Abstract
Background:
Primary high-risk human papillomavirus (hr-HPV) testing of self-collected cervico-vaginal swabs could increase cervical cancer screening coverage, although triage strategies are needed to reduce unnecessary colposcopies. We evaluated the use of extended hr-HPV genotyping of self-collected samples for cervical cancer screening.
Methods:
We recruited women age 25-65 years at two colposcopy clinics in North Carolina between 11/2016-01/2019, and obtained self-collected cervico-vaginal samples, provider-collected cervical samples, and cervical biopsies from all enrolled women. Self- and provider-collected samples were tested for 14 hr-HPV genotypes using the Onclarity assay (Becton Dickinson, Sparks, MD). We calculated hr-HPV genotype-specific prevalence and assessed agreement between results in self- and provider-collected samples. We ranked the hr-HPV genotypes according to their positive predictive value (PPV) for the detection of cervical intraepithelial neoplasia grade 2 or higher (CIN2+).
Results:
Three-hundred-and-fourteen women participated (median age: 36 years); 85 women (27%) had CIN2+. More women tested positive for any hr-HPV on self-collected (76%) than on provider-collected samples (70%; p=0.009) with type-specific agreement ranging from substantial to almost perfect. HPV-16 was the most common genotype in self-collected (27%) and provider-collected samples (20%), and HPV-16 prevalence was higher in self- than provider-collected samples (p<0.001). In self- and provider-collected samples, HPV-16 had the highest PPV for CIN2+ detection.
Conclusion:
Overall sensitivity for CIN2+ detection was similar for both sample types, but the higher HPV-16 prevalence in self-collected samples could result in increased colposcopy referral rates.
Impact:
Additional molecular markers might be helpful to improve the triage of women who are hr-HPV-positive on self-collected samples.
Introduction
Almost all cervical cancer cases are caused by oncogenic high-risk (hr) human papillomavirus (HPV) types.1 However, not all hr-HPV types have the same potential for causing progression to cervical cancer.2–5 In 2018, the United States Preventive Services Task Force (USPSTF) proposed primary hr-HPV testing or a combination of Pap and hr-HPV testing (co-testing) every five years for women ages 30 to 65 years.6 Current guidelines of the American Society for Colposcopy and Cervical Pathology (ASCCP) recommend immediate referral to colposcopy for women who are cytology negative for intraepithelial lesion or malignancy (NILM) if they are infected with hr-HPV genotypes 16 or 18, irrespective of HPV vaccination status.7 This screening algorithm was designed because HPV-16 and HPV-18 are the two most common types associated with cervical cancer, accounting for approximately 60% and 10-15% of cervical cancers, respectively.3,8 HPV types 31, 33, 45, 52, and 58 combined account for another 19% of cervical cancer cases.9 However, the introduction of HPV vaccinations has led to a shift in the HPV genotype distributions with lower HPV-16/18 prevalence among younger immunized women as compared with unimmunized women.10,11 Extended hr-HPV genotyping beyond HPV-16/18 could serve as a triage strategy and aid in optimizing the detection of clinically important high-grade cervical dysplasia while simultaneously minimizing unnecessary colposcopy referrals for cases of transient or non-progressing HPV infection.5,12–14
Most cervical cancer cases occur in under-screened women.15–17 Main barriers to care include limited access to costly screening, lack of transportation, and personal reasons including embarrassment, fear of finding cancer, and anxiety about undergoing a pelvic examination.18 Many of these barriers could be addressed by accurate and low-cost hr-HPV self-sampling strategies. Most women prefer self-collected cervico-vaginal brushes/swabs over provider-collected samples for hr-HPV testing.19,20 However, self-collection for hr-HPV testing is not yet FDA approved for clinical use.
Many different HPV tests exist, but currently, only the cobas 4800 assay (Roche Diagnostics, Indianapolis, IN) and the Onclarity assay (Becton Dickinson, Sparks, MD) are FDA-approved with a clinical indication for primary hr-HPV screening. The cobas assay provides individual hr-HPV results for HPV-16 and 18, although not for the other hr-HPV types. The Onclarity assay has the potential to provide extended genotyping results by simultaneously detecting DNA from 14 hr-HPV types – six types are individually genotyped (HPV-16, 18, 31, 45, 51, and 52) and the remaining eight types in three groups (33/58, 56/59/66, and 33/39/68).21
In this study, we evaluated the potential use of extended hr-HPV genotyping of self-collected cervico-vaginal samples for cervical cancer screening. Specifically, using the Onclarity assay, we compared the hr-HPV type distributions in self- and provider-collected samples stratified by cervical lesion grade, assessed agreement between self- and provider-collected samples for hr-HPV positivity, and computed positive predictive values (PPVs) of different hr-HPV genotypes for the detection of cervical intraepithelial neoplasia grade 2 or higher (CIN2+).
Methods
Study population
Between November 2016 and January 2019, we recruited a convenience sample of women ages 25 to 65 years attending colposcopy clinics at either the University of North Carolina (UNC) Women’s Hospital or Duke University Hospital for one of the following reasons: i) abnormal cytology results, ii) infection with HPV-16 or 18, iii) persistent infection with other hr-HPV genotypes, or iv) treatment for CIN2+. In addition, we invited women to participate in the study if they were NILM on cytology, but positive for hr-HPV genotypes other than 16 or 18 at their routine screening. This group was referred to as “research only,” because immediate referral for colposcopy is currently not recommended for these women.7
Potentially eligible women were selected through a review of electronic medical records and were contacted via phone or during their clinic visit to ask if they would participate. Women were excluded from participation if they were pregnant or had had their cervix removed; additionally, women in the “research only” group were excluded if they were taking blood thinners, or if the enrollment date was not within three months of their original hr-HPV diagnosis. Women were not asked to abstain from sexual intercourse before the study visit. Written informed consent was obtained from each eligible woman willing to participate, and the study was approved by the Institutional Review Boards of UNC and of Duke University.
Sample collection
During the clinic visit participating women received detailed verbal and written instructions concerning the study procedures in either English or Spanish. Women self-collected a cervico-vaginal sample by inserting a Viba brush (Rovers Medical Devices BV, The Netherlands) to the top of the vaginal canal, rotating five times, removing it, and releasing the brush head into a vial prefilled with 6 mL of preservative liquid-based cytology media (ThinPrep, Hologic Inc., Bedford, Mass.). A urine pregnancy test was performed when clinically indicated.
Next, women underwent a pelvic examination during which the clinical provider collected a cervical scraping with two 360 degree turns in a clockwise fashion of a brush-like cervical cell collector (Wallach Papette, Wallach Surgical Devices, Trumbull, CT). The provider-collected cervical sample was preserved in a standard 20 mL vial of ThinPrep media for subsequent hr-HPV testing. Colposcopy was performed on all participating women, following cervical treatment with 3-5% acetic acid (followed by Lugol’s iodine at the Duke site), according to standard clinical procedures. Directed biopsies were taken from visible cervical lesions, and an endocervical curettage (ECC) was performed if the transformation zone or the limits of a lesion near the cervical os could not be fully visualized. If no cervical lesions were observable, one random biopsy at the 12 o’clock position of the cervix was taken and an ECC was performed. Loop electrosurgical excision procedure (LEEP) was performed when clinically indicated. At the end of the visit, women received a gift card for their participation in the study.
Sample processing and laboratory analyses
All samples were placed in a cooler with frozen gel packs within 10 minutes of sample collection and kept cool until they could be further aliquoted the same day. The self- and provider-collected samples were vortexed for 10-30 seconds and 0.5 mL of each were transferred to separate BD molecular tubes containing 1.7 mL of an HPV diluent buffer. Initially, samples were not vortexed. When this laboratory error was discovered, samples were re-aliquoted after vortexing them for 10-30 seconds. However, 69 provider-collected samples and 3 self-collected samples were not available for re-aliquoting and, therefore, had to be excluded from the analysis. The tubes are fitted with a pierceable cap to facilitate automated sample processing on the BD Viper™ LT System. The tubes were stored at −20°C until shipment to BD for hr-HPV testing using the Onclarity assay (Becton Dickinson, Sparks, MD). The staff who performed the hr-HPV testing at BD did not have access to any clinical information including cervical histology results of the participants. The BD Onclarity assay uses PCR and nucleic acid hybridization to detect DNA of 14 hr-HPV genotypes. Six hr-HPV genotypes (16, 18, 31, 45, 51, and 52) are individually genotyped, and the remaining eight are identified in three groups (33/58, 56/59/66, and 35/39/68). Self- and provider-collected specimens were processed using the standard liquid-based cytology workflow on the BD Viper™ LT System. We used the following established PCR cycle thresholds (Ct) for both self- and provider-collected samples: ≤ 38.3 for HPV-16, and ≤ 34.2 for all other hrHPV genotypes. The remaining provider-collected cervical sample was sent to the UNC cytopathology laboratory for liquid-based ThinPrep cytological analysis, if clinically indicated, or otherwise stored at −20°C.
Cervical biopsies from participating women who underwent a colposcopy as part of their scheduled clinical appointment were sent to UNC or Duke Hospital Surgical Pathology Laboratory for histological evaluation per standard clinical procedures. Pathologists had access to clinical information captured in the electronic medical records but were unaware of the study hr-HPV test results. Women who underwent a clinically indicated colposcopy were informed of their histological results by the clinical team per standard of care. Biopsies taken from women in the “research only” group, who underwent the colposcopy for study purposes, were histologically analyzed by a gynecologic pathologist (O’Connor) at the UNC translational pathology laboratory who did not have access to any additional clinical information. Women in the “research-only” group were contacted with histological results by the study team after review by a practicing gynecologist (Rahangdale, Knittel). If CIN2+ was detected, the women were referred to further treatment as per standard of care.
Statistical analyses
We included women with valid hr-HPV test results on both self- and provider-collected samples and valid cervical histology results. We compared socio-demographic characteristics and CIN2+ status between eligible women with and without matched self- and provider-collected samples using Fisher’s exact test. We calculated hr-HPV genotype-specific prevalence stratified by cervical lesion grade and compared genotype-specific prevalence between self- and provider-collected samples using the McNemar’s test. We computed the unweighted Cohen’s kappa and its 95% confidence interval (CI) to assess agreement beyond chance between hr-HPV results in self- and provider-collected samples. We calculated Cohen’s kappa overall, stratified by cervical lesion grade, and for specific hr-HPV genotypes. We used the following interpretation for Cohen’s kappa: ≤ 0, no agreement; 0.01-0.20, slight agreement; 0.21-0.40, fair agreement; 0.41-0.60, moderate agreement; 0.61-0.80, substantial agreement; and 0.81-1.00, almost perfect agreement.23 We ranked the hr-HPV genotypes detected in self- and provider-collected samples according to their PPV for the detection of CIN2+ from the highest to the lowest PPV excluding women with multiple-type infections. In a sensitivity analysis, we ranked the genotypes based on both single- and multiple-type infections. For this analysis, PPVs were calculated after excluding women who had one of the previously ranked genotypes. We computed cumulative sensitivity and specificity for the detection of CIN2+ based on the sequential addition of genotypes according to the obtained order. In a second sensitivity analysis, we ranked the genotypes based on their PPVs for detection of CIN3+. Analyses were done using SAS/STAT® (SAS Institute Inc., Cary, NC, USA), Stata 15 (College Station, TX: StataCorp LLC), and R software (R Foundation for Statistical Computing, Vienna, Austria).
In total, 434 women (363 with an indication for colposcopy, and 71 in the “research only” group) were enrolled in the study (Figure 1). Of those, 413 women had valid histology results and were available for analysis. However, 70 provider-collected samples and 33 self-collected samples had hr-HPV results that were either invalid or unavailable. Thus, the analyses reported here are based on data from 314 women with valid hr-HPV results for both the self-collected cervico-vaginal and the provider-collected cervical samples. Baseline characteristics such as age, race/ethnicity, marital status, education, health insurance, and smoking status, as well as CIN2+ status did not significantly differ (Supplementary table S1) between eligible women with (N = 314) and without matched samples (N = 99).
Figure 1:
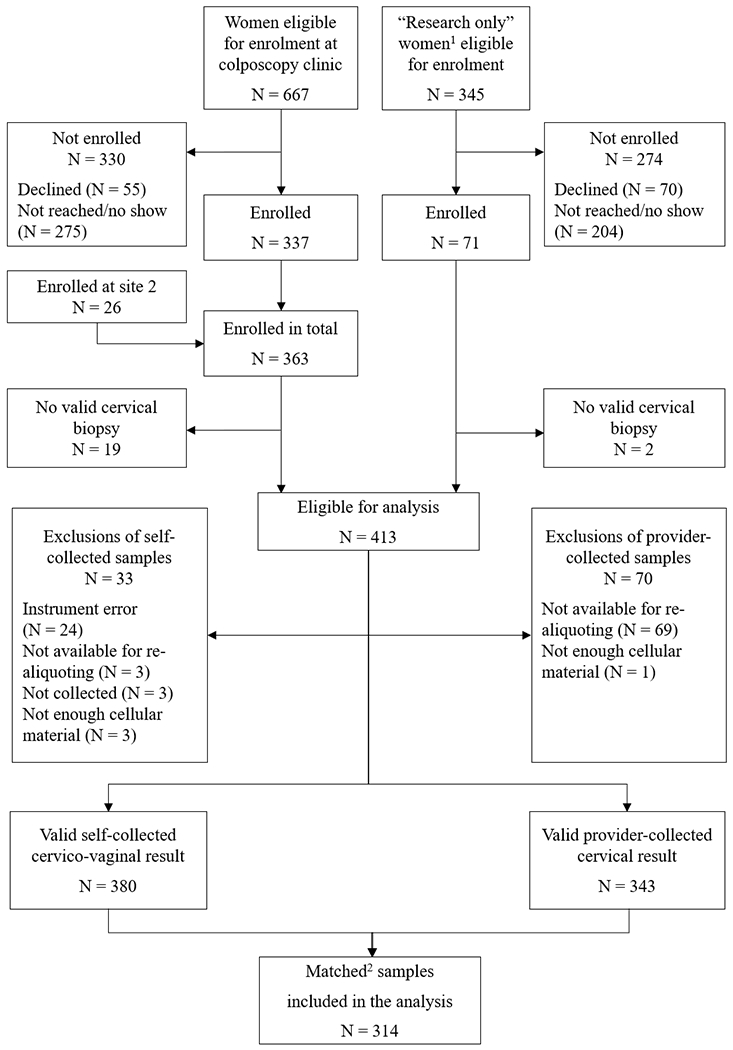
Study Flow diagram. 1“Research only” women were negative for intraepithelial lesion or malignancy on cytology, but positive for hr-HPV genotypes other than 16 or 18. 2Matched self-collected and provider-collected samples available for the same participant
Results
Study population
The median age of the 314 participating women was 36 years (interquartile range [IQR] 31-45 years). The study population was ethnically and racially diverse with 38% non-Hispanic White (N = 120), 29% Hispanic (N = 92), 26% non-Hispanic Black (N = 82), and 6% women with other racial identities (N = 20; Table 1). Eighty-five (27%) women were diagnosed with CIN2+.
Table 1:
Characteristics of and high-risk (hr)-HPV positivity on self- or provider-collected samples among 314 study participants*
| Overall N = 314 | Self-collection hr-HPV-positive† N = 239 |
Provider-collection hr-HPV-positive† N = 220 |
|
|---|---|---|---|
| n (%) | n (%) | n (%) | |
| Age groups [years] | |||
| 25 – 29 | 56 (18%) | 46 (19%) | 45 (20%) |
| 30 – 39 | 135 (43%) | 110 (46%) | 98 (45%) |
| 40 – 49 | 69 (22%) | 45 (19%) | 43 (20%) |
| 50 – 65 | 54 (17%) | 38 (16%) | 34 (15%) |
| Race and Ethnicity | |||
| Hispanic | 92 (29%) | 69 (29%) | 72 (33%) |
| Non-Hispanic Black | 82 (26%) | 66 (28%) | 51 (23%) |
| Non-Hispanic White | 120 (38%) | 88 (37%) | 83 (38%) |
| Other‡ | 20 (6%) | 16 (7%) | 14 (6%) |
| Marital Status | |||
| Married or living with a partner | 119 (39%) | 87 (38%) | 85 (40%) |
| Divorced, separated, widowed | 82 (27%) | 64 (28%) | 56 (26%) |
| Single/never married | 101 (33%) | 80 (35%) | 71 (33%) |
| Missing | 12 | 8 | 8 |
| Education | |||
| Elementary school or less | 27 (9%) | 20 (9%) | 20 (9%) |
| High school | 93 (31%) | 68 (29%) | 69 (33%) |
| Some college | 106 (35%) | 80 (35%) | 69 (33%) |
| College graduate | 77 (25%) | 63 (27%) | 54 (25%) |
| Missing | 11 | 8 | 8 |
| Health insurance | |||
| Private insurance | 120 (39%) | 91 (38%) | 80 (37%) |
| Medicaid/Medicare/TRICARE | 71 (23%) | 52 (22%) | 43 (20%) |
| None/Uninsured | 120 (39%) | 94 (40%) | 95 (44%) |
| Missing | 3 | 2 | 2 |
| Current Smoker | |||
| Yes | 72 (23%) | 55 (23%) | 53 (25%) |
| No | 238 (77%) | 180 (77%) | 163 (75%) |
| Missing | 4 | 4 | 4 |
Study population includes 250 women (80%) with a clinical indication for colposcopy and 64 women (20%) who underwent the colposcopy for study purposes.
Positive for any of the 14 high-risk (hr) HPV types detected by the Onclarity assay
Includes Asian (10), American Indian/Alaskan Native (5), Native Hawaiian/Other Pacific Islander (1), Black Indian (1), Mediterranean (1), not further specified (2).
High-risk HPV prevalence
Overall, more women tested positive for any hr-HPV on self-collected samples (N = 239, 76%) than on provider-collected samples (N = 220, 70%; p-value = 0.009; Figure 2A). Multiple-type hr-HPV infections were also more common in self-collected samples (N = 76, 24%) than in provider-collected samples (N = 44, 14%; p-value < 0.001). HPV-16 was the most common hr-HPV type detected in self-collected (N = 85, 27%) and in provider-collected samples (N = 62, 20%). Prevalence of HPV-16 was significantly higher in self-collected cervico-vaginal samples than in provider-collected cervical samples (p-value < 0.001). Especially, prevalence of multiple-type HPV-16 infections was higher in self-collected versus provider-collected samples (Table 2). When we restricted the analysis to women with CIN2+ or CIN3+, HPV prevalence did not significantly differ between self-collected cervico-vaginal samples and provider-collected cervical samples (Figure 2B, Table 2). Among women with a histological diagnosis of <CIN2, the pattern of a higher hr-HPV prevalence in self-collected than provider-collected samples was observed across all hr-HPV genotypes, although differences by collection site reached statistical significance only for any hr-HPV (p-value = 0.004), HPV-16 (p-value < 0.001), HPV-33/58 (p-value = 0.046), and HPV-35/39/68 (p-value = 0.008). Supplementary tables S2 and S3 show prevalence of hr-HPV genotypes in self- and provider-collected samples among all 85 CIN2+ cases, stratified by race/ethnicity and age, respectively.
Figure 2:
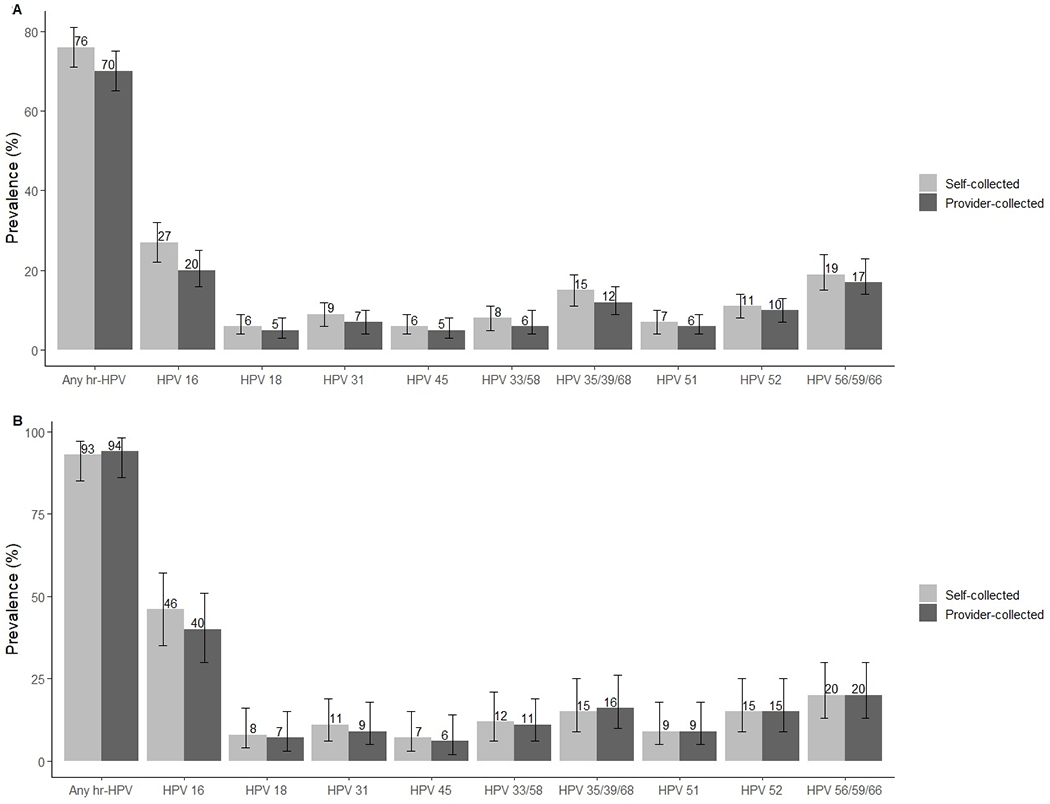
Prevalence of any hr-HPV and specific genotypes with 95% confidence intervals in all (Panel A) self-collected cervico-vaginal and provider-collected cervical samples and among CIN2+ cases (Panel B)
Table 2:
Counts and prevalence of high-risk (hr)-HPV types, stratified by cervical lesion grade, sample collection method, and infection type (single- versus multiple-type)
| hr-HPV and infection type | <CIN2† (N = 229) | CIN2+‡ (N = 85) | CIN3+§∥ (N = 49) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Self | Provider | p-value¶ | Self | Provider | p-value¶ | Self | Provider | p-value¶ | |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | ||||
| Any hr-HPV* | 160 (70%) | 140 (61%) | 0.004 | 79 (93%) | 80 (94%) | 0.65 | 47 (96%) | 47 (96%) | 1.00 |
| Single | 115 (50%) | 120 (52%) | 48 (56%) | 56 (66%) | 26 (53%) | 31 (63%) | |||
| Multiple | 45 (20%) | 20 (9%) | 31 (36%) | 24 (28%) | 21 (43%) | 16 (33%) | |||
| 16 | 46 (20%) | 28 (12%) | <0.001 | 39 (46%) | 34 (40%) | 0.13 | 29 (59%) | 24 (49%) | 0.06 |
| Single | 25 (11%) | 21 (9%) | 22 (26%) | 21 (25%) | 16 (33%) | 14 (29%) | |||
| Multiple | 21 (9%) | 7 (3%) | 17 (20%) | 13 (15%) | 13 (27%) | 10 (20%) | |||
| 18 | 11 (5%) | 9 (4%) | 0.41 | 7 (8%) | 6 (7%) | 0.56 | 4 (8%) | 4 (8%) | 1.00 |
| Single | 6 (3%) | 7 (3%) | 2 (2%) | 0 (0%) | 0 (0%) | 0 (0%) | |||
| Multiple | 5 (2%) | 2 (1%) | 5 (6%) | 6 (7%) | 4 (8%) | 4 (8%) | |||
| 31 | 18 (8%) | 13 (6%) | 0.10 | 9 (11%) | 8 (9%) | 0.32 | 4 (8%) | 4 (8%) | 1.00 |
| Single | 7 (3%) | 8 (3%) | 5 (6%) | 6 (7%) | 2 (4%) | 3 (6%) | |||
| Multiple | 11 (5%) | 5 (2%) | 4 (5%) | 2 (2%) | 2 (4%) | 1 (2%) | |||
| 45 | 13 (6%) | 11 (5%) | 0.41 | 6 (7%) | 5 (6%) | 0.56 | 4 (8%) | 3 (6%) | 0.32 |
| Single | 10 (4%) | 10 (4%) | 2 (2%) | 2 (2%) | 1 (2%) | 2 (4%) | |||
| Multiple | 3 (1%) | 1 (<1%) | 4 (5%) | 3 (4%) | 3 (6%) | 1 (2%) | |||
| 33/58 | 15 (7%) | 11 (5%) | 0.046 | 10 (12%) | 9 (11%) | 0.32 | 6 (12%) | 6 (12%) | 1.00 |
| Single | 11 (5%) | 10 (4%) | 5 (6%) | 7 (8%) | 2 (4%) | 4 (8%) | |||
| Multiple | 4 (2%) | 1 (<1%) | 5 (6%) | 2 (2%) | 4 (8%) | 2 (4%) | |||
| 35/39/68 | 34 (15%) | 23 (10%) | 0.008 | 13 (15%) | 14 (16%) | 0.56 | 5 (10%) | 6 (12%) | 0.32 |
| Single | 19 (8%) | 17 (7%) | 4 (5%) | 8 (9%) | 1 (2%) | 2 (4%) | |||
| Multiple | 15 (7%) | 6 (3%) | 9 (11%) | 6 (7%) | 4 (8%) | 4 (8%) | |||
| 51 | 13 (6%) | 11 (5%) | 0.16 | 8 (9%) | 8 (9%) | 1.00 | 4 (8%) | 4 (8%) | 1.00 |
| Single | 3 (1%) | 8 (3%) | 2 (2%) | 2 (2%) | 1 (2%) | 1 (2%) | |||
| Multiple | 10 (4%) | 3 (1%) | 6 (7%) | 6 (7%) | 3 (6%) | 3 (6%) | |||
| 52 | 20 (9%) | 17 (7%) | 0.26 | 13 (15%) | 13 (15%) | 1.00 | 9 (18%) | 8 (16%) | 0.32 |
| Single | 12 (5%) | 11 (5%) | 4 (5%) | 6 (7%) | 3 (6%) | 4 (8%) | |||
| Multiple | 8 (3%) | 6 (3%) | 9 (11%) | 7 (8%) | 6 (12%) | 4 (8%) | |||
| 56/59/66 | 43 (19%) | 40 (17%) | 0.37 | 17 (20%) | 17 (20%) | 1.00 | 10 (20%) | 10 (20%) | 1.00 |
| Single | 22 (10%) | 28 (12%) | 2 (2%) | 4 (5%) | 0 (0%) | 1 (2%) | |||
| Multiple | 21 (9%) | 12 (5%) | 15 (18%) | 13 (15%) | 10 (20%) | 9 (18%) | |||
hr-HPV: Positive for any of the 14 high-risk HPV types detected by the Onclarity assay
<CIN2: Less than cervical intraepithelial neoplasia grade 2
xCIN2+: Cervical intraepithelial neoplasia grade 2 or higher
CIN3+: Cervical intraepithelial neoplasia grade 3 or higher
CIN2/3 cases were analyzed as CIN3+ cases
p-value: From McNemar’s test which assessed difference between the hr-HPV prevalence in self- and provider-collected samples for a given HPV type or group
Agreement between hr-HPV results in self- and provider-collected samples
Overall agreement between self- and provider-collected samples for any hr-HPV was 83% (Supplementary table S4); agreement beyond chance was moderate with a Cohen’s kappa value of 0.57 (95% CI 0.47-0.67). Type-specific agreement beyond chance among all included women ranged from substantial (0.69; 95% CI 0.60-0.79) for HPV-16 to almost perfect (0.95; 95% CI 0.87-1.00) for HPV-51 (Figure 3A). When we restricted the analysis to women with CIN2+, agreement beyond chance remained moderate overall (kappa: 0.51, 95% CI 0.14-0.89) and ranged from substantial to perfect for specific hr-HPV genotypes (Figure 3B). Among the 85 CIN2+ cases, 31 cases were positive for HPV-16 on both sample types, 8 cases on self-collected specimens only, and 3 cases on provider-collected specimens only.
Figure 3:
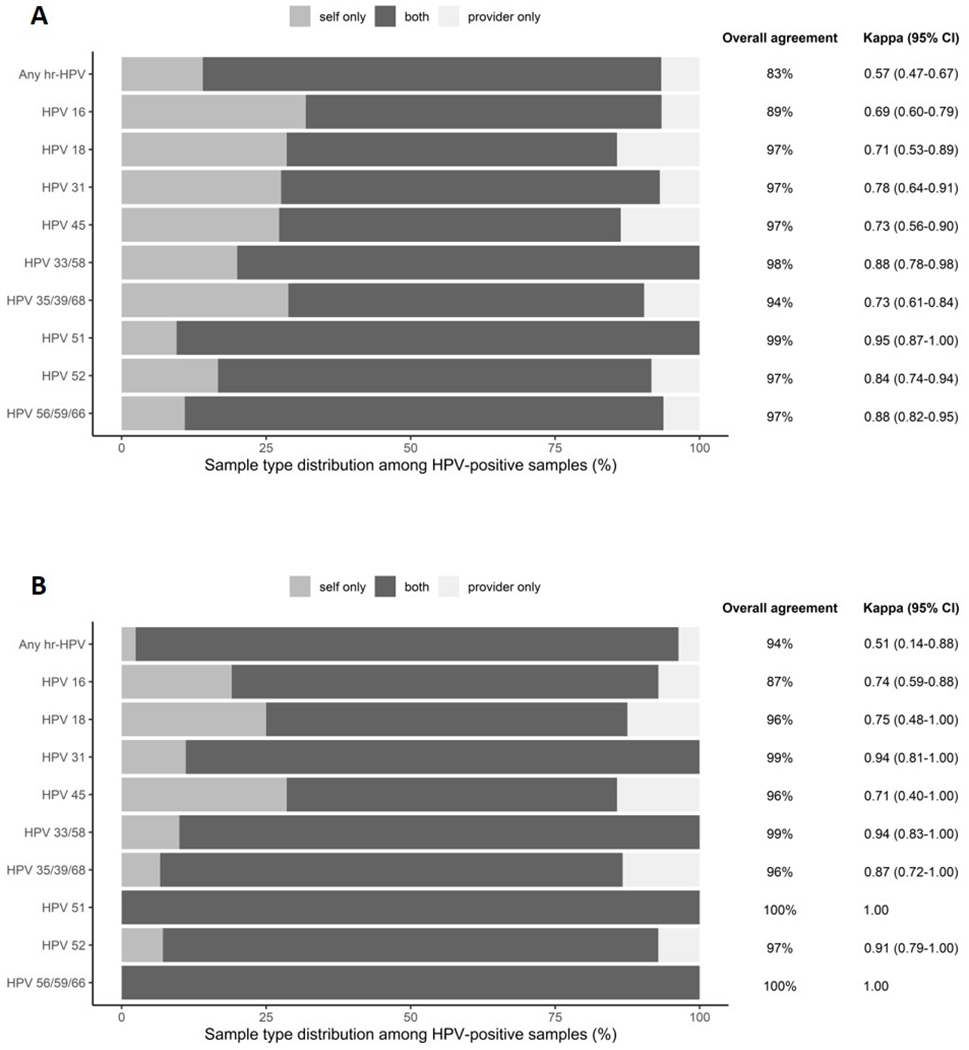
HPV type-specific agreement between all (Panel A) self-collected cervico-vaginal and provider-collected cervical samples and among CIN2+ cases (Panel B).
Cumulative sensitivity and specificity of different hr-HPV genotypes for CIN2+ detection
In both self- and provider-collected samples, positivity for HPV-16 had the highest PPV for the detection of CIN2+ when considering single-type infections only (Table 3). However, the subsequent order of hr-HPV genotypes selected to maximize the PPV among the remaining samples varied slightly between self- and provider-collection. In provider-collected samples, the order was HPV-16, 31, 33/58, 52, 35/39/68, 51, 45, 56/59/66, and 18. In self-collected samples, the order was HPV-16, 31, 51, 33/58, 52, 18, 35/39/68, 45, and 56/59/66. Positivity for HPV-16 alone had a sensitivity of 34.4% in provider-collected samples and 40.7% in self-collected samples for the detection of CIN2+; specificity was 90.0% and 86.4%, respectively. By adding hr-HPV genotypes to the algorithm in the determined order, the cumulative sensitivity for detection of CIN2+ increased to 91.8% in provider-collected and 88.9% in self-collected specimens when all 14 hr-HPV were included (Table 3). Cumulative specificity decreased to 42.6% in provider-collected and 37.5% in self-collected samples when an algorithm based on all 14 hr-HPV genotypes was used.
Table 3:
Classification of hr-HPV genotypes according to positive predictive value for CIN2+, excluding women with multiple type infections
| hr-HPV Type | Cumulative | ||||
|---|---|---|---|---|---|
| N at risk | CIN2+/hr-HPV+ | PPV | Sensitivity | Specificity | |
| Provider-collected samples | |||||
| HPV-16 | 270 | 21/42 | 50.0% | 34.4% | 90.0% |
| HPV-31 | 228 | 6/14 | 42.9% | 44.3% | 86.1% |
| HPV-33/58 | 214 | 7/17 | 41.2% | 55.7% | 81.3% |
| HPV-52 | 197 | 6/17 | 35.3% | 65.6% | 76.1% |
| HPV-35/39/68 | 180 | 8/25 | 32.0% | 78.7% | 67.9% |
| HPV-51 | 155 | 2/10 | 20.0% | 82.0% | 64.1% |
| HPV-45 | 145 | 2/12 | 16.7% | 85.2% | 59.3% |
| HPV-56/59/66 | 133 | 4/32 | 12.5% | 91.8% | 45.9% |
| HPV-18 | 101 | 0/7 | 0.0% | 91.8% | 42.6% |
| Self-collected samples | |||||
| HPV-16 | 238 | 22/47 | 46.8% | 40.7% | 86.4% |
| HPV-31 | 191 | 5/12 | 41.7% | 50.0% | 82.6% |
| HPV-51 | 186 | 2/5 | 40.0% | 53.7% | 81.0% |
| HPV-33/58 | 170 | 5/16 | 31.3% | 63.0% | 75.0% |
| HPV-52 | 154 | 4/16 | 25.0% | 70.4% | 68.5% |
| HPV-18 | 146 | 2/8 | 25.0% | 74.1% | 65.2% |
| HPV-35/39/68 | 123 | 4/23 | 17.4% | 81.5% | 54.9% |
| HPV-45 | 111 | 2/12 | 16.7% | 85.2% | 49.5% |
| HPV-56/59/66 | 87 | 2/24 | 8.3% | 88.9% | 37.5% |
CIN2+; cervical intraepithelial neoplasia grade 2 or more severe; hr-HPV, high-risk human papillomavirus; PPV, positive predictive value.
When including women with multiple-type infections, we found that HPV-16, 33/58, 51 and 31 had the highest PPVs for CIN2+ detection in both provider- and self-collected samples (Supplementary table S5). In provider-collected samples, the order was HPV-16, 33/58, 51, 31, 52, 35/39/68, 45, 56/59/66, and 18. In self-collected samples, the order was HPV-16, 51, 33/58, 31, 52, 18, 56/59/66, 45, and 35/39/68. Cumulative sensitivity based on all 14 hr-HPV genotypes for single- and multiple-type infections combined was 94.1% in provider-collected samples and 92.9% in self-collected samples. For the detection of CIN3+, HPV-16, 33/58, and 52 had the highest PPVs in both provider- and self-collected samples based on single- and multiple-type infections combined (Supplementary table S6).
Discussion
Among women with histologically confirmed CIN2+ included in our study, the prevalence of hr-HPV in provider-collected (94%) and self-collected samples (93%) was similar. Among all 314 participants, however, more women tested positive for any hr-HPV on self-collected cervico-vaginal samples (76%) than on provider-collected cervical samples (70%). Overall agreement between self- and provider-collected samples was moderate (kappa = 0.57), but type-specific agreement ranged from substantial (e.g. HPV-16) to almost perfect (e.g. HPV-51). Based on single-type infections only, HPV-16 and 31 had the highest PPVs for CIN2+ in both self- and provider-collected samples.
HPV prevalence was somewhat higher in self-collected cervico-vaginal compared to provider-collected cervical samples in this study, which is consistent with some studies,24–27 although other studies have shown comparable28,29 or slightly lower HPV prevalence in self-collected samples.30,31 When summarized in a systematic review and meta-analysis, however, HPV prevalence in self- and provider-collected samples seems to be similar.32 In line with others,28,33 we also found a higher prevalence of multiple-type infections in self-collected cervico-vaginal as compared to provider-collected cervical samples. The sampling order could have played a role in finding a higher HPV prevalence in self-collected samples, as the sample obtained first might have yielded more exfoliated cells and, therefore, more HPV DNA, than the second sample. The sequence of sample collection in our study was not randomized with self-collection always occurring before provider-collection. However, a randomized clinical trial found that hr-HPV detection was not affected by the sequence in which self- and provider-collection were performed.34 We provided participating women with comprehensive self-collection instructions in English and Spanish, but incorrect performance of sample self-collection might still have affected the hr-HPV test results obtained for self-collected samples. Although, in an acceptability study based on the same study population, we found that women generally thought that the self-collection was easy to perform.35 Furthermore, in our study the self-collected sample was approximately 3.3-fold more concentrated than the provider-collected sample (resuspended in 6mL versus 20 mL ThinPrep), which might also have contributed to the higher HPV prevalence in self-collected samples. The unequal sample concentrations may have affected genotypes differentially, given that the Ct cut-off to determine positivity was higher for HPV-16 (≤38.3) than for all other hr-HPV genotypes (≤34.2). However, we used the same established Ct cut-offs for both self- and provider-collected samples. Another potential explanation for the higher HPV prevalence in self-collected samples is that vaginal HPV infections may be acquired earlier in time than cervical infections,36 and that not all vaginal HPV infections will go on to infect the cervix.
In line with other studies,25,28 we have shown good type-specific agreement, indicating that HPV detected in cervico-vaginal samples are representative of cervical HPV infections. High type-specific agreement is essential for self-sampling to be a valid alternative to provider-based hr-HPV testing for cervical cancer screening, as the ultimate goal is to prevent cervical disease. Our finding that only few HPV-16 infections among CIN2+ cases detected in provider-collected cervical samples were missed by the self-collected samples is reassuring, as this is the hr-HPV genotype most commonly associated with cervical cancer.3,8 However, the higher observed HPV-16 prevalence in self- compared to provider-collected samples would also result in higher colposcopy referral rates. Additional triage strategies should be considered to avoid unnecessary referrals and overtreatment among women who are positive for HPV-16 on self-collected cervico-vaginal samples but do not have CIN2+.
A recent meta-analysis showed that PCR-based hr-HPV assays were similarly accurate for the detection of CIN2+ in self- and in provider-collected samples indicating that self-collected samples can substitute for provider-collected samples to reach under-screened women.37 In high-risk study participants enriched for CIN2+ outcomes, the meta-analysis reported sensitivities of approximately 90% and specificities of approximately 50% for PCR-based hr-HPV testing in self- and provider-collected samples for the detection of CIN2+.37 In our study, we found similar sensitivities for CIN2+ detection of approximately 90%, but lower specificities of 30-40%. Our specificity estimates increased substantially to approximately 85% when we used a primary screening algorithm based on HPV-16 only, consistent with other studies,38,39 although our sensitivity estimates dropped to 40-46% using this strategy. We and others40,41 found HPV-33, 16, and 31 to be among the genotypes with the highest PPVs for CIN2+ detection. This is also in line with studies that identified HPV-16 and 31 as the genotypes with the highest risk for CIN2+ among women with normal11 or low-grade cervical cytology.14 HPV 51 had a relatively high PPV for CIN2+ detection in our study, especially when multiple-type infections were included in a sensitivity analysis, but was considered intermediate risk for CIN2+ in other studies.40,41 Of note, one of these studies was cross-sectional,40 like ours, whereas the other used histological findings obtained over a 3-year follow-up period.41 Interestingly, HPV-18 and 45, two hr-HPV genotypes commonly associated with cervical cancer,3,8,9 ranked low in our ordering of genotypes based on PPVs for CIN2+. This might be partly attributable to a more rapid trajectory for progression given infection with these two HPV types.42 Alternatively, HPV-18 and 45, both of which belong to the alpha-7 HPV species, are associated with glandular lesions and cervical adenocarcinoma.43 These endocervical lesions are harder to detect by conventional colposcopy, and thus, some of these lesions might have been missed in our study. However, given that we performed an ECC if the transformation zone or the limits of a lesion near the cervical os could not be fully visualized the risk of missing endocervical lesions is low in our study. Furthermore, although provider-collection is more likely to sample from the endocervix and thus, detect glandular lesions, than self-collection, HPV-18 and 45 prevalence and the genotype ranking appeared to be similar in self- and provider-collected samples. In general, our genotype ranking should be interpreted cautiously due to the relatively low number of samples positive for specific genotypes such as HPV-51, 18, and 45 and, therefore, the limited precision of our estimates for these genotypes.
The majority of cervical cancer cases occur in under-screened women.15–17 Primary hr-HPV testing on self-collected cervico-vaginal samples may help increase cervical cancer screening coverage, as women generally prefer self-collection over provider-collection for hr-HPV testing.19,20 However, most hr-HPV infections clear within one year, and only a small proportion develops into cervical cancer.44,45 Therefore, a triage strategy is needed to reduce the number of unnecessary colposcopy referrals among cases of transient or non-progressing hr-HPV infection identified through primary hr-HPV testing. Biomarkers for triage of hr-HPV infections can be broadly categorized into morphological and molecular biomarkers.46 Extended hr-HPV genotyping beyond HPV-16/18 could serve as a molecular triage strategy.5,12–14,46 We found type-specific agreement between self- and provider-collected samples to range from substantial to almost perfect, indicating that extended hr-HPV genotyping algorithms identified in provider-collected samples may also be valid for self-collected samples. For HPV-16 we found a significantly higher prevalence in self- versus provider-collected samples, which may result in higher colposcopy referral rates when self-collected samples are used. However, when we restricted the analysis to women with CIN2+ or CIN3+, hr-HPV prevalence did not significantly differ between self-collected cervico-vaginal samples and provider-collected cervical samples. DNA-based extended hr-HPV genotyping tests are unable to discriminate between transient and persistent hr-HPV infections at a cross-sectional timepoint.46 Therefore, it is likely that a combination of biomarkers involved in different stages of the cervical carcinogenesis are required to obtain adequate clinical sensitivity and specificity. Novel molecular biomarker including DNA methylation and cell cycle markers may become particularly relevant as HPV vaccination coverage increases on the population-level, resulting in changes of hr-HPV genotype distribution among screen-eligible women and decreases in type-specific PPVs for CIN2+.47 Other triage methods such as machine-learning based automated visual examination of cervical images may also be useful.48 Further studies are needed to evaluate extended hr-HPV testing strategies in self-collected samples among vaccinated and unvaccinated primary screening populations, to quantify the effect of sample collection method on colposcopy referral rates, and to assess the performance of other molecular biomarkers for triage of hr-HPV positive women.
To our knowledge, this is the first study to compare hr-HPV prevalence and agreement between self- and provider-collected samples at the same time point using the Onclarity assay. All women included in the analysis provided both a self-collected cervico-vaginal and a provider-collected cervical sample for testing. Testing was done under standardized conditions in the same laboratory, and the staff performing the hr-HPV testing was unaware of the related clinical and histopathological information. We also have to acknowledge several limitations. Women in our study self-collected cervico-vaginal samples at the clinic, although the target setting for a future rollout would likely be home-based sample self-collection.37 Nevertheless, our results are likely to also apply to that setting, as a recent study showed comparable detection of CIN2+ in home- and clinic-collected self-samples.49 Our study population mostly consisted of women with abnormal screening results referred for colposcopy, and our findings might not necessarily be generalizable to a primary screening population. Importantly, however, we found agreement of hr-HPV test results between self- and provider-collected samples to be similar among women with and without advanced cervical disease. We did not collect individual-level data on HPV vaccination status but given the median age of our study population was 36 years and HPV vaccination was introduced in the U.S. in 2006, most included women are expected to have not been vaccinated. Thus, our findings are not generalizable to a population of vaccinated women. Extended hr-HPV genotyping will remain important to assess non-vaccine genotypes and their associated type-specific risk of high-grade cervical precancer and cancer.
To conclude, HPV 16 had the highest PPV for detection of CIN2+ in both self-collected cervico-vaginal and provider-collected cervical samples and overall sensitivity for CIN2+ detection was similar for both sample types. However, HPV-16 prevalence was significantly higher in self- versus provider-collected samples, which could result in increased colposcopy referral rates and overtreatment. In the future, additional molecular marker such as DNA methylation might be helpful to improve the triage of women positive for hr-HPV on self-collected samples.
Supplementary Material
Acknowledgements:
We are grateful to all women who participated in this research project. We also thank Johana Bravo, Elena DiRosa, and Sara Smith for the day-to-day coordination of this study including the recruitment of participants and data collection. Furthermore, we thank the UNC Center for AIDS Research HIV/STD Laboratory Core for processing, storage, and clinical trial support.
Source of Funding: This research was supported by National Institutes of Health (NIH) grants U54 CA156733 and 5R01CA183891 to J.S. Smith and the UNC Center for AIDS Research (CFAR), an NIH-funded program (P30 AI50410). E. Rohner was supported by a grant from the Swiss Cancer Research foundation (BIL KFS-4423-02-2018). Specimen transport tubes and HPV testing were donated by BD, self-collection brushes were donated by Rovers Medical Devices BV, and cytology media was donated by Hologic Corporation.
Footnotes
Conflict of Interest: J.S. Smith has received research grants, supply donations, and consultancies; served on paid advisory boards; and/or been a paid speaker for Arbor Vita, Becton Dickenson Corporation, Hologic, Rovers Medical Devices, and Trovagene in the past 5 years. J.A.E. Nelson has received financial support from Hologic for work on a different study.
IRB status: Approved, UNC IRB# 15-2872 and Duke IRB# Pro00083075.
Data availability: The data that support the findings of this study are available on request from the corresponding author, in accordance to the NIH guidelines on data sharing, as proposed in our original grant submission.
References
- 1.Walboomers JMM, Jacobs MV., Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999; 189: 12–9. [DOI] [PubMed] [Google Scholar]
- 2.Smith JS, Lindsay L, Hoots B, et al. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer 2007; 121: 621–32. [DOI] [PubMed] [Google Scholar]
- 3.Muñoz N, Bosch FX, de Sanjosé S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 2003; 348: 518–27. [DOI] [PubMed] [Google Scholar]
- 4.Schiffman M, Clifford G, Buonaguro FM. Classification of weakly carcinogenic human papillomavirus types: addressing the limits of epidemiology at the borderline. Infect Agent Cancer 2009; 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schiffman M, Hyun N, Raine-Bennett TR, et al. A cohort study of cervical screening using partial HPV typing and cytology triage. Int J Cancer 2016; 139: 2606–15. [DOI] [PubMed] [Google Scholar]
- 6.Curry SJ, Krist AH, Owens DK, et al. Screening for Cervical Cancer. JAMA 2018; 320: 674. [DOI] [PubMed] [Google Scholar]
- 7.Saslow D, Solomon D, Lawson HW, et al. American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. CA Cancer J Clin 2012; 62: 147–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Sanjose S, Quint WG, Alemany L, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol 2010; 11: 1048–56. [DOI] [PubMed] [Google Scholar]
- 9.Serrano B, Alemany L, Tous S, et al. Potential impact of a nine-valent vaccine in human papillomavirus related cervical disease. Infect Agent Cancer 2012; 7: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatia R, Kavanagh K, Cubie HA, et al. Use of HPV testing for cervical screening in vaccinated women - Insights from the SHEVa (Scottish HPV Prevalence in Vaccinated Women) study. Int J Cancer 2016; 138: 2922–31. [DOI] [PubMed] [Google Scholar]
- 11.Drolet M, Bénard É, Pérez N, et al. Population-level impact and herd effects following the introduction of human papillomavirus vaccination programmes: updated systematic review and meta-analysis. Lancet 2019; 394: 497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan P, Howell-Jones R, Li N, et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer 2012; 131: 2349–59. [DOI] [PubMed] [Google Scholar]
- 13.Stoler MH, Wright TC, Parvu V, Yanson K, Cooper CK, Andrews J. Stratified risk of high-grade cervical disease using onclarity HPV extended genotyping in women, ≥25 years of age, with NILM cytology. Gynecol Oncol 2019; 153: 26–33. [DOI] [PubMed] [Google Scholar]
- 14.Wright TC, Stoler MH, Parvu V, Yanson K, Cooper C, Andrews J. Risk detection for high-grade cervical disease using Onclarity HPV extended genotyping in women, ≥21 years of age, with ASC-US or LSIL cytology. Gynecol Oncol 2019; 154: 360–7. [DOI] [PubMed] [Google Scholar]
- 15.Spence AR, Goggin P, Franco EL. Process of care failures in invasive cervical cancer: Systematic review and meta-analysis. Prev Med 2007; 45: 93–106. [DOI] [PubMed] [Google Scholar]
- 16.Pruitt SL, Werner CL, Borton EK, et al. Cervical Cancer Burden and Opportunities for Prevention in a Safety-net Healthcare System. Cancer Epidemiol Biomarkers Prev 2018; 27: 1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrae B, Kemetli L, Sparen P, et al. Screening-Preventable Cervical Cancer Risks: Evidence From a Nationwide Audit in Sweden. J Natl Cancer Inst 2008; 100: 622–9. [DOI] [PubMed] [Google Scholar]
- 18.Akinlotan M, Bolin JN, Helduser J, Ojinnaka C, Lichorad A, McClellan D. Cervical Cancer Screening Barriers and Risk Factor Knowledge Among Uninsured Women. J Community Health 2017; 42: 770–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asciutto KC, Ernstson A, Forslund O, Borgfeldt C. Self-sampling with HPV mRNA analyses from vagina and urine compared with cervical samples. J Clin Virol 2018; 101: 69–73. [DOI] [PubMed] [Google Scholar]
- 20.Snijders PJF, Verhoef VMJ, Arbyn M, et al. High-risk HPV testing on self-sampled versus clinician-collected specimens: A review on the clinical accuracy and impact on population attendance in cervical cancer screening. Int J Cancer 2013; 132: 2223–36. [DOI] [PubMed] [Google Scholar]
- 21.Ejegod DM, Junge J, Franzmann M, et al. Clinical and analytical performance of the BD OnclarityTM HPV assay for detection of CIN2+ lesions on SurePath samples. Papillomavirus Res 2016; 2: 31–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper DM, Longacre MR, Noll WW, Belloni DR, Cole BF. Factors affecting the detection rate of human papillomavirus. Ann Fam Med 2003; 1: 221–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McHugh ML. Interrater reliability: the kappa statistic. Biochem medica 2012; 22: 276–82. [PMC free article] [PubMed] [Google Scholar]
- 24.de Campos KLM, Machado AP, de Almeida FG, et al. Good agreements between self and clinician-collected specimens for the detection of human papillomavirus in Brazilian patients. Mem Inst Oswaldo Cruz 2014; 109: 352–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerigo H, Coutlée F, Franco EL, Brassard P. Dry self-sampling versus provider-sampling of cervicovaginal specimens for human papillomavirus detection in the Inuit population of Nunavik, Quebec. J Med Screen 2012; 19: 42–8. [DOI] [PubMed] [Google Scholar]
- 26.Ajenifuja OK, Ikeri NZ, Adeteye OV, Banjo AA. Comparison between self sampling and provider collected samples for Human Papillomavirus (HPV) Deoxyribonucleic acid (DNA) testing in a Nigerian facility. Pan Afr Med J 2018; 30: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorenzi AT, Fregnani JHTG, Possati-Resende JC, Neto CS, Villa LL, Longatto-Filho A. Self-collection for high-risk HPV detection in Brazilian women using the careHPVTM test. Gynecol Oncol 2013; 131: 131–4. [DOI] [PubMed] [Google Scholar]
- 28.Dijkstra MG, Heideman DAM, van Kemenade FJ, et al. Brush-based self-sampling in combination with GP5+/6+-PCR-based hrHPV testing: high concordance with physician-taken cervical scrapes for HPV genotyping and detection of high-grade CIN. J Clin Virol 2012; 54: 147–51. [DOI] [PubMed] [Google Scholar]
- 29.Polman NJ, Ebisch RMF, Heideman DAM, et al. Performance of human papillomavirus testing on self-collected versus clinician-collected samples for the detection of cervical intraepithelial neoplasia of grade 2 or worse: a randomised, paired screen-positive, non-inferiority trial. Lancet Oncol 2019; 20: 229–238. [DOI] [PubMed] [Google Scholar]
- 30.Lorenzato FR, Singer A, Ho L, et al. Human papillomavirus detection for cervical cancer prevention with polymerase chain reaction in self-collected samples. Am J Obstet Gynecol 2002; 186: 962–8. [DOI] [PubMed] [Google Scholar]
- 31.Ortiz AP, Romaguera J, Pérez CM, et al. Human papillomavirus infection in women in Puerto Rico: Agreement between physician-collected and self-collected anogenital specimens. J Low Genit Tract Dis 2013; 17: 210–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petignat P, Faltin DL, Bruchim I, Tramèr MR, Franco EL, Coutlée F. Are self-collected samples comparable to physician-collected cervical specimens for human papillomavirus DNA testing? A systematic review and meta-analysis. Gynecol Oncol 2007; 105: 530–5. [DOI] [PubMed] [Google Scholar]
- 33.Bhatla N, Dar L, Patro AR, et al. Can human papillomavirus DNA testing of self-collected vaginal samples compare with physician-collected cervical samples and cytology for cervical cancer screening in developing countries? Cancer Epidemiol 2009; 33: 446–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harper DM, Noll WW, Belloni DR, Cole BF. Randomized clinical trial of PCR-determined human papillomavirus detection methods : Self-sampling versus clinician-directed – Biologic concordance and women ‘ s preferences. Am J Obstet Gynecol 2002; 186: 365–73. [DOI] [PubMed] [Google Scholar]
- 35.Rohner E, McGuire FH, Liu Y, et al. Racial and Ethnic Differences in Acceptability of Urine and Cervico-Vaginal Sample Self-Collection for HPV-Based Cervical Cancer Screening. J Women’s Health 2020; 29: 971–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winer RL, Lee S- K, Hughes JP, Adam DE, Kiviat NB, Koutsky LA. Genital Human Papillomavirus Infection: Incidence and Risk Factors in a Cohort of Female University Students. Am J Epidemiol 2003; 157: 218–26. [DOI] [PubMed] [Google Scholar]
- 37.Arbyn M, Smith SB, Temin S, Sultana F, Castle P, Collaboration on Self-Sampling and HPV Testing. Detecting cervical precancer and reaching underscreened women by using HPV testing on self samples: updated meta-analyses. BMJ 2018; 363: k4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gage JC, Schiffman M, Solomon D, et al. Risk of precancer determined by HPV genotype combinations in women with minor cytologic abnormalities. Cancer Epidemiol Biomarkers Prev 2013; 22: 1095–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu L, Benoy I, Cuschieri K, Poljak M, Bonde J, Arbyn M. Accuracy of genotyping for HPV16 and 18 to triage women with low-grade squamous intraepithelial lesions: a pooled analysis of VALGENT studies. Expert Rev Mol Diagn 2019; 19: 543–51. [DOI] [PubMed] [Google Scholar]
- 40.Cuzick J, Ho L, Terry G, et al. Individual detection of 14 high risk human papilloma virus genotypes by the PapType test for the prediction of high grade cervical lesions. J Clin Virol 2014; 60: 44–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adcock R, Cuzick J, Hunt WC, McDonald RM, Wheeler CM, New Mexico HPV Pap Registry Steering Committee. Role of HPV Genotype, Multiple Infections, and Viral Load on the Risk of High-Grade Cervical Neoplasia. Cancer Epidemiol Biomarkers Prev 2019; 28: 1816–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herrero R, Hildesheim A, Bratti C, et al. Population-based study of human papillomavirus infection and cervical neoplasia in rural Costa Rica. J Natl Cancer Inst 2000; 92: 464–74. [DOI] [PubMed] [Google Scholar]
- 43.Clifford G, Franceschi S. Members of the human papillomavirus type 18 family (alpha-7 species) share a common association with adenocarcinoma of the cervix. Int J Cancer 2007; 122: 1684–5. [DOI] [PubMed] [Google Scholar]
- 44.Ho GYF, Bierman R, Beardsley L, Chang CJ, Burk RD. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med 1998; 338: 423–8. [DOI] [PubMed] [Google Scholar]
- 45.Rositch AF, Koshiol J, Hudgens MG, et al. Patterns of persistent genital human papillomavirus infection among women worldwide: A literature review and meta-analysis. Int J Cancer 2013; 133: 1271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ebisch RMF, Siebers AG, Bosgraaf RP, Massuger LFAG, Bekkers RLM, Melchers WJG. Triage of high-risk HPV positive women in cervical cancer screening. Expert Rev Anticancer Ther 2016; 16: 1073–85. [DOI] [PubMed] [Google Scholar]
- 47.Cuschieri K, Ronco G, Lorincz A, et al. Eurogin roadmap 2017: Triage strategies for the management of HPV-positive women in cervical screening programs. Int J Cancer 2018; 143: 735–45. [DOI] [PubMed] [Google Scholar]
- 48.Hu L, Bell D, Antani S, et al. An Observational Study of Deep Learning and Automated Evaluation of Cervical Images for Cancer Screening. J Natl Cancer Inst 2019; 111: 923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Des Marais AC, Zhao Y, Hobbs MM, et al. Home Self-Collection by Mail to Test for Human Papillomavirus and Sexually Transmitted Infections. Obstet Gynecol 2018; 132: 1412–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


