Abstract
In this study, we consider admixed populations through their expected heterozygosity, a measure of genetic diversity. A population is termed admixed if its members possess recent ancestry from two or more separate sources. As a result of the fusion of source populations with different genetic variants, admixed populations can exhibit high levels of genetic diversity, reflecting contributions of their multiple ancestral groups. For a model of an admixed population derived from K source populations, we obtain a relationship between its heterozygosity and its proportions of admixture from the various source populations. We show that the heterozygosity of the admixed population is at least as great as that of the least heterozygous source population, and that it potentially exceeds the heterozygosities of all of the source populations. The admixture proportions that maximize the heterozygosity possible for an admixed population formed from a specified set of source populations are also obtained under specific conditions. We examine the special case of K = 2 source populations in detail, characterizing the maximal admixture in terms of the heterozygosities of the two source populations and the value of FST between them. In this case, the heterozygosity of the admixed population exceeds the maximal heterozygosity of the source groups if the divergence between them, measured by FST, is large enough, namely above a certain bound that is a function of the heterozygosities of the source groups. We present applications to simulated data as well as to data from human admixture scenarios, providing results useful for interpreting the properties of genetic variability in admixed populations.
Keywords: Admixture, allele frequencies, heterozygosity, population genetics
1. Introduction
Admixed populations are populations that possess ancestry from multiple source groups. They result from the fusion of populations that have long been separated, in processes such as long-distance migration and hybrid-zone formation at population boundaries.
Several features of ancestry and allele frequencies are characteristic of admixed populations (Chakraborty, 1986; Long, 1991; Verdu & Rosenberg, 2011; Gravel, 2012). In an admixed population, the values of allele frequencies are typically intermediate between those of the various sources. Unlike in a mixture that pools individuals taken from separate populations, in an admixed population, alleles from different sources cooccur within individuals. The contributions from the source populations are each large enough that most members of an admixed population have ancestry in more than one source group.
In admixed populations, the history of mating among populations is recent enough that time has not yet eroded differences among admixed individuals in their relative proportions of ancestry. This feature of high levels of variability in admixture proportions has been central to studies of admixed populations. Investigations of such phenomena as the timing and contributions of the source populations (Verdu & Rosenberg, 2011; Gravel, 2012), the effect of admixture levels on assortative mating patterns (Risch et al., 2009; Zou et al., 2015), and the genetic basis of traits in admixed populations (Buerkle & Lexer, 2008; Zhu et al., 2008) all make use of variation in levels of admixture levels across admixed individuals.
A second aspect of variability in admixed populations is potentially of interest: the variability of alleles as captured by genetic diversity measures. The effect of admixture in contributing to increased genetic diversity, however, is not simple. For example, in a study of the genetics of populations founded by relatively small groups, Mooney et al. (2018) examined genetic diversity in admixed and non-admixed populations, some of which were regarded as founder populations. Mooney et al. (2018) observed that genetic diversity was relatively high in multiple admixed populations of Latin America. This pattern was observed even for populations that, on the basis of small population size and past history of isolation, might have been expected to have relatively low levels of genetic diversity.
Here, to deepen understanding of the relationship between admixture and genetic variability, we focus in admixed populations on levels of genetic diversity computed from allele frequencies, rather than on variability among individuals in admixture proportions. For a model of an admixed population with K source groups, we derive a relationship between genetic diversity, as measured by heterozygosity, and proportions of admixture drawn from the various source populations. The model is the same model we have previously used to examine the genetic differentiation between admixed populations and their source groups, as measured by FST (Boca & Rosenberg, 2011). We show that for all values of the admixture contributions from the source populations, the heterozygosity of the admixed population is greater than or equal to the smallest of the source population heterozygosities. We further examine the maximal values of the heterozygosity of the admixed population over the space of possible admixture proportions. We consider in more detail special cases with K = 2 and K = 3 source populations, providing explicit results for K = 2 in terms of relatively few parameters. Finally, we use simulations and example analyses from human population data to illustrate the mathematical results.
2. Notation and model
We consider a model with K ⩾ 2 source populations and an admixed population arising from these sources. A single polymorphic locus is considered, with J ⩾ 2 alleles, such that each of the J alleles appears in at least one of the K source populations.
In Sections 2.1, 2.2, and 2.3, respectively, we define the expected heterozygosity and the fixation index, and we provide a result about relationships between fixation indices and heterozygosities. In Section 2.4, we introduce the admixture model. Notation is summarized in Table 1.
Table 1:
Notation
| Type of quantity | Symbol | Description |
|---|---|---|
| Indices | j = 1,..., J | Index over alleles |
| k = 1,..., K | Index over source populations | |
| Allele frequencies | pkj | Frequency of allelic type j in population k |
|
J × 1 vector of allele frequencies for population k J × K matrix of allele frequencies in the source populations |
||
| Frequency of allelic type j in the admixed population | ||
| Admixture fractions | γk | Admixture fraction for population k |
| γ | K × 1 vector of admixture fractions | |
| Heterozygosities | Hk | Heterozygosity for population k; probability that two alleles drawn from population k differ in type |
| Hadm | Heterozygosity for the admixed population | |
| Ckℓ | Probability that an allele drawn from population k and an allele drawn from population ℓ differ in type | |
| Fixation index | Fkℓ | Fixation index FST between populations k and ℓ |
2.1. Expected heterozygosity
The expected heterozygosity is a measure of genetic diversity, giving the probability that two alleles randomly drawn from a population differ in type.
Definition 1. The expected heterozygosity in a population for a given locus with J distinct alleles is defined as , where pj is the frequency of allelic type j.
We denote by pkj the frequency of allelic type j, 1 ⩽ j ⩽ J, in source population k, 1 ⩽ k ⩽ K, with 0 ⩽ pkj ⩽ 1. We denote by Hk the expected heterozygosity of source population k at a locus. We have 0 ⩽ Hk < 1, with Hk = 0 if and only if source population k has only a single allelic type of nonzero frequency. For fixed J, the maximal value of Hk is , attained when all J alleles have the same frequency, namely (Reddy & Rosenberg, 2012, Lemma 4). We refer to expected heterozygosity simply as heterozygosity.
2.2. Fixation index
The fixation index FST is a measure of genetic divergence among a set of subpopulations. In its general form, it is computed from HS, the mean of the heterozygosities of the subpopulations, and HT, the heterozygosity of a population formed by pooling the subpopulations into a single “total” population.
Definition 2. The fixation index, FST is defined as FST = (HT − HS)/HT, where HT is the heterozygosity of the total population and HS is the mean heterozygosity across subpopulations.
The fixation index can be regarded as a measure of genetic divergence between two populations, with Fkℓ denoting the value of FST between source populations k and ℓ. For its calculation, the two subpopulations have the same contribution to the overall population, so that they are weighted equally in producing the total population. We assume that when pooled together, the two subpopulations produce a polymorphic population. In other words, for each (k, ℓ), we disallow the case in which there is some allelic type 1 ⩽ j ⩽ J for which pkj = pℓj = 1. Our assumption that pooling any two populations produces a polymorphic population avoids a denominator of 0 in the formula for Fkℓ.
For this pairwise scenario, HS = (Hk + Hℓ)/2, , and
| (1) |
We can observe by the Cauchy-Schwarz inequality that 0 ⩽ Fkℓ ⩽ 1, with Fkℓ = 0 requiring pkj = pℓj for all j. Fkℓ = 1 requires HS = Hk = Hℓ = 0.
2.3. The fixation index in relation to the heterozygosities
We will need a result on the relationship between the fixation index for source populations k and ℓ, Fkℓ, and the heterozygosities of those source populations, Hk and Hℓ. We first introduce a quantity, Ckℓ, the probability that, when randomly drawing one allele from population k and one allele from population ℓ, the two alleles differ in type. For population k, let denote a J × 1 column vector of its allele frequencies. Ckℓ can then be written as 1 minus the dot product of the allele frequency vectors of populations k and ℓ:
| (2) |
This quantity is a generalization of heterozygosity to two populations, as Hk = Ckk. Because we exclude the case in which populations k and ℓ are fixed for the same allelic type, Ckℓ strictly exceeds 0, so that 0 < Ckℓ ⩽ 1. The upper bound of 1 is achieved if populations k and ℓ share no allelic types in common.
We can rewrite eq. 1 as
| (3) |
If Fkℓ < 1, then we can solve for Ckℓ:
| (4) |
Recall that Fkℓ = 1 implies Hk = Hℓ = 0, so that populations k and ℓ each have only a single allelic type with nonzero frequency. We have excluded the case in which the two populations are fixed for the same allelic type; hence, they must be fixed for different allelic types, and Ckℓ = 1 in eq. 2.
We have previously shown by the Cauchy-Schwarz inequality that (Mehta et al., 2019, eq. 7). Equality in the lower bound requires pkj = pℓj for all j, and hence Hk = Hℓ. Rewriting this inequality with eq. 4, we obtain the allowable space of Fkℓ given Hk, Hℓ ∈ [0, 1):
| (5) |
The lower limit is achieved if and only if the two populations k and ℓ are identical, with Hk = Hℓ and pkj = pℓj for all j. The upper limit is achieved if and only if populations k and ℓ share no allelic types in common. This result adds to the understanding of constraints on FST placed by genetic diversity (Nagylaki, 1998; Hedrick, 1999; Long & Kittles, 2003; Rosenberg et al., 2003; Hedrick, 2005; Boca & Rosenberg, 2011; Maruki et al., 2012; Jakobsson et al., 2013; Edge & Rosenberg, 2014; Alcala & Rosenberg, 2017, 2019; Mehta et al., 2019). We use the allowable region to constrain our examples to permissible values of (Hk, Hℓ, FST).
Appendix A of Mehta et al. (2019) shows that given Hk and Hℓ in [0, 1), if the number of distinct alleles J is not fixed, then we can choose allele frequency vectors and such that each Ckℓ value in is achievable. The lower bound is achievable only if Hk = Hℓ. Hence, each value in the interval in eq. 5 for FST is also achievable by some pair and , the lower bound only if Hk = Hℓ.
2.4. Admixture model
We use an admixture model that describes current patterns of variation in an admixed population, rather than mechanistic dynamics. This model follows a commonly used approach, treating allele frequencies in the admixed population as linear combinations of those of the source populations (e.g. Pritchard et al., 2000; Boca & Rosenberg, 2011).
In our K-source-population model, K ⩾ 2, we follow Section 2.2 in assuming that no two populations are fixed for the same allelic type. We now make a stronger assumption that no two populations are identical, so that for each (k, ℓ), some j exists for which pkj ≠ pℓj. Further, it is convenient to assume that no source population can have its vector of allele frequencies written as the linear combination of vectors of allele frequencies of other source populations; otherwise, an admixed population would not have a unique representation as a linear combination of sources. We thus assume that not only are no two source populations identical, no source can be described as an admixture of two or more of the other sources.
Note that the assumption that no population is a linear combination of the others also excludes linear combinations with one or more negative coefficients. Because the maximal number of vectors of length J that can be linearly independent is J, the linear independence assumption implies J ⩾ K. A succinct way of describing the assumption is that if we define the J×K matrix of allele frequencies in the source populations,
| (6) |
then we assume that P has rank K.
For the admixed population generated from the K source populations, we denote by γk the admixture fraction for source population k; for each k with 1 ⩽ k ⩽ K, fraction γk of the ancestry of the admixed population, 0 ⩽ γk ⩽ 1, derives from source k. We denote by γ the K×1 column vector of admixture fractions. This vector lies in the simplex ΔK−1, the set of all vectors of K nonnegative entries with .
The frequency of allele j in the admixed population is denoted . By the linear combination assumption,
| (7) |
In the special case that for each K, the admixed population is equivalent to the “pooled population” used in defining the fixation index FST among the K populations.
3. General case: K source populations
Our goal is to study the heterozygosity of the admixed population. Using Definition 1 with eq. 7, we compute the heterozygosity for the admixed population, which we denote by Hadm:
| (8) |
The heterozygosity of the admixed population can be written in terms of the heterozygosities of the source populations and the dot products of the allele frequencies. Using eq. 4 in eq. 8, we have:
| (9) |
| (10) |
The last simplification can be made only for Fkℓ ≠ 1; if Fkℓ = 1, then eq. 9 is used, or, as noted after eq. 4, (Hk + Hℓ)(1 + Fkℓ)/(1 − Fkℓ) is understood to equal 2.
With the formula for Hadm established, we now explore how Hadm varies in relation to the admixture fractions γ. Given the allele frequencies P, we determine the range of Hadm over the space of possible values of γ. We write Hm for the smallest heterozygosity among the source populations, , and HM for the largest heterozygosity among the source populations, .
3.1. Minimum of Hadm in terms of the ancestry proportions
For the minimum of Hadm over vectors (γ1, γ2, …, γK), we can immediately observe from the form of eq. 10 that for a fixed set of source population allele frequencies P, Hadm is minimized as a function of the admixture fractions when the admixed population consists of only one of the source populations.
Proposition 3. The minimum of Hadm as a function of the ancestry proportions γ is , the smallest heterozygosity among the source populations, and it is obtained when the admixed population consists solely of that source population.
Proof. To obtain this result, we use eq. 10 and the fact that Hk ⩾ Hm for all k:
Because equality is achieved when γm = 1 and γk = 0 for all k ≠ m, we have shown that the minimal value of Hadm as a function of the ancestry proportions is Hm. □
The result finds that nonzero admixture inflates heterozygosity at least above the level seen in the least heterozygous source. It applies whether or not H1, H2, …, HK are mutually distinct. If two or more of H1, H2, …, HK are tied for the minimal heterozygosity Hm, then the minimum of Hadm is achieved at each vector associated with complete ancestry from one of the minimally heterozygous populations.
A consequence of Proposition 3 is that if all K populations have the same heterozygosity Hm—for example, in cases where the different alleles have distinct frequencies and each population has an allele frequency vector that is a permutation of the vectors for the other populations—then Hadm > Hm for all ancestry vectors γ with two or more nonzero entries. In particular, note that Fkℓ > 0 for each (k, ℓ), k ≠ ℓ, by the assumption that each pair of source populations has distinct allele frequencies. Hence, (Hk +Hℓ)(1+ Fkℓ)/(1 − Fkℓ) > 2Hm for each (k, ℓ), k ≠ ℓ. Because at least one product γkγℓ is positive, the inequality is strict for at least one (k, ℓ), so that . This same reasoning shows that if two or more populations are tied with heterozygosity Hm, then Hadm > Hm for each γ with two or more nonzero entries.
We note that the result for all γ ∈ ΔK−1 in Proposition 3 can be quickly obtained from the classic Wahlund principle, by which the heterozygosity of a population formed by mixing populations 1, 2, …, K, with proportion γk of the mixed population taken from population k, 0 ⩽ γk ⩽ 1, is greater than or equal to the mean of the K population heterozygosities (e.g. Rosenberg & Calabrese, 2004, Theorem 2). The heterozygosity of the population mixture in the setting of the Wahlund principle is the same as the heterozygosity of the admixed population in our scenario. Thus, in our notation, setting for all k, the Wahlund principle gives . Because the mean is greater than or equal to the minimum , it immediately follows that .
3.2. Maximum of Hadm in terms of the ancestry proportions
To obtain the maximum of Hadm over the space of values of γ, we write eq. 9 as a quadratic form:
Here, γ′ represents the transpose of the column vector γ and A is the K × K symmetric matrix with the Hk on the diagonal and the Ckℓ off the diagonal:
| (11) |
where P is the J × K allele frequency matrix (eq. 6) and 1 is a K × 1 vector of ones.
Maximizing Hadm in terms of γ is equivalent to finding subject to 1′γ = 1. We denote by γarg max the location of the maximal value of Hadm. We first observe that γarg max is sometimes interior to the simplex, and that it sometimes lies at a vertex. In other words, for a fixed set of sources, a population nontrivially admixed among the sources can sometimes have a higher heterozygosity than all of the sources, but sometimes, no population admixed among the sources has higher heterozygosity than all the sources.
Proposition 4. Consider the case of K source populations, K ⩾ 2.
(i) There exists some collection of source population allele frequencies P and some collection of admixture proportions γ for which the heterozygosity of the admixed population exceeds the heterozygosity HM of the most heterozygous source population.
(ii) There exists some collection of source population allele frequencies P for which no collection of admixture proportions γ produces an admixed population with heterozygosity greater than the heterozygosity HM of the most heterozygous source population.
Proof. (i) Consider K populations, each with different allele frequencies, but identical heterozygosity: for k ≠ ℓ but Hk = H for k = 1, 2, …, K. Suppose that a locus has K + 1 distinct alleles, and that the allele frequencies are , . By eq. 9, , which is minimized if and only if or for some k. The minimal value of Hadm is thus , all other values of the admixture proportions resulting in .
(ii) Consider K populations and a locus with K distinct alleles. Suppose that the number of distinct alleles at the locus is k for population k, with . Hence, and, in particular, H1 < … < HK. We show that Hadm ⩽ HK irrespective of γ.
By eq. 9,
By the Cauchy-Schwarz inequality:
Thus, . □
The proof is constructive, exhibiting example source groups for which specific features are obtained. In part (i), each source has an allele that is not present in the other sources, and a nontrivially admixed population—which possesses all of these private alleles—is necessarily more heterozygous than each source. For part (ii), we have a sequence of increasingly heterozygous source populations, each with one additional allele, and no population admixed among them is more heterozygous than the most heterozygous source. Other constructive examples are possible, with, for example, low heterozygosities but distinct alleles across populations generating additional examples along the lines of Proposition 4i.
Note that it is trivial to see that in general, : the K source populations simply correspond to the K vertices of the simplex. This result that the maximal Hadm is at least is great as the heterozygosity of the most heterozygous source population immediately implies .
Having established that the maximum can be at a vertex or an interior point of the simplex—a trivial admixed population consisting only of a single source population, or a population admixed among all the sources—we now provide a general theorem. The theorem gives the location of the maximum when it lies in the interior of ΔK−1, rather than on the boundary, assuming a condition applies on the allele frequencies. The proof is in Appendix 1, making use of a general constrained quadratic optimization procedure.
Theorem 5. Suppose that 1′(P′P)−11 ≠ 1. Suppose also that . Then the maximum of Hadm as a function of the ancestry proportions γ ∈ ΔK−1 is attained at γarg max = γ*, where:
The maximum is equal to:
If , then γarg max lies on the boundary of the set {γ : 1′γ = 1 and γ ∈ ΔK−1}.
The “boundary” of a set R is the set of points in R for which a neighborhood around them always contains both points in R and points in the complement of R. For simplex ΔK−1, the boundary includes all points for which at least one of the K coordinates is 0, with the vertices occurring at locations where all of the coordinates except one are 0.
The following corollary, also proven in Appendix 1, further describes the possible locations of the maximal Hadm. Note that if the maximum is not at γ∗, then it lies at a point that has some elements equal to 0, the nonzero subvector having a similar form to γ∗, but in a lower number of dimensions. Thus, the maximum can occur in a scenario in which the admixture involves only a strict subset of the source populations.
Consider a nonempty subset . Define by the matrix that has diagonal terms Hk for each and off-diagonal terms Ckℓ for each distinct . Additionally, denote by the matrix consisting of the columns of P corresponding to the subset . contains the allele frequencies for the source populations in .
Corollary 6. Suppose that for all nonempty . Then the maximum of Hadm as a function of the ancestry proportions γ ∈ ΔK−1 is attained at a point that has nonzero elements for some nonempty subset of the source populations . The nonzero subvector of ancestry proportions at the location of the maximum is equal to .
In particular, note that γarg max = γ* corresponds to : all source populations contribute nonzero admixture fractions. The K vertices of the simplex ΔK−1 correspond to the cases of , at which only one source population contributes. has 2K − 1 nonempty subsets, each representing a distinct collection of source populations.
4. K = 2 source populations
With general results established for the case of arbitrary K, we now focus on the simplest case, with K = 2 source populations contributing to the admixed population.
We continue to exclude the scenario in which the allele frequencies for the two source populations are identical, so that we assume . Noting that γ2 = 1 − γ1, we can consider Hadm in terms of a single admixture coefficient γ1, the admixture fraction of the first population, with γ1 ∈ [0, 1]. Using eqs. 9 and 10 with this substitution, we obtain:
| (12) |
| (13) |
| (14) |
In particular, we note from eq. 13 that Hadm is increasing as a function of F12.
From eq. 14, we can see that Hadm is concave down in γ1. We have . By Definition 1 and eq. 2, . Because for at least one choice of j, and hence . By symmetry, Hadm is also concave down in γ2.
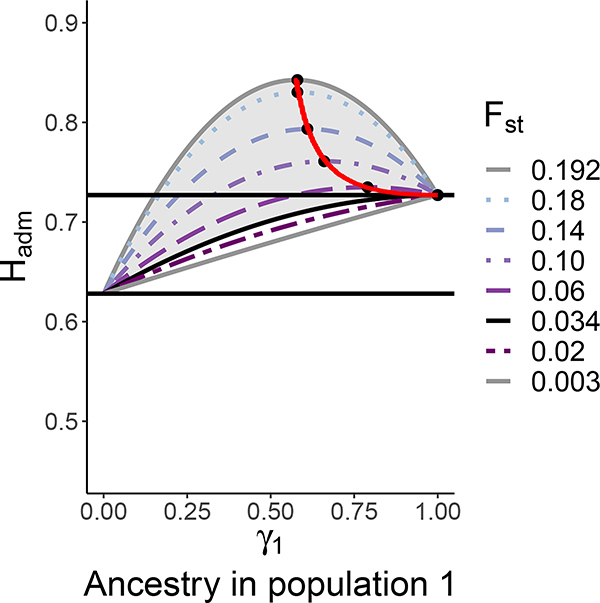
To illustrate eq. 13, for H1 and H2 fixed, Figure 1 plots the concave-down Hadm as a function of γ1 for a variety of values of F12. We observe that for each value of F12 considered, the minimum of Hadm occurs at (γ1, γ2) = (0, 1), reflecting the result of Proposition 3 that the minimum occurs when the admixed population consists solely of the less heterozygous source population. In accord with the fact that in eq. 13, Hadm increases for fixed H1, H2, and γ1 with increasing F12, the value at the maximum increases with increasing F12. The location of the maximum lies at a value of , decreasing with increasing F12. This location has a pattern where for larger values of F12, it lies interior to the unit interval, and for smaller values of F12, it occurs when the admixed population consists solely of the more heterozygous source population. We now consider this pattern in more detail.
Figure 1:
Hadm versus γ1 for fixed values of H1 and H2. We choose H1 = 0.727 and H2 = 0.628; the horizontal lines represent Hadm = H1 and Hadm = H2. Eq. 13 is plotted for multiple values of F12, considering the allowable range of F12 values in [0.003,0.192] as specified by eq. 5. The red curve, which plots (γ1, Hadm) in terms of H1, H2, and F12 in the form of eqs. 24 and 25, indicates the maxima of Hadm as F12 varies, with black dots specifying the maxima for the specific plotted values of F12. The shaded region corresponds to the region where , as specified by Corollary 10; the value F12 ≈ 0.034 gives the boundary of this region. The values chosen for H1 and H2 are, respectively, the mean heterozygosities across 8 European and 29 Native American populations, based on population-wise estimates in Table S20 of Pemberton et al. (2013). The value of γ1 can be viewed as the fraction of European ancestry in an admixed population and 1 − γ1 can be considered the fraction of Native American ancestry.
4.1. Minimum and maximum of Hadm in terms of the ancestry proportions
Applying the results from Section 3.1 on the minimum and maximum of Hadm as a function of γ, by Proposition 3, Hadm has minimum min{H1, H2}. The maximum can occur in one of three locations.
Proposition 7. Consider two source populations with distinct allele frequencies, p1 ≠ p2. As a function of γ1, Hadm is maximized at , where takes one of three forms.
(i) If H1 < C12 and H2 < C12, then satisfies
| (15) |
and Hadm has maximum equal to
| (16) |
(ii) If H1 < C12 and H2 ⩾ C12, then and Hadm has maximum H2.
(iii) If H1 ⩾ C12 and H2 < C12, then and Hadm has maximum H1.
An elementary proof appears in Appendix 2. The locations specified in Proposition 7 accord with Theorem 5 and Corollary 6. For K = 2, the result of Theorem 5 gives , where
The locations in Corollary 6 are and , and and .
We now give two corollaries of Proposition 7, providing more features of the maximal Hadm for specific cases. Proofs appear in Appendix 2. In accord with the observation in Figure 1 that the maximal Hadm lies at a value of in an example with H1 ⩾ H2, Corollary 8 demonstrates if and only if H1 ⩾ H2.
Corollary 8. Consider two source populations with distinct allele frequencies, . As a function of γ1, Hadm is maximized at if and only if H1 ⩾ H2.
A second corollary is that the maximal Hadm is always at least as great as HT.
Corollary 9. Consider two source populations with distinct allele frequencies, . Then , with equality occurring if H1 = H2.
We can also succinctly describe the region where lies interior to (0, 1).
Corollary 10. Consider two source populations with distinct allele frequencies, . lies in (0, 1) if and only if the following inequality holds:
| (17) |
This corollary is proven in Appendix 2. Note that if H1 + H2 is fixed, then the right-hand side of eq. 17 increases with |H1 −H2|, from a minimum of 0 when H1 = H2 to a maximum of as |H1 −H2| approaches H1+H2. Thus, in accord with the observation in Section 3.1 that Hadm > H for all nontrivial admixtures of equal-heterozygosity source populations, the maximal Hadm exceeds max{H1, H2} over a broader range of F12 values if |H1−H2| is small rather than large. Moreover, if , then eq. 17 necessarily holds. Hence, irrespective of H1 and H2, if the source populations are distant enough that , then the maximal heterozygosity exceeds the heterozygosities of the source populations.
4.2. Special case of J = 2 alleles
For K = 2 sources, when the locus has only J = 2 allelic types, further simplifications are possible, as results can be stated in terms of frequencies of one specific allele. We substitute p12 = 1 − p11 and p22 = 1 − p21.
Proposition 11. Consider two source populations with distinct allele frequencies, . For a biallelic locus, Hadm is maximized at , where takes one of three forms.
(i) If or , then satisfies
| (18) |
and Hadm has maximum equal to
| (19) |
(ii) If or , then and Hadm has maximum H2.
(iii) If or , then and Hadm has maximum H1.
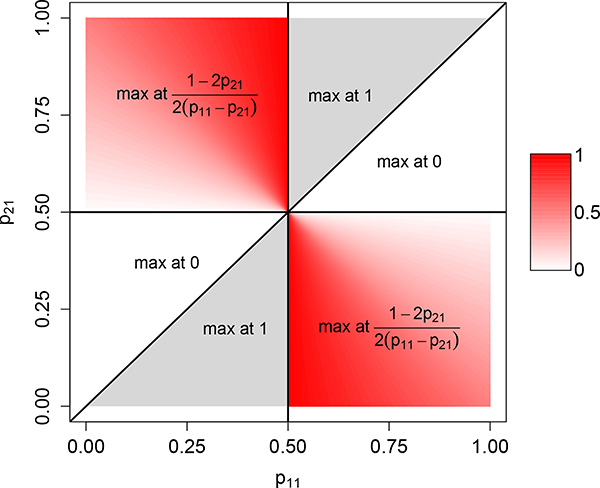
The result is proven in Appendix 2. The unit square representing possible values of the location of the maximum appears in Figure 2. It has six nonoverlapping regions: in Proposition 11, each of the three cases generates two disjoint subsets of [0,1]2. A smooth gradient exists for regions in case (i). However, an abrupt transition occurs at the line p21 = p11 between case-(ii) regions where and case-(iii) regions where . Note that the p21 = p11 line, where the two populations have equal allele frequencies, is disallowed.
Figure 2:
The admixture coefficient γ1 that maximizes Hadm in the case of K = 2 source populations and J = 2 allelic types. The plot shows the unit square for (p11, p21). In the red regions, the maximizing value of γ1 lies in (0,1), whereas in the white and gray regions, it lies on one or the other boundary. The figure depicts the result of Proposition 11.
5. Simulations
We illustrate properties of Hadm by simulating population sets for different values of K and J. Given a value of K, we generated allele frequency vectors for the K source populations from independent and identically distributed symmetric multivariate J-dimensional Dirichlet distributions with a common concentration parameter α = 1. This distribution corresponds to a uniform distribution on the simplex ΔJ−1. A number of mathematical results can be obtained in this Dirichlet setting; these appear in Appendix 3.
First, for K = 2 and K = 3, we assessed the probability that the maximal Hadm over possible admixture vectors γ occurs interior to the simplex ΔK−1, rather than on its boundary. This computation gives the probability that the heterozygosity-maximizing admixture vector contains nonzero contributions from all K source populations. We considered 2 ⩽ J ⩽ 30 for K = 2 and 3 ⩽ J ⩽ 30 for K = 3, recalling the condition J ⩾ K for the K allele frequency vectors to be linearly independent.
For each (K, J), we ran 10,000 simulation replicates. In each replicate, to determine the location of the maximum, we applied Theorem 5 and Corollary 6 to identify the locations specified for each choice of the nonempty subset of the K populations with nonzero allele frequencies. Among these 2K−1 locations, excluding those outside the simplex ΔK−1, we identified the point with the largest Hadm. Note that in each replicate, we observed that the condition of Corollary 6 was satisfied for each .
Figure 3 finds that, for both K = 2 and K = 3, the maximum of Hadm is increasingly likely to be in the interior of the simplex as the number of distinct alleles, J, increases. For K = 3, we also observe that the probability that Hadm is maximized on an edge, corresponding to nonzero contributions from two of three sources, exceeds the probability that it is maximized at a vertex, with only one contributing source.
Figure 3:
Location of the maximum of Hadm in simulation replicates. (A) K = 2. (B) K = 3. The location γarg max can be in the interior of the simplex ΔK−1, corresponding to nontrivial admixture of all source groups, or on the boundary of the simplex. For K = 3, it can be on an edge, corresponding to admixture of two of three source populations, and for both K = 2 and K = 3, it can be at a vertex, corresponding to membership in only one source population. For each (K, J), points plotted are based on 10,000 simulations with independently and identically distributed Dirichlet-(1, 1, …, 1) distributions for the allele frequency vectors in the K populations.
Next, we assessed the probability in a scenario in which both the allele frequency vectors pk and the admixture fractions γ were chosen from independent Dirichlet distributions. We simulated the pk as before, additionally simulating γ from a K-dimensional symmetric Dirichlet-(1, 1, …, 1) distribution. For each (K, J) with K = 2, 3, 4, 5 and J = 2, 3, …, 30, we simulated 50,000 replicate populations. Note that here, unlike in Section 2.4, we impose no restrictions on linear combinations of allele frequency vectors from the source populations, so that it is not necessarily true that J ⩾ K.
The fraction of replicates with appears in Figure 4. We see that this fraction increases with K: for an admixture involving more populations, the probability is larger that the admixed population exceeds all source populations in heterozygosity. This probability also increases with J.
Figure 4:
The fraction of simulation replicates for which Hadm > max{H1, …, HK}, for various values of K and J. For each (K, J), points plotted are based on 50,000 simulation replicates with independently and identically distributed Dirichlet-(1, 1, …, 1) distribitions for the allele frequency vectors in the K populations, and a Dirichlet-(1, 1, …, 1) distribution for the admixture coefficient vector γ.
For (K, J) = (2, 2), Proposition 17 in Appendix 3 obtains the probability analytically, . Following this result, the K = 2 curve in Figure 4 begins near (2, 0.307).
Figure 5 provides further detail on Hadm in the K = 2 case by graphing Hadm versus γ1 for 10 simulation replicates chosen at random for each of three values of J. The figure illustrates that Hadm is a concave-down quadratic polynomial in γ1, as in eq. 14. Averaging across replicates, by examining the figure panels from left to right, we can also observe that increases as a function of J, as in Corollary 16 of Appendix 3. For J = 2, as in Proposition 11, the possible values of Hadm at the maximum are H1, H2, and .
Figure 5:
Hadm versus γ1 for 10 simulation replicates for K = 2 source populations, for each of three values of the number of allelic types J. For each replicate, allele frequency vectors in the two populations are simulated according to Dirichlet-(1, 1, …, 1) distributions, and Hadm is plotted as a function of γ1 according to eq. 8. The maximum of Hadm is indicated by a black circle in each replicate. The red dashed lines represent the expected values of Hadm according to Corollary 16 in Appendix 3.
6. Application to data
Next, we illustrate the mathematical results using data from human populations. As multiallelic loci satisfy J ⩾ K with both K = 2 and K = 3, we focus on a multiallelic data example. First, we begin with a simpler biallelic data set whose set of individuals overlaps with the multiallelic data set, illustrating our maximal heterozygosity results in the case of K = 2 source populations. For both data sets, we treat allele frequencies, heterozygosities, and FST values computed from the data as parametric values rather than estimates.
6.1. Biallelic loci: K = 2 source populations
We consider the single-nucleotide polymorphism (SNP) data of Li et al. (2008), as employed by Pemberton et al. (2012) in phased form with no missing data. In this data set, which contains 640,034 autosomal SNPs, we consider Europeans and Native Americans as putative source populations for an admixed population, considering the 156 Europeans and 63 Native Americans in the data. We drop from consideration the 32,989 SNPs with identical allele frequencies in the two populations; 32,888 of these are monomorphic.
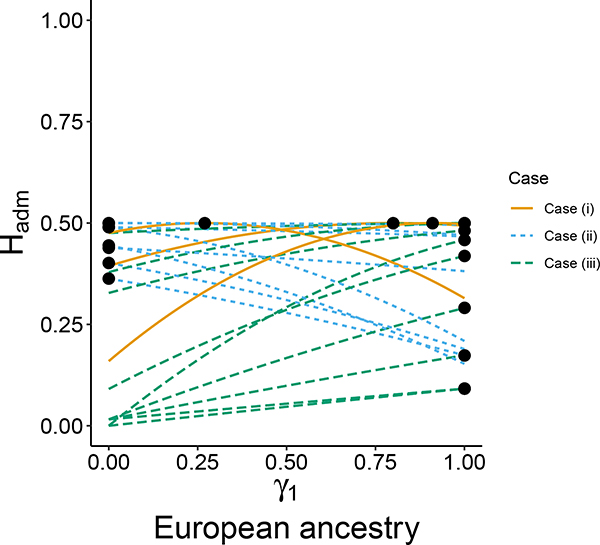
We select 20 loci at random from the data set for illustration. Treating γ1 as the fraction of European ancestry in an admixed population and 1 − γ1 as the fraction of Native American ancestry, for each locus, the plot for Hadm versus γ1 appears in Figure 6. Following Proposition 3, the minimum of Hadm lies either at γ1 = 0 or at γ1 = 1 for all loci. For 3 of the 20 loci, the maximum lies in the interior of the unit interval (case (i) of Proposition 11); 8 loci have the maximum at γ1 = 0, representing membership in the less heterozygous Native American population (case (ii)); and 9 loci have the maximum at γ1 = 1, representing membership in the more heterozygous European population (case (iii)). Following Proposition 11i, at each locus for which the maximum lies in the interior, the maximum is equal to .
Figure 6:
Hadm versus γ1 for 20 random biallelic loci from Pemberton et al. (2012). The two source populations providing the allele frequencies are the European and Native American populations, with γ1 corresponding to membership in the European population. Hadm is plotted according to eq. 8. Circles indicate the location of the maximum along each curve. Different colors and line types correspond to the three cases in Proposition 11 for the location of the maximal Hadm.
Examining all 607,045 loci, 19% have the maximum in the interior, 27% at γ1=0, and 54% at γ1 = 1. That more loci have the maximum at γ1 = 1 than γ1 = 0 is expected from the fact that European populations generally have greater heterozygosity than Native American populations (e.g. Pemberton et al., 2013).
6.2. Multiallelic loci: K = 2 source populations
For our multiallelic data set, we follow Boca & Rosenberg (2011) in considering data from Wang et al. (2008) on 678 microsatellite loci typed in 160 Europeans, 463 Native Americans, 123 Africans, and 249 individuals from admixed Mestizo populations. To represent Mestizo populations under our model, we use Europeans and Native Americans as source populations in the K = 2 case, also including Africans for K = 3.
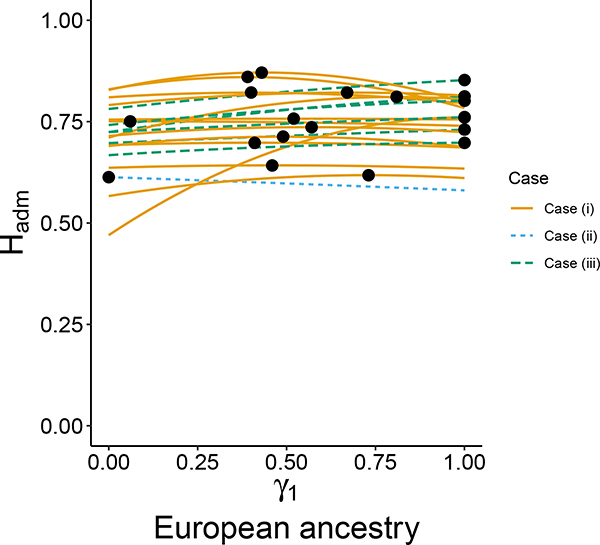
As we did in the biallelic data set, we select 20 loci at random from Wang et al. (2008), choosing the same loci as in Boca & Rosenberg (2011). Again treating γ1 as the fraction of European ancestry and 1 − γ1 as the fraction of Native American ancestry in an admixed population, for each locus, the plot for Hadm versus γ1 appears in Figure 7. Comparing Figures 7 and 6, we see that the maximum of Hadm lies in the interior of the unit interval for γ1 more often for the multiallelic than for the biallelic loci. Indeed, examining all 678 loci, 53% have the maximum in the interior—a greater number than for the SNPs. The fraction with the maximum at γ1 = 1 is 39%, and 8% have the maximum at γ1 = 0.
Figure 7:
Hadm versus γ1 for 20 random multiallelic loci from Wang et al. (2008). The two source populations providing the allele frequencies are the European and Native American populations, with γ1 corresponding to membership in the European population. Hadm is plotted according to eq. 8. Circles indicate the location of the maximum along each curve. Different colors and line types correspond to the three cases in Proposition 7 for the location of the maximal Hadm.
The Dirichlet model in Corollary 16 in Appendix 3 and Figures 3 and 5 predicts a dependence of the location of the maximum on the number of distinct alleles of a locus, with the probability that the maximum lies in the interior increasing with the number of distinct alleles. The multiallelic data produce a trend in the same direction as this prediction. The mean numbers of distinct alleles are 9.36, 10.40, and 10.75, for the loci with at 0, 1, and in (0, 1), respectively (one-way ANOVA, P = 0.008, F test, 2 df). The mean number of distinct alleles for the loci with the maximum on either boundary is 10.24, smaller than the mean of 10.74 for those with the mean in the interior (P = 0.03, two-tailed t test).
6.3. Comparison of predicted Hadm to observed Hadm
We next compare predicted and observed Hadm values for the 678 loci for the admixed Mestizo population. In this approach, we used estimated locus-wise values of γ1 in the Mestizo population together with locus-wise heterozygosities in the European and Native American populations to “predict” locus-wise Mestizo heterozygosities. The prediction is compared to the observed heterozygosity value to examine if our formulas for the heterozygosity of an admixed population are reflected in actual heterozygosities in an admixed group.
This computation follows a similar computation of Boca & Rosenberg (2011). The estimated admixture fractions, computed for the same data, are taken from Schroeder et al. (2009), who obtained them by a maximum likelihood approach (Millar, 1987) that does not take into account source population heterozygosities. Using these estimates, locus-wise heterozygosity estimates in the source populations, and locus-wise FST values calculated from allele frequencies in the source populations, we predicted Hadm with eq. 13.
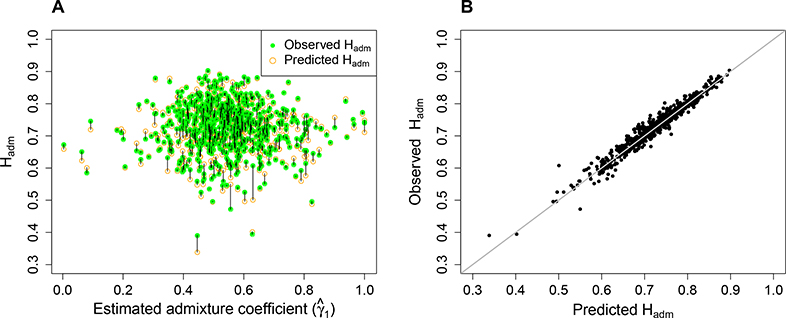
The predicted and observed Hadm values for individual loci are compared in Figure 8. In general, the observation closely matches the prediction (Figure 8A), with the correlation between the observed and predicted Hadm values equaling 0.978 (Figure 8B). For 56% of the 678 loci, the prediction provides an underestimate of the observed value.
Figure 8:
Predicted and observed Hadm. (A) The predicted and observed Hadm values for an admixed Mestizo population are plotted against the locus-wise estimated European admixture fraction in the Mestizo population, estimated by maximum likelihood. The prediction is based on eq. 8, using European and Native American allele frequencies estimated from Wang et al. (2008) as and , respectively, together with the maximum likelihood estimate of γ1. The observation is based on Hadm computed from Definition 1, inserting estimated allele frequencies from Wang et al. (2008) for the Mestizo population. (B) The observed Hadm value is plotted against the predicted Hadm value. The identity line is shown in gray. In both panels, each point represents one of the 678 loci used. The correlation coefficient between the predicted and observed Hadm values is 0.978.
6.4. K = 3 source populations
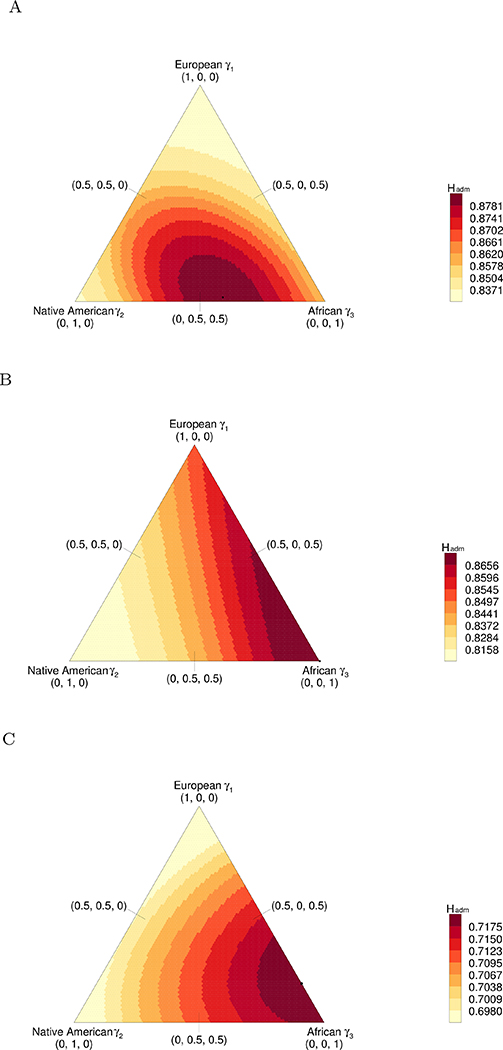
We now consider the European, Native American, and African populations as the source populations, using γ1 for the proportion of European ancestry, γ2 for Native American ancestry, and γ3 for African ancestry. We select 3 loci for illustration, choosing the same ones as in a similar analysis of Boca & Rosenberg (2011).
Plots for Hadm over the unit simplex for (γ1, γ2, γ3) appear in Figure 9. Each plot depicts Hadm as a function of (γ1, γ2, γ3) for a specific locus. The three panels show the possible locations of the maximal value of Hadm: in the first panel, the maximum lies in the interior of the simplex; in the second panel, at a vertex, and in the third panel, on an edge.
Figure 9:
Hadm versus (γ1, γ2, γ3) for three loci. The loci are from Wang et al. (2008) and have 14, 14, and 8 distinct alleles, respectively. The value of Hadm is computed from eq. 8. Black circles indicate the maximum Hadm. (A) Locus D2S1399: the maximum lies in the interior of the region. (B) Locus GATA101G01: the maximum lies at the (0, 0, 1) vertex. (C) Locus GATA146D07: the maximum lies on the γ2 = 0 edge.
Considering all 678 loci, 15% have the maximum in the interior of the region, with γ1 > 0, γ2 > 0, and γ3 > 0. The fractions with the maximum on an edge are 20% for a maximum on the edge with γ1 = 0, 26% on the γ2 = 0 edge, and 5% on the γ3 = 0 edge. The fractions with the maximum at a vertex are 27% for the vertex (0, 0, 1), 2% for (0, 1, 0), and 5% for (1, 0, 0). The observations that (0, 0, 1) is the vertex with the largest number of maxima and (1, 0, 1) is the edge with the most maxima accord with the fact that African populations have generally higher heterozygosity than European populations, which in turn have higher heterozygosity than Native American populations (e.g. Pemberton et al., 2013).
7. Discussion
We have considered the heterozygosity Hadm of an admixed population in terms of the admixture fractions of the source populations, and their heterozygosities and FST values at a locus. We have derived formulas describing Hadm in relation to these quantities (eqs. 8–10). In particular, we showed that Hadm is minimized over the set of possible admixture coefficient vectors when the admixed population consists of only one of the source populations (Proposition 3): an admixed population is at least as heterozygous as the least heterozygous source population. The maximal Hadm is more complicated, as its heterozygosity can either exceed or equal that of the most heterozygous source population (Proposition 4).
In studying the possible locations of the maximal Hadm for a fixed set of source populations, we found that the maximum can lie either in the interior of the region describing the allowable values of the admixture fractions—in which case all source populations contribute to the admixed population—or on the boundary, where one or more source populations does not contribute to the admixed population (Propositions 4–6, Figures 1–3). Simulations under a Dirichlet model for allele frequencies suggest that the maximal value of Hadm lies with increasing frequency in the interior of the allowable region as K and J increase (Figure 4).
For K = 2 source populations, we obtained further results, in particular showing that Hadm is a concave-down quadratic polynomial in the admixture coefficient γ1 (eqs. 12–14). We obtained an analytical expression for the maximal heterozygosity of an admixture of a specific pair of source populations in terms of H1, H2, and the FST value between the two populations (Proposition 7). For fixed values of H1, H2, and the admixture fraction γ1, Hadm is increasing as a function of FST (eq. 13, Figure 1). If H1 > H2, then the admixture fraction in source population 1 that maximizes Hadm is greater than (Proposition 7), meaning that at the maximal heterozygosity of the admixed population, the contribution of the more heterozygous source population exceeds that of the less heterozygous one. Interestingly, for the K = 2 case with J = 2 allelic types, if the location of the maximal value lies in (0, 1), then heterozygosity at the maximum is always (Proposition 11 and Figure 5): irrespective of the allele frequencies of the source populations, a linear combination (γ1, γ2) always exists so that the admixed population has frequencies of for both alleles.
For K = 2 source populations, a key result is that the maximal value of Hadm exceeds the larger of the two source population heterozygosities if and only if FST exceeds a bound defined by those heterozygosities (Corollary 10). Thus, with all other quantities equal, combining source populations that are more rather than less divergent is more likely to lead to an admixed population with heterozygosity exceeding those of the source populations. To obtain this result, it was important to utilize bounds on FST that constrain its values within a possibly narrow region of the unit interval, particularly for high-heterozygosity loci.
In multiallelic human data, we observed that for heterozygosities and FST values for putative sources of Mestizo populations, the maximal Hadm was more likely to be in the interior of the unit simplex or on an edge rather than at a vertex (Figures 7 and 9). This result indicates that the heterozygosities and FST values of these populations lie in a parameter range for which admixed populations are frequently more heterozygous than all their source populations. Examining heterozygosities of 267 worldwide populations in Table S20 of Pemberton et al. (2013), the 13 Mestizo populations all have heterozygosities exceeding all 29 Native American populations, and 4 have heterozygosities exceeding all 8 European populations. Interestingly, the 10 most heterozygous populations among the 267 include all five admixed populations involving a source population from the high-heterozygosity region of Africa: a Cape Mixed Ancestry group from South Africa, and four African-American populations. Thus, our mathematical results predicting that admixed populations often exceed all their source populations in heterozygosity are reflected in admixed human groups.
For K = 2, our model successfully predicted the heterozygosities in an admixed population from the source population heterozygosities, FST between the source populations, and the estimated admixture coefficient (Figure 8). Because Hadm is not necessarily monotonic in γ1, however, the reverse problem of using Hadm to estimate γ1 is problematic—unlike for the monotonically varying FST between an admixed population and one of the source populations (Boca & Rosenberg, 2011, Theorem 3). Given Hadm, source population heterozygosities H1 and H2, and FST between the source populations, two solutions to eq. 13 might exist for γ1—so that although Hadm can be predicted from γ1, it is inadvisable to proceed in the reverse direction to estimate γ1 from the heterozygosity of an admixed population.
We note that we have assumed J ⩾ K: the number of alleles is greater than or equal to the number of populations. While the results are suited to biallelic markers for K = 2, they apply primarily to multiallelic markers. Thus, in addition to the microsatellite loci we have used, we can use them with haplotype loci, for which each distinct haplotype over a length of genome is regarded as a separate allele (Mehta et al., 2019), and haplotype clusters, for which haplotypes are grouped into a fixed number of clusters and each individual is assigned a haplotype cluster membership at each site in the genome (San Lucas et al., 2012).
Our approach has followed the study of FST and admixture from Boca & Rosenberg (2011), and it shares similar limitations. The model assumes source population allele frequencies are known rather than estimated, and it considers population-level rather than individual-level admixture. It relies on patterns of variation from a single time point and does not incorporate mechanistic admixture processes or a bottleneck at the founding of the admixed population; strong genetic drift since the onset of admixture might interfere with the linear combination assumption for allele frequencies in the admixed population. Despite these limitations, the observed Hadm values and those predicted under our model are correlated in the Mestizo example (Figure 8B), indicating that the model captures key features relevant to the relationship between admixture and heterozygosity. Thus, the empirical results suggest that assessing this relationship in the mathematical formulations we have presented can be useful for understanding the genetics of admixed populations.
Acknowledgments.
Rohan Mehta provided assistance with the SNP data. We thank two reviewers for comments on the manuscript. Support was provided by NIH grant HG005855 and NSF grant BCS-1515127.
Appendix 1. Proofs for arbitrary K: Theorem 5 and Corollary 6
For the proof of Theorem 5, we first show (i) that P′P and A are both invertible under the conditions stated in the theorem, and that:
We then (ii) use constrained optimization via Lagrange multipliers to obtain the maximum of γ′Aγ subject to 1′γ = 1. This step consists of the first-derivative test to find a stationary point, coupled with the second-derivative test, in Lemma 12, to show that the stationary point defines a local maximum. Finally, we (iii) show that this means that the overall maximum is either at the local maximum γ∗ as described in the statement of the theorem or on the boundary of the set {γ : 1′γ = 1 and γ ∈ ΔK−1}.
Proof of Theorem 5 (i) Because P is a J × K matrix with column rank K, K × K matrix P′P is positive definite. As a positive definite matrix, P′P is invertible and (P′P)−1 is also positive definite (Graybill, 1976, pp. 21–22).
To show that A = 11′ − P′P is invertible, we use the Sherman-Morrison formula for the inverse of a rank-one update of an invertible matrix (Horn & Johnson, 2012, pp. 18–19). This formula states that for an invertible square n × n matrix X and n × 1 column vectors y and z, X + yz′ is invertible if and only if 1 + z′X−1y ≠ 0, with:
Because we assumed 1′(P′P)−11 ≠ 1, the Sherman-Morrison formula applies with −(P′P) in the role of X, and K × 1 column vectors 1 in the role of y and z. A has inverse:
| (20) |
Left-multiplying by 1′ and right-multiplying by 1, we obtain
Because (P′P)−1 is positive definite, 1′(P′P)−11 > 0 by definition, and because 1′(P′P)−11 ≠ 1 by assumption, we conclude that is always defined.
(ii) To maximize γ′Aγ subject to 1′γ = 1, we use Lagrange multipliers. Let f(γ) = γ′Aγ, and let g(γ) = 1′γ. The Lagrange function is defined as:
Denoting by 0 is a column vector of length K, we solve a system of equations for γ and λ,
| (21) |
Eq. 21 includes K equations for 1 ⩽ k ⩽ K.
A is symmetric, so we have
For the derivatives of the Lagrange function, we have:
Setting the derivatives with respect to γ to 0 leads to:
Hence, the solution for γ is:
Because γ′Aγ is a differentiable function of γ, its maximum on ΔK−1 can occur either on the boundary or at a critical point. The following lemma shows that the critical point is a local maximum.
Lemma 12. The critical point is a local maximum of Hadm seen as a function of γ on ΔK−1, under the conditions stated in Theorem 5.
Proof. To show that γ∗ is a local maximum, we use the second-derivative test for constrained optimization (e.g. Magnus & Neudecker, 2007, p. 155). This test considers the bordered Hessian matrix, representing the matrix of second derivatives of the Lagrange function Λ with respect to λ and the components of γ:
We must consider the principal minors—determinants of matrices in the upper-left corner—of F. We denote the upper-left corner matrix of size r × r of F by Fr, for r = 2, 3, …, K. The principal minors are the det(Fr). Using the definition of A from eq. 11, we obtain
A sufficient condition for the critical point to be a local maximum is for (−1)r det(Fr) > 0 for each r (Magnus & Neudecker, 2007, p. 155). We now show that this condition is satisfied.
Using the fact that multiplying a row or column of a matrix by a scalar multiplies the determinant by that scalar, we multiply rows 2 through r + 1 by −1 and get
Using the fact that adding a multiple of a row or column to another row does not change the determinant, we add −2 times the first column to each of the remaining columns. We also multiply the first column by −1. We then have
| (22) |
where Mr is the r × r matrix consisting of the upper-left corner of matrix P′P, and is the column vector of length r consisting of 1s.
We now apply a result for the determinant of partitioned matrices (Graybill, 1976, pp. 19–20). If W is invertible, then
Applying this result to eq. 22, we obtain
Because P′P is positive definite, Mr is also positive definite. To demonstrate this result, note that because x′P′Px > 0 for each nonzero column vector x, x′P′Px > 0 for each nonzero x with xk = 0 for k > r. Because Mr is positive definite, det(Mr) > 0 and is also positive definite, leading to . We conclude
so that the critical point is the location of a local maximum. □
Concluding the proof of Theorem 5. Returning to part (iii) of the proof, following Lemma 12, if is interior to the simplex ΔK−1, then Hadm is maximal at γ = γ∗, with maximum . This value is the reciprocal of the sum of the elements of A−1. If γ∗ is not interior to ΔK−1, then the maximum lies on the boundary of ΔK−1.
Finally, we note that by using eq. 20. □
Proof of Corollary 6. In Theorem 5, the maximum of Hadm occurs either in the interior of the simplex ΔK−1 or on its boundary, {γ : 1′γ = 1 and γ ∈ ΔK−1}.
The boundary of the simplex is the union of K faces, which are themselves (K − 2)-simplices. If the maximum lies on the boundary of ΔK−1, then without loss of generality, we can permute the labels of the source populations so that γK = 0.
We drop column K from matrix P and apply Theorem 5 with this new J ×(K −1) matrix, P{1,…,K−1}, which has rank K − 1. By assumption, .
We then apply Theorem 5 to P{1,…,K−1}. The maximum of Hadm occurs either at the point , where , or on the boundary of the set {γ : 1′γ = 1 and γ ∈ ΔK−2}.
We repeat this method of descent, decrementing the dimension (and permuting population labels without loss of generality) until we reach the case of only two source populations. A final application of Theorem 5 then finds that Hadm is maximized either interior to the 1-simplex—the line connecting vertices (1, 0) and (0, 1)—or at one of these vertices. □
Appendix 2. Proofs for K = 2: Propositions 7–11
Proof of Proposition 7. We maximize the quadratic polynomial in eqs. 12–14 over γ ∈ [0, 1]. The maximum occurs at the unique critical point or on the boundary of the interval.
Setting the derivative of eq. 14 with respect to γ1 to 0, we find that the critical point is
| (23) |
Because the leading coefficient of eq. 14 is negative for , the critical point is a maximum. Hence, if (C12 − H2)/[2(C12 − HS)] ∈ (0, 1), then the maximum of Hadm on the interval [0, 1] lies at γ1 = (C12 − H2)/[2(C12 − HS)]. Otherwise, the maximum lies either at γ1 = 0, in which case it equals H2, or at γ1 = 1, in which case it equals H1.
The conditions describing the location of the maximum can be written in terms of H1, H2, and C12. Because the denominator of in eq. 23 is always positive for (Section 4), becomes equivalent to C12 > H1 and C12 > H2, the former inequality arising from the condition and the latter from the condition .
If the requirement C12 > H1 and C12 > H2 for fails, then the maximum occurs on the boundary of the unit interval. We have Hadm(0) = H2 and Hadm(1) = H1. Thus, the maximum lies at γ1 = 0 if H2 > H1 and at γ1 = 1 if H1 > H2.
If C12 > H1 and C12 > H2 do not both hold, then one of them must hold, as we showed in Section 4 that 2C12 > H1 + H2. Combining the fact that either C12 > H1 or C12 > H2 holds with the observation that H2 > H1 leads to a maximum at γ1 = 0 and H1 > H2 leads to a maximum at γ1 = 1, we complete the characterization of the three cases.
Note that the three cases in the statement of the proposition capture all possible values of (H1, H2, C12). By the Cauchy-Schwarz inequality, (1 − C12)2 ⩽ (1 − H1)(1 − H2), with equality requiring . Hence, with assumed, either 1 − C12 < 1 − H1 and 1 − C12 ⩾ 1 − H2 (case (ii)), 1 − C12 < 1 − H2 and 1 − C12 ⩾ 1 − H1 (case (iii)), or both 1 − C12 < 1 − H1 and 1 − C12 < 1 − H2 (case (i)).
Alternative expressions in terms of H1, H2, and F12 can be derived by noting that , and C12 = HS(1 + F12)/(1 − F12), the latter simply restating eq. 4 (recalling C12 = 1 for F12 = 1). Thus, we have
| (24) |
| (25) |
Another formulation uses the heterozygosity of a population formed by equal admixture of populations 1 and 2, or HT. Because F12 = 1−HS/HT by eq. 1, F12/(1−F12) = (HT −HS)/HS. Using this relationship in eqs. 24 and 25,
□
Proof of Corollary 8. Suppose H1 ⩾ H2. If case (i) from Proposition 7 applies, then because HT > HS, . Case (ii) cannot apply because H1 < C12, H2 ⩾ C12, and H1 ⩾ H2 cannot hold simultaneously. In case (iii), . For the reverse direction, if H1 < H2 and case (i) or case (ii) applies, then . Case (iii) cannot apply because H1 ⩾ C12, H2 < C12, and H1 < H2 cannot hold simultaneously. □
Proof of Corollary 9. First, we see that in case (i) of Proposition 7. In case (ii), H2 > HT = (H1 + H2 + 2C12)/4 because H2 > H1 and H2 ⩾ C12. In case (iii), H1 > HT because H1 > H2 and H1 ⩾ C12. Note that if H1 = H2, then case (i) applies, producing . □
Proof of Corollary 10. We restate the condition 0 < (C12 − H2)/[2(C12 − HS)] < 1 as
Subtracting from both sides and multiplying by 2, an equivalent condition is
or, equivalently, . We rearrange this last expression to obtain the desired result. □
Proof of Proposition 11. We apply Proposition 7 with J = 2. Substituting p12 = 1 − p11 and p22 = 1 − p21 in eqs. 15 and 16, we obtain C12 −H2 = (p11 −p21)(1−2p21), C12 −H1 = (p21 −p11)(1−2p11), C12 −HS = (p11 − p21)2, and . Thus, because p11 = p21 is not permitted, the quantities in eqs. 15 and 16 reduce to those of eqs. 18 and 19, respectively.
To complete the application of Proposition 7 to K = 2, note that case (i) of Proposition 7 occurs when (p11 − p21)(1 − 2p21) > 0 and (p21 − p11)(1 − 2p11) > 0. The first of this pair of inequalities requires both p11 − p21 > 0 and 1 − 2p21 > 0, so that p11 > p21 and , or both p11 − p21 < 0 and 1 − 2p21 < 0, so that p11 < p21 and . The second inequality requires both p21 − p11 > 0 and 1 − 2p11 > 0, so that p21 > p11 and , or both p21 − p11 < 0 and 1 − 2p11 < 0, so that p21 < p11 and . Thus, the conditions of case (i) of Proposition 7 obtain if and only if or .
Similarly, using the expressions for H1, H2, and C12 when K = 2, the conditions of case (ii) of Proposition 7 are equivalent to or . The conditions of case (iii) are equivalent to or . □
Appendix 3: Dirichlet model for allele frequencies
We first provide results concerning Hadm in the case that the K source populations have independently and identically distributed (IID) allele frequency vectors. Next, we specify these IID vectors to be Dirichlet distributions.
IID allele frequency vectors
We begin by examining the expected values of Hk and Hadm.
Proposition 13. Suppose the allele frequency vectors are independently and identically distributed for 1 ⩽ k ⩽ K. Then .
Proof. We use eq. 8:
Using the IID assumption and simplifying by noting that , we have
from which the result follows. □
An immediate corollary of Proposition 13 is that Hadm has expectation greater than or equal to the expectation of the heterozygosity of each of the source populations.
Corollary 14. Suppose the allele frequency vectors are independently and identically distributed for 1 ⩽ k ⩽ K. Then .
A second corollary results from the Cauchy-Schwarz inequality, by which , with equality if and only if .
Corollary 15. Suppose the allele frequency vectors are independently and identically distributed for 1 ⩽ k ⩽ K. Considering all admixture vectors is maximized at , and has maximal value .
IID allele frequency vectors from a symmetric Dirichlet distribution
We now further assume that the independently and identically distributed allele frequency vectors follow a symmetric multivariate Dirichlet distribution. This distribution is frequently used for allele frequency distributions (Balding & Nichols, 1995; Pritchard et al., 2000; Huelsenbeck & Andolfatto, 2007), and it is a natural probability distribution to assume for allelic types with the same marginal distributions.
The J-dimensional Dirichlet-(α1, α2, …, αJ) distribution is defined over the open unit (J − 1)-simplex ΔJ−1 and has concentration parameters αj > 0. The means and variances for the individual allele frequencies are (Lange, 1997; Kotz et al., 2000, chapter 49):
where .
The symmetric Dirichlet distribution assumes , leading to:
Making these substitutions in Proposition 13, we obtain the expectation of Hadm under the assumption that the allele frequency vectors follow independent Dirichlet distributions.
Corollary 16. Suppose the allele frequency vectors are independently and identically distributed for 1 ⩽ k ⩽ K, all with symmetric multivariate Dirichlet distributions with concentration parameter . Then
This corollary implies that both and are increasing functions of J and .
The next proposition considers the special case of K = 2 and J = 2, further specifying a uniform distribution for γ1.
Proposition 17. Consider K = 2 and J = 2. Suppose that the values of p11 and p21 are independently chosen from a uniform-[0,1] distribution. Suppose also that γ1 is also chosen from a uniform-[0, 1] distribution. Then .
Proof. Using Proposition 11, we identify the regions of the unit square for (p11, p21) in which . These regions are and .
Within those regions, we must determine the portion of the unit interval for γ1 in which Hadm(γ1) > max{H1, H2}. Hadm(γ1) is a quadratic function of γ1. We ignore the set of zero volume with H1 = H2. In the regions for (p11, p21) in which and H2 > H1, the interval for γ1 in which is . In the regions for (p11, p21) in which and H1 > H2, the interval for γ1 in which is .
The desired probability is the volume within the unit cube for (p11, p21, γ1) of the regions in which Hadm(γ1) > max{H1, H2}. The volume is
□
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- Alcala N and Rosenberg NA 2017. Mathematical constraints on FST: biallelic markers in arbitrarily many populations, Genetics 206, 1581–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcala N and Rosenberg NA 2019. G′ST, Jost’s D, and FST are similarly constrained by allele frequencies: a mathematical, simulation, and empirical study, Mol. Ecol 28, 1624–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balding DJ and Nichols RA 1995. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity, Genetics 96, 3–12. [DOI] [PubMed] [Google Scholar]
- Boca SM and Rosenberg NA 2011. Mathematical properties of Fst between admixed populations and their parental source populations, Theor. Pop. Biol 80, 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerkle CA and Lexer C 2008. Admixture as the basis for genetic mapping, Trends Ecol. Evol 23, 686–694. [DOI] [PubMed] [Google Scholar]
- Chakraborty R 1986. Gene admixture in human populations: Models and predictions, Yrbk. Phys. Anthropol 29, 1–43. [Google Scholar]
- Edge MD and Rosenberg NA 2014. Upper bounds on FST in terms of the frequency of the most frequent allele and total homozygosity: the case of a specified number of alleles, Theor. Pop. Biol 97, 20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S 2012. Population genetics models of local ancestry, Genetics 191, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybill FA 1976. “Theory and application of the linear model”, Duxbury, Pacific Grove, CA. [Google Scholar]
- Hedrick PW 1999. Perspective: highly variable loci and their interpretation in evolution and conservation, Evolution 53, 313–318. [DOI] [PubMed] [Google Scholar]
- Hedrick PW 2005. A standardized genetic differentiation measure, Evolution 59, 1633–1638. [PubMed] [Google Scholar]
- Horn RA and Johnson CR 2012. “Matrix analysis”, Cambridge University Press, New York, NY. [Google Scholar]
- Huelsenbeck JP and Andolfatto P 2007. Inference of population structure under a Dirichlet process model, Genetics 175, 1787–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Edge MD, and Rosenberg NA 2013. The relationship between FST and the frequency of the most frequent allele, Genetics 193, 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotz S, Balakrishnan N, and Johnson NL 2000. “Continuous Multivariate Distributions. Volume 1: Models and Applications”, Wiley, New York. [Google Scholar]
- Lange K 1997. “Mathematical and Statistical Methods for Genetic Analysis”, Springer, New York. [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, and Myers RM 2008. Worldwide human relationships inferred from genome-wide patterns of variation, Science 319, 1100–1104. [DOI] [PubMed] [Google Scholar]
- Long JC 1991. The genetic structure of admixed populations, Genetics 127, 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC and Kittles RA 2003. Human genetic diversity and the nonexistence of biological races, Hum. Biol. 75, 449–471. [DOI] [PubMed] [Google Scholar]
- Magnus JR and Neudecker H 2007. “Matrix differential calculus with applications in statistics and econometrics”, John Wiley & Sons, Chichester, UK, 3rd edition. [Google Scholar]
- Maruki T, Kumar S, and Kim Y 2012. Purifying selection modulates the estimates of population differentiation and confounds genome-wide comparisons across single-nucleotide polymorphisms, Mol. Biol. Evol. 29, 3617–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta RS, Feder AF, Boca SM, and Rosenberg NA 2019. The relationship between haplotype-based FST and haplotype length, Genetics 213, 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar RB 1987. Maximum likelihood estimation of mixed stock fishery composition, Can. J. Fish. Aquat. Sci 44, 583–590. [Google Scholar]
- Mooney JA, Huber CD, Service S, Sul JH, Marsden CD, Zhang Z, Sabatti C, Ruiz-Linares A, Bedoya G, Costa Rica/Colombia Consortium for Genetic Investigation of Bipolar Endophenotypes, Freimer N, and Lohmueller KE 2018. Understanding the hidden complexity of Latin American population isolates, Am. J. Hum. Genet 103, 707–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki T 1998. Fixation indices in subdivided populations, Genetics 148, 1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, and Li JZ 2012. Genomic patterns of homozygosity in worldwide human populations, Am. J. Hum. Genet 91, 275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ, DeGiorgio M, and Rosenberg NA 2013. Population structure in a comprehensive genomic data set on human microsatellite variation, G3: Genes, Genomes, Genetics 3, 891–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, and Donnelly P 2000. Inference of population structure using multilocus genotype data, Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SB and Rosenberg NA 2012. Refining the relationship between homozygosity and the frequency of the most frequent allele, J. Math. Biol 64, 87–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Choudhry S, Via M, Basu A, Sebro R, Eng C, Beckman K, Thyne S, Chapela R, Rodriguez-Santana JR, Rodriguez-Cintron W, Avila PC, Ziv E, and Burchard EG 2009. Ancestry-related assortative mating in Latino populations, Genome Biol. 10, R132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA and Calabrese PP 2004. Polyploid and multilocus extensions of the Wahlund inequality, Theor. Pop. Biol 66, 381–391. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Li LM, Ward R, and Pritchard JK 2003. Informativeness of genetic markers for inference of ancestry, Am. J. Hum. Genet 73, 1402–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Lucas FA, Rosenberg NA, and Scheet P 2012. Haploscope: a tool for the graphical display of haplotype structure in populations, Genet. Epidemiol 35, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KB, Jakobsson M, Crawford MH, Schurr TG, Boca SM, Conrad DF, Tito RY, Osipova LP, Tarskaia LA, Zhadanov SI, Wall JD, Pritchard JK, Malhi RS, Smith DG, and Rosenberg NA 2009. Haplotypic background of a private allele at high frequency in the Americas, Mol. Biol. Evol 26, 995–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu P and Rosenberg NA 2011. A general mechanistic model for admixture histories of hybrid populations, Genetics 189, 1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ray N, Rojas W, Parra MV, Bedoya G, Gallo C, Poletti G, Mazzotti G, Hill K, Hurtado AM, Camrena B, Nicolini H, Klitz W, Barrantes R, Molina JA, Freimer NB, Bortolini MC, Salzano FM, Petzl-Erler ML, Tsuneto LT, Dipierri JE, Alfaro EL, Bailliet G, Bianchi NO, Llop E, Rothhammer F, Excoffier L, and Ruiz-Linares A 2008. Geographic patterns of genome admixture in Latin American Mestizos, PLoS Genet. 4, e1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Tang H, and Risch N 2008. Admixture mapping and the role of population structure for localizing disease genes, Adv. Genet 60, 547–569. [DOI] [PubMed] [Google Scholar]
- Zou JY, Park DS, Burchard EG, Torgerson DG, Pino-Yanes M, Song YS, Sankararaman S, Halperin E, and Zaitlen N 2015. Genetic and socioeconomic study of mate choice in Latinos reveals novel assortment patterns, Proc. Natl. Acad. Sci. USA 112, 13621–13626. [DOI] [PMC free article] [PubMed] [Google Scholar]