Abstract
Purpose:
The use of genomic sequencing (GS) in military settings poses unique considerations, including the potential for GS to impact service members’ careers. The MilSeq Project investigated the use of GS in clinical care of active-duty Airmen in the United States Air Force (USAF).
Methods:
We assessed perceived risks, benefits, and attitudes toward use of GS in the USAF among patient-participants (n=93) and health-care provider-participants (HCPs) (n=12) prior to receiving or disclosing GS results.
Results:
Participants agreed that there are health benefits associated with GS (90% patients, 75% HCPs), though more HCPs (75%) than patients (40%) agreed that there are risks (p=.048). The majority of both groups (67% HCPs, 77% patients) agreed that they trust the USAF with genetic information, but far fewer agreed that genetic information should be used to make decisions about deployment (5% patients, 17% HCPs) or duty assignments (3% patients, 17% HCPs). Despite their hesitancy, patients were supportive of the USAF testing for non-disease traits that could impact their duty performance. Eighty-seven percent of patients did not think their GS results would influence their career.
Conclusion:
Results suggest favorable attitudes toward the use of GS in the USAF when not used for deployment or assignment decisions.
Keywords: Genomics, Military, ELSI, Exome Sequencing, Healthy Populations
INTRODUCTION
Advances in genomic sequencing (GS) technologies have allowed researchers and clinicians to leverage a wealth of genomic information in diagnostic and precision medicine applications, garnering considerable interest for GS.1 From diagnosing medical conditions to tailoring treatments for many diseases, particularly cancer, based on molecular signatures of a patient’s tissues, there is much promise in the putative utility of GS, which has gained traction in the wider practice of civilian medicine.2–4 Given the growing applications of GS and its increasing affordability, the United States military has been investigating the feasibility, ethics, and challenges of integrating GS into military settings.5–7 Issues surrounding privacy and the potential for discrimination present unique challenges in the military, where fitness for specific duties and perceived mission-readiness are often important considerations.5,8–10
The collection and use of genetic information is not new in the military. Upon enlistment, all military service members are required to provide a blood sample in case genetic information is needed to help identify remains.8 Some branches of the military also regularly screen for some genetic conditions, including sickle cell trait and variants associated the metabolism of certain malaria medications, and make duty assignment decisions based on this information to reduce risk to service members.11–14 Additionally, scientific advisors to the military have highlighted the potential for GS to identify people with traits that would be of particular relevance to military performance and cost saving,7 and recent research has identified genetic variants associated with such phenotypes, including cognitive ability, endurance, tolerance of various conditions (e.g. sleep deprivation, dehydration, extreme temperature, and high altitude), and susceptibility to post-traumatic stress disorder.15–22 Whether and how to use screening information for such traits may pose new ethical considerations.
While targeted genetic screening raises concerns of its own, GS has the potential to raise additional issues. For instance, GS may reveal unanticipated or incidental genetic findings,23 though scholars believe there is only a small likelihood of such findings affecting service member job security.6,8 Additionally, issues surrounding genetic privacy and discrimination in the military are compounded by the fact that the employment protections afforded by the Genetic Information Non-Discrimination Act (GINA)24 do not apply to the military, though there are other provisions that govern the use of genetic information in the military healthcare system that offer many similar protections.6,11,25 Finally, there are challenges associated with preparing health-care providers (HCPs) who lack specialized training in genetics to disclose genetic testing results and counsel patients, an issue relevant to military health-care systems where geneticists may not be readily available.26,27
Despite ongoing discussions about the unique considerations concerning GS in the military, few studies have explored the views of active duty service members and military HCPs.27,28 The MilSeq Project is a pilot study exploring the integration of GS into the clinical care of Airmen in the U.S. Air Force (USAF). Here we report perceived risks, benefits, and utility of GS, and attitudes toward its use in the USAF among MilSeq patient-participants and HCP participants prior to receiving or disclosing GS results. Our findings contribute much needed empirical data on service members’ attitudes toward the use of GS in an active-duty military setting.
PATIENTS AND METHODS
Ethics Statement
The MilSeq Project was approved by the 59th Medical Wing Institutional Review Board (IRB). The Baylor College of Medicine IRB and the Partners HealthCare Human Research Committee (IRB of record for Brigham and Women’s Hospital) ceded review via reliance protocols. Study details may be publicly accessed at ClinicalTrials.gov with MilSeq identifier: NCT03276637. The voluntary, fully informed consent of the subjects used in this research was obtained as required by 32 CFR 219 and AFI 40-402, Protection of Human Subjects in Biomedical and Behavioral Research.
Study Design
The MilSeq Project enrolled ostensibly healthy, active-duty patients (n=93) and active-duty HCPs (n=12) from the USAF. HCP-participants (hereinafter “HCPs”), who were also Airmen, were recruited in person and by group announcement. HCPs attended a 3-hour genetics education session with a genetic counselor29 and completed pre- and post-education surveys.
Patient-participants (hereinafter “patients”) were recruited in person in USAF health clinics, and via email, social media advertisements, and word of mouth. Patients completed an electronic survey pre-enrollment, which assessed their interest in undergoing GS. Patients who indicated interest in receiving GS provided their contact information and were invited to a consent session for GS with a MilSeq Project genetic counselor. HCPs returned GS results directly to patients, and all results were included in the patient’s medical record. This paper focuses on attitudes toward GS, including perceived risks, benefits, and utility, prior to receiving or disclosing GS results collected via patients’ pre-enrollment and the HCPs’ pre-education surveys.
Measures
Participants’ perceived risks and benefits of GS, concerns, and attitudes toward the use of GS in the military were assessed using ten items with five-point Likert scale response options from 1 (“strongly disagree”) to 5 (“strongly agree”) with 3 as a neutral midpoint. To measure perceived utility, participants were asked to rate how useful they felt GS was for managing health now and managing health in the future on a ten-point scale from 1 (“not at all useful”) to 10 (“extremely useful”). Attitudes toward USAF testing for mental health risks and nondisease traits that could affect duty performance were measured using 15 items answered on a five-point Likert scale anchored by 1 (“definitely not”) and 5 (“definitely yes”) with 3 (“I don’t know”) as the midpoint. Participants were informed that the MilSeq Project would not test for these mental health risks and nondisease traits and that the genetic basis for most of these traits is not yet well established. General health was assessed using the first item of the Veterans RAND 12 Item Health Survey (VR-12).[30, 31] Patients’ genetic knowledge was assessed using 11 true–false items.[32] HCPs’ genetic knowledge was assessed using 14 multiple-choice items developed using published and novel items, including vignettes based on the HCP education session.[29, 32–35]
Data Analysis
Descriptive statistics, including means, standard deviations, and proportions, were calculated to evaluate participants’ characteristics. The total number of correct genetic knowledge questions were summed for each participant. Since the data for each measure were not normally distributed, non-parametric Mann-Whitney U tests were used to compare the perceived risks and benefits, concerns, attitudes, and perceived current and future utility among patients versus HCPs. Wilcoxon signed-rank tests were used to assess whether there were differences in perceived current versus future utility of GS among patients as well as HCPs. A p-value less than 0.05 was considered statistically significant. All statistical calculations were performed using R statistical software (R Foundation for Statistical Computing, Vienna, Austria).
Participants who agreed or strongly agreed that there were risks or benefits of GS were asked to describe those risks and benefits in an open-ended response. A coding team experienced in qualitative research methods and overseen by a qualitative research methods expert (SP) conducted thematic analysis on these responses using a consensus coding approach.
RESULTS AND DISCUSSION
Participant Characteristics
Ninety-three patients and 12 HCPs completed surveys at baseline (pre-enrollment and pre-education). All patients indicated interest in receiving GS. Patients’ and HCPs’ characteristics are reported in Tables 1 and 2. Patients were 52% male, 69% non-Hispanic White, and the majority were college graduates or higher (61%) with an average age of 34 years old. Half (52.7%) of the patients indicated that they worked in a health-related field. Of the 12 HCPs, 8 were male (66.7%), and half were non-Hispanic White. The majority of the HCPs had been in practice for less than 10 years (83.4%) and only one reported previous training in genetics beyond the typical medical school curriculum.
Table 1.
Patient Characteristics
| Characteristic – N (%) unless otherwise noted | N=93 |
|---|---|
| Age (n=87) | |
| Mean in years (SD) | 33.3 (±8.2) |
| Gender (n=93) | |
| Male | 48 (52%) |
| Female | 45 (48%) |
| Race/Ethnicitya (n=93) | |
| Hispanic or Latino | 17 (18%) |
| Non-Hispanic White | 64 (69%) |
| Non-Hispanic Otherb | 13 (14%) |
| Prefer Not to Answer | 2 (2%) |
| Education (n=93) | |
| Did not graduate from college | 36 (39%) |
| College graduate or higher | 57 (61%) |
| Annual Household Income (n=93) | |
| ≤ $99,999 | 64 (69%) |
| ≥ $100,000 | 29 (31%) |
| Genetic Knowledge Score (n=93) | |
| Mean percentage correct (SD) | 91% (±10%) |
| General Self-Rated Healthc (n=93) | |
| Very good to excellent health | 67 (72%) |
3 participants selected Non-Hispanic White and another race
Non-Hispanic Other includes Black or African American, Asian, and Other
General self-rated health was asked on a 5-point scale from Poor (1) to Excellent (5), with Good (3) as the midpoint
Table 2.
Health Care Provider Characteristics
| Characteristic – N (%) unless otherwise noted | N=12 |
|---|---|
| Age (n=11) | |
| Mean in years (SD) | 37.9 (±9.3) |
| Gender (n=12) | |
| Male | 8 (67%) |
| Female | 4 (33%) |
| Race/Ethnicity (n=12) | |
| Non-Hispanic White | 6 (50%) |
| Non-Hispanic Othera | 6 (50%) |
| Years in Practice (n=12) | |
| 1-10 | 10 (83%) |
| 21-30 | 2 (17%) |
| Genetics Training (n=12) | |
| No | 11 (92%) |
| Yes | 1 (8%) |
| Genetic Knowledge Score (n=12) | |
| Mean percentage correct (SD) | 71% (±10%) |
Non-Hispanic Other includes Black or African American and Asian
Benefits, Risks, and Perceived Utility of Genomic Sequencing
Overall, participants agreed that there are health benefits associated with GS, though more (90%) patients than HCPs (75%) agreed (median patients =5 (strongly agreed), HCP =4 (agreed); p=.002). In response to open-ended questions, the main themes cited by both patients and HCPs related to the health benefits of GS included the potential for risk assessment, early detection and prevention, and precision medicine. Patients also reported the potential for GS to increase their feelings of agency by giving them a greater sense of control and enabling proactive health choices, the impact this information could have on their family and family planning, and the sense that they were contributing to research as benefits.
More HCPs (75%) agreed that there are risks associated with GS compared to patients (40%) (median patients =3, HCP =4, p=0.048). When asked to describe those risks, the most common theme among patients and HCPs was the risk of negative psychological impact of GS results and potential for discrimination, including insurance and employment discrimination. Both groups reported concerns about the likelihood of receiving results of unclear significance and the information’s potential to prompt unnecessary medical interventions.
Aligned with these findings, more patients (82%) than HCPs (42%) reported that the benefits of GS outweigh the risks (median patients =4, HCP =3, p<0.001). Additionally, more HCPs (64%) than patients (16%) agreed that they were concerned about the potential for discrimination if patients’ genetic information is not protected (median patients =3, HCP =4, p=0.002), though patients were largely unsure, with 44% selecting neither agree nor disagree.
With regard to utility, patients rated the current utility of genomic information higher than HCPs did (median patient =8, HCP =5.5, p=0.003), though patients and HCPs rated the future utility of GS similarly (median patient =9, HCP =8.5, p=0.176). Both groups rated the utility of GS for managing health significantly higher in the future compared to now (both p<0.001).
Use of Genomic Sequencing in the Air Force
We also explored participants’ attitudes toward the use of GS specifically in the USAF (Figure 1). The majority of both groups reported that they trust the USAF with their genetic information (67% of HCPs and 77% of patients). When asked if the USAF should require GS for all Airmen, however, only 17% of patients and 25% of HCPs agreed. Participants who agreed that the military should require GS (n=16) cited the potential to improve the health of individuals and the military population’s fitness as a whole and to strengthen the military healthcare system and research. Patients and HCPs who disagreed that testing should be required believed that undergoing GS should be a personal decision and questioned both the military health system’s logistical readiness for the implementation of GS and individuals’ psychological preparedness and willingness to receive the information.
Figure 1.
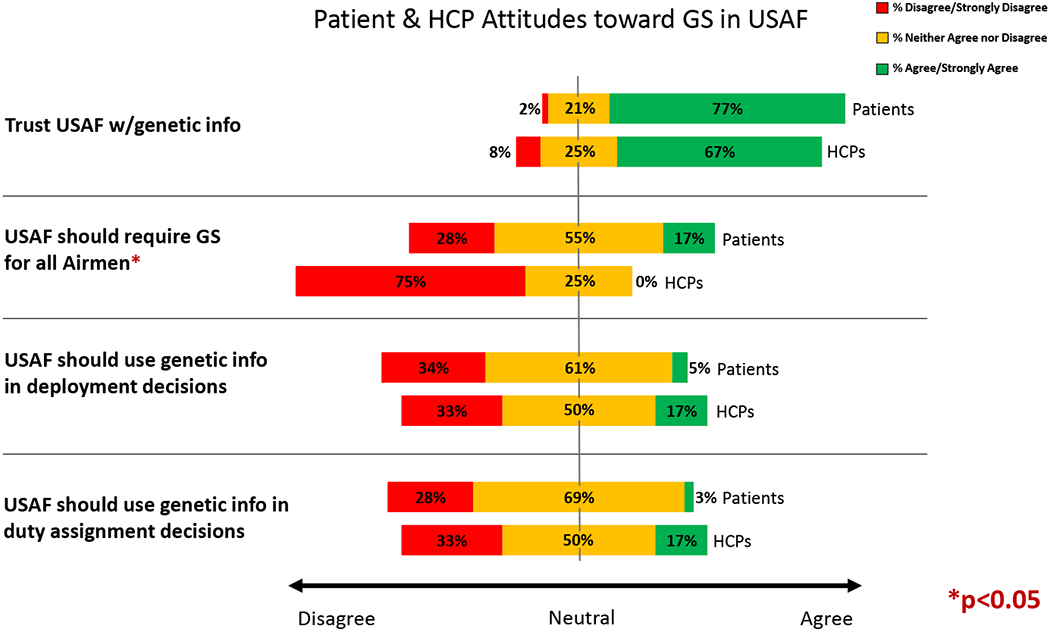
Patient and HCP Attitudes toward GS in the USAF
[Genome sequencing] costs a lot of money to taxpayers and the [Department of Defense] in general spends too much money as it is. Also, what I read about genomic sequencing leads me to believe this research is new and it comes with a lot of controversy. Does the [Air Force Medical Service] want to take on that scrutiny? This information could be potentially damaging to a person’s psychological well-being, without the disease or affliction ever coming to fruition.
–Patient
Because some patients don’t want to know their genetic make-up - it is their right not to know.
–HCP
Participants were generally ambivalent or negative when asked whether genetic information should be used to make decisions about deployment (61% of patients ambivalent and 34% negative; 50% of HCPs ambivalent and 33% negative) or duty assignments (69% of patients ambivalent and 28% negative; 50% of HCPs ambivalent and 33% negative). A minority of patients and HCPs saw potential to use this information to “avoid undue risk.” As one HCP said, “It depends on the findings. If a deployed setting could be the environmental factor in a genetically predisposed person, then deployment should be avoided.”
Most HCPs and patients, however, expressed ambivalence about using genetic information for deployment decisions due to uncertainty regarding its applicability.
How would it be helpful? What genetic problems have historically been a problem on deployment? None that I’m aware of.
–HCP
Genetics do not equal phenotype. The Air Force wouldn’t decide to deploy me because my parents were both expert marksmen, I don’t expect them to make decisions based just on genomics.
—Patient
Additionally, they thought that acting on this information could lead to issues including discrimination or unnecessary logistical burdens.
There’s no relevance of genetic information to deploying. A person is capable of deploying regardless. Using genetics to determine qualified deployers would limit the available manning and put more of a burden on certain members to deploy often.
–Patient
Eighty-seven percent of patients did not think their GS results would influence their career, and only 13% agreed that they were concerned that their GS results could affect their career. In comparison, a third of HCPs (33%) agreed that their patients’ careers could be affected by their GS results, while the majority (58%) neither agreed nor disagreed.
Mental health and duty performance trait testing
We asked participants whether they thought the USAF should routinely test Airmen for mental health risks and hypothetical traits that could impact duty performance, including tolerance for extreme conditions and personal traits, to assess potential acceptance of this testing (Table 3). It was explicitly stated that the MilSeq Project would not be testing for any of these and that the genetic bases for these traits are not yet well established. A majority of patients indicated support for the USAF to probably or definitely test for all listed conditions and traits. Those supported by the most patients were testing for injury recovery, tolerance for sleep deprivation, and physical endurance. Though still a majority for each, fewer patients supported testing for impulsivity, tolerance for starvation, and risk of substance abuse. HCPs were less receptive to such testing, with only 5 of the 15 traits and conditions supported by a majority of HCPs. The traits supported by the most HCPs were tolerance for sleep deprivation, tolerance for dehydration, and tolerance for extreme environments. The traits supported by the least HCPs were speed, physical endurance, and tolerance for pain.
Table 3.
Patient and HCP attitudes toward hypothetical implementation of routine mental health and nondisease trait testing in the USAF
| Trait | Yes | No | I donť know | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | HCP | Patient | HCP | Patient | HCP | ||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | ||
| Tolerance for Extreme Conditions | Extreme environments | 62 | 67% | 8 | 67% | 13 | 14% | 2 | 17% | 18 | 19% | 2 | 17% |
| Sleep deprivation | 71 | 76% | 8 | 67% | 8 | 9% | 8 | 17% | 14 | 15% | 8 | 17% | |
| Starvation | 53 | 57% | 6 | 50% | 21 | 23% | 2 | 17% | 19 | 20% | 4 | 33% | |
| Dehydration | 67 | 72% | 8 | 67% | 11 | 12% | 2 | 17% | 15 | 16% | 2 | 17% | |
| Personal Traits | Tolerance for pain | 57 | 62% | 4 | 33% | 16 | 17% | 2 | 17% | 19 | 21% | 6 | 50% |
| Injury recovery | 75 | 81% | 6 | 50% | 6 | 6% | 4 | 33% | 12 | 13% | 2 | 17% | |
| Strength | 63 | 68% | 5 | 42% | 8 | 9% | 4 | 33% | 21 | 23% | 3 | 25% | |
| Speed | 62 | 67% | 4 | 33% | 9 | 10% | 4 | 33% | 21 | 23% | 4 | 33% | |
| Physical endurance | 70 | 75% | 4 | 33% | 7 | 8% | 4 | 33% | 16 | 17% | 4 | 33% | |
| Intelligence | 65 | 70% | 4 | 33% | 12 | 13% | 5 | 42% | 16 | 17% | 3 | 25% | |
| Impulsivity | 48 | 52% | 7 | 58% | 18 | 19% | 1 | 8% | 27 | 29% | 4 | 33% | |
| Mental Health Risks | PTSD | 61 | 66% | 6 | 50% | 14 | 15% | 0 | 0% | 18 | 19% | 6 | 50% |
| Depression | 64 | 69% | 6 | 50% | 12 | 13% | 0 | 0% | 17 | 18% | 6 | 50% | |
| Anxiety | 65 | 70% | 5 | 42% | 11 | 12% | 1 | 8% | 17 | 18% | 6 | 50% | |
| Substance Abuse | 58 | 62% | 7 | 58% | 17 | 18% | 2 | 17% | 18 | 19% | 3 | 25% | |
Discussion
Overall, both patients and HCPs in the USAF were generally positive toward genomic sequencing, agreeing that there are health benefits associated with GS, and that there is the potential for GS information to be useful for managing one’s health, particularly in the future. Patients were slightly more optimistic than HCPs, perceiving less risk and more benefit, but they did identify the potential for genetic discrimination as a risk of GS. Patients also identified more types of benefits associated with GS than HCPs did. The way participants balance these perceived risks and benefits may underlie their general positive attitudes toward GS and patients’ willingness to undergo sequencing. It seems patients felt that the potential benefits, including benefits to their own health and the health of their families, outweighed the risks. HCPs, conversely, were less confident that the benefits of GS outweigh the risks to their patients.
Despite general optimism and relatively high trust in the Air Force, a strong majority of both patients and HCPs were ambivalent or negative toward making GS mandatory in the USAF, and toward the use of genetic information for job-specific decisions including deployment and duty assignments. This is particularly interesting since most (n=75) of the pre-enrollment survey participants went on to enroll in the GS portion of the study within the context of the USAF healthcare system, in which GS results were entered into the patients’ medical records. The reason for the seeming disconnect may be that very few patients actually thought that their results would influence their career or expressed any concern about that, possibly related to the fact that they rated themselves as generally healthy. USAF policy prohibits any action based on a genetic variant if there is no accompanying phenotype.6 The potential for GS information to harm service members’ careers has been highlighted in the literature as one of the major unique issues associated with integration of GS into military settings,6,9,10,27 but our patient participants were largely unconcerned about this. Similarly, only one-third of HCPs in this study thought their patients’ careers could be impacted by GS. These findings contradict those from a survey of genetics providers who care for active-duty military personnel, where 71% of providers were more concerned about the genetic privacy of their active-duty patients than their civilian patients and reported that they had had patients who were concerned about how genetic testing could impact their careers.28 It is worth noting that nearly all the providers surveyed in that study were civilians and the majority had not received any training in military healthcare, while the HCPs in our study were all active-duty military personnel practicing within the USAF healthcare system. It is possible that those providers’ lack of knowledge and familiarity with military healthcare drove their concern. Other research, however, has found that experience with and knowledge of military rules related to genetics varies widely by specialty even among HCPs working within a military healthcare facility.27
Additionally, despite their hesitancy toward the use of genetic information for duty-specific purposes, patients were largely supportive of the USAF testing for non-disease traits that could impact their duty performance and, consequently, their careers. HCPs were less supportive than patients, but a majority still supported testing for most of the tolerance for extreme conditions traits. In 2010, a group of scientific advisors to the military encouraged the use of genomic technologies to identify individuals with propensity for traits that would be of particular interest to the military, either for performance or cost saving purposes.7 Others have argued that genetic testing for traits associated with military-relevant abilities could exacerbate existing disparities.36 Though the genetic bases for many of the traits we asked about have not been definitively identified, recent research has made progress on some of them, making the potential to screen for such genotypes increasingly possible. Though patients were hypothetically supportive of such testing, their reluctance overall to use genetic information for duty-specific purposes points to unease or uncertainty regarding how this information could be used. It is unclear what uses patients had in mind when responding to the trait testing questions, if not for use in duty-specific decisions. Patients may have underestimated the likelihood that genetic propensity for traits would be used for such purposes if the USAF tested for them. Future research that further explores this tension should be used to inform any trait-screening program.
Of note, for questions about universal genomic screening in the USAF and use of genomic information to inform deployment and duty assignments, many patients and HCPs chose neutral midpoints when available as response options, indicating that they were largely unsure of or ambivalent toward how GS should be used in this setting, despite their general optimism and trust and lack of concern about career impact. This may be due to lack of experience with GS or lack of knowledge about how such information could be used. It may also be that though patients felt it was unlikely their own GS results would include a career-impacting finding, particularly since they rated themselves as generally healthy, they may have been concerned that their colleagues could be negatively impacted, or that such screening could negatively impact military recruitment by screening out a larger portion of the population. This ambivalence toward how GS should be used may have important implications for its integration in military settings. Additional education on how genetic information could impact military careers could help military personnel make informed decisions about GS if offered. A robust informed consent procedure for GS could help ensure that military personnel are aware of the potential uses of this information before agreeing to undergo any such screening.
Participants in this study self-selected for participation, and as such, their attitudes may not be representative of the larger United States Air Force population. Those who were uninterested in participating in the MilSeq Project may have more critical opinions of genomic sequencing. About half of our patient participants reported working in a health-related field, which may have influenced their perspectives toward GS, though it is unclear whether that would lead to more positive or more critical attitudes. Further, no pilots chose to enroll in this study, which may be a reflection of our recruitment strategy and potentially non-representative clinic populations, or may be due to pilots’ concerns about how genetic information could impact their careers. Our patient sample was also largely non-Hispanic White, and thus not representative of the active duty forces. Due to our sampling strategy, we are also unable to determine our response rate since it is unknown how many people viewed our social media advertisements or heard of the study through word of mouth, thereby decreasing our ability to speak to the representativeness of our sample. Additionally, the data presented here are from surveys completed at enrollment, and thus do not reflect attitudes in response to receiving or disclosing genomic information. Future work will examine data from follow-up surveys to investigate participants’ attitudes post-disclosure of genomic results. Despite these limitations, this is one of the first studies of Airmen’s attitudes toward the use of GS in an active duty military setting and thus contributes much needed empirical evidence that can help shape future practice and policy around the use of GS in the military.
As genomic sequencing is increasingly integrated into different clinical care settings, population-specific considerations need to be taken into account. This study is a first step toward exploring the use of genomic sequencing in a military setting. Our findings suggest that Airmen may be receptive to the incorporation of GS in a military setting, provided use limitations are in place. Ongoing work will explore whether attitudes change after return of GS results to assess impact of providing this information to ostensibly healthy Airmen.
ACKNOWLEDGMENTS
This material is based on research sponsored by Air Force Medical Support Agency, Research and Acquisition Directorate AFMSA/SG5 under cooperative agreement number FA8650-17-2-6704. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright notation thereon. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of Air Force Medical Support Agency, Research and Acquisition Directorate AFMSA/SG5 or the U.S. Government.
This work was also supported by K01 HG009173/HG/NHGRI NIH HHS/United States.
We thank Steven Dicker for his invaluable input on our hypothetical trait testing questions.
Robert C. Green MD, MPH receives compensation for advising the following companies: AIA, Applied Therapeutics, Grail, Humanity, SavvySherpa, Wamberg Advisors; and is co-founder of Genome Medical, Inc, a technology and services company providing genetics expertise to patients, providers, employers and care systems. Emily Sirotich, MS is a Board Member of the Canadian Arthritis Patient Alliance, a patient run, volunteer-based organization whose activities are largely supported by independent grants from pharmaceutical companies.
Footnotes
CONFLICT OF INTEREST NOTIFICATION PAGE
The remaining authors do not have any commercial association that might pose, create or create the appearance of a conflict of interest with the information presented in the submitted manuscript.
References
- 1.Biesecker LG & Green RC Diagnostic clinical genome and exome sequencing. N. Engl. J. Med 370, 2418–2425 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Aronson SJ & Rehm HL Building the foundation for genomics in precision medicine. Nature 526, 336–342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins FS & Varmus H A new initiative on precision medicine. N. Engl. J. Med 372, 793–795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakagawa H, Wardell CP, Furuta M, Taniguchi H & Fujimoto A Cancer whole-genome sequencing: present and future. Oncogene 34, 5943–5950 (2015). [DOI] [PubMed] [Google Scholar]
- 5.De Castro MJ & Turner CE Military genomics: a perspective on the successes and challenges of genomic medicine in the Armed Services. Mol. Genet. Genomic Med 5, 617–620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Castro M et al. Genomic medicine in the military. Npj Genomic Med. 1, 15008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.JASON, The MITRE Corporation. The $100 Genome: Implications for the DOD. (2010). [Google Scholar]
- 8.Mehlman MJ & Li TY Ethical, legal, social, and policy issues in the use of genomic technology by the U.S. Military. J. Law Biosci 1, 244–280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helm BM, Langley K, Spangler BB & Schrier Vergano SA Military Health Care Dilemmas and Genetic Discrimination: A Family’s Experience with Whole Exome Sequencing. Narrat. Inq. Bioeth 5, 179–186 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Hellwig LD et al. Return of secondary findings in genomic sequencing: Military implications. Mol. Genet. Genomic Med 7, e00483 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baruch S & Hudson K Civilian and military genetics: nondiscrimination policy in a post-GINA world. Am. J. Hum. Genet 83, 435–444 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webber BJ & Witkop CT Screening for sickle-cell trait at accession to the United States military. Mil. Med 179, 1184–1189 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Kark JA, Posey DM, Schumacher HR & Ruehle CJ Sickle-cell trait as a risk factor for sudden death in physical training. N. Engl. J. Med 317, 781–787 (1987). [DOI] [PubMed] [Google Scholar]
- 14.O’Connor FG et al. ACSM and CHAMP summit on sickle cell trait: mitigating risks for warfighters and athletes. Med. Sci. Sports Exerc 44, 2045–2056 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Banerjee SB, Morrison FG & Ressler KJ Genetic approaches for the study of PTSD: Advances and challenges. Neurosci. Lett 649, 139–146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Coso J et al. More than a ‘speed gene’: ACTN3 R577X genotype, trainability, muscle damage, and the risk for injuries. Eur. J. Appl. Physiol 119, 49–60 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Bruenig D et al. Nitric oxide pathway genes (NOS1AP and NOS1) are involved in PTSD severity, depression, anxiety, stress and resilience. Gene 625, 42–48 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Costa-Mattioli M et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 129, 195–206 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czeisler CA Medical and Genetic Differences in the Adverse Impact of Sleep Loss on Performance: Ethical Considerations for the Medical Profession. Trans. Am. Clin. Climatol. Assoc 120, 249–285 (2009). [PMC free article] [PubMed] [Google Scholar]
- 20.Bray MS et al. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med. Sci. Sports Exerc 41, 35–73 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Varley I, Patel S, Williams AG & Hennis PJ The current use, and opinions of elite athletes and support staff in relation to genetic testing in elite sport within the UK. Biol. Sport 35, 13–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dlamini N et al. Mutations in RYR1 are a common cause of exertional myalgia and rhabdomyolysis. Neuromuscul. Disord. NMD 23, 540–548 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Dorschner MO et al. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am. J. Hum. Genet 93, 631–640 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genetic Information Nondiscrimination Act of 2008. vol. 122 Stat. 881 (2008). [Google Scholar]
- 25.Genetic Discrimination. Genome.gov https://www.genome.gov/about-genomics/policy-issues/Genetic-Discrimination. [Google Scholar]
- 26.Vassy JL, Korf BR & Green RC How to know when physicians are ready for genomic medicine. Sci. Transl. Med 7, 287fs19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellwig LD, Turner C & O’Neill SC Patient-centered care and genomic medicine: A qualitative provider study in the military health system. J. Genet. Couns 28, 940–949 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Jacober J Counseling the Active-Duty Military Population: Knowledge, Attitudes, and Experience of Clinical Genetics Providers. (Brandeis University, 2019). [Google Scholar]
- 29.Maxwell MD et al. Educating military primary health-care providers in genomic medicine: lessons learned from the MilSeq Project. Genet. Med 1–8 (2020) doi: 10.1038/s41436-020-0865-7. [DOI] [PubMed] [Google Scholar]
- 30.DeSalvo KB, Bloser N, Reynolds K, He J & Muntner P Mortality Prediction with a Single General Self-Rated Health Question. J. Gen. Intern. Med 21, 267–275 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selim AJ et al. Updated U.S. population standard for the Veterans RAND 12-item Health Survey (VR-12). Qual. Life Res. Int. J. Qual. Life Asp. Treat. Care Rehabil 18, 43–52 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Kaphingst KA et al. Effects of informed consent for individual genome sequencing on relevant knowledge. Clin. Genet 82, 408–415 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vassy JL et al. The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials 15, 85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowling BV et al. Development and evaluation of a genetics literacy assessment instrument for undergraduates. Genetics 178, 15–22 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Harvard Personal Genome Project (PGP) – enabling participant-driven science. https://pgp.med.harvard.edu/. [Google Scholar]
- 36.Roberts JL ‘Good soldiers are made, not born’⊥: the dangers of medicalizing ability in the military use of genetics. J. Law Biosci 2, 92–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]


