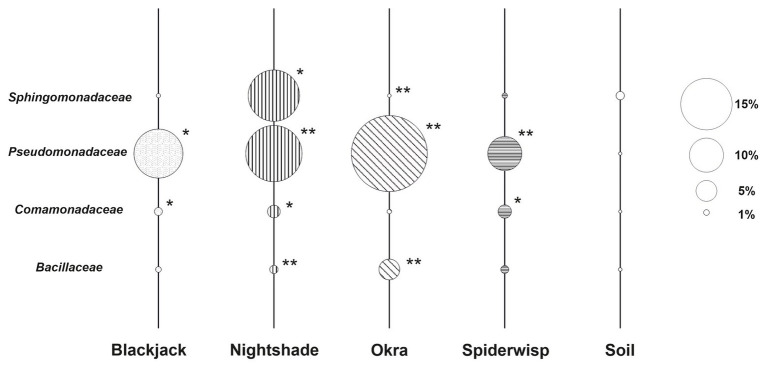
The distribution and abundance of bacterial families comprising taxa with high antagonistic activity toward fungal phytopathogens (Bacillaceae and Sphingomonadaceae) within the microbiome of the leafy green crops were compared in order to highlight possible links to the robustness of the plant host. Bacterial families that were isolated from Ugandan tomato and were shown to comprise nematicidal effects to plant-pathogenic nematodes in earlier studies (Wolfgang et al., 2019) were included. Most families were significantly enriched in plants compared to soil (Supplementary Table 4), although plant-specific differences were observed (Figure 7); while Pseudomonadaceae was the most abundant antagonistic family across all plant species with the highest relative abundance in okra, Sphingomonadaceae were higher abundant in soil except for nightshade. Sphingomonadaceae account for 9.6% (range 0.6–21.9) relative abundance in nightshade. Additionally, abundances of Sphingomonadaceae significantly differ within nightshade, with a higher relative abundance in rhizosphere and phyllosphere than in root-endosphere (Supplementary Table 5). Bacterial communities of nightshade consisted of the highest share of antagonistic families (31.2%), followed by okra (25.5%), blackjack (16.3%), and spiderwisp (14.8%). Within soil, antagonistic families comprised only 4.5% of all recorded microorganisms.
Figure 7.
Relative abundance of antagonistic families of bacteria associated with leafy green vegetables. The diameter of the bubble represents the abundance of each family within the microbiome of each leafy green crop and soil. Soil is used as a reference. Families, which were found to produce nematicidal VOCs in Ugandan tomatoes (Wolfgang et al., 2019) – namely Comamonadaceae and Pseudomonadaceae – are included. Asterisks indicate significant differences in relative abundance compared to soil based on LEfSe (*p ≤ 0.05; **p ≤ 0.001).
Discussion
Research on plant-associated microbiomes in tropical regions is still in its beginning. Many factors known to influence crop-associated bacterial communities, e.g., soil quality, plants life cycle, or agronomic practices (Philippot et al., 2013), were not addressed in this study. Nevertheless, this study is a first step for understanding microbial communities in crops that are usually understudied in tropical agricultural research, but have a high relevance to local people. When investigating four locally popular leafy green vegetables (blackjack, nightshade, okra, and spiderwisp) in Uganda, we identified a microbiome that has both common and specific components between plant species. The general taxonomic composition was comparable with many other plant and crop species as well as the abundance of microbes (Bulgarelli et al., 2013; Berg et al., 2016). The rhizosphere was confirmed as the microbial hot spot for plants (Berg et al., 2006; Berg and Smalla, 2009) as well as the rhizosphere effect, which describes the selective enrichment visible in the composition of the microbiota (Foster et al., 1983; Buée et al., 2009). Interesting specific components were also observed. For instance, the microbiota of indigenous leafy greens were characterized by: (I) an unusually large core microbiome with only minor differences between plant species; (II) a high diversity of bacteria and archaea forming a network of potentially copiotrophic bacteria and oligotrophic archaea; and (III) a high proportion (15–31%) of potential plant beneficial microbes. The latter were identified in our culture collection, and can be potentially employed as biologically-based options for protection against stresses.
Leafy Green Vegetables Harbor Common Bacteria With Copiotrophic Lifestyle
The diversity and community structure of bacteria and archaea in four leafy greens was found to be microhabitat-specific, rather than plant genotype-specific. The extent of the impact of numerous variables (e.g., plant genotype, plant organ, habitat, developmental stage, and soil quality) is a persistent question in microbial ecology across studies. However, in studies focused on natural vegetation in particular, the plant genotype seem to be the most important factors to determine plant-associated bacterial communities, followed by soil traits (Berg and Smalla, 2009; Bulgarelli et al., 2013). Recent studies revealed a decrease in diversity of crop-associated microbial communities through breeding practices (Cardinale et al., 2015; Mendes et al., 2019). The less pronounced impact of the plant genotype can be explained by the life strategy of plants. Three (blackjack, nightshade, and spiderwisp) of the four leafy greens in the current study were naturally occurring, and in general are ubiquitous, mostly invasive, produce many seeds and are, therefore, categorized as r-strategists, which often have a copiotrophic lifestyle (Andrews and Harris, 1986). This life strategy might also affect the composition of their associated microorganisms, which may have even co-evolved with them (Cordovez et al., 2019). For example, invasive plants, such as cheatgrass (Bromus tectorum L.), knapweed (Centaurea stoebe L.), and leafy spurge (Euphorbia esula L.), enrich copiotrophic bacteria in their associated rhizosphere (Gibbons et al., 2017). In our study, the microbiome associated with the leafy greens was neither specific nor depended on the plant genotype. We found that the most abundant bacterial phyla followed the same copiotrophic life strategy as their host, such as Proteobacteria, Firmicutes, Bacteriodetes, and Actinobacteria (Ho et al., 2017). In contrast, the archaeal phyla are considered to be oligotrophic, especially Thaumarchaeota (Uksa et al., 2015; Youssef et al., 2015). This indicates high substrate specificity and supports the assumption of a niche-colonization by archaea, including their role as followers of bacteria. Interestingly, a rhizosphere effect was also observed for archaea, especially for nightshade. The enrichment may depend on quality and composition of the root exudates, as demonstrated for archaea in tomato plants (Simon et al., 2005; Taffner et al., 2020). Altogether, copiotrophic bacteria and oligotrophic archaea appear to form a potent trophic network on leafy greens, which would warrant further investigation.
High Microbial Diversity of Leafy Green Vegetables Compared to Cultivated Crops
The four leafy greens studied all showed high and relatively similar values for Shannon’s indices, with H-values ranging from 5.31 and 4.51 (okra) to 6.24 and 4.95 (nightshade), for bacteria and archaea, respectively. When comparing the Shannon’s indices in bacterial phyllosphere communities [ranging from H(okra) = 4.4 to H(nightshade) = 5.74], the diversity of the leafy greens in this study is considerably higher than in some commercially cultivated leafy greens, such as spinach [Spinacia oleracea L.; H(spinach) = 3.15, Lopez-Velasco et al., 2013]. This also applies for rhizosphere: maize (Zea mays, L.) rhizosphere displayed a distinctly lower alpha diversity [H(maize) = 3.42; García-Salamanca et al., 2013] than the diversity of the rhizosphere of the leafy greens [H(mean_rhizosphere) = 6.91]. Further, the archaeal diversity in rhizosphere of leafy greens tested was found to be higher [H(mean_rhizosphere) = 4.51–4.95] than in other cultivated crops, such as rice [Oryza sativa, L., H(rice) = 4.08–4.43], Barbados nut [Jatropha curcas, L., H(barbados_nut) = 3.16] and tomato [H(tomato) = 3.4; Lee et al., 2015; Dubey et al., 2016]. This large disparity in microbial diversity is attributed to the overbreeding of our main crops (Pérez-Jaramillo et al., 2016), whereas natural leafy greens have received much less attention and remain less intensively bred, having only recently attracted interest in agriculture. Further, agricultural practices affect microbial diversity. Comparing organic farming with conventional intensive farming, significant differences in the microbiome of maize, melon (Citrullus lanatus Thunb.), pepper (Capsicum annuum L.), and tomato, as well as the soil, were prominent (Hartmann et al., 2015; Xia et al., 2015). Given the large difference in diversity indices between the uncultivated leafy greens from Uganda with intensively cultivated field crops, we could assume that highly focused breeding programs, as well as intensive agricultural practices have led to a reduction and loss of diversity in the microbiome of these crops (Pérez-Jaramillo et al., 2016). Naturally occurring vegetables, such as leafy greens, have a high microbial diversity, which is directly correlated with healthier, more robust plants that are less vulnerable to pathogenic outbreaks (Berg et al., 2016).
High Proportion of Plant-Beneficial Bacteria of the Microbiome in Leafy Green Vegetables
A broad range of the taxa recovered from the core microbiome of leafy greens are well-known plant growth promoters, such as members of Enterobacteriaceae and Pseudomonadaceae, which occur frequently on leafy greens (Hayat et al., 2010). They are also known to be antagonistic against phytopathogenic fungi, either through competition or production of antimicrobial metabolites (Haas and Défago, 2005). However, Enterobacteriaceae also include human enteric pathogens, some of which, through occupation of crops via roots enter human digestive systems and have been associated with stimulating immune responses or acting as a “natural vaccination” as opposed a pathogen (Brandl, 2006; Berg et al., 2015). Further, Actinobacteria and Proteobacteria species were broadly distributed throughout the microbiome of the four leafy greens, both of which have previously been associated with host plant protection against fungal infections (Mendes et al., 2011). Besides the dominant families mentioned, some of the less common members of the microbiome showed growth promoting properties. Bacillaceae species such as Bacillus subtilis, B. amyloliquefaciens and B. cereus, and Oxalobacteraceae species such as Herbaspirillum seropedicae are known for supporting plant growth (Hayat et al., 2010), as well as members of the families Xanthomonadaceae, Paenibacillaceae, Sphingobacteriaceae, Chitinophagaceae, and Alcaligenaceae (Berg, 2009; Yang et al., 2014). One interesting fact is the relatively high abundance of Sphingomonadaceae in nightshade rhizosphere and phyllosphere (Figures 1, 7), compared to the other leafy greens. A high relative abundance of Sphingomonadaceae was frequently measured in other Solanum-associated communities, namely in rhizosphere (12%), root endosphere (5%), fruits (12–24%), and flowers (2–12%) of tomato (Allard et al., 2016; dataset of Wolfgang et al., 2019), rhizosphere of potatoes (Solanum tuberosum L., Pfeiffer et al., 2017), and rhizosphere of eggplant (Solanum melongena L., Li et al., 2019). Sphingomonadaceae comprise members with remarkable biotechnological potential, for instance degraders of aromatic or metalorganic compounds (Asaf et al., 2020). The high relative abundance of Sphingomonadaceae may be attributable to the diverse secondary metabolites (e.g., alkaloids) found in Solanaceae. However, further studies have to confirm the connection between Sphingomonadaceae and Solanum.
The archaeal community was clearly dominated by Thaumarchaeota, which are common colonizers of leafy greens, such as arugula (Taffner et al., 2019). This phylum consists mostly of ammonia oxidizing archaea (AOA), which are important for nitrogen cycling (Francis et al., 2007), and therefore, for supporting nutrient supplies to the plant. Further, recent studies show that archaea have the potential to directly support plant growth via auxin biosynthesis, a plant growth hormone (Taffner et al., 2018). Besides Thaumarchaeota, methanogens of the phylum Euryarchaeota were observed. These are also common in plants, colonizing anoxic niches in the rhizosphere, such as on maize or arugula (Chelius and Triplett, 2001; Taffner et al., 2019). However, there was a high relative abundance of taxonomically unassigned archaeal features, although an up-to-date established pipeline was used for the bioinformatic analysis. This limitation is well-known for archaea, especially in novel, less studied habitats such as Uganda, and is mainly due to poorly defined reference databases. We can conclude therefore that the core microbiome of leafy greens contained several taxa with the potential to support plant growth and protection against pathogenic fungi, and thereby contribute to the robustness and health of plant hosts.
Promising Key Species for Future Biocontrol Agents
In the core microbiome of the leafy greens, we identified Bacillus spp. and Sphingomonas spp. playing a pivotal role in suppressing the key pathogenic fungi B. cinerea, F. oxysporum, F. verticillioides, S. rolfsii, and V. dahliae. Bacillus spp. have previously been shown to produce antimicrobial compounds, such as mycosubtilin and lipopeptides produced by B. subtilis (Leclère et al., 2005), or the antagonistic compound bacillomycin by B. amyloliquefaciens (Mülner et al., 2020). Sphingomonas are mainly known for their ability to degrade refractory contaminants, but have also been reported to be antagonistic against bacteria and fungi (White et al., 1996; Innerebner et al., 2011). These highly effective antagonists further showed resistance to abiotic stresses and plant-growth promotion capabilities in the current study. One isolate of Bacillus was able to tolerate high levels of hydrogen peroxide, which is a major abiotic stress factor for plants. Further, all isolates could grow under saline conditions up to 10%. Salinity reduces water-uptake efficiency and photosynthesis rate in plants, but microorganisms capable of dealing with such osmotic stress may confer resistance in plants to salt stress (Mayak et al., 2004). Furthermore, episodic drying and re-wetting of soil causes fluctuations in the soil’s water potential and challenges microbes. We showed that all our selected isolates were highly resistant to desiccation. Effective consortia of biological control agents, therefore, should include bacteria that support plant growth in addition to antagonistic species. In our study, priming of tomato seeds with Bacillus isolates resulted in significant plant-growth promotion of up to 70%, whereas Sphingomonas isolates did not show any effect. However, Sphingomonas are known to promote plant growth by producing gibberellic acids (GAs) and indole acetic acid (IAA), improving crop productivity, which have also been reported for Bacillus spp. (Khan et al., 2014). Although Bacillus strains have previously been reported as solubilizers of inorganic phosphate (Hayat et al., 2010), we could not identify phosphate solubilizers among our isolates. This provides confirmation of the high specificity of plant beneficial traits at strain level (Berg, 2009). The antagonistic and plant-growth-promoting characteristics of the Bacillus and Sphingomonas isolates tested make them promising candidates for their application as biological control agents against fungal infections and for increasing robustness and plant health in Ugandan agriculture.
Conclusion
In our study, we found a unique, diverse, and robust microbiome occurring on natural leafy green vegetables in Uganda. Blackjack, okra, nightshade, and spiderwisp harbored microbes with strong antagonistic activities against pathogenic fungi, as well as promoting plant growth and demonstrating properties to enable host plants to withstand abiotic stresses. Six isolates in particular, assigned to the families Sphingomonadaceae and Bacillaceae, proved to be promising key-candidates for future sustainable biocontrol agents, toward supporting crop production in smallholder production systems in Sub-Saharan Africa. The biocontrol approach provides a more environmentally sustainable opportunity to produce crops and reduce or even replace excessive pesticide use. Identification of microbial isolates that are indigenous and adapted to African smallholder production systems will enable the development of technologies to support smallholders and improve human and environmental health.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ebi.ac.uk/ena, PRJEB39392.
Author Contributions
JT, DC, AW, and GB performed the sampling. JT and OL conducted experiments in the laboratory and drafted the manuscript. JT, OL, and AW performed bioinformatics analyses. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank their project partners, namely the Austrian Development Agency (ADA), the International Institute of Tropical Agriculture (IITA, Kenia, part of CGIAR), Makerere University (Kampala, Uganda), Volunteer Efforts for Development Concern (VEDCO, Uganda), and Addis Ababa University (Ethiopia); Kyadondo County Headquarters (Buganda kingdom, Uganda), Expedito Olimi Kaboyo, Doreen Nampamya, and Fassil Assefa for help during the sampling process; and Lea Gibitz-Lambert, Monika Schneider-Trampitsch, Tobija Glawogger, and Isabella Wrolli for support in the laboratory.
Funding. This work was undertaken as a part of the research project “IITA–Healthy seedling systems for a safer, more productive vegetables in East Africa” (F37139), funded by the Austrian Development Agency (ADA) to DC and GB.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.585690/full#supplementary-material
References
- al-Saleh I. A. (1994). Pesticides: a review article. J. Environ. Pathol. Toxicol. Oncol. 13, 151–161. PMID: [PubMed] [Google Scholar]
- Allard S. M., Walsh C. S., Wallis A. E., Ottesen A. R., Brown E. W., Micallef S. A. (2016). Solanum lycopersicum (tomato) hosts robust phyllosphere and rhizo-sphere bacterial communities when grown in soil amended with variousorganic and synthetic fertilizers. Sci. Total Environ. 573, 555–563. 10.1016/j.scitotenv.2016.08.157, PMID: [DOI] [PubMed] [Google Scholar]
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. 10.1111/j.1442-9993.2001.01070.pp.x [DOI] [Google Scholar]
- Andrews J. H., Harris R. F. (1986). “R‐ and K-selection and microbial ecology” in Advances in Microbial Ecology. Vol 9 ed. Marshall K. C. (Boston, MA: Springer; ), 99–147. [Google Scholar]
- Asaf S., Numan M., Khan A. L., Al-Harrasi A. (2020). Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 40, 138–152. 10.1080/07388551.2019.1709793, PMID: [DOI] [PubMed] [Google Scholar]
- Bartolome A. P., Villaseñor I. M., Yang W. -C. (2013). Bidens pilosa L. (Asteraceae): botanical properties, traditional uses, phytochemistry, and pharmacology. Evid. Based Complement. Alternat. Med. 2013:340215. 10.1155/2013/340215, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S. F., Wagg C., van der Heijden M. G. A. (2016). An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evol. 31, 440–452. 10.1016/j.tree.2016.02.016, PMID: [DOI] [PubMed] [Google Scholar]
- Berg G. (2009). Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. 10.1007/s00253-009-2092-7, PMID: [DOI] [PubMed] [Google Scholar]
- Berg G., Erlacher A., Grube M. (2015). “The edible plant microbiome: importance and health issues” in Principles of plant-microbe interactions. ed. Lugtenberg B. (Cham: Springer International Publishing; ), 419–426. [Google Scholar]
- Berg G., Opelt K., Zachow C., Lottmann J., Götz M., Costa R., et al. (2006). The rhizosphere effect on bacteria antagonistic towards the pathogenic fungus Verticillium differs depending on plant species and site. FEMS Microbiol. Ecol. 56, 250–261. 10.1111/j.1574-6941.2005.00025.x, PMID: [DOI] [PubMed] [Google Scholar]
- Berg G., Rybakova D., Grube M., Köberl M. (2016). The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67, 995–1002. 10.1093/jxb/erv466, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G., Smalla K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. 10.1111/j.1574-6941.2009.00654.x, PMID: [DOI] [PubMed] [Google Scholar]
- Bokulich N. A., Kaehler B. D., Rideout J. R., Dillon M., Bolyen E., Knight R. A., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with qiime 2’s q2-feature-classifier plugin. Microbiome 6:90. 10.1186/s40168-018-0470-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. 10.1038/s41587-019-0209-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragina A., Berg C., Cardinale M., Shcherbakov A., Chebotar V., Berg G. (2012). Sphagnum mosses harbor highly specific bacterial diversity during their whole lifecycle. ISME J. 6, 802–813. 10.1038/ismej.2011.151, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl M. T. (2006). Fitness of human enteric pathogens on plants and implications for food safety. Annu. Rev. Phytopathol. 44, 367–392. 10.1146/annurev.phyto.44.070505.143359, PMID: [DOI] [PubMed] [Google Scholar]
- Buée M., De Boer W., Martin F., van Overbeek L., Jurkevitch E. (2009). The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321, 189–212. 10.1007/s11104-009-9991-3 [DOI] [Google Scholar]
- Bulgarelli D., Schlaeppi K., Spaepen S., van Themaat E. V. L., Schulze-Lefert P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. 10.1146/annurev-arplant-050312-120106, PMID: [DOI] [PubMed] [Google Scholar]
- Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). Dada2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. 10.1038/nmeth.3869, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. 10.1038/nmeth.f.303, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108, 4516–4522. 10.1073/pnas.1000080107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale M., Grube M., Erlacher A., Quehenberger J., Berg G. (2015). Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environ. Microbiol. 17, 239–252. 10.1111/1462-2920.12686, PMID: [DOI] [PubMed] [Google Scholar]
- Cernansky R. (2015). The rise of Africa’s super vegetables. Nature 522, 146–148. 10.1038/522146a, PMID: [DOI] [PubMed] [Google Scholar]
- Cernava T., Aschenbrenner I. A., Grube M., Liebminger S., Berg G. (2015). A novel assay for the detection of bioactive volatiles evaluated by screening of lichen-associated bacteria. Front. Microbiol. 6:398. 10.3389/fmicb.2015.00398, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelius M. K., Triplett E. W. (2001). The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41, 252–263. 10.1007/s002480000087, PMID: [DOI] [PubMed] [Google Scholar]
- Chong J., Liu P., Zhou G., Xia J. (2020). Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 15, 799–821. 10.1038/s41596-019-0264-1, PMID: [DOI] [PubMed] [Google Scholar]
- CIAT; BFS/USAID (2017). Climate-smart agriculture in Uganda. CSA Country profiles for Africa Series. International Center for Tropical Agriculture (CIAT); Bureau for Food Security, United States Agency for International Development (BFS/USAID), Washington, D.C. 22 p., Available at: https://climateknowledgeportal.worldbank.org/sites/default/files/2019-06/UGANDA_CSA_Profile.pdf (Accessed June 30, 2020)
- Cordovez V., Dini-Andreote F., Carrión V. J., Raaijmakers J. M. (2019). Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol. 73, 69–88. 10.1146/annurev-micro-090817-062524, PMID: [DOI] [PubMed] [Google Scholar]
- Coyne D., Abberton M., Adetonah S., Ayodele M., Cortada-Gonzales L., Gbaguidi L., et al. (2019). “Making integrated pest management (IPM) work in sub-Saharan Africa” in Critical issues in plant health: 50 years of research in African agriculture. eds. Neuenschwander P., Tamò M. (Cambridge, UK: Burleigh Dodds Science Publishing; ), 397–414. [Google Scholar]
- Dean R., Van Kan J. A. L., Pretorious Z. A., Hammond-Kosack K. E., Di Pietro A., Spanu P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. 10.1111/j.1364-3703.2011.00783.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey G., Kollah B., Gour V. K., Shukla A. K., Mohanty S. R. (2016). Diversity of bacteria and archaea in the rhizosphere of bioenergy crop Jatropha curcas. 3 Biotech 6:257. 10.1007/s13205-016-0546-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. 10.1016/0006-3207(92)91201-3 [DOI] [Google Scholar]
- Foster R. C., Rovira A. D., Cock T. W. (1983). Ultrastructure of the root-soil interface. ed. Paul St. (MN: American Phytophathological Society; ). [Google Scholar]
- Francis C. A., Beman J. M., Kuypers M. M. M. (2007). New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1, 19–27. 10.1038/ismej.2007.8, PMID: [DOI] [PubMed] [Google Scholar]
- García-Salamanca A., Molina-Henares M. A., van Dillewijn P., Solano J., Pizarro-Tobías P., Roca A., et al. (2013). Bacterial diversity in the rhizosphere of maize and the surrounding carbonate-rich bulk soil. Microb. Biotechnol. 6, 36–44. 10.1111/j.1751-7915.2012.00358.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons S. M., Lekberg Y., Mummey D. L., Sangwan N., Ramsey P. W., Gilbert J. A. (2017). Invasive plants rapidly reshape soil properties in a grassland ecosystem. mSystems 2:e00178–16. 10.1128/msystems.00178-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D., Défago G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. 10.1038/nrmicro1129, PMID: [DOI] [PubMed] [Google Scholar]
- Hamady M., Walker J. J., Harris J. K., Gold N. J., Knight R. (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5, 235–237. 10.1038/nmeth.1184, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardoim P. R., van Overbeek L. S., Berg G., Pirttilä A. M., Compant S., Campisano A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. 10.1128/MMBR.00050-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M., Frey B., Mayer J., Mäder P., Widmer F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. 10.1038/ismej.2014.210, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayat R., Ali S., Amara U., Khalid R., Ahmed I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. 10.1007/s13213-010-0117-1 [DOI] [Google Scholar]
- Ho A., di Lonardo D. P., Bodelier P. L. E. (2017). Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 93:fix006. 10.1093/femsec/fix006 [DOI] [PubMed] [Google Scholar]
- Innerebner G., Knief C., Vorholt J. A. (2011). Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 77, 3202–3220. 10.1128/AEM.00133-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- James B., Atcha-Ahowé C., Godonou I., Baimey H., Goergen H., Sikirou R., et al. (2010). Integrated pest management in vegetable production: A guide for extension workers in West Africa. Ibadan, Nigeria: International Institute of Tropical Agriculture. [Google Scholar]
- Katoh K., Standley D. M. (2013). Mafft multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. L., Waqas M., Kang S.-M., Al-Harrasi A., Hussain J., Al-Rawahi A., et al. (2014). Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 52, 689–695. 10.1007/s12275-014-4002-7, PMID: [DOI] [PubMed] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. 10.1093/nar/gks808, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Dagnoko S., Haougui A., Ratnadass A., Pasternak N., Kouamé C. (2010). Okra (Abelmoschus spp.) in West and Central Africa: potential and progress on its improvement. Afr. J. Agric. Res. 5, 3590–3598. 10.5897/AJAR10.839 [DOI] [Google Scholar]
- Leclère V., Béchet M., Adam A., Guez J. -S., Wathelet B., Ongena M., et al. (2005). Mycosubtilin overproduction by Bacillus subtilis BBG100 enhances the organism’s antagonistic and biocontrol activities. Appl. Environ. Microbiol. 71, 4577–4584. 10.1128/AEM.71.8.4577-4584.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Jeong S. E., Kim P. J., Madsen E. L., Jeon C. O. (2015). High resolution depth distribution of bacteria, archaea, methanotrophs, and methanogens in the bulk and rhizosphere soils of a flooded rice paddy. Front. Microbiol. 6:639. 10.3389/fmicb.2015.00639, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire G., Mnif W., Mauvais P., Balaguer P., Rahmani R. (2006). Activation of alpha‐ and beta-estrogen receptors by persistent pesticides in reporter cell lines. Life Sci. 79, 1160–1169. 10.1016/j.lfs.2006.03.023, PMID: [DOI] [PubMed] [Google Scholar]
- Li H., Ding X., Chen C., Zheng X., Han H., Li C., et al. (2019). Enrichment of phosphate solubilizing bacteria during late developmental stages of eggplant (Solanum melongena L.). FEMS Microbiol. Ecol. 3:fiz023. 10.1093/femsec/fiz023, PMID: [DOI] [PubMed] [Google Scholar]
- Lopez-Velasco G., Carder P. A., Welbaum G. E., Ponder M. A. (2013). Diversity of the spinach (Spinacia oleracea) spermosphere and phyllosphere bacterial communities. FEMS Microbiol. Lett. 346, 146–154. 10.1111/1574-6968.12216, PMID: [DOI] [PubMed] [Google Scholar]
- Lozupone C. A., Hamady M., Kelley S. T., Knight R. (2007). Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. 10.1128/AEM.01996-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B., Kamilova F. (2009). Plant-growth-promoting Rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. 10.1146/annurev.micro.62.081307.162918, PMID: [DOI] [PubMed] [Google Scholar]
- Lundberg D. S., Yourstone S., Mieczkowski P., Jones C. D., Dangl J. L. (2013). Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10, 999–1002. 10.1038/nmeth.2634, PMID: [DOI] [PubMed] [Google Scholar]
- Mariotte P., Mehrabi Z., Bezemer T. M., De Deyn G. B., Kulmatiski A., Drigo B., et al. (2018). Plant-soil feedback: bridging natural and agricultural sciences. Trends Ecol. Evol. 33, 129–142. 10.1146/annurev.micro.62.081307.162918, PMID: [DOI] [PubMed] [Google Scholar]
- Mayak S., Tirosh T., Glick B. R. (2004). Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 42, 565–572. 10.1016/j.plaphy.2004.05.009, PMID: [DOI] [PubMed] [Google Scholar]
- Mendes L. W., de Chaves M. G., de Cassia Fonseca M., Mendes R., Raaijmakers J. M., Tsai S. M. (2019). Resistance breeding of common bean shapes the physiology of the rhizosphere microbiome. Front. Microbiol. 10:2252. 10.3389/fmicb.2019.02252, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R., Kruijt M., de Bruijn I., Dekkers E., van der Voort M., Schneider J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. 10.1126/science.1203980, PMID: [DOI] [PubMed] [Google Scholar]
- Moissl-Eichinger C., Huber H. (2011). Archaeal symbionts and parasites. Curr. Opin. Microbiol. 14, 364–370. 10.1016/j.mib.2011.04.016, PMID: [DOI] [PubMed] [Google Scholar]
- Moissl-Eichinger C., Pausan M., Taffner J., Berg G., Bang C., Schmitz R. A. (2018). Archaea are interactive components of complex microbiomes. Trends Microbiol. 26, 70–85. 10.1016/j.tim.2017.07.004, PMID: [DOI] [PubMed] [Google Scholar]
- Mülner P., Schwar E., Dietel K., Junge H., Herfort S., Weydmann M., et al. (2020). Profiling for bioactive peptides and volatiles of plant growth promoting strains of the bacillus subtilis complex of industrial relevance. Front. microbiol. 11:1432. 10.3389/fmicb.2020.01432, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango C. M., Kunyanga C. N., Ontita E. G., Narla R. D., Kimenju J. W. (2013). Current status on production and utilization of spider plant (Cleome gynandra L.) an underutilized leafy vegetable in Kenya. Genet. Resour. Crop. Evol. 60, 2183–2189. 10.1007/s10722-013-0036-7, PMID: 28826642 [DOI] [Google Scholar]
- Pérez-Jaramillo J. E., Mendes R., Raaijmakers J. M. (2016). Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 90, 635–644. 10.1007/s11103-015-0337-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer S., Mitter B., Oswald A., Schloter-Hai B., Schloter M., Declerck S., et al. (2017). Rhizosphere microbiomes of potato cultivated in the high Andes show stable and dynamic core microbiomes with different responses to plant development. FEMS Microbiol. Ecol. 93:fiw242. 10.1093/femsec/fiw242, PMID: [DOI] [PubMed] [Google Scholar]
- Philippot L., Raaijmakers J. M., Lemanceau P., van der Putten W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. 10.1038/nrmicro3109, PMID: [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010). Fasttree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. 10.1093/nar/gks1219, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademaker J. L. W., de Bruijn F. J. (1997). “Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer assisted pattern analysis” in DNA markers: Protocols, applications and overviews. eds. Caetano-Anollés G., Gresshoff P. M. (Hoboken, NJ: Wiley & Sons; ), 151–171. [Google Scholar]
- Ronoh R., Linde M., Winkelmann T., Abukutsa-Onyango M., Dinssa F. F., Debener T. (2019). Morphological characterization, genetic diversity and population structure of African nightshades (section Solanum L.). Genet. Resour. Crop. Evol. 66, 105–120. 10.1007/s10722-018-0700-z [DOI] [Google Scholar]
- Rosenstock L., Keifer M., Daniell W. E., McConnell R., Claypoole K. (1991). Chronic central nervous system effects of acute organophosphate pesticide intoxication. Lancet 338, 223–227. 10.1016/0140-6736(91)90356-T, PMID: [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. 10.1101/gr.1239303, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora R. A., Terry E. R., Vlek P. L. G., Chitja J. (2019). Transforming agriculture in Southern Africa: Constraints, technologies, policies and processes. Milton, UK: Routledge publisher, 324. [Google Scholar]
- Simon H. M., Jahn C. E., Bergerud L. T., Sliwinski M. K., Weimer P. J., Willis D. K., et al. (2005). Cultivation of mesophilic soil crenarchaeotes in enrichment cultures from plant roots. Appl. Environ. Microbiol. 71, 4751–4760. 10.1128/AEM.71.8.4751-4760.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taffner J., Bergna A., Cernava T., Berg G. (2020). Tomato-associated archaea show a cultivar-specific rhizosphere effect but an unspecific transmission by seeds. Phytobiomes J. 4, 133–141. 10.1094/PBIOMES-01-20-0017-R [DOI] [Google Scholar]
- Taffner J., Cernava T., Erlacher A., Berg G. (2019). Novel insights into plant-associated archaea and their functioning in arugula (Eruca sativa Mill.). J. Adv. Res. 19, 39–48. 10.1016/j.jare.2019.04.008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taffner J., Erlacher A., Bragina A., Berg C., Moissl-Eichinger C., Berg G. (2018). What is the role of archaea in plants? New insights from the vegetation of alpine bogs. mSphere 3:e00122–18. 10.1128/mSphere.00122-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uksa M., Schloter M., Endesfelder D., Kublik S., Engel M., Kautz T., et al. (2015). Prokaryotes in subsoil-evidence for a strong spatial separation of different phyla by analysing co-occurrence networks. Front. Microbiol. 6:1269. 10.3389/fmicb.2015.01269, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenkoornhuyse P., Quaiser A., Duhamel M., Le Van A., Dufresne A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. 10.1111/nph.13312, PMID: [DOI] [PubMed] [Google Scholar]
- Vanlauwe B., Coyne D., Gockowski J., Hauser S., Huising J., Masso C., et al. (2014). Sustainable intensification and the African smallholder farmer. Curr. Opin. Environ. Sustain. 8, 15–22. 10.1016/j.cosust.2014.06.001 [DOI] [Google Scholar]
- Vázquez-Baeza Y., Pirrung M., Gonzalez A., Knight R. (2013). Emperor: a tool for visualizing high-throughput microbial community data. Gigascience 2:16. 10.1186/2047-217X-2-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswarlu B., Shanker A. K., Shanker C., Maheswari M. (eds.) (2012). Crop stress and its management: Perspectives and strategies. Dordrecht: Springer. [Google Scholar]
- Walters W., Hyde E. R., Berg-Lyons D., Ackermann G., Humphrey G., Parada A., et al. (2016). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009–15. 10.1128/mSystems.00009-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D. C., Sutton S. D., Ringelberg D. B. (1996). The genus Sphingomonas: physiology and ecology. Curr. Opin. Biotechnol. 7, 301–306. 10.1016/S0958-1669(96)80034-6, PMID: [DOI] [PubMed] [Google Scholar]
- Wolfgang A., Taffner J., Guimarães R. A., Coyne D., Berg G. (2019). Novel strategies for soil-borne diseases: exploiting the microbiome and volatile-based mechanisms toward controlling Meloidogyne-based disease complexes. Front. Microbiol. 10:1296. 10.3389/fmicb.2019.01296, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., DeBolt S., Dreyer J., Scott D., Williams M. A. (2015). Characterization of culturable bacterial endophytes and their capacity to promote plant growth from plants grown using organic or conventional practices. Front. Plant Sci. 6:490. 10.3389/fpls.2015.00490, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Zhang X., Cao Z., Zhao K., Wang S., Chen M., et al. (2014). Growth-promoting Sphingomonas paucimobilis ZJSH1 associated with Dendrobium officinale through phytohormone production and nitrogen fixation. Microb. Biotechnol. 7, 611–620. 10.1111/1751-7915.12148, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh Y. K., Dennis P. G., Paungfoo-Lonhienne C., Weber L., Brackin R., Ragan M. A., et al. (2017). Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat. Commun. 8:215. 10.1038/s41467-017-00262-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef N. H., Rinke C., Stepanauskas R., Farag I., Woyke T., Elshahed M. S. (2015). Insights into the metabolism, lifestyle and putative evolutionary history of the novel archaeal phylum ‘Diapherotrites’. ISME J. 9, 447–460. 10.1038/ismej.2014.141, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachow C., Müller H., Tilcher R., Donat C., Berg G. (2013). Catch the best: novel screening strategy to select stress protecting agents for crop plants. Agronomy 3, 794–815. 10.3390/agronomy3040794 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ebi.ac.uk/ena, PRJEB39392.