Abstract
Background:
Genome-wide association studies (GWAS) have identified several single nucleotide polymorphisms (SNPs) associated with pancreatic cancer. No studies yet have attempted to replicate these SNPs in US minority populations. We aimed to replicate the associations of 31 GWAS-identified SNPs with pancreatic cancer and build and test a polygenic risk score (PRS) for pancreatic cancer in an ethnically diverse population.
Methods:
We evaluated 31 risk variants in the Multiethnic Cohort and the Southern Community Cohort Study. We included 691 pancreatic ductal adenocarcinoma (PDAC) cases and 13,778 controls from African-American, Japanese-American, Latino, Native Hawaiian, and white participants. We tested the association between each SNP and PDAC, established a PRS using the 31 SNPs and tested the association between the score and PDAC risk.
Results:
Eleven of the 31 SNPs were replicated in the multiethnic sample. The PRS was associated with PDAC risk [OR top vs. middle quintile = 2.25 (95% CI: 1.73, 2.92)]. Notably, the PRS was associated with PDAC risk in all ethnic groups except Native Hawaiian (OR per risk allele ranged from 1.33 in Native Hawaiians to 1.91 in African Americans; P heterogeneity=0.12).
Conclusions:
This is the first study to replicate 11 of the 31 GWAS-identified risk variants for pancreatic cancer in multiethnic populations, including African Americans, Japanese Americans and Latinos. Our results also suggest a potential utility of PRS with GWAS-identified risk variants for the identification of individuals at increased risk for PDAC across multiple ethnic groups.
Impact:
PRS can potentially be used to stratify pancreatic cancer risk across multiple ethnic groups.
Keywords: Pancreatic Cancer, PDAC, Genetics, Replication, Polygenic Risk Score
Introduction
Pancreatic cancer is the fourth leading cause of cancer deaths in the United States with over 56,000 new cases and 45,000 deaths in 2019 (1). By 2030, pancreatic cancer is projected to be the second leading cause of cancer-related death (2). Diagnosis at a late stage is common due to lack of symptoms at early stage of disease and regular forms of screening (3). These characteristics result in a 5-year survival of only 9% (1), emphasizing the importance of primary prevention strategies for this disease.
Pancreatic cancer incidence differs by ethnicity. African Americans experience 1.36 times the rate of pancreatic cancer (10.4 per 100,000) relative to non-Hispanic whites (7.7) (4). Differences in incidence rates are observed across other ethnic groups [Hispanic (7.1), Japanese (8.1), Asian/Pacific Islander (6.2 per 100,000)]. In the Multiethnic Cohort (MEC), the incidence rates of pancreatic cancer are notably higher among Native Hawaiians (1.8 times that of whites), followed by African Americans and Japanese Americans (1.3-1.4 times that of whites) (5). Epidemiologic studies have associated body mass index (BMI) (5–7), type 2 diabetes (5,8,9), diet patterns (10,11) and smoking (5,12) with pancreatic cancer. In the MEC, ~20% of pancreatic cancer can be attributed to these factors (13).
Common genetic variants have been associated with pancreatic cancer risk in genome-wide association studies (GWAS) (14–22). So far, these GWAS have identified 31 risk variants for pancreatic cancer. Twenty-two were identified by the PanScan and the PanC4 studies, composed of populations of primarily European-ancestry (14–16,19,20,22). Of the remaining variants, four were discovered in Japanese and five in Chinese (17,18,21).
The associations between GWAS variants and pancreatic cancer have yet to be examined in other ethnic groups, especially in high-risk African Americans and other minority populations. Few single nucleotide polymorphisms (SNPs) identified in European ancestry replicated in Asian samples. In Chinese, Wang et al. replicated 4 SNPs identified in GWAS and pathway analysis in Europeans, Chinese, and Japanese (23). Among Japanese, Nakatochi et al. has replicated 13 GWAS-significant and suggestive loci, discovered in Europeans, Japanese, and Chinese (24); Ueno et al. has replicated one European ancestry loci in Japanese (25). Similarly, there is limited cross-ethnic replications among Europeans (16,26), with only one Japanese-identified SNP replicated in Europeans (16). Additionally, three other Asian GWAS have also reported on replication of GWAS identified SNPs in their samples (17,18,21). Lack of replication of pancreatic cancer-associated SNPs across ethnic groups may be due to low minor allele frequencies, monomorphic loci, and differences of linkage disequilibrium of tagging SNPs between ethnic groups. Identifying the association of these SNPs with pancreatic cancer in a multiethnic population, and in ethnic-specific analyses, will help us identify the value of these SNPs for disease prediction in an admixed sample.
In this study, we assessed the transportability of prior GWAS findings in an ethnically diverse population and examined how these variants contribute to pancreatic cancer risk across populations. We first attempted to replicate the 31 GWAS-significant risk variants in the MEC and the Southern Community Cohort Study (SCCS). Using the 31 SNPs, we then built a multiethnic polygenic risk score (PRS) and assessed its association with pancreatic cancer risk.
Materials and Methods
Study Population.
This study included case-control samples within the MEC and SCCS. Information on recruitment, characteristics, and case ascertainment in the MEC and SCCS has been described (27,28) (Supplementary Methods Text 1). Briefly, the MEC is a population-based prospective cohort study initiated between 1993 and 1996 to investigate cancer etiology. The MEC consists of over 215,000 men and women from Los Angeles County and Hawaii who were 45 to 75 years old at enrollment and from these racial/ethnic groups: African Americans, Japanese Americans, Latinos, Native Hawaiians and whites. The SCCS was initiated in 2002 to investigate sources of racial disparities in cancer and chronic disease. The SCCS participants were mainly African Americans and whites between the ages of 40 and 79 who resided in one of 12 US southern states. At baseline, the MEC and SCCS gathered detailed information on demographics, lifestyle, diet, anthropometry, reproductive history, and medical history. In both cohorts, cancer cases were identified through annual linkage to state cancer registries. Pancreatic cancer cases were defined as primary invasive pancreatic cancer with pancreatic ductal adenocarcinoma (PDAC) histology (ICD-O-3 code C25). Controls were selected by matching to incident cases based on age, sex, and ethnicity. For the MEC, we also added eligible controls (without PDAC) with genotype data from prior GWAS. We conducted all analyses with the original cases and matched controls then with added controls. We present the results using the added controls since the effect estimates were similar between analyses and we had improved statistical power.
Genotyping, Quality Control and Genotype Imputation.
Samples were genotyped using the Multi-Ethnic Genotyping Array (MEGA) chip (Illumina, San Diego, CA), which was developed to ensure genome-wide coverage of variants down to 1% frequency in non-European ancestry populations. Samples underwent an intensive quality control process including SNP call-rate filtering, sample call rate filtering, concordance checks of inter- and intra-plate controls, removal of redundant or discordant variants based on location and call rates, removal of SNPs with race-specific allele frequency differences over 25% in comparison to 1000G phase 3 race-specific estimates (Supplementary Figure 1). Following QC, 932,530 SNPs, 691 cases and 13,778 controls were used for imputation. The sample was stratified based on self-reported ethnicity, then imputed using Minimac3, ShapeIT v2, and the cosmopolitan 1000 Genomes Project reference panel (Phase 3 v5).
Statistical Analysis.
Participants with missing covariate values or with implausible values for age, sex, diabetes, and body mass index (BMI in kg/m2) were removed from analysis. Related samples (first- and second-degree relatives) were identified using KING software for robust relationship inference then removed based on a kinship coefficient of 0.0884 or greater (Supplementary Figure 2) (29).
SNP and PDAC associations were examined using logistic regression, adjusting for age at sample collection, sex, study, BMI, diabetes, and population stratification using principal components (PC 1-6). PCs were estimated using PLINK and a set of >50,000 independent SNPs (30). Most global ancestry variation among the five ethnic groups was captured in the first six PCs (Supplementary Figure 3). Measures of association were reported on the ratio scale along with corresponding likelihood ratio test (LRT) p-values. As a sensitivity analysis, we estimated multiethnic associations by meta-analyzing ethnic-specific results using both fixed effect and random effects models. We present multiethnic pooled results since there was no effect heterogeneity between the pooled multiethnic analysis and the meta-analyses. All SNPs were modeled as log odds of PDAC per risk allele (0, 1, 2). A log-odds weighted PRS was estimated for each participant by multiplying the multiethnic log-odds for each of the 31 SNPs by the number of risk alleles at the given loci, then summing all values. This PRS took the following form: PRS = β1x1+β2x2+ … . βkxk+βnxn. In this algorithm, β1 is the log-odds ratio for risk of PDAC associated with a per allele increase in risk for a given SNP in our replication analysis. xk is the number of risk alleles an individual has for the corresponding SNP (0, 1, or 2). We additionally conducted sensitivity analyses using the following alternative weighting methods: external weights, external weights only using 22 SNPs from European studies, unweighted, ethnic-specific internal weights, and multiethnic weights from a meta-analysis of ethnic-specific associations from the replication, using both a random-effects and a fixed-effect.
Logistic regression was used to estimate the log odds of PDAC based on binned percentiles ([1%-20%], (20%-40%], (40%-60%], (60%-80%], (80%-100%]) generated using the PRS distribution among controls within each ethnic group, except for the multiethnic analysis which used the control distribution from all groups combined. Log-odds of PDAC in each percentile were compared to the mid-quantile category (40%-60%). The PRS was also modeled continuously, after standardizing the score to the ethnic-specific interquartile range (IQR) among controls, except for the multiethnic analysis which used all controls. The replication and PRS analyses were stratified by ethnicity. P <0.05 was used to determine statistical significance. Analysis was conducted using R 3.5.0 (31).
Results
Sample Characteristics.
The final analytical sample included 691 PDAC cases and 13,778 controls (518 cases and 13,426 controls from the MEC; 173 cases and 352 controls from the SCCS). Most cases were African American (230 cases/5,235 controls), followed by Japanese American (181 cases/3,285 controls), white (132 cases/570 controls), Latino (105 cases/ 2,935 controls), and Native Hawaiian (43 cases/1,753 controls) (Table 1). SCCS samples were younger, had a higher prevalence of diabetes, and a higher mean BMI than MEC participants. Diabetes was common among SCCS African Americans (32.4% of cases) and MEC Native Hawaiians (25.6% of cases). A large portion of the sample was overweight or obese. SCCS African-American and white cases had a mean BMI of 31.4 kg/m2. MEC Japanese Americans had the lowest mean BMI (24.8 kg/m2 among cases).
Table 1:
Characteristics of pancreatic cancer cases and controls
| Case | Control | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | Age | Female n (%) | Diabetes n (%) | Mean BMI | n | Age | Female n (%) | Diabetes n (%) | Mean BMI | |
| MEC | ||||||||||
| African American | 94 | 71.3 | 56 (59.6) | 8 (8.5) | 28.0 | 4961 | 69.1 | 3091 (62.3) | 757 (15.3) | 28.8 |
| Latino | 105 | 69.5 | 45 (42.9) | 21 (20.0) | 28.4 | 2935 | 67.0 | 1600 (54.5) | 211 (7.2) | 27.8 |
| Japanese American | 181 | 70.9 | 103 (56.9) | 16 (8.8) | 24.8 | 3285 | 69.0 | 1541 (46.9) | 761 (23.2) | 25.4 |
| White | 95 | 69.2 | 42 (44.2) | 0 (0) | 26.4 | 492 | 60.2 | 234 (47.6) | 2 (0.4) | 25.1 |
| Native Hawaiian | 43 | 67.0 | 21 (48.8) | 11 (25.6) | 29.6 | 1753 | 65.4 | 968 (55.2) | 237 (13.5) | 28.6 |
| SCCS | ||||||||||
| African American | 136 | 57.6 | 70 (51.5) | 44 (32.4) | 29.4 | 274 | 57.5 | 141 (51.5) | 69 (25.2) | 30.0 |
| White | 37 | 56.8 | 19 (51.4) | 10 (27.0) | 29.4 | 78 | 57.3 | 39 (50.0) | 13 (16.7) | 28.7 |
MEC = Multiethnic Cohort; SCCS = Southern Community Cohort Study; BMI = Body Mass Index
SNP Frequencies.
All SNPs, except rs78193826 and rs35226131 had a minor allele frequency (MAF) >0.05 in the multiethnic sample. Multiple SNPs were rare in ethnic-specific groups (Supplementary Table 1), and all had similar risk allele frequencies to what is reported in prior studies (Supplementary Figure 4). Among cases or controls combined, there were 2 SNPs in the multiethnic sample with a MAF <0.05, 3 in whites, 3 in African Americans, 6 in Japanese Americans, 2 in Latinos, and 3 in Native Hawaiians. When considering MAF <0.01, there were 0 SNPs in the multiethnic sample, 2 in whites, 2 in African Americans, 2 in Japanese Americans, 1 in Latinos, and 1 in Native Hawaiians.
Replication Analysis.
11 of the 31 SNPs were replicated at P <0.05, with consistent direction of association with that observed in the literature (Figure 1; Figure 2; Supplementary Table 1). Of the replicating SNPS, 10 were discovered in Europeans (rs505922, rs6971499, rs4795218, rs10094872, rs401681, rs7190458, rs7214041, rs1517037, rs13303010, rs9543325) and one (rs1547374) in Chinese. Replicating SNPs had a similar mean effect size to what is reported in the literature (mean log-odds of replicating SNPs from literature: 0.20; mean log-odds of replicating SNPs in multiethnic sample: 0.17). Of these 11 replicated SNPs, 8 were statistically significant in at least one ethnic group after filtering out SNPs with an MAF <0.05 in cases or controls. Four replicated in African Americans, three in whites, one in Japanese Americans, one in Latinos, and one in Native Hawaiians, at P <0.05. Within the set of 11 replicating SNPs in the multiethnic sample, with MAF >0.05 among cases or controls, we assessed directional consistency of SNP associations with the literature. Among whites, 9/11 SNPs had consistent direction of effect; 11/11 among African Americans, 8/9 among Japanese Americans, 10/11 among Latinos, 7/11 among Native Hawaiians.
Figure 1.
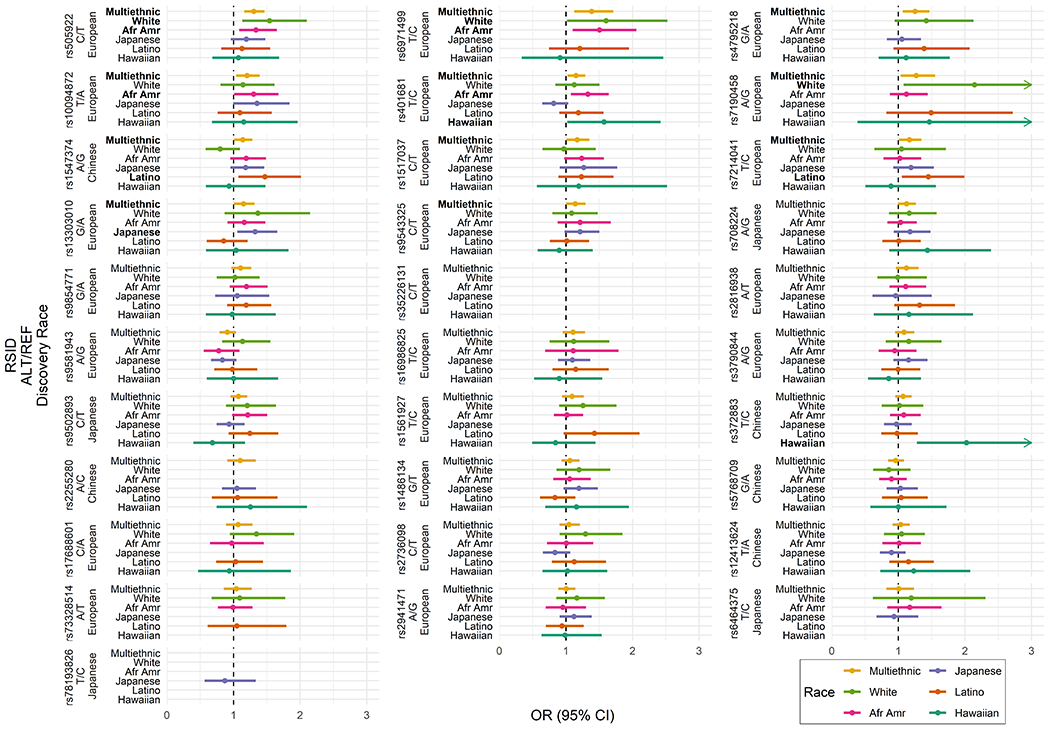
Multiethnic and ethnic-specific replication analysis results for 31 SNPs identified in prior GWAS of pancreatic cancer in European, Chinese, and Japanese ancestry. Results shown on the odds ratio (OR) scale with corresponding 95% confidence intervals (CIs) and ordered from lowest to highest p-values from multiethnic replication analysis. Only SNPs with minor allele frequency > 0.05 are shown. RSID = Reference SNP cluster ID; ALT = Alternative (risk) allele; REF = Reference allele.
Figure 2.
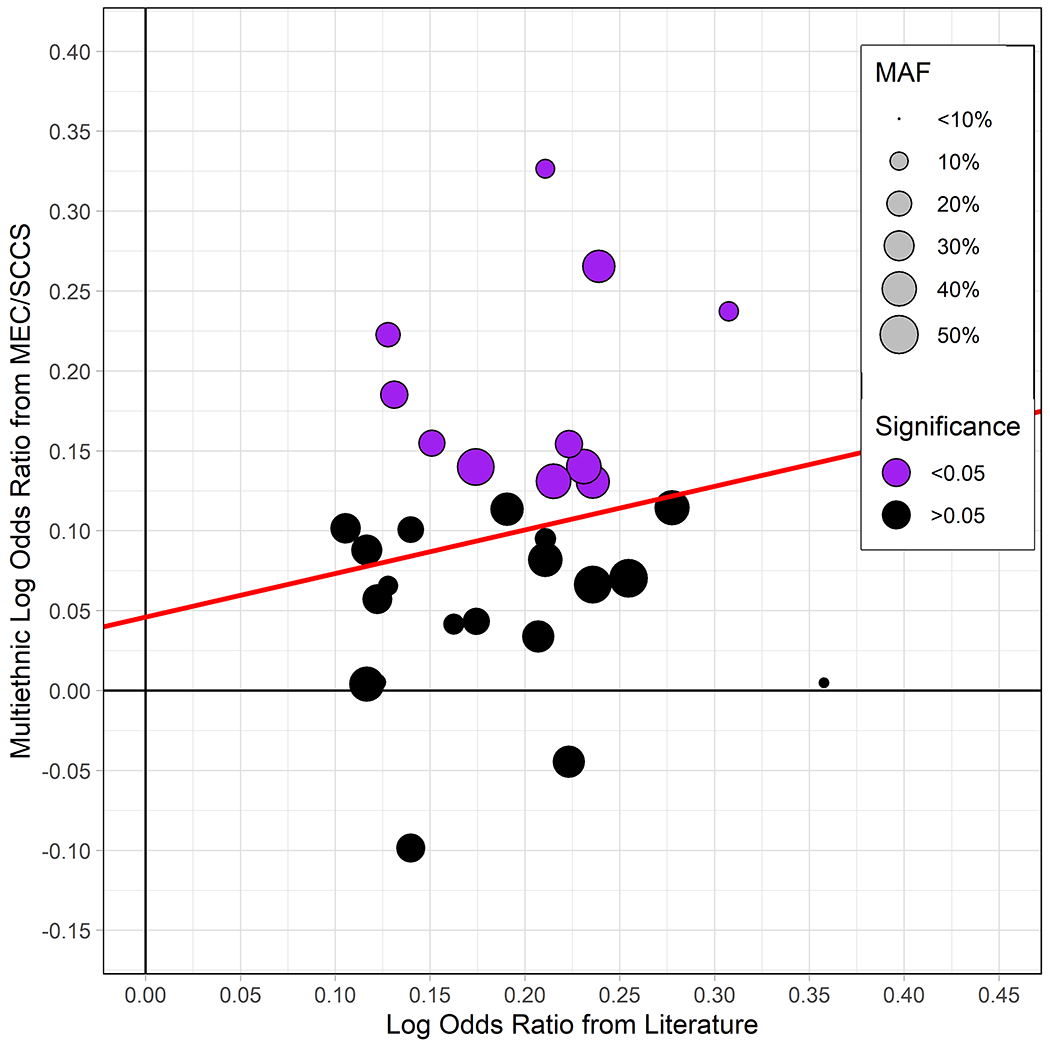
Comparison between 31 replicating SNPs from multiethnic replication analysis and most recent GWAS results on the log OR scale. Point size corresponds to minor allele frequency (MAF) among controls in the replication analysis. The red line represents, MAF-weighted least squares fit. One point removed from figure due to extreme replication result (rs2816938, OR = 0.69). MEC = Multiethnic Cohort; SCCS = Southern Community Cohort Study.
Of the 20 SNPs not replicating in the multiethnic sample with an MAF >0.05, one was replicated in Native Hawaiians (rs372883; discovered in Chinese), with consistent direction of association to that observed in the literature. Within this set of non-replicating SNPs in the multiethnic sample, with ethnic-specific MAF >0.05, we assessed directional consistency of associations in each race/ethnic group with the literature. Among whites, 15/17 SNPs had consistent direction of effect; 10/17 among African Americans, 8/16 among Japanese-Americans, 12/17 among Latinos, 7/16 among Native Hawaiians.
Polygenic Risk Score.
We estimated a genetic risk score using the multiethnic effect estimates as the weight for each SNP. When comparing PRS distributions, we observed a significant difference in risk scores by case status, where cases had 0.13 higher mean PRS than controls (P<0.001). In the multiethnic sample, those in the (80%-100%] risk score group had OR=2.25 (95% CI: 1.73, 2.92) for PDAC relative to the reference group (Figure 3; Supplementary Figure 5). The IQR-standardized PRS was significantly associated with PDAC (OR per IQR increase=1.94; 95% CI: 1.70, 2.22; P LRT=4.092 x 10−17). The PRS was significantly associated with PDAC risk in African Americans (OR per IQR increase=1.91; 95% CI: 1.55, 2.35; P LRT: 3.121 x 10−8), Japanese Americans (OR=1.46 1.19, 1.79; P LRT: 0.003), Latinos (OR=1.65; 95%CI 1.26, 2.17; P LRT: 0.013 and whites (OR=1.85; 95% CI: 1.38, 2.46; P LRT: 2.937 x 10−4). There was no significant ethnic heterogeneity of the association between PRS and PDAC in the continuous model (P heterogeneity=0.12). We observed similar results when using alternative weighting schemes for the multiethnic analysis (Supplementary Figures 6–11).
Figure 3.
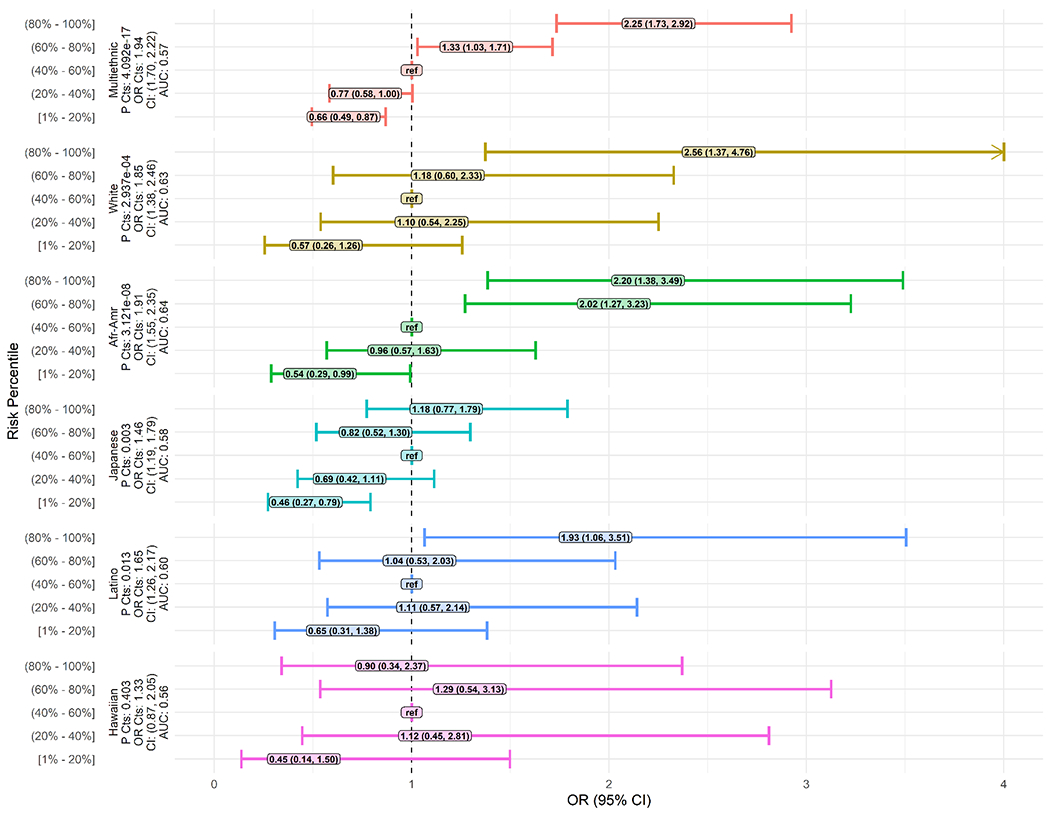
Multiethnic and ethnic-specific polygenic risk score odds ratios (ORs) and 95% confidence intervals (CIs). Weights used from multiethnic replication analysis. Multiethnic analysis used binned risk score percentile groups from the complete, multiethnic, sample among controls. Ethnic-specific analysis used binned risk score percentile groups from the control ethnic-specific risk score distribution among controls. ref = Reference category used in binned regression analysis; P Cts = P-value from continuous polygenic risk score model.
Discussion
We investigated the association of GWAS-identified SNPs with pancreatic cancer risk in an ethnically diverse population. Of the 31 SNPs tested, we replicated 11 in the multiethnic sample at an alpha of 0.05. In comparison to prior replication attempts across racial groups (16,23,24), we found a number of SNPs identified in Europeans and Asians to be associated with PDAC in a US multiethnic sample. Furthermore, we showed the potential utility of PRS with GWAS-identified risk variants for the identification of individuals at increased risk for PDAC across multiple ethnic groups.
Of the SNPs tested, the 3 least common SNPs in our multiethnic sample (MAF <0.1 among controls), were not replicated, but 60% of the most common SNPs in our sample (MAF >0.4) were replicated. This highlights a possible pattern between replication and allele frequencies in our multiethnic sample. Although most SNPs have been identified in GWAS of European ancestry (14–16,19,20,22), only 3 of these SNPs were replicated in our white population (14,20), the group which most similarly reflects European ancestry. This limited replication likely results from the small number of whites relative to the other ethnic groups in our study.
The most significant replicating SNP (rs505922 in ABO) was the first SNP identified to be associated with PDAC (14). This association was significant in whites and African Americans. In our study, this SNP was not replicated in Japanese Americans, however it was directionally consistent with a similar effect size (OR=1.19; 95% CI: 0.96, 1.48) to prior Japanese studies (ORs range 1.11 - 1.36) (18,24). The next three most significant replicating SNPs in our multiethnic sample (rs6971499, rs4795218, rs10094872; discovered in European ancestry) have not been replicated across any race/ethnicity to our knowledge. Following these, rs401681 (CLPTM1L) has been replicated in Chinese (21,23). The study that reports replication has a similar effect to ours (Wang et al. OR=1.39; 95% CI: 1.11, 1.74; MEC/SCCS OR=1.15; 95% CI 1.03, 1.29), however, we observed significant heterogeneity of association by ethnic group (P heterogeneity= 0.01). Another frequent cross-ethnic replication from prior studies, rs9543325 (KLF5), was replicated in our multiethnic sample. This SNP was associated with PDAC in Japanese and Chinese (17,18,21,24) with a larger effect estimate than in our multiethnic analysis and an estimate similar to our Japanese-American sample. In contrast, the single Chinese-discovered SNP (rs1547374; TFF1) (21), which was replicated in our multiethnic sample and in the Latino subset, was not replicated in prior studies of European (16), Chinese (23), and Japanese samples (24). Lastly, rs3790844 and rs3790843 in NR5A2 have been associated with pancreatic cancer in Europeans (16,32) and Japanese (18,25), however we did not replicate this finding. The effect estimates in MEC whites and Japanese Americans were most similar to those reported in European (16) and in Japanese ancestry (18,25).
Reason for lack of replication between studies is likely due to limited sample size. The size of our case group is less than half the number of cases included in the first European ancestry GWAS and is only around 7% the size of the most recent European ancestry GWAS (14,16). It is likely that the limited number of cases in our study, relative to what is seen in pancreatic cancer GWAS, resulted in a lower statistical power than required for replication of additional SNPs. A second likely factor limiting replication in our study is racial heterogeneity. Across ethnic groups, differing linkage-disequilibrium structures can lead to the tagging SNPs not being in association with the true causal SNP, resulting in lack of replication (33).
We built and tested a multiethnic genetic risk score using the previously identified 31-GWAS SNPs. The associations between PRS and PDAC risk from the multiethnic and ethnic-specific continuous models were statistically significant, except in Native Hawaiians. In the ethnic analysis, a monotonic pattern between categorical PRS and PDAC risk was clearest in African Americans. Two studies have reported a PRS analysis for PDAC (16,24). In the most recent study, Klein et al. used the 22 SNPs identified in European ancestry to estimate a weighted PRS using their results and found a strong association with PDAC (16). In an earlier study, Nakatochi et al. first attempted to replicate 61 GWAS-identified SNPs (both significant and suggestive) then used the 8 replicating SNPs in stepwise regression to select five independent SNPs for use in a PRS (24). They observed significant associations between the extreme PRS categories and PDAC risk.
Consistent with findings from Klein et al. and the Japanese ancestry PRS in Nakatoshi et al. (16,24), we observed an association between PRS and PDAC risk in whites and Japanese Americans. In our sensitivity analysis using weights from Klein et al, we found similar performance of the PRS, with three of the four risk quantile groups differing from the reference. Both previous studies used ethnic-specific weights in their analysis which might provide a better fit in a large study. In our main analysis, we used multiethnic weights to uniformly weight SNPs across ethnicities. This was done so differences in PRS-PDAC association reflects case-control differences and not discrepancies in ethnic-specific weights which can be highly variable due to small sample sizes within some ethnic groups.
There are several strengths and limitations to this study. This is the first replication study of pancreatic cancer risk variants in a multiethnic population. Our ethnic-specific analysis is the first to produce replication estimates and show transportability of GWAS findings for multiple ethnic groups, including African Americans who have notably high pancreatic cancer incidence, yet have not been studied in the context of genetics. We leveraged existing MEC GWAS data to boost sample size which improved power needed to replicate multiple SNPs. Finally, we showed that multiethnic estimates for SNPs known to be associated with pancreatic cancer perform better than expected in both a multiethnic and ethnic-stratified PRS analysis. Limitations include our relatively small number of cases in comparison to what was included in previous GWAS, which may be responsible for some SNPs not replicating in our sample. We stratified the replication and PRS analysis by self-reported ethnicity. As observed in the principal component figures, there can be considerable variation of global ancestry within these groups.
In conclusion, we successfully replicated 11 of the 31 GWAS-identified loci in a multiethnic population. These replications provide evidence for the importance of these SNPs in understanding genetic pancreatic cancer risk in an admixed population and in understudied ethnic groups. We showed a potential value of PRS with GWAS-identified variants for the identification of individuals at increased risk for PDAC across multiple ethnic groups. Currently there is no routine screening recommended for PDAC, and thus PRS may be useful in identifying a subgroup of high-risk individuals who may benefit the most from screening with endoscopic ultrasound or MRI. Furthermore, with known modifiable risk factors (i.e. smoking, excess weight, diabetes) for PDAC, PRS may be useful for prioritizing individuals for targeted health and lifestyle-related interventions.
Supplementary Material
Acknowledgments
Grant support: The National Cancer Institute at the National Institutes of Health (R01CA209798 to VWS, U01CA164973 to LLM, U01CA202979 to WJB and U01HG007397 to CAH). VWS is supported by an American Cancer Society Research Scholar Grant (RSG-16-250-01-CPHPS).
Footnotes
Conflicts of Interest: none.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: A Cancer Journal for Clinicians 2019;69(1):7–34 doi 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Research 2014;74(11):2913–21 doi 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Kardosh A, Lichtensztajn DY, Gubens MA, Kunz PL, Fisher GA, Clarke CA. Long-Term Survivors of Pancreatic Cancer: A California Population-Based Study. Pancreas 2018;47(8):958–66 doi 10.1097/MPA.0000000000001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Zhang J, Deapen D, Stern MC, Sipin A, Pandol SJ, et al. Differences in Pancreatic Cancer Incidence Rates and Temporal Trends Across Asian Subpopulations in California (1988-2015). Pancreas 2019;48(7):931–3 doi 10.1097/MPA.0000000000001337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang BZ, Stram DO, Marchand LL, Haiman CA, Wilkens LR, Pandol SJ, et al. Interethnic differences in pancreatic cancer incidence and risk factors: The Multiethnic Cohort. Cancer Medicine 2019;0(0) doi 10.1002/cam4.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arslan AA, Helzlsouer KJ, Kooperberg C, Shu X-O, Steplowski E, Bueno-de-Mesquita HB, et al. Anthropometric measures, body mass index, and pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan). Archives of Internal Medicine 2010;170(9):791–802 doi 10.1001/archinternmed.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aune D, Greenwood DC, Chan DSM, Vieira R, Vieira AR, Navarro Rosenblatt DA, et al. Body mass index, abdominal fatness and pancreatic cancer risk: a systematic review and non-linear dose-response meta-analysis of prospective studies. Annals of Oncology: Official Journal of the European Society for Medical Oncology 2012;23(4):843–52 doi 10.1093/annonc/mdr398. [DOI] [PubMed] [Google Scholar]
- 8.Batabyal P, Vander Hoorn S, Christophi C, Nikfarjam M. Association of diabetes mellitus and pancreatic adenocarcinoma: a meta-analysis of 88 studies. Annals of Surgical Oncology 2014;21(7):2453–62 doi 10.1245/s10434-014-3625-6. [DOI] [PubMed] [Google Scholar]
- 9.Bosetti C, Rosato V, Li D, Silverman D, Petersen GM, Bracci PM, et al. Diabetes, antidiabetic medications, and pancreatic cancer risk: an analysis from the International Pancreatic Cancer Case-Control Consortium. Annals of Oncology: Official Journal of the European Society for Medical Oncology 2014;25(10):2065–72 doi 10.1093/annonc/mdu276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arem H, Reedy J, Sampson J, Jiao L, Hollenbeck AR, Risch H, et al. The Healthy Eating Index 2005 and risk for pancreatic cancer in the NIH-AARP study. Journal of the National Cancer Institute 2013;105(17):1298–305 doi 10.1093/jnci/djt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan JM, Gong Z, Holly EA, Bracci PM. Dietary patterns and risk of pancreatic cancer in a large population-based case-control study in the San Francisco Bay Area. Nutrition and Cancer 2013;65(1):157–64 doi 10.1080/01635581.2012.725502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch SM, Vrieling A, Lubin JH, Kraft P, Mendelsohn JB, Hartge P, et al. Cigarette Smoking and Pancreatic Cancer: A Pooled Analysis From the Pancreatic Cancer Cohort Consortium. American journal of epidemiology 2009;170(4):403–13 doi 10.1093/aje/kwp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang BZ, Stram DO, Le Marchand L, Haiman CA, Wilkens LR, Pandol SJ, et al. Interethnic differences in pancreatic cancer incidence and risk factors: The Multiethnic Cohort. Cancer Med 2019;8(7):3592–603 doi 10.1002/cam4.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nature Genetics 2009;41(9):986–90 doi 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Childs EJ, Mocci E, Campa D, Bracci PM, Gallinger S, Goggins M, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nature Genetics 2015;47(8):911–6 doi 10.1038/ng.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein AP, Wolpin BM, Risch HA, Stolzenberg-Solomon RZ, Mocci E, Zhang M, et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nature Communications 2018;9(1) doi 10.1038/s41467-018-02942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y, Nakatochi M, Ito H, Kamatani Y, Inoko A, Sakamoto H, et al. Genome-wide association meta-analysis identifies novel GP2 gene risk variants for pancreatic cancer in the Japanese population. bioRxiv 2018:498659 doi 10.1101/498659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Low S-K, Kuchiba A, Zembutsu H, Saito A, Takahashi A, Kubo M, et al. Genome-Wide Association Study of Pancreatic Cancer in Japanese Population. PLOS ONE 2010;5(7):e11824 doi 10.1371/journal.pone.0011824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nature Genetics 2010;42(3):224–8 doi 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nature Genetics 2014;46(9):994–1000 doi 10.1038/ng.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C, Miao X, Huang L, Che X, Jiang G, Yu D, et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nature Genetics 2011;44(1):62–6 doi 10.1038/ng.1020. [DOI] [PubMed] [Google Scholar]
- 22.Zhang M, Wang Z, Obazee O, Jia J, Childs EJ, Hoskins J, et al. Three new pancreatic cancer susceptibility signals identified on chromosomes 1q32.1, 5p15.33 and 8q24.21. Oncotarget 2016;7(41):66328–43 doi 10.18632/oncotarget.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Lin X, Na R, Jiang D, Zhang P, Li J, et al. An evaluation study of reported pancreatic adenocarcinoma risk-associated SNPs from genome-wide association studies in Chinese population. Pancreatology: official journal of the International Association of Pancreatology (IAP) [et al] 2017;17(6):931–5 doi 10.1016/j.pan.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Nakatochi M, Lin Y, Ito H, Hara K, Kinoshita F, Kobayashi Y, et al. Prediction model for pancreatic cancer risk in the general Japanese population. PloS One 2018;13(9):e0203386 doi 10.1371/journal.pone.0203386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueno M, Ohkawa S, Morimoto M, Ishii H, Matsuyama M, Kuruma S, et al. Genome-wide association study-identified SNPs (rs3790844, rs3790843) in the NR5A2 gene and risk of pancreatic cancer in Japanese. Scientific Reports 2015;5:17018 doi 10.1038/srep17018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campa D, Rizzato C, Bauer AS, Werner J, Capurso G, Costello E, et al. Lack of replication of seven pancreatic cancer susceptibility loci identified in two Asian populations. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology 2013;22(2):320–3 doi 10.1158/1055-9965.EPI-12-1182. [DOI] [PubMed] [Google Scholar]
- 27.Kolonel LN, Henderson BE, Hankin JH, Nomura AMY, Wilkens LR, Pike MC, et al. A Multiethnic Cohort in Hawaii and Los Angeles: Baseline Characteristics. American journal of epidemiology 2000;151:346–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Signorello LB, Hargreaves MK, Steinwandel MD, Zheng W, Cai Q, Schlundt DG, et al. Southern community cohort study: establishing a cohort to investigate health disparities. Journal of the National Medical Association 2005;97(7):972–9. [PMC free article] [PubMed] [Google Scholar]
- 29.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M. Robust relationship inference in genome-wide association studies. Bioinformatics (Oxford, England) 2010;26(22):2867–73 doi 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S, Chang C. PLINK 2.
- 31.R: A Language and Environment for Statistical Computing. 3.5.1. Vienna, Austria: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 32.Chen Q, Yuan H, Shi G-D, Wu Y, Liu D-F, Lin Y-T, et al. Association between NR5A2 and the risk of pancreatic cancer, especially among Caucasians: a meta-analysis of case-control studies. OncoTargets and Therapy 2018;11:2709–23 doi 10.2147/OTT.S157759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marigorta UM, Navarro A. High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants. PLOS Genetics 2013;9(6):e1003566 doi 10.1371/journal.pgen.1003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


