Summary
Hepatic Tank-Binding Kinase 1 (TBK1) activity is increased during obesity, and administration of a TBK1 inhibitor reduces fatty liver. Surprisingly, liver-specific TBK1 knockout in mice produces fatty liver by reducing fatty acid oxidation. TBK1 functions as a scaffolding protein to localize acyl-CoA synthetase long-chain family member 1 (ACSL1) to mitochondria, which generates acyl-CoAs that are channeled for β-oxidation. TBK1 is induced during fasting, and maintained in the unphosphorylated, inactive state, enabling its high affinity binding to ACSL1 in mitochondria. In TBK1-deficient liver, ACSL1 is shifted to the endoplasmic reticulum to promote fatty acid re-esterification in lieu of oxidation in response to fasting, which accelerates hepatic lipid accumulation. The impaired fatty acid oxidation in TBK1-deficient hepatocytes is rescued by expression of kinase-dead TBK1. Thus, TBK1 operates as a rheostat to direct the fate of fatty acids in hepatocytes, supporting oxidation when inactive during fasting, and promoting re-esterification when activated during obesity.
Keywords: Tank-binding kinase 1 (TBK1), Acyl-CoA synthetase long chain family member 1 (ACSL1), Hepatic lipid metabolism, Fasting, Mitochondria, β-oxidation, Re-esterification, Non-alcoholic fatty liver disease (NAFLD)
eTOC blurb
Huh et al. demonstrate that TBK1 is necessary for fasting-induced fat oxidation in liver via channeling substrates to mitochondria. While obesity increases the kinase activity of TBK1, fasting reduces its activity, which elevates the inactive TBK1 to support more efficient fat oxidation. This pathway may contribute to the mechanism by which TBK1 inhibitors improve fatty liver.
Graphical Abstract

Introduction
Non-alcoholic fatty liver disease (NAFLD) is a complication of obesity, often associated with type 2 diabetes (T2D), dyslipidemia, and hypertension (Anstee et al., 2013, Byrne and Targher, 2015, Yki-Järvinen, 2014). NAFLD encompasses a broad spectrum of symptoms from simple hepatic steatosis to non-alcoholic steatohepatitis (NASH) and cirrhosis, which can lead to hepatocellular carcinoma (HCC) and liver failure (Anstee et al., 2013, Pais et al., 2016). NASH will soon become the leading cause of liver transplant and liver-associated death in the US (Estes et al., 2018, Friedman et al., 2018). Despite years of attention, the underlying mechanisms and links between lipid homeostasis, inflammation and hepatocellular death remain unresolved. Notwithstanding this ensuing public health crisis, there are no approved treatments.
Levels of hepatic fat are largely dictated by the uptake of free fatty acids (FFAs) from white adipose tissue (Donnelly et al., 2005) and de novo lipogenesis (Jiang et al., 2005, Koo, 2013), offset by β-oxidation of fatty acids (Lee et al., 2016, McGarry and Brown, 1997) and VLDL secretion (Schonfeld, 2003). NAFLD ensues when fatty acid oxidation and lipid export fail to compensate for increased lipid uptake and synthesis. Once incorporated into hepatocytes, fatty acids can undergo either oxidation or re-esterification into glycerolipids and phospholipids. In either case, the first step is conversion into acyl-CoA, which is catalyzed by the acyl-CoA synthetases (ACS), such as acyl-CoA synthetase long chain family member 1 (ACSL1) (Coleman et al., 2000). Hepatic ACSL1 is reported to localize both in mitochondria and endoplasmic reticulum (ER) (Li et al., 2009). When generated in the mitochondrial outer membrane by ACSL1, acyl-CoAs are destined for β-oxidation; acyl-CoA is converted to acylcarnitine by CPT1, which translocates into the mitochondrial matrix and converted back to acyl-CoA by CPT2 (Coleman, 2019). Acyl-CoA is then metabolized to acetyl-CoA via acyl-CoA dehydrogenases (VLCAD, LCAD, and MCAD) and the other fatty acid-degrading enzymes. Although the fate of acyl-CoA produced by ER-localized ACSL1 is not clearly understood, it is likely involved in lipid synthesis, since the other key lipid synthetic enzymes (GPAT and DGAT) are localized in ER (Coleman, 2019, Tang et al., 2018). Confirming the importance of this enzyme, ACSL1 variants have emerged in several GWAS studies for type 2 diabetes, both in European and South Asian populations (Manichaikul et al., 2016, Scott et al., 2017). The processes that determine the different metabolic fates of acyl-CoAs are not well understood, but clearly depend on localization of the specific pools, and thus the localization of ACSL1.
Both genetic and dietary forms of obesity are accompanied by low-grade inflammation in adipose and liver tissue (Lumeng et al., 2007, Olefsky and Glass, 2010, Saltiel and Olefsky, 2017). Rodent models of obesity display chronic inflammation, and a strong correlation exists between insulin resistance and inflammation in patients (Blackburn et al., 2006, Festa et al., 2002). Inflammation is also a key component of the transition of NAFLD to NASH (Schuster et al., 2018). Obesity-associated inflammation is accompanied by macrophage switching from a Type 2 to Type 1 polarization state (Lumeng et al., 2007, Lumeng et al., 2008, Schenk et al., 2008, Huh et al., 2014), and activation of other immune cells (Feuerer et al., 2009, Winer et al., 2009, Huh et al., 2017). Obesity also increases recruited hepatic macrophage infiltration, along with the local production of inflammatory chemokines and cytokines from both macrophages and hepatocytes (Obstfeld et al., 2010).
We have explored the role of the noncanonical IKKs, TBK1 and IKKε as effectors of inflammatory signaling in adipocytes and hepatocytes (Mowers et al., 2013, Reilly et al., 2013, Reilly et al., 2015, Zhao et al., 2018). TBK1 is thought to play an essential role in immune responses that mediate the induction of type I interferons in response to Toll-like receptors and viral infection (Fitzgerald et al., 2003, McWhirter et al., 2004, Zhao, 2013), and its activity is increased in obese liver due to the preponderance of inflammatory signals (Reilly et al., 2013). Moreover, a placebo-controlled, 40 patient clinical trial in obese patients with T2D demonstrated that treatment with the TBK1/IKKε inhibitor amlexanox produced a significant 0.5% reduction in HbA1c, with an approximate 20% reduction in hepatic fat in responders (Oral et al., 2017).
We recently reported that TBK1 can phosphorylate and inactivate AMPK to repress energy expenditure in adipocytes (Zhao et al., 2018). However, the pathways by which TBK1 regulates hepatic energy metabolism remain uncertain. Here we report the surprising discovery that TBK1 impacts lipid metabolism via binding to the key rate-limiting enzyme ACSL1. In the fasted state, TBK1 expression is induced, but remains inactive, and can serve as a molecular scaffold to localize ACSL1 to the mitochondrial outer membrane, thus facilitating fatty acid β-oxidation. In the absence of TBK1, fasting-stimulated ACSL1 localization to mitochondria is blunted, driving the localization of the enzyme to the ER for fatty acid re-esterification. These findings suggest that TBK1 occupies a unique place in metabolic regulation at the intersection of two crucial pathways, serving to balance ACSL1 localization in response to nutritional status so that acyl-CoAs can be generated for both oxidation and re-esterification. Thus, TBK1 serves as a key rheostat to direct the fate of fatty acids in hepatocytes, and controls the activity of a major regulator of steatosis.
Results
Hepatic TBK1 mRNA expression is induced by fasting and its activity increases upon high fat diet
While the regulation of hepatic metabolism has been intensively investigated, our understanding of the pathways that control energy balance remains incomplete. We have focused our attention on inflammatory signaling activated in response to overnutrition, particularly NFκB (Chiang et al., 2009). To understand the effectors of this pathway, we investigated the noncanonical IκB kinase TBK1. While knockout and inhibitor studies revealed the role of this kinase as a link between obesity, insulin resistance and reduced energy expenditure in adipocytes (Chiang et al., 2009, Reilly et al., 2013, Zhao et al., 2018), its role in hepatocytes remains unknown.
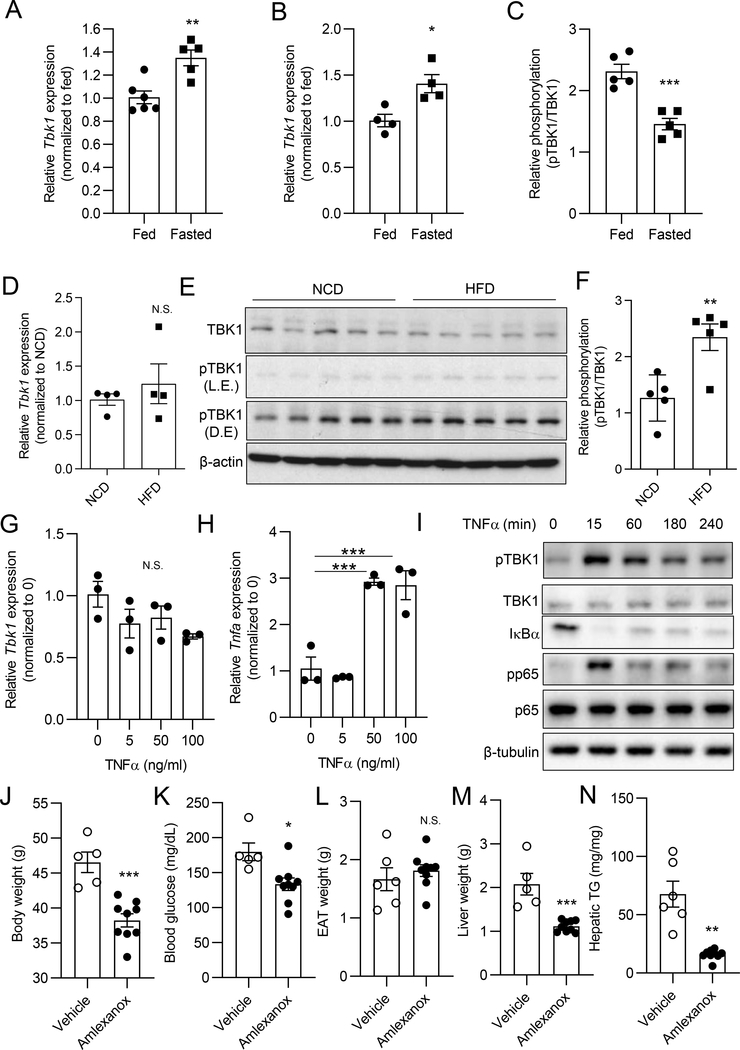
To characterize the regulation of TBK1 in hepatocytes, we examined mRNA and protein expression during both fasting and high fat diet (HFD) feeding. As we previously shown in adipocytes (Zhao et al., 2018), Tbk1 mRNA levels in liver were modestly but significantly induced by 16 h fasted mice compared to ad libitum-fed mice (Fig. 1A). To examine whether fasting-induced Tbk1 mRNA expression is derived from hepatocytes rather than other cell types in liver, we isolated primary hepatocytes from ad libitum-fed mice and 16 h fasted mice. Hepatocyte Tbk1 mRNA expression was induced in fasted compared with ad libitum-fed mice (Fig. 1B). TBK1 protein levels were slightly but significantly increased by fasting in liver tissues (Fig. S1A, S1B). However, TBK1 Ser172 phosphorylation, representative of enzyme activity, was significantly reduced in fasted mice (Fig. 1C, Fig. S1C).
Figure 1. Hepatic TBK1 mRNA expression is induced by fasting and its activity increases upon HFD.
(A) Tbk1 expression in liver tissues from ad libitum-fed or 16 h fasted mice. n = 5–6 mice per group. (B) Tbk1 expression in primary hepatocytes from ad libitum-fed or 16 h fasted mice. n = 4 mice per group. (C) Quantified amount of pTBK1 (Ser172) and TBK1 in liver tissues from ad libitum-fed or 16 h fasted mice. Band density quantified from immunoblot (IB) shown in Fig. S1C. n = 5 mice per group. (D) Tbk1 expression in liver tissues from NCD or 8 weeks of HFD-fed mice. n = 4 mice per group. (E) IB analysis of TBK1 and pTBK1(Ser172) in livers from NCD or 8 weeks of HFD-fed mice. n = 5 mice per group. L.E., light exposure; D.E, dark exposure. (F) Quantified ratio of TBK1 and pTBK1(Ser172) in liver tissues from NCD or HFD-fed mice. n = 5 mice per group. (G) Tbk1 expression in primary hepatocytes treated with vehicle (DMSO) or TNFα for 24 h. n = 3 replicates per group. (H) Tnfa expression in primary hepatocytes treated with vehicle (DMSO) or TNFα for 24 h. n = 3 replicates per group. (I) IB analysis of pTBK1 (S172), TBK1, IκBα, pp65 (S536), and p65 in primary hepatocytes treated with vehicle or TNFα (50 ng/ml) for the indicated time. (J-N) 8 days of amlexanox treatment in diet-induced obese (DIO) mice. n = 5 mice for vehicle and 9 mice for amlexanox group. (J) Body weight after amlexanox treatment. (K) Blood glucose levels. (L) Epididymal adipose tissue (EAT) weight. (M) Liver weight. (N) Hepatic triglyceride (TG) levels (mg/g tissue). * p < 0.05, **p < 0.01, ***p < 0.001, N.S. no significance; p > 0.05. (Student’s t test for A, B, C, D, F, J, K, L, M, and N, One-way ANOVA for G and H). All data in the figure are shown as the mean ± SEM. See also Figure S1 and S2.
Tbk1 mRNA expression was comparable in liver tissues from normal chow diet (NCD) or HFD-fed mice (Fig. 1D). On the other hand, TBK1 Ser172 phosphorylation was slightly but significantly increased in liver from HFD-fed mice (Fig. 1E and 1F) (Kishore et al., 2002). To explore the mechanism of this effect, we treated primary hepatocytes with TNFα; while there was no increase in Tbk1 mRNA expression, other inflammatory genes, including Tnfα itself, were significantly induced (Fig. 1G and 1H). However, TBK1 Ser172 phosphorylation was stimulated by acute TNFα treatment (Fig. 1I). We confirmed TNFα signaling in this experiment by demonstrating reduced levels of IκB (due to degradation) and elevated phosphorylation of p65. These data are consistent with our initial evaluations (Chiang et al., 2009), and indicate that the transcriptional regulation of TBK1 in hepatocytes differs from that seen in adipocytes (Zhao et al., 2018), in which the gene is markedly induced by TNFα treatment, NFκB activation and obesity. However, inflammatory pathways can nevertheless stimulate the phosphorylation of TBK1 in hepatocytes, accounting for increased kinase activity in obesity.
Inhibition of TBK1 kinase activity reverses NAFLD and NASH
In previous studies we demonstrated that treatment with the TBK1/IKKε inhibitor amlexanox for 4 weeks or longer improves obesity, NAFLD and diabetes in both genetic and dietary rodent models of obesity (Oral et al., 2017, Reilly et al., 2013). The beneficial effects of amlexanox on NAFLD has also been reported by others (He et al., 2019). To investigate the impact of amlexanox on liver during the early phase of treatment, we administered the drug to mice with established diet-induced obesity by gavage for 8 days. Amlexanox treatment resulted in a 17 % loss of body weight and 26 % reduction in blood glucose levels (Fig. 1J and 1K), although epididymal adipose tissue (EAT) weight was unaffected (Fig. 1L). However, there was a major reduction in liver weight of 41 %, with an overall reduction in hepatic lipid accumulation of 76 % (Fig. 1M and 1N). To check whether amlexanox inhibits TBK1 activity in liver, we tested TBK1 phosphorylation upon 4 hours of treatment. We observed increased phosphorylation of TBK1 at Ser172, which reflects reduced feedback regulation of TBK1 phosphorylation due to inhibition of kinase activity (Fig. S2A and S2B) (Zhao et al., 2018). mRNA levels of lipogenesis-related genes from RNAseq data (GSE57054) (Reilly et al., 2015), were significantly down-regulated by amlexanox treatment, which likely contributes to improved fatty liver produced by the drug (Fig. S2C). We also examined mice fed a methionine/choline-deficient/high fat diet (MCD/HFD) to induce NASH. Treatment of these mice with amlexanox lowered levels of hepatic lipid accumulation and liver weight, also reversing the diet-induced increase in circulating levels of ALT (Fig. S2D, S2E, S2F), demonstrating that inhibition of TBK1 kinase activity improves fatty liver and NASH.
Liver-specific TBK1 deficiency increases hepatic lipid accumulation in response to fasting
To investigate the specific role of hepatic TBK1 in liver, we generated hepatocyte-specific TBK1 knockout (LTKO) mice by crossing albumin-cre transgenic and TBK1-loxp mice. Tbk1 mRNA and protein levels were significantly reduced in liver tissue from LTKO compared to flox mice (Fig. 2A, Fig. 6A). Upon NCD feeding, body weights were comparable between the genotypes (Fig. 2B and Fig. S3A). Since TBK1 is known to reduce energy expenditure in adipocytes via inhibitory phosphorylation of AMPK (Zhao et al., 2018), we explored the response to 16 h fasting. The body weight loss produced by a 16 h fast was comparable between two genotypes (Fig. S3B), as were serum FGF21 levels (Fig. S3C) and liver weight (Fig. S3D, S3E), indicating that hepatic TBK1 deficiency does not globally impair the fasting response. We also analyzed inflammatory gene expression in TBK1-deficient liver tissues. Although there was increased mRNA expression of the related protein kinase Ikbke in LTKO liver, other inflammatory genes such as Tnfα, Cd11b, Ccl5 were not different between the genotypes (Fig. 2C). However, H&E staining revealed that LTKO mice have a significantly higher lipid accumulation in liver, particularly in the fasted state (Fig. 2D). The levels of hepatic triglycerides and cholesterol were markedly elevated in LTKO mice upon fasting (Fig. 2E and 2F). Together, these data indicate that TBK1 deficiency surprisingly increases hepatic lipid accumulation in NCD-fed mice without affecting inflammatory status.
Figure 2. Liver-specific TBK1 deficiency increases hepatic lipid accumulation in response to fasting.
(A~F) NCD-fed flox mice and LTKO mice. (A) Tbk1 mRNA expression in liver tissues from flox or LTKO mice. n = 5–6 mice per group. ***p < 0.001. (Student’s t test). (B) Body weight of 16 week-old NCD-fed mice. n = 11 mice per group. (C) Inflammatory gene expression in liver tissues. n = 4–7 mice per group. (D) Representative images of H&E-stained livers from flox and LTKO mice in ad libitum-fed or 16 h fasted. Scale bar, 50 μm. (E) Hepatic triglyceride (TG) content (mg/g tissue) of NCD-fed mice. n = 5–6 mice per group. (F) Hepatic cholesterol content (mg/g tissue) of NCD-fed mice. n = 6 mice for each group. (G~L) 8 weeks of HFD-fed flox and LTKO mice. (G) Body weight change during HFD feeding. n = 6–9 mice per group. (H) Liver weight of HFD-fed flox and LTKO mice in ad libitum-fed or 16 h fasted. n = 5–7 mice per group. (I) Inflammatory gene expression in liver tissues. n = 6–8 mice per group. (J) Representative images of H&E-stained livers from HFD-fed flox and LTKO mice in ad libitum-fed or 16 h fasted. Scale bars, 100 μm. (K) Hepatic triglyceride (TG) content (mg/g tissue) of HFD-fed mice. n = 6–9 mice per group. (L) Hepatic cholesterol content (mg/g tissue) of HFD-fed mice. n = 6–9 mice per group. For data in (C), (E), (F), (H), (I), (K), and (L), * p < 0.05, **p < 0.01, ***p < 0.001 for flox compared to LTKO in same feeding condition and #p < 0.05, ##p < 0.01, ###p < 0.001 for fed compared to fasted group in same genotype (two-way ANOVA followed Holm-Sidak’s multiple comparisons test). All data in the figure are shown as the mean ± SEM. See also Figure S3.
Figure 6. The fasting-stimulated mitochondrial localization of ACSL1 is impaired in LTKO liver.
(A) The protein levels of ACSL1, ACSL4, and TBK1 in liver tissues from NCD-fed flox mice or LTKO mice. n = 4–8 mice per group. (B) Quantified levels of ACSL1 and ACSL4 expression in livers. (C) The expression of ACSL1 and TBK1 in mitochondrial and microsomal fraction of liver tissues from NCD-fed flox or LTKO mice. Ponceau band was used for loading control. n = 5 mice per group. (D) Quantified level of ACSL1 (normalized with ponceau band) in mitochondria. (E) Quantified level of ACSL1 (normalized with ponceau band) in microsome. (F) Quantified level of TBK1 (normalized with ponceau band) in mitochondria of flox mice. (G) Quantified level of TBK1 (normalized with ponceau band) in microsome of flox mice. (H-J) Re-esterification activity in primary hepatocytes from flox or LTKO mice upon [U-14C] palmitate. n = 3 replicates per group. (H) Autoradiography image for triglyceride (TG), diacylglyceride (DG) and polar lipid (PL). (I) Quantified band density of TG. (J) Quantified ratio of TG/PL. For data in (B), (D), and (E), *p < 0.05, **p < 0.01 for flox compared to LTKO in same feeding condition, #p < 0.05, ##p < 0.01 for fed compared to fasting in each genotype, two-way ANOVA followed Holm-Sidak’s multiple comparisons test was analyzed. Student t test was analyzed for (F), (G), (I), and (J). All data in the figure are shown as the mean ± SEM. See also Figure S6A and S6B.
We fed 60 % HFD to flox and LTKO mice for 8 weeks, and analyzed metabolic status after ad libitum-feeding and 16 h fasting. Body weight and fasting-dependent weight loss were comparable between the genotypes (Fig. 2G, S3F and S3G), nor were there differences in serum FGF21 levels upon fasting (Fig. S3H). Surprisingly, absolute liver weight and liver weight as a percentage of total body weight were increased in fasted LTKO compared with fasted flox mice (Fig. 2H, S3I), the opposite effect of administering a TBK1 inhibitor. mRNA expression of inflammatory genes encoding Tnfα, Ccl2, Ccl5, Cd11b were increased in LTKO mice in response to fasting (Fig. 2I). H&E staining of liver revealed significantly more lipid droplets in LTKO mice, especially upon fasting (Fig. 2J). Moreover, LTKO mice had increased hepatic triglyceride in fasting conditions (Fig. 2K) without a difference in cholesterol levels (Fig. 2L). Thus, unlike what is observed after treatment of mice with the TBK1 inhibitor amlexanox, HFD-mediated hepatic steatosis is exacerbated in TBK1 deficient mice, especially upon fasting.
Loss of TBK1 suppresses fasting-induced fatty acid oxidation in liver
Hepatic lipid levels are maintained by the equilibrium of anabolic and catabolic metabolism under normal fasting and feeding cycles. To determine the mechanism whereby TBK1 deficiency leads to increased lipid accumulation, we assessed both anabolic and catabolic processes. Expression levels of all lipid metabolism-related genes were comparable between genotypes in NCD-fed mice (Fig. S4A, S4B, S4C). Upon HFD feeding, the mRNA levels of genes encoding proteins involved in lipid uptake, such as Cd36, Fatp2, and Fatp5 and lipogenesis, such as Srebf1, Fasn, and Acaca, were also not different between LTKO or flox littermates during either ad libitum-feeding or fasting (Fig. 3A, 3B). Although Cpt2 was modestly lower in LTKO compared with flox mice upon fasting, other fatty acid oxidation-related mRNAs were comparable between the genotypes (Fig. 3C).
Figure 3. Loss of TBK1 suppresses fasting-induced fatty acid oxidation in liver.
(A-C) mRNA expression of fatty acid uptake-related genes (A), lipogenesis genes (B), and fatty acid oxidation genes (C) in the liver tissues from HFD-fed flox and LTKO mice in ad libitum-fed or 16 h fasted group. n = 6–9 mice per group. (D) Hepatic lipid secretion analysis with HFD-fed flox and LTKO mice. n = 6 mice for flox and 9 mice for LTKO. (E, F) De novo lipogenesis activity in livers from HFD-fed flox and LTKO mice. n = 8 mice for flox and n = 4 mice for LTKO. N.S., no significance (Student t-test). (G) Fatty acid uptake in primary hepatocytes from NCD-fed flox or LTKO mice. n = 8 replicates per group. (H, I) Serum levels of β-hydroxybutyrate (BOH) (H) and total ketone bodies (TKB) (I) in HFD-fed flox or LTKO mice upon 16 h fasting. n = 5–6 mice per group. *p < 0.05 (Student t-test). (J) 14C-palmitate oxidation activity in primary hepatocytes from flox or LTKO mice. n = 6 replicates per group. ***p < 0.001 (Student t-test). (K) Complete oxidation rate by 14C-palmitate. n = 6 replicates per group. **p < 0.01 (Student t test). For the data in (A), (B), (C), (D), and (G), *p < 0.05 for flox compared to LTKO in same feeding condition, #p < 0.05, ##p < 0.01 for fed compared to fasting in each genotype, N.S.; no significance between genotypes; p > 0.05 (two-way ANOVA followed Holm-Sidak’s multiple comparisons test). All data in the figure are shown as the mean ± SEM. See also Figure S4.
To assess VLDL secretion, we injected poloxamer-407, an inhibitor of peripheral lipid uptake, into HFD-fed flox and LTKO mice, followed by measurement of plasma triglyceride levels during 3 hours. Plasma TGs were not different between the genotypes (Fig. 3D), suggesting that lipid secretion cannot account for triglyceride accumulation in TBK1-deficient liver. We then compared de novo lipogenesis in HFD-fed flox and LTKO mice by administration of D2O-labeled water in vivo. Levels of newly synthesized stearate and palmitate were also comparable between the genotypes (Fig. 3E and 3F). Moreover, fatty acid uptake assayed with fluorescence-labeled fatty acids in primary hepatocytes from flox and LTKO mice was comparable between two groups (Fig. 3G).
Because lipid uptake and synthesis appeared to be similar between the genotypes under all conditions, we assayed hepatic fatty acid oxidation in vivo by measuring the levels of serum ketone bodies in HFD-fed flox and LTKO mice after a 16 h fast. Interestingly, the levels of ketone bodies were significantly lower in HFD-fed LTKO compared with flox mice (Fig. 3H and 3I), suggesting that TBK1 deficiency reduces fatty acid oxidation. To explore further this possibility, [1-14C]-labeled palmitate was added to primary hepatocytes isolated from NCD-fed flox and LTKO mice, followed by measurement of 14C-labeled CO2 and acid soluble metabolites (ASM). As shown in Fig. 3J, TBK1-deficient hepatocytes had significantly lower palmitate oxidation activity. On the other hand, the complete oxidation rate (the ratio of CO2 versus ASM) was even higher in LTKO hepatocytes (Fig. 3K), indicating that TBK1 is necessary for fatty acid oxidation upstream of the TCA cycle. Since both fatty acid uptake and oxidation are activated in liver during fasting, these data indicate that TBK1 is necessary for catabolism of fatty acids during fasting, and further suggest that hepatic TBK1 expression is induced under these conditions to regulate fatty acid oxidation.
LTKO mice have increased mitochondrial number and size
One possible explanation for decreased fatty acid oxidation is that TBK1 deficiency produces a loss of mitochondria, or perhaps a fundamental defect in mitochondrial function. Because the mitochondrion is the main organelle responsible for fatty acid oxidation, we investigated whether TBK1-deficient liver exhibits a defect in mitochondrial number or function. The relative mitochondrial DNA normalized by nuclear DNA was significantly higher in liver tissues of LTKO mice compared with flox mice fed either NCD or HFD (Fig. 4A and 4B). Since we previously demonstrated that mitochondrial biogenesis was elevated through AMPK activation in TBK1-deficient adipocytes (Zhao et al., 2018), we checked mitochondrial biogenesis-related gene expression in livers of flox and LTKO mice. The mRNA levels of Ppargc1a, Ndufs7, Cox5b, Cox8a, and Atp5d were comparable between the genotypes in both NCD and HFD-fed mice (Fig. 4C, 4D). However, electron microscopy revealed significantly increased mitochondrial size in LTKO compared with flox mice after fasting (Fig. 4E, 4F). Even though it was not statistically significant, mitochondria exhibited a greater length (3.5 μm~4.99 μm) in LTKO compared to flox liver (Fig. 4G). In addition, the number of mitochondria was elevated in fasted LTKO mice compared with fasted flox mice (Fig. 4H). However, despite increased mitochondrial number, fatty acid oxidation was dramatically impaired, indicating that the difference in mitochondrial number cannot account for excessive hepatic lipid accumulation in LTKO mice.
Figure 4. LTKO mice have increased mitochondria number and size.
(A) The mitochondrial DNA copy number in livers from NCD-fed flox and LTKO mice after 16 h fasting. n = 5–6 mice per group. (B) The mitochondrial DNA copy number in livers from HFD-fed flox and LTKO mice after 16 h fasting. n = 5–6 mice per group. (C) The mRNA levels of mitochondrial biogenesis-related genes from NCD-fed flox and LTKO mice in 16 h fasting. n = 6–7 mice per group. (D) The mRNA levels of mitochondrial biogenesis-related genes from HFD-fed flox and LTKO mice in 16 h fasting. n = 5–6 mice per group. (E) TEM photographs of liver tissues from HFD-fed flox and LTKO mice after 16 h fasting. Scale bar, 2 μm. KC, Kupffer cell; LD, lipid droplet; Mc, mitochondria; Nuc, nucleus; SD, space of Disse or perisinusoidal space; SS, sinusoidal space. (F) Distribution of mitochondrial area. (G) Distribution of mitochondrial length. (H) Mitochondria number per area. For the data in (A), (B), (C), (D), and (H) *p < 0.05, **p < 0.01 (Student t test). For (F) and (G), Kolmogorov-Smimov test was analyzed. N.S., no significance. All data in the figure are shown as the mean ± SEM.
Acyl-CoA synthetase activity is impaired in hepatic mitochondria of LTKO mice
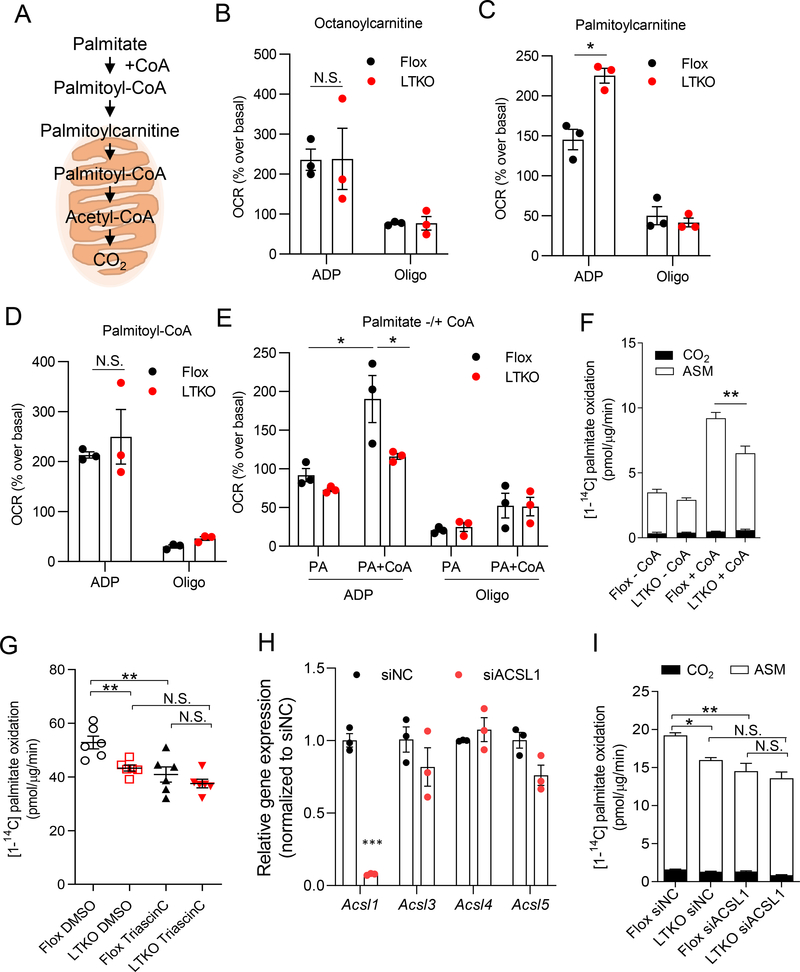
To explore how TBK1 deficiency reduces fatty acid oxidation in hepatocytes, mitochondria were isolated from flox and LTKO mice after 16 h fasting, followed by measurement of oxygen consumption rate (OCR) in the presence of different substrates (Fig. 5A). We compared the ADP-stimulated (State 3) and oligomycin-induced rate (State 4) between the genotypes. No differences were detected with octanoylcarnitine as substrate, whereas OCR was greater in LTKO liver with palmitoylcarnitine (Fig. 5B and 5C), demonstrating that the observed changes in morphology in LTKO mice do not influence mitochondrial function. Additionally, use of palmitoyl-CoA as substrate for OCR revealed no differences between the genotypes (Fig. 5D). There were no differences in OCR levels between genotypes in oligomycin-induced rate using any of these substrates. These data indicate that mitochondria from LTKO mice are fully functional, and further that there is no defect in the activity of CPT1, or in downstream pathways for fatty acid oxidation. However, addition to isolated mitochondria of palmitate plus CoA revealed a substantial defect in TBK1 KO mitochondria, suggesting a defect in the formation of acyl-CoAs (Fig. 5E).
Figure 5. Acyl-CoA synthetase activity is impaired in hepatic mitochondria of LTKO mice.
(A) The substrates for fatty acid oxidation. (B-E) ADP-stimulated and oligomycin-induced oxygen consumption rate (OCR) of isolated hepatic mitochondria from NCD-fed flox or LTKO mice with various substrates including octanoylcarnitine (B), palmitoylcarnitine (C), palmitoyl-CoA (D), and palmitate with (+) or without (−) coenzyme A (CoA) (E). n =3 mice per genotype and n = 6 replicates per mouse. Each dot indicates the mean value for each mouse. For data in (B), (C), (D) and (E), *p < 0.05, N.S., no significance (two-way ANOVA followed Holm-Sidak’s multiple comparisons test). (F) 14C-palmitate oxidation rate in the PMP (plasma membrane permeanilizer, 3 nM)-treated primary hepatocytes from flox or LTKO mice with or without CoA. n = 6 replicates per group. (G) Fatty acid oxidation rate in primary hepatocytes from flox or LTKO mice upon 5 μM triascin C treatment (pretreatment for 30 min and during 3 hours of oxidation assay). n = 6 replicates per group. (H) The mRNA levels of ACSL isoforms in primary hepatocytes by ACSL1 siRNA transfection. n = 3 replicates per group. siNC, siRNA for negative control. ***p < 0.001 (Student t test). (I) Fatty acid oxidation rate in siACSL1 transfected primary hepatocytes from flox or LTKO mice. n = 6 replicates per group. For data in (F), (G), and (I), *p < 0.05, **p < 0.01, N.S., no significance, (two-way ANOVA followed Holm-Sidak’s multiple comparisons test). All data in the figure are shown as the mean ± SEM. See also Figure S5.
The conversion of palmitate to palmitoyl-CoA is catalyzed in liver mitochondria by long-chain acyl-CoA synthetases (ACSL). Reduced oxygen consumption with palmitate plus CoA as substrate in KO cells thus suggests that TBK1 is required for ACSL activity in mitochondria. To confirm these findings, we added [1-14C] labeled palmitate with or without CoA, and directly measured 14C-palmitate oxidation in permeabilized hepatocytes to provide these substrates directly to mitochondria. As shown in Figure 5F, palmitate oxidation was dramatically increased by CoA addition in control cells, while TBK1-deficient hepatocytes were significantly less affected.
ACSL1 is involved in TBK1-mediated fatty acid oxidation
To understand the molecular interactions of TBK1 in mitochondria, we overexpressed FLAG-tagged WT and kinase-inactive (K38A) TBK1 (Fitzgerald et al., 2003) in primary hepatocytes, and then precipitated TBK1-bound proteins with anti-FLAG antibodies, followed by mass-spectrometry analysis. Among the TBK1-interacting proteins, we identified acyl-CoA synthetase long chain family member 1 (ACSL1), which is known to regulate lipid metabolism in liver (Fig. S5) (Li et al., 2009). To further evaluate whether the reduced fatty acid oxidation observed in TBK1 KO hepatocytes is mediated by a defect in ACSL1 activity, cells were incubated with the ACSL inhibitor triascin C prior to addition of [1-14C] palmitate and measurement of 14C-labeled CO2 and ASM. Triascin C reduced palmitate oxidation activity in flox hepatocytes, but this inhibitory effect was blunted in LTKO hepatocytes (Fig. 5G). We also transfected flox and LTKO hepatocytes with siRNA against ACSL1, followed by assay of palmitate oxidation. Treatment of cells with ACSL1 siRNA reduced the expression of ACSL1, but not other ACSL isoforms (Fig. 5H). Fatty acid oxidation was modestly, but significantly diminished by ACSL1 knockdown in WT hepatocytes (Fig. 5I). Importantly, the inhibitory effects of TBK1 deletion on fatty acid oxidation were eliminated in cells transfected with ACSL1 siRNA, demonstrating that TBK1-mediated regulation of hepatic fatty acid oxidation operates through the regulation of ACSL1. Taken together, these data suggest that TBK1 is required for long-chain acyl-CoA synthesis in hepatic mitochondria by supporting ACSL1 activity.
The fasting-stimulated mitochondrial localization of ACSL1 is impaired in LTKO liver
How does TBK1 influence ACSL1 activity? ACSL1 has been shown to be upregulated by fasting in the liver (Mashek et al., 2006). We confirmed this finding, demonstrating that mRNA and protein levels of ACSL1 were increased by fasting in both flox and LTKO mice, while ACSL4 was unchanged (Fig. 6A, 6B, S4D). Since TBK1 forms a complex with ACSL1 in hepatocytes, we wondered whether TBK1 might control the subcellular localization of ACSL1 in response to nutritional state, thereby influencing the fate of fatty acids. We thus performed subcellular fractionation of liver tissues into crude mitochondria and ER-enriched microsomes, and checked the protein levels of TBK1 and ACSL1. We first examined TBK1 protein levels in mitochondrial, microsomal, and cytosolic fractions from liver tissues, revealing TBK1 in all fractions (Fig. S6A). Assessment of ACSL1 levels in mitochondria and microsomes from the liver tissues of NCD-fed flox and LTKO mice upon ad libitum feeding and 16 h fasting revealed that ACSL1 was increased in the mitochondrial fraction upon 16 h fasting in flox mice; however, LTKO mice showed less ACSL1 enrichment in mitochondria (Fig. 6C and 6D). Conversely, LTKO mice had more microsomal ACSL1 protein compared to flox mice upon fasting (Fig. 6C, 6E). Furthermore, TBK1 localization in mitochondria was significantly increased by fasting in flox mice (Fig. 6C, 6F), while microsomal TBK1 was not altered (Fig. 6C, 6G). To check the purity of each fraction, we compared the protein levels of TOM20 and cytochrome C for mitochondrial proteins, and calreticulin for ER proteins (Fig. S6B). Although there was some contamination between the fractions, TOM20 and calreticulin were significantly enriched in mitochondria and microsomes, respectively. These data suggest that fasting increases ACSL1 expression and localization in mitochondria, and further that the presence of TBK1 is required for this fasting-dependent localization shift.
The acyl-CoAs produced by ACSL1 can be directed towards oxidation or storage via re-esterification. Since TBK1 appears to influence ACSL1 localization towards mitochondria and away from ER, we assayed fatty acid re-esterification in hepatocytes derived from flox and LTKO mice. [U-14C] palmitate was incubated with hepatocytes for 2 hours, and the lipid fraction was extracted and separated by thin layer chromatography (TLC). 14C-incorporation into triglycerides was markedly increased in TBK1 KO hepatocytes (Fig. 6H, 6I, 6J). These data indicate that TBK1 acts as a scaffolding protein, directing ACSL1 to mitochondria to promote the beta-oxidation of fatty acids in lieu of re-esterification.
Inactive TBK1 has a higher binding affinity for ACSL1
Because TBK1 inhibitors exert an effect opposite to that of knockout of the kinase, we wondered whether the scaffolding function of TBK1 was regulated by the phosphorylation state of TBK1. Endogenous ACSL1 co-precipitated in hepatocytes with both WT TBK1 and its K38A kinase-inactive mutant that does not undergo autophosphorylation (Fig. 7A). We confirmed the interaction between the two proteins by reciprocal co-IP in HEK293 cells overexpressing both TBK1 and ACSL1 (Fig. 7B). More importantly, kinase-dead TBK1 (K38A mutant) interacted with much higher affinity, suggesting that the interaction of TBK1 with ACSL1 does not rely on the catalytic activity of the protein kinase. Furthermore, the S172A TBK1 mutant, which cannot undergo phosphorylation in the site crucial for TBK1 activity, also showed a higher binding affinity to ACSL1 than did WT TBK1. These data reveal that phosphorylation of TBK1 reduces its affinity for ACSL1, suggesting that activation of the kinase might inhibit its scaffolding function.
Figure 7. Inactive TBK1 has a higher binding affinity for ACSL1.
(A) IP analysis with overexpressed TBK1 WT or TBK1 K38A and endogenous ACSL1 of primary hepatocytes. (B) IP analysis with overexpressed ACSL1 and TBK1 (WT, K38A, S172A) in HEK293 cell line. (C) IP analysis with overexpressed ACSL1 and each domain deleted TBK1 mutants (Full length, ΔULD, and ΔC-terminal). (D) The level of GST and pTBK1 (Ser172) in each purified GST-TBK1 from sf9 cells for in vitro binding assay. (E) In vitro binding assay with full length TBK1 (WT), C-terminal truncated TBK1 (ΔC WT), C-terminal truncated TBK1 with K38A mutation (ΔC K38A), and C-terminal truncated TBK1 with S172A mutation (ΔC S172A). n = 3 replicates per group. (F) Quantified level of GST normalized by flag (ACSL1). *p < 0.05 (Student t-test). (G) Cayman 10576 (20 μM, pre-treatment for 30 min and 3 hours of fatty acid oxidation assay) increases fatty acid oxidation in primary hepatocytes. n = 6 replicates per group. **p < 0.01 (Student t test). (H) Fatty acid oxidation rate in TBK1 WT or TBK1 K38A overexpressed hepatocytes from LTKO mice. n = 6 replicates per group. *p < 0.05, (two-way ANOVA followed Holm-Sidak’s multiple comparisons test). All IP analysis were replicated in more than three experiments. All data in the figure are shown as the mean ± SEM. See also Figure S6C and S7.
TBK1 contains a kinase domain, ubiquitin-like domain and C-terminal domain. To explore the domain in TBK1 required for ACSL1 binding, we utilized a co-immunoprecipitation assay by expressing HA-tagged ACSL1 with myc-tagged full length murine TBK1, ubiquitin-like domain (304–382) deleted (ΔULD) TBK1, and C-terminal deleted (ΔC-ter) TBK1, which has only the kinase domain and ULD (1–382) (Goncalves et al., 2011). We could not assess a mutant with the kinase domain deleted, as this construct was too unstable for cellular expression. Interestingly, TBK1 mutants in which either ULD or C-terminal domains were deleted interacted strongly with ACSL1, suggesting that the major region for ACSL1 binding lies within the kinase domain (Fig. 7C). We also assessed the binding affinity between ACSL1 and each mouse TBK1 mutant with an in vitro binding assay. Full-length mouse TBK1(mTBK1), C-terminal truncated mTBK1 (1–657), C-terminal truncated mTBK1 with the K38A mutation, and C-terminal truncated mTBK1 with the S172A mutation were purified from sf9 insect cells transfected by baculovirus (Fig. 7D) (Shu et al., 2013). As shown in figure 7E and 7F, both the K38A and S172A mutants exhibited a higher binding affinity for ACSL1 compared to WT TBK1. These data indicate that binding of ACSL1 occurs in the kinase domain of TBK1, and is negatively regulated by autophosphorylation.
Interestingly, mass spectrometry analyses revealed no phosphorylation of ACSL1 by TBK1 overexpression in HEK 293 cell lines, indicating that despite strong binding, ACSL1 is not a substrate of the TBK1 kinase (data not shown). These data, together with the enhanced binding of the non-phosphorylated mutant form of the kinase, indicate that TBK1 phosphorylation is likely to provide a negative signal for ACSL1 binding, such that binding at the mitochondria would be greater in the fasted state when TBK1 phosphorylation is low, and reduced in the obese state when TBK1 phosphorylation is increased, leading to the suppressed translocation of ACSL1 to the mitochondria. To test this hypothesis, we analyzed fatty acid oxidation activity in primary hepatocytes after addition of the TBK1 kinase inhibitor Cayman 10576. As shown in figure 7G, the fatty acid oxidation rate was significantly increased by addition of the TBK1 kinase inhibitor. The rate of palmitate oxidation was also assessed in primary WT or TBK1 KO hepatocytes overexpressing either WT or kinase-inactive TBK1. Overexpression of the K38A mutant increased fatty acid oxidation more efficiently than did WT TBK1 (Fig. 7H). We note that TBK1 WT and the kinase inactive K38A mutant showed comparable levels of mitochondrial localization in hepatocytes (Fig. S6C). These data suggest that the scaffolding activity of TBK1, especially in its inactive form, is responsible for supporting fatty acid oxidation. These findings demonstrate that TBK1 occupies a unique place in metabolic regulation at the intersection of two crucial pathways, serving as a rheostat that senses nutritional status to balance ACSL1 localization so that long-chain acyl-CoAs can be generated for both oxidation and re-esterification (Fig. S7).
Discussion
In previous studies we demonstrated that administration of the TBK1/IKKε inhibitor amlexanox to genetic or dietary mouse models of obesity reduced weight, insulin resistance, inflammation and fatty liver (Reilly et al., 2013). We show here that the drug reduces hepatic expression of lipogenic enzymes in HFD-fed mice, while also reducing steatosis and NASH in a methionine/choline-deficient murine model. Amlexanox inhibits the kinase activity of TBK1 and IKKε in both liver and fat cells, as well as in immune cells in both liver and adipose tissue, leading to increased catabolic pathways that enhance energy expenditure and repress energy storage (Reilly et al., 2013). However, because increasing energy expenditure in adipose tissue has the potential to indirectly reduce liver fat, it was not possible to determine whether the drug has direct effects on liver. To resolve this question, we conditionally deleted TBK1 from hepatocytes by crossing floxed-TBK1 with albumin-Cre mice. Surprisingly, the resulting liver-specific KO (LTKO) mice exhibited increased steatosis, which was primarily observed in the fasting state in both normal weight and obese mice. To explore the mechanism of this unexpected finding, we performed a series of metabolic studies to assess hepatic lipid metabolism. While differences in fatty acid uptake, de novo lipogenesis and hepatic lipid secretion could not account for the phenotype, we found that fatty acid oxidation was dramatically reduced in KO mice. Interestingly, this decrease could be fully rescued with a kinase-inactive mutant of TBK1, indicating that this activity of TBK1 does not rely on protein phosphorylation, but rather on a scaffolding activity of the protein. This kinase-independent activity of TBK1 was attributed to binding of the enzyme to ACSL1, the rate-limiting enzyme in production of acyl-CoA.
The fate of long-chain acyl-CoA in cells depends on the localization of ACSL1 (Coleman, 2019, Li et al., 2015). When localized to the outer mitochondrial membrane, ACSL1 generates long-chain acyl-CoAs for β-oxidation in mitochondria, and fatty acid oxidation was reduced in liver-specific ACSL1 KO mice (Li et al., 2009). Although it has not yet been clearly evaluated, it is likely that ER-localized ACSL1 generates substrates for re-esterification of fatty acids into glycerolipids. Our data indicate that TBK1 serves as a scaffold to recruit ACSL1 to the outer mitochondrial membrane in the fasted state, channeling acyl-CoAs to mitochondria for oxidation. Indeed, the phenotype of LTKO mice was most pronounced during fasting. Moreover, fasting increased not only ACSL1, but also TBK1 gene expression; both mRNA and protein were elevated after a fasting. Fasting also shifted ACSL1 to mitochondrial fractions, and this fasting-induced distribution was not seen in LTKO mice. Our in vitro binding studies revealed that inactive TBK1 binds with much higher affinity to ACSL1 compared to the fully active form of the kinase. Moreover, when induced during fasting, TBK1 remains inactive and capable of recruiting ACSL1 to mitochondria, supporting fatty acid oxidation. Thus, these data suggest that TBK1 functions to scaffold ACSL1 to mitochondria more effectively when in the inactive, unphosphorylated state, and is less effective when activated during obesity, resulting in less ACSL1 in mitochondria and hence more fatty acid re-esterification.
These findings demonstrate that TBK1 occupies a unique place in metabolic regulation at the intersection of two crucial pathways, serving to balance ACSL1 localization so that acyl-CoAs can be generated for both oxidation and re-esterification. During fasting, TBK1 is induced, but remains in the inactive state, supporting fatty acid oxidation by scaffolding ACSL1 in mitochondria. In obesity, TBK1 is activated via phosphorylation, which reduces its binding to ACSL1, shifting the enzyme from mitochondria to the ER. This produces a net reduction in fatty acid oxidation while increasing fatty acid re-esterification. Because binding appears to be less efficient when TBK1 is active, it remains possible that a portion of the beneficial effects of TBK1 inhibitors might be to stabilize the hepatic TBK1/ACSL1 complex, increasing fatty acid β-oxidation. Future studies will focus on understanding the potential therapeutic value of manipulating this interaction.
Limitations of Study
Our study demonstrates the role of TBK1 in the regulation of fatty acid oxidation in hepatocytes via stimulating the mitochondrial localization of ACSL1. To investigate the mechanism of how TBK1 mediates fatty acid oxidation, we isolated the hepatic mitochondrial fraction from flox and LTKO mice by using a differential centrifugation method. However, this mitochondrial fraction (also called crude mitochondria) includes both pure mitochondria and mitochondria-associated membrane (MAM), which is known as a part of the ER membrane structure. Given that the mitochondrial fraction is more enriched in crude mitochondria than is MAM, mitochondria would be the major contributor accounting for our data. Nonetheless, because it has been reported that MAM plays a role in lipid metabolism, the function of TBK1 in the regulation of ACSL1 localization in MAM, or transition of the enzyme between organelles remains elusive.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alan Saltiel (asaltiel@ucsd.edu)
Materials availability
All unique reagents generated in this study are available from Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
This study did not generate/analyze dataset.
Experimental Model and Subject Details
Animals
All mice used were on the C57BL/6J background. For 4 hours and 8 days of amlexanox studies with DIO mice, C57BL6/J mice purchased from the Jackson Laboratory were fed 60 % HFD (Research Diets Inc.) for 12 weeks, then 25 mg/kg body weight of amlexanox (Abcam) was administered by daily oral gavage. For 8 days of amlexanox treatment experiment, mice were fasted for 16 h before sacrifice and tissue collection. For amlexanox administration to MCD-induced NASH mice, C57BL6/J mice were fed with MCD diet (Research Diets Inc.) for 7 weeks. Starting after 4 weeks of MCD feeding, 25 mg/kg of amlexanox was administered by daily oral gavage for 3 weeks. Liver-specific TBK1 knockout (LTKO) mice were generated by crossing TBK1f/f (flox) mice (Marchlik et al., 2010) with albumin-cre mice (Jackson Laboratory). TBK1f/f mice were bred with TBK1f/f Cre/+ mice to generate TBK1f/f (flox) and TBK1f/fCre/+ (LTKO) mice as littermates. Male mice were analyzed for all experiments. 60 % HFD (Research Diets Inc.) was fed to 8-week-old flox or LTKO mice for 8 weeks. NCD-fed mice were analyzed at 16 weeks of age. Mice in the overnight fasting group were fasted for 16 hours. During metabolic studies, ear tag numbers were used to identify animals. Mice were housed in a specific pathogen-free facility with 12 hour light and 12 hour dark cycles and given free access to food and water, except during fasting periods. All animal use was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, San Diego.
Mouse primary hepatocyte isolation
Mouse primary hepatocytes were isolated from 8-week-old NCD-fed flox or LTKO male mice. Briefly, mice were fasted for 16 hours (except the mice for ad libitum-fed group in Fig. 1B) and anesthetized with isoflurane. Mice were infused through the inferior vena cava with 25 ml of perfusion buffer (138 mM NaCl, 5.4 mM KCl, 0.6 mM NaH2PO4.H2O, 0.8 mM Na2HPO4, 10 mM HEPES, 4.2 mM NaHCO3, 0.5 mM EGTA, 5mM glucose, pH 7.4) for 3 min and followed by 25 ml of digestion buffer (40 μg/ml of Liberase TM (Roche), 138 mM NaCl, 5.4 mM KCl, 0.6 mM NaH2PO4.H2O, 0.8 mM Na2HPO4, 10 mM HEPES, 4.2 mM NaHCO3, 5 mM CaCl2, pH 7.4) for 3 min by using peristaltic pump. Liver tissues were washed with 25 ml of perfusion buffer for 3 min after digestion. Excised liver were minced and centrifuged at 50 x g for 1 min. Dead cells were excluded by removing the floating cells after centrifugation at 100 x g for 10 min in 36 % percoll solution (Sigma-Aldrich). Cells were resuspended with William’s medium E (Life Technologies) supplemented with 10 % of fetal bovine serum (FBS), GlutaMax (Life Technologies), and 1 % penicillin/streptomycin to make 2.5×105 cells/ml. 0.5 ml, 1 ml, 2 ml of cells were plated in 24-well, 12-well, and 6-well collagen-coated plates. After 4–6 h incubation for cell attachment, the media was replaced with fresh media. For TNFα treatment experiment (Fig. 1G–I), human TNFα (BioLegend) was treated to flox hepatocytes in 10% FBS and GlutaMax supplemented William’s medium E. All experiments with primary hepatocytes were performed within 48 hours after isolation.
qRT-PCR
For liver tissues and primary hepatocytes, RNA was isolated by using TRIzol reagent (Life Technologies). 3 μg of RNA was used for reverse-transcription PCR to generate cDNA with Maxima First Strand cDNA Synthesis kits (Thermo Scientific). The expression level of mRNA was analyzed by using ΔΔCt real-time PCR with Power SYBR Green PCR Master Mix (Life Technologies). Cyclophilin was used as endogenous control gene. Primer sequences are listed in supplemental table 1.
Histology
Liver tissue was harvested and fixed in 10 % formalin. Paraffin-embedding and sectioning for H&E staining was completed at the UCSD histology core.
Hepatic lipid measurement
Frozen liver tissues (50 mg) was homogenized in 1 ml of homogenization buffer (50 mM pH 8.0 Tris, 5 mM EDTA, 30 mM Mannitol). 200 μl of lysates were mixed with 5 μl of 10 M KOH and 800 μl of chloroform:methanol (2:1) mixture and vortexed. After incubating for 5 min at room temperature, lipid fractions in the lower phase after 10 min centrifugation at 13000 rpm were transferred and completely dried in the fume hood. 200 μl of 5 % Triton X-100 solution was used to solubilize the lipids. Triglyceride and cholesterol levels were determined by using triglyceride (Thermo Fisher Scientific) and cholesterol reagents (Thermo Fisher Scientific), respectively. Lipid amount was normalized to liver weight.
Lipid secretion assay
Hepatic TG secretion rate was assessed as described in previous studies (Millar et al., 2005). In brief, 8-weeks of HFD-fed flox and LTKO mice were fasted for 4 hours before basal (time = 0) blood collection, then poloxamer-407 (Sigma-Aldrich) was injected intraperitoneally at a dose of 1 mg/g body weight. Blood was then collected at 1, 2, and 3 hours after injection. Plasma was used to measure triglycerides were measured using triglyceride reagent (Thermo Fisher Scientific).
In Vivo de novo lipogenesis assay
8-week-old HFD-fed mice were intraperitoneally injected with 0.035 ml (per gram of body weight) D2O (Sigma-Aldrich) in 0.9 % NaCl and were allowed 24 h of labeling with drinking water containing 8 % enriched D2O. After 4 h of fasting, mice were sacrificed, and the plasma and liver were collected. For mass spectrometry, plasma and liver were analyzed as described in previous studies (Svensson et al., 2016). The 2H labeling of water from samples or standards was determined via deuterium acetone exchange. 5 μl of sample or standard was reacted with 4 μl of 10 N NaOH and 4 μl of a 5% (v/v) solution of acetone in acetonitrile for 24 hours. Acetone was extracted by the addition of 600 μl chloroform and 0.5 g Na2SO4 followed by vigorous mixing. 100 μl of the chloroform was then collected to a GCMS vial. Acetone was analyzed using an Agilent DB-35MS column (30 m 3 0.25mm i.d. 3 0.25 mm, Agilent J&W Scientific) installed in an Agilent 7890A gas chromatograph (GC) interfaced with an Agilent 5975C mass spectrometer (MS) with temperature program as follows: 60 °C initial, increase by 20 °C/min to 100 °C, increase by 50 °C/min to 220 °C, and hold for 1 min. The split ratio was 40:1 with a helium flow of 1 ml/min. Acetone eluted at approximately 1.5 min. The mass spectrometer was operated in the electron impact mode (70 eV). The mass ions 58–60 were quantified. Given that up to six deuterium atoms can transfer to acetone depending on 2H2O enrichment, mole-percent enrichment was calculated and compared to a standard curve to determine plasma 2H2O enrichment. Total fatty acids were extracted from tissues and plasma using a bligh and dyer based methanol/chloroform/water extraction with [U-2H31] C16:0 (d31-C16:0) as an internal standard. Briefly, 500 μl MeOH, 500 μl CHCL3, 200 μl H2O and fatty acid isotope internal standards were added to weighed pre-ground tissue. This was vortexed for 10 minutes followed by centrifugation at 10,000 g for 5 min. The lower chloroform phase was dried and then derivatized to form fatty acid methyl esters (FAMEs) via adding 500 μl 2% H2SO4 and incubation at 50 °C for 2 hours. FAMEs were extracted with 100 μl saturated salt solution and 500 μl hexane and these were analyzed using a Select FAME column (100 m x 0.25mm i.d.) installed in an Agilent 7890A GC interfaced with an Agilent 5975C MS with following temperature setting: 80 °C initial, increase by 20 °C/min to 170 °C, increased by 1 °C/min to 204 °C, then 20 °C/min to 250 °C and held for 10 min. Calculation of the fraction of newly synthesized fatty acids (FNS) was based on the protocol previously described (Lee et al., 2000) where FNS is determined by the following equation:
Where ME is the average number of deuterium atoms incorporated per molecule (ME =1 x m1 + 2 x m2 +3 x m3 …), p is the deuterium enrichment in water and n is the maximum number of hydrogen atoms from water incorporated per molecule. N was calculated using the equation:
Where q is the fraction of hydrogen atoms and p + q = 1. The molar amount of newly synthesized fatty acids was determined by the following equation:
Fatty acid uptake assay
Isolated primary hepatocytes from NCD-fed flox and LTKO mice were plated in 96-well-black plates with collaged-coated clear flat bottom. After 6 h incubation for attachment, fatty acid uptake assay was performed using free fatty acid uptake assay kits (Abcam). Fluorescence was measured by TECAN infinite 200 Pro.
Fatty acid oxidation assay
We modified the published fatty acid oxidation assay protocol(Huynh et al., 2014). Attached primary hepatocytes were incubated in serum-free William E medium for 16 h and then treated with fatty acid oxidation media (0.3 % fatty acid free BSA (Akron Biotechnology)/100 μM palmitate (Sigma-Aldrich) /0.4 μCi [1-14C] palmitate (Perkin Elmer) /1mM carnitine (Bachem Americas INC) in William’s medium E) in 24-well plates for 3 hours at 37°C. 400 μl of the media was added to acidification vials, which have filter paper under the cap and 200 μl of 1 M perchloric acid in the tube. After 1 hour, captured 14CO2 and acid-soluble metabolites (ASM) were used to measure radioactivity for fatty acid oxidation rates, and the CO2/ASM ratio was calculated for complete oxidation rate. The cells on the plates were lysed with 0.1 M HCl to quantify protein concentrations.
Measurement of mitochondrial DNA (mtDNA) copy number
Mitochondrial DNA copy number was analyzed as described in previous studies (Quiros et al., 2017). 20 mg of liver tissue was added to 600 ul of lysis buffer (0.2 mg/ml Proteinase K (Roche), 100 mM NaCl, 10 mM EDTA, 0.5 % SDS, 20 mM Tris-HCl (pH7.4)) and incubated overnight at 55 °C. 100 μg/ml of RNase A (Roche Diagnostics) was added and samples were incubated at 37 °C for 30 min. Samples were mixed with 250 μl of 7.5 M ammonium acetate and 600 μl of isopropyl alcohol and then centrifuged at 15000 x g for 10 min at 4 °C. Pellets were washed with 70% ethanol, dried, and solubilized in TE buffer. qRT-PCR was used to measure relative numbers of copies of mtDNA and nuclear DNA. Mitochondrial DNA quantities were analyzed with mitochondrial DNA specific sequences and normalized with nuclear DNA sequences (GAPDH or HK2). Primer sequences are listed in supplemental table 2.
Transmission electron microscopy for hepatic mitochondria morphology
For mitochondrial morphology analysis, 8-week HFD-fed flox and LTKO mice were used after 16h fasting. Deeply anesthetized mice were flushed with a pre-warmed (37 °C) calcium and magnesium free buffer containing DPBS, 10 mM HEPES, 0.2 mM EGTA, 0.2 % BSA, 5 mM glucose and 10 mM KCl for 2 min followed by perfusion with fresh and pre-warmed fixative buffer (2.5 % glutaraldehyde, 2 % paraformaldehyde in 0.15 M cacodylate buffer) for 3 min using a peristaltic pump through the left ventricle (Pasqua et al., 2016). Excised liver tissues were sliced at 2 mm x 2 mm x 2 mm cube and put in fixative overnight (2 h at room temperature and 12 h at 4 °C), post-fixed in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer for 1 h on ice, and stained en bloc with 2 % uranyl acetate for 1 h on ice. The stained tissues were dehydrated in ethanol (20–100%) and embedded with Durcupan (Sigma-Aldrich). Ultrathin (50–60 nm) sections were post-stained with uranyl acetate and lead stain. Samples were viewed using a JEOL JEM1400-plus TEM (JEOL) and photographed using a Gatan OneView digital camera with 4k x 4k resolution. Morphometric measurements of mitochondrial area and length were done using the free-hand tool of ImageJ by manually tracing around the mitochondria as described previously (Pasqua et al., 2016). For quantification of mitochondria area and length, 616 mitochondria for flox and 606 mitochondria for LTKO were analyzed. Cytoplasmic area (excluding sinusoidal spaces and lipid droplet areas) was calculated using ImageJ software. Mitochondrial numbers were calculated by placing the EM micrographs onto a grid with 10 horizontal x 10 vertical square grids. Mitochondrial numbers were then divided by the area and was expressed as mitochondrial number/10 μm2 area. For quantification of mitochondria number, 42 images per group were analyzed.
Measurement of liver mitochondrial oxygen consumption rate
For seahorse experiment, liver tissues from 16 week-old NCD-fed flox and LTKO mice were collected after 16 h fasting. Liver mitochondria was isolated as described in previous studies (Rogers et al., 2011). Briefly, livers were minced in mitochondrial isolation buffer (MSHE, 70 mM sucrose, 210 mM mannitol, 5 mM HEPES, 1mM EGTA, pH 7.2) including 2% fatty acid-free BSA by stroking 5 times with a Teflon glass homogenizer. The homogenate was centrifuged at 800 x g for 10 min and the supernatant was centrifuged at 8000 x g for 10 min to pellet the mitochondrial fraction. After removing the light mitochondrial layer, the pellet was completely resuspended with 2% BSA /MSHE buffer and was centrifuged at 8000 x g to wash mitochondria pellet. Mitochondria pellet was resuspended with 200 μl of mitochondrial assay solution (MAS, 70 mM sucrose, 220 mM mannitol, KH2PO4 5 mM, MgCl2 5 mM, HEPES 2mM, EGTA 1 mM, pH 7.2 adjusted with 1 M KOH) contained with 2 % fatty acid-free BSA. All steps for mitochondria isolation were performed on ice. 3 ug of mitochondria per well (for 96 well plate) was used. For substrates, 40 μM octanoyl carnitine (Sigma-Aldrich), 40 μM palmitoyl carnitine (Sigma-Aldrich), 40 μM palmitoyl-CoA (Sigma-Aldrich) were added to MAS buffer with 1 mM malate and 1 mM carnitine. 20 μM of BSA-conjugated palmitate (Agilent Technologies) with or without 0.5 mM of CoA (Sigma-Aldrich) was also added to MAS buffer (with 1 mM malate, 5 mM ATP, 1mM carnitine, 8 mM MgCl2) to determine ACSL activity, which is modified from previously published paper (Füllekrug and Poppelreuther, 2016). Calibrated cartridges were used for analysis using the seahorse XF96 analyzer. 4 mM ADP and 2 μM oligomycin were treated via injection ports at 5 min and 10 min, respectively. OCR were measured at 0.8 sec for baseline, 7 min for ADP, and 12 min for oligomycin. OCR levels were expressed as a percentages of the baseline oxygen consumption (100 %) for each group.
Fatty acid oxidation assay with permeabilized primary hepatocyte
Primary hepatocytes from flox or LTKO mice were used for assay after 16 h incubation with serum free William’s medium E. 3 nM of XF Plasma Membrane Permeabilizer (XF PMP) (Agilent Technologies) and 4 mM ADP contained MAS buffer was used for base media. 1 mM malate, 5 mM ATP, 1 mM carnitine, 8 mM MgCl2, 0.3 % fatty acid free BSA /100 μM palmitate/0.4 μCi [1-14C] palmitate with or without 0.5 mM of CoA were treated as substrates. After 30 min incubation at 37 °C, we performed same steps for fatty acid oxidation assay as we previously described.
Subcellular fractionation for western blot
To conduct subcellular fractionation of liver tissues, we used the isolation method as we used for mitochondrial isolation for seahorse analysis. For microsome fraction, we used a modified protocol previously published (Schenkman and Cinti, 1978). The microsomal fraction was collected by centrifuging mitochondrial pellet at 8000 x g, transferring the supernatant to new tubes containing 8 mM of CaCl2, and incubating on a rotating platform in the cold room for 10 min. Samples were then centrifuged at 22000 x g for 30 min to yield microsomal pellets. The mitochondrial and microsomal pellets were resuspended in protease and phosphatase inhibitor and 1 mM DTT contained RIPA buffer and sonicated for 10 sec pulses at an output power of 2 to prepare western blot samples.
Re-esterification assay
Primary hepatocytes from flox and LTKO mice were plated in 24-well plates. On the following day, cells were incubated in serum-free William’s medium E with 0.3% fatty acid free BSA (Akron Biotechnology) /100 μM palmitate (Sigma-Aldrich) / 0.4 μCi [U-14C] palmitate (Perkin Elmer) for 2 hours at 37 °C. After PBS washing, lipids were extracted using hexane:isopropanol mixture (3:2). Dried lipid was solubilized with 10 μl of chloroform and then used for thin layer chromatography (TLC). Before running the TLC plate, the loading chamber was filled with 100 ml of hexane:diethy ether:acetic acid mixture (80:20:1). Lipid extracts, as well as lipid standard for monoglyceride, diglyceride, and triglyceride (Sigma-Aldrich), were loaded on TLC plates (EMD Millipore) and separated in chamber. Plate was dried for 5–10 min (until all solvents evaporated), transferred to an exposure cassette (GE Healthcare), and exposed to autoradiography film. ImageJ software was used to quantify the band density of autoradiography.
Site-directed mutagenesis
ACSL1-HA and flag-TBK1 S172A were generated by using CloneAmp Hifi DNA PCR premix (Takara) with 0.1 μg ACSL1-myc/flag (Origene) and flag-TBK1 WT plasmid, respectively. PCR products were treated with DpnI (NEB) to remove circular double stranded templates from the reaction. After agarose gel purification, eluted DNA was ligated by using In-Fusion HD enzyme premix (Takara) and transformed into Stellar Competent Cells (Clontech).
Transfection
HEK293 cells were transfected with a combination of pCMV6 Entry mouse ACSL1-myc/flag, pCMV6 Entry mouse ACSL1-HA, pcDNA-flag-human TBK1 WT, pcDNA-flag-human TBK1 K38A, pcDNA-flag-human TBK1 S172A plasmids, 6myc-mouse TBK1 full length, 6myc-mouse TBK1 ULD deleted, and 6myc-mouse TBK1 C-terminal deleted plasmids as described in each experiment, using Lipofectamine 3000 (Life Technologies). Primary hepatocytes were transfected with pcDNA-flag-human TBK1 WT or pcDNA-flag-human TBK1 K38A by using Lipofectamine LTX (Life Technologies). GFP vector was used to normalize the transfected DNA amount in control (mock) group as well as to check transfection efficiency. Lipofectamine RNAiMAX (Life Technologies) was used to transfect negative control siRNA or ACSL1 siRNA (5 pmol per well of 24 well plate) to primary hepatocytes. Cells were harvested after 40 hours of transfection for further assays.
Immunoprecipitation
For HEK293 cells, 500 μg of lysates were incubated with anti-c-Myc (Thermo Scientific Pierce) or anti-HA (Thermo Scientific Pierce) magnetic beads overnight at 4 °C on rotating shakers. The beads were rinsed three times for 10 min each in washing buffer (TBS added with 0.05% Tween-20 and 0.35 M NaCl) and eluted with SDS buffer. For primary hepatocytes, 500 μg of protein lysate were incubated with anti-flag M2 affinity gel (Sigma) overnight at 4 °C to pull down the protein complex with TBK1. After washing three times for 10 min each in washing buffer (TBS added with 0.05% Tween-20 and 0.35 M NaCl), samples were used for mass spectrometry. LC-MS/MS was performed by the Biomolecular and Proteomics Mass Spectrometry Facility at the University of California, San Diego.
Western blot
Cells or liver tissues were lysed in RIPA buffer (25 mM Tris/HCl pH7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) with 1 mM DTT and protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). After centrifuging the lysates at 14000 x g for 15 min, the supernatant was used for protein quantification using BCA protein assay kits (Pierce).
In Vitro TBK1-ACSL1 binding assay
Full-length, C-terminal truncated (1–657) WT, C-terminal truncated with K38A mutant, and C-terminal truncated S172A mutant mouse TBK1 in pAcGHLTc baculovirus transfer vectors (gifted from Pingwei Li) (Shu et al., 2013) were mixed with baculovirus DNA (AB Vector). This DNA mixture was transfected to sf9 cells with profectine (AB Vector). After 72 h incubation, recombinant baculoviruses were harvested. The recombinant baculoviruses were treated to sf9 cells for 72 h and infected sf9 cells were used for GST-TBK1 protein purification. PBS-washed cell pellet was lysed with lysis buffer (50 mM Tris pH 7.7, 300 mM NaCl, 5% glycerol, 0.1% TritonX100, protease and phosphatase inhibitor cocktail) and centrifuged at 20000 rpm for 30 min. The supernatant was used to purify GST-TBK1 by using GST Spin Purification Kits (Pierce). Eluted proteins were filtrated with spin columns (Abcam) to remove glutathione and resuspended with binding buffer (25 mM HEPES, 100 mM NaCl, 0.01 % Triton X-100, and 5% glycerol) (Lapetina and Gil-Henn, 2017). After GST protein quantification by Coomassie blue staining, 40 ng of each GST-TBK1 was incubated with ACSL1, which was overexpressed in HEK293 cells and bound to anti-flag magnetic beads (Sigma-Aldrich) for 2 hours at 4 °C with end-over-end rotation. Beads were washed 4 times and SDS buffer was added to prepare samples for western blotting.
Quantification and Statistical analysis
Averaged values are shown as mean ± SEM. Statistical analyses were performed with Prism software version 8.4.3 (GraphPad Software).Comparisons of two groups were made by conducting Student’s t-tests. For more than two groups, we evaluated the data with one-way ANOVA or two-way ANOVA (Holm-Sidak’s multiple comparisons) test. For mitochondria morphology data, Kolmogorov-Smimov test was used. Differences were considered statistically significant if p < 0.05 (* or #); p < 0.01 (** or ##) or p < 0.001 (*** or ###). The statistical methods of each experiment are indicated in the figure legends. Data from animal experiments were collected in a blinded fashion. All in vitro experiments were replicated at least three independent times.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-ACSL1 | Cell Signaling Technology | Cat#9189S; RRID:AB_10891616 |
| Rabbit monoclonal anti-ACSL4 | Abcam | Cat#Ab155282; RRID:AB_2714020 |
| Rabbit monoclonal anti-beta-actin | Cell Signaling Technology | Cat#4970S; RRID:AB_2223172 |
| Rabbit monoclonal anti-Calreticulin | Abcam | Cat#Ab92516; RRID:AB_10562796 |
| Rabbit polyclonal anti-Cytochrome C | Cell Signaling Technology | Cat#4272; RRID:AB_2090454 |
| Rabbit monoclonal anti-GST | Cell Signaling Technology | Cat#2625S; RRID:AB_490796 |
| Rabbit monoclonal anti-HA | Cell Signaling Technology | Cat#3724; RRID:AB_1549585 |
| Rabbit monoclonal anti-IκBα | Cell Signaling Technology | Cat#4812S; RRID:AB_10694416 |
| Rabbit polyclonal anti-MTCO2 | Abcam | Cat#198286; RRID:AB_2861364 |
| Rabbit monoclonal anti-Myc | Cell Signaling Technology | Cat#2278; RRID:AB_490778 |
| Rabbit monoclonal anti-p65 | Cell Signaling Technology | Cat#8242T; RRID:AB_10859369 |
| Rabbit monoclonal anti-p-p65 (S536) | Cell Signaling Technology | Cat#3033S; RRID:AB_331284 |
| Rabbit polyclonal anti-TBK1 | Cell Signaling Technology | Cat#3031S; RRID:AB_2199749 |
| Rabbit monoclonal anti-pTBK1 (Ser172) | Cell Signaling Technology | Cat#5483S; RRID:AB_10693472 |
| Rabbit monoclonal anti-Tom20 | Cell Signaling Technology | Cat#42406; RRID:AB_2687663 |
| Anti-Flag M2 affinity gel | Sigma-Aldrich | Cat#A2220; RRID:AB_10063035 |
| Anti-Flag M2 magnetic beads | Sigma-Aldrich | Cat#M8823; RRID:AB_2637089 |
| Anti-HA magnetic bead | Thermo Scientific Pierce | Cat#88837; RRID:AB_2861399 |
| Anti-Myc magnetic bead | Thermo Scientific Pierce | Cat#88843; RRID:AB_2861398 |
| Bacterial and Virus Strains | ||
| Stellar Competent Cells | Clontech | Cat#636763 |
| GST-Full length TBK1 baculovirus | Shu et al., 2013 | N/A |
| GST-C-terminal truncated TBK1 baculovirus | Shu et al., 2013 | N/A |
| GST-C-terminal truncated TBK1 K38A baculovirus | Shu et al., 2013 | N/A |
| GST-C-terminal truncated TBK1 K38A baculovirus | Shu et al., 2013 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Amlexanox | Abcam | Cat#Ab142825 |
| Carnitine | Bachem Americas INC | Cat#F2700.0025 |
| Cayman 10576 | Cayman | Cat#10011249 |
| Coenzyme A sodium salt hydrate | Sigma-Aldrich | Cat#C3144 |
| Deuterium oxide (D2O) | Sigma-Aldrich | Cat#151882 |
| Fatty acid free BSA | Akron Biotechnology | Cat#AK8909–0100 |
| Liberase TM | Roche | Cat#5401127001 |
| Lipid standard, MG, DG, TG mix | Sigma-Aldrich | Cat#1787–1AMP |
| Octanoyl carnitine | Sigma-Aldrich | Cat#50892 |
| Palmitate-BSA | Agilent Technologies | Cat#102720–100 |
| 1-[C14]-palmitic acid | Perkin Elmer | Cat#NEC075H050UC |
| U-[C14]-palmitic acid | Perkin Elmer | Cat#NEC534050UC |
| Palmitoylcarnitine | Sigma-Aldrich | Cat#P1645 |
| Palmitoyl CoA | Sigma-Aldrich | Cat#P9716 |
| Percoll | Sigma-Aldrich | Cat#GE17089101 |
| Plasma Membrane Permeabilizer (PMP) | Agilent Technologies | Cat#102504–100 |
| Poloxamer 407 purified | Sigma-Aldrich | Cat#16758 |
| Protease and phosphatase inhibitor | Thermo Fisher Scientific | Cat#78444 |
| Proteinase K | Roche | Cat#03115836001 |
| Recombinant human TNFα | BioLegend | Cat#570106 |
| RNase A | Roche Diagnostics | Cat#10109169001 |
| Sodium palmitate | Sigma-Aldrich | Cat#P9767 |
| Triascin C | Enzo Life Sciences | Cat#BML-EI218–0100 |
| Critical Commercial Assays | ||
| BCA protein assay kits | Pierce | Cat#23227 |
| Cholesterol reagent | Thermo Fisher | Cat#TR13421 |
| CloneAmp Hifi DNA PCR premix | Takara | Cat#639298 |
| Free fatty acid uptake assay kit | Abcam | Cat#ab176768 |
| GST protein purification kit | Pierce | Cat#16107 |
| In-Fusion HD enzyme premix | Takara | Cat#639690 |
| Ketone body assay kit | Abnova | Cat#89028362 |
| Lipofectamine LTX | Life Technologies | Cat#A12621 |
| Lipofectamine p3000 | Life Technologies | Cat#L3000015 |
| Lipofectamine RNAiMAX | Life Technologies | Cat#13778500 |
| Profectine | AB vector | Cat#T10 |
| Maxima First Strand cDNA Synthesis kit | Thermo Scientific | Cat#K1642 |
| Power SYBR Green PCR Master Mix | Life Technologies | Cat#4367659 |
| Triglyceride reagent | Thermo Fisher | Cat#TR22421 |
| TRIzol reagent | Life Technologies | Cat#15596018 |
| Experimental Models: Cell Lines | ||
| HEK293 | ATCC | Cat#CRL-1573 RRID:CVCL_0045 |
| sf9 cells | Gibco | Cat#B82501 RRID:CVCL_0549 |
| Experimental Models: Organisms/Strains | ||
| Mouse: TBK1 f/f | Marchlik et al., 2010 | N/A |
| Mouse: Albumin Cre
B6.Cg-Speer6-ps1Tg(Alb-cre)21Mgn/J |
Jackson Laboratory | Cat#003574; RRID:IMSR_JAX:003574 |
| Mouse: C57BL/6J | Jackson Laboratory | Cat#000664; RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| Mouse ACSL1 siRNA | Dharmacon | Cat#14081 |
| Negative control siRNA | Integrated DNA Technologies | Cat#51–01–14–04 |
| Primers for Q-PCR, see Table S1, S2 | ||
| Recombinant DNA | ||
| pcDNA-Flag-human TBK1 | Fitzgerald et al., 2003 | N/A |
| pcDNA-Flag-human TBK1 K38A | Fitzgerald et al., 2003 | N/A |
| pcDNA-Flag-human TBK1 S172A | This paper | N/A |
| 6myc-mTBK1 WT | Goncalves et al., 2011 | N/A |
| 6myc-mTBK1 ULD deleted | Goncalves et al., 2011 | N/A |
| 6myc-mTBK1 C-terminal deleted | Goncalves et al., 2011 | N/A |
| pCMV6 Entry mACSL1-myc/flag | Origene | Cat#MR210117 |
| pCMV6 Entry mACSL1-HA | This paper | N/A |
| Baculovirus genomic vector DNA | AB vector | Cat#A1 |
| pAcGHLTc-full length TBK1 | Shu et al., 2013 | N/A |
| pAcGHLTc-C-terminal truncated TBK1 | Shu et al., 2013 | N/A |
| pAcGHLTc-C-terminal truncated TBK1 K38A | Shu et al., 2013 | N/A |
| pAcGHLTc-C-terminal truncated TBK1 S172A | Shu et al., 2013 | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.net/Welcome |
| GraphPad Prism 8.4.3 | GraphPad Software | https://www.graphpad.com/ |
| BioRender (for Graphical Abstract) | BioRender | https://www.biorender.com/ |
| Other | ||
| High Fat Diet (60% kcal% fat) | Research Diet, Inc. | Cat#D12492 |
| Methionine/Choline-deficient diet | Research Diet, Inc. | Cat#A06071302 |
| William’s medium E | Life Technologies | Cat#12551–032 |
| GlutaMax | Life Technologies | Cat#35050–061 |
| Spin columns | Abcam | Cat#ab93349 |
| TLC plate | EMD Millipore | Cat#1.05553.0001 |
Context and Significance
Abnormal lipid accumulation can accelerate the development of non-alcoholic fatty liver diseases in obesity. Researchers at the University of California, San Diego and their colleagues show that chemical inhibitors of the enzyme TBK1 can reverse diet-induced fatty liver. Surprisingly, in liver-specific TBK1 deficient mice, fatty liver is worsened, due to reduced fatty acid oxidation. Further studies revealed that TBK1 serves as a scaffold for the enzyme ACSL1.
TBK1 is required for the fasting-stimulated localization of ACSL1 to mitochondria, providing acyl-CoAs for oxidation, while reducing localization of the enzyme to the ER for fatty acid re-esterification. While obesity elevates active TBK1, fasting increases inactive TBK1 which can rescue the activity of fatty acid oxidation. TBK1 thus senses nutritional status to determine the fate of fatty acids, perhaps explaining the mechanism by which TBK1 inhibitors improve fatty liver.
Highlights
Hepatic TBK1 activity is reduced by fasting while elevated by obesity
Liver-specific TBK1 KO mice have increased liver lipid due to less fat oxidation
Fasting-stimulated mitochondrial localization of ACSL1 is impaired in TBK1 KO mice
Inactive TBK1 rescues fatty acid oxidation, suggesting a scaffolding function
Acknowledgments
We thank members of Saltiel laboratory for helpful discussion. We thank Dr. Kate Fitzgerald for sharing TBK1 floxed mice. We thank Dr. Giulio Superti-Furga for sharing domain deleted TBK1 plasmids. We thank Dr. Pingwei Li for sharing GST-TBK1 plasmids for in vitro protein binding assays. We thank Dr. Majid Ghassemian and the Biomolecular and Proteomics Mass Spectrometry Facility for mass spectrometry analysis. We thank Dr. Alexander Andreyev for helpful discussions about seahorse experiments. We thank the UCSD histology core for tissue sectioning and staining. This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education 2018R1A6A3A03012174 to J.Y.H., and US National Institutes of Health grants 1K01DK105075 and R03DK118195 to S.M.R., R01NS087611 to A.N.M, VA Merit Review Grant (I01 BX003934) for S.K.M., P30 DK063491, R01DK117551, 1R01DK125820 and R01DK076906 to A.R.S.
Footnotes
Declaration of Interests
A.R.S. is a founder of Elgia Therapeutics and named inventor on several patent applications that include amlexanox. The other authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- ANSTEE QM, TARGHER G & DAY CP 2013. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol, 10, 330–44. [DOI] [PubMed] [Google Scholar]
- BLACKBURN P, DESPRÉS JP, LAMARCHE B, TREMBLAY A, BERGERON J, LEMIEUX I & COUILLARD C 2006. Postprandial variations of plasma inflammatory markers in abdominally obese men. Obesity (Silver Spring), 14, 1747–54. [DOI] [PubMed] [Google Scholar]
- BYRNE CD & TARGHER G 2015. NAFLD: a multisystem disease. J Hepatol, 62, S47–64. [DOI] [PubMed] [Google Scholar]
- CHIANG SH, BAZUINE M, LUMENG CN, GELETKA LM, MOWERS J, WHITE NM, MA JT, ZHOU J, QI N, WESTCOTT D, DELPROPOSTO JB, BLACKWELL TS, YULL FE & SALTIEL AR 2009. The protein kinase IKKepsilon regulates energy balance in obese mice. Cell, 138, 961–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLEMAN RA 2019. It takes a village: channeling fatty acid metabolism and triacylglycerol formation via protein interactomes. J Lipid Res, 60, 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLEMAN RA, LEWIN TM & MUOIO DM 2000. Physiological and nutritional regulation of enzymes of triacylglycerol synthesis. Annu Rev Nutr, 20, 77–103. [DOI] [PubMed] [Google Scholar]
- DONNELLY KL, SMITH CI, SCHWARZENBERG SJ, JESSURUN J, BOLDT MD & PARKS EJ 2005. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest, 115, 1343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESTES C, RAZAVI H, LOOMBA R, YOUNOSSI Z & SANYAL AJ 2018. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology, 67, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FESTA A, D’AGOSTINO R, TRACY RP, HAFFNER SM & STUDY IRA 2002. Elevated levels of acute-phase proteins and plasminogen activator inhibitor-1 predict the development of type 2 diabetes: the insulin resistance atherosclerosis study. Diabetes, 51, 1131–7. [DOI] [PubMed] [Google Scholar]
- FEUERER M, HERRERO L, CIPOLLETTA D, NAAZ A, WONG J, NAYER A, LEE J, GOLDFINE AB, BENOIST C, SHOELSON S & MATHIS D 2009. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med, 15, 930–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FITZGERALD KA, MCWHIRTER SM, FAIA KL, ROWE DC, LATZ E, GOLENBOCK DT, COYLE AJ, LIAO SM & MANIATIS T 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol, 4, 491–6. [DOI] [PubMed] [Google Scholar]
- FRIEDMAN SL, NEUSCHWANDER-TETRI BA, RINELLA M & SANYAL AJ 2018. Mechanisms of NAFLD development and therapeutic strategies. Nat Med, 24, 908–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FÜLLEKRUG J & POPPELREUTHER M 2016. Measurement of Long-Chain Fatty Acyl-CoA Synthetase Activity. Methods Mol Biol, 1376, 43–53. [DOI] [PubMed] [Google Scholar]
- GONCALVES A, BÜRCKSTÜMMER T, DIXIT E, SCHEICHER R, GÓRNA MW, KARAYEL E, SUGAR C, STUKALOV A, BERG T, KRALOVICS R, PLANYAVSKY M, BENNETT KL, COLINGE J & SUPERTI-FURGA G 2011. Functional dissection of the TBK1 molecular network. PLoS One, 6, e23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HE Q, XIA X, YAO K, ZENG J, WANG W, WU Q, TANG R & ZOU X 2019. Amlexanox reversed non-alcoholic fatty liver disease through IKKε inhibition of hepatic stellate cell. Life Sci, 239, 117010. [DOI] [PubMed] [Google Scholar]
- HUH JY, PARK J, KIM JI, PARK YJ, LEE YK & KIM JB 2017. Deletion of CD1d in Adipocytes Aggravates Adipose Tissue Inflammation and Insulin Resistance in Obesity. Diabetes, 66, 835–847. [DOI] [PubMed] [Google Scholar]
- HUH JY, PARK YJ, HAM M & KIM JB 2014. Crosstalk between adipocytes and immune cells in adipose tissue inflammation and metabolic dysregulation in obesity. Mol Cells, 37, 365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUYNH FK, GREEN MF, KOVES TR & HIRSCHEY MD 2014. Measurement of fatty acid oxidation rates in animal tissues and cell lines. Methods Enzymol, 542, 391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG G, LI Z, LIU F, ELLSWORTH K, DALLAS-YANG Q, WU M, RONAN J, ESAU C, MURPHY C, SZALKOWSKI D, BERGERON R, DOEBBER T & ZHANG BB 2005. Prevention of obesity in mice by antisense oligonucleotide inhibitors of stearoyl-CoA desaturase-1. J Clin Invest, 115, 1030–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KISHORE N, HUYNH QK, MATHIALAGAN S, HALL T, ROUW S, CREELY D, LANGE G, CAROLL J, REITZ B, DONNELLY A, BODDUPALLI H, COMBS RG, KRETZMER K & TRIPP CS 2002. IKK-i and TBK-1 are enzymatically distinct from the homologous enzyme IKK-2: comparative analysis of recombinant human IKK-i, TBK-1, and IKK-2. J Biol Chem, 277, 13840–7. [DOI] [PubMed] [Google Scholar]
- KOO SH 2013. Nonalcoholic fatty liver disease: molecular mechanisms for the hepatic steatosis. Clin Mol Hepatol, 19, 210–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAPETINA S & GIL-HENN H 2017. A guide to simple, direct, and quantitative. J Biol Methods, 4, e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE J, CHOI J, SCAFIDI S & WOLFGANG MJ 2016. Hepatic Fatty Acid Oxidation Restrains Systemic Catabolism during Starvation. Cell Rep, 16, 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE WN, BASSILIAN S, LIM S & BOROS LG 2000. Loss of regulation of lipogenesis in the Zucker diabetic (ZDF) rat. Am J Physiol Endocrinol Metab, 279, E425–32. [DOI] [PubMed] [Google Scholar]
- LI LO, ELLIS JM, PAICH HA, WANG S, GONG N, ALTSHULLER G, THRESHER RJ, KOVES TR, WATKINS SM, MUOIO DM, CLINE GW, SHULMAN GI & COLEMAN RA 2009. Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and beta-oxidation and alters phospholipid fatty acid composition. J Biol Chem, 284, 27816–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI LO, GREVENGOED TJ, PAUL DS, ILKAYEVA O, KOVES TR, PASCUAL F, NEWGARD CB, MUOIO DM & COLEMAN RA 2015. Compartmentalized acyl-CoA metabolism in skeletal muscle regulates systemic glucose homeostasis. Diabetes, 64, 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUMENG CN, BODZIN JL & SALTIEL AR 2007. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest, 117, 175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUMENG CN, DELPROPOSTO JB, WESTCOTT DJ & SALTIEL AR 2008. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes, 57, 3239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANICHAIKUL A, WANG XQ, ZHAO W, WOJCZYNSKI MK, SIEBENTHALL K, STAMATOYANNOPOULOS JA, SALEHEEN D, BORECKI IB, REILLY MP, RICH SS & BORNFELDT KE 2016. Genetic association of long-chain acyl-CoA synthetase 1 variants with fasting glucose, diabetes, and subclinical atherosclerosis. J Lipid Res, 57, 433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARCHLIK E, THAKKER P, CARLSON T, JIANG Z, RYAN M, MARUSIC S, GOUTAGNY N, KUANG W, ASKEW GR, ROBERTS V, BENOIT S, ZHOU T, LING V, PFEIFER R, STEDMAN N, FITZGERALD KA, LIN LL & HALL JP 2010. Mice lacking Tbk1 activity exhibit immune cell infiltrates in multiple tissues and increased susceptibility to LPS-induced lethality. J Leukoc Biol, 88, 1171–80. [DOI] [PubMed] [Google Scholar]
- MASHEK DG, LI LO & COLEMAN RA 2006. Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. J Lipid Res, 47, 2004–10. [DOI] [PubMed] [Google Scholar]
- MCGARRY JD & BROWN NF 1997. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur J Biochem, 244, 1–14. [DOI] [PubMed] [Google Scholar]
- MCWHIRTER SM, FITZGERALD KA, ROSAINS J, ROWE DC, GOLENBOCK DT & MANIATIS T 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci U S A, 101, 233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLAR JS, CROMLEY DA, MCCOY MG, RADER DJ & BILLHEIMER JT 2005. Determining hepatic triglyceride production in mice: comparison of poloxamer 407 with Triton WR-1339. J Lipid Res, 46, 2023–8. [DOI] [PubMed] [Google Scholar]
- MOWERS J, UHM M, REILLY SM, SIMON J, LETO D, CHIANG SH, CHANG L & SALTIEL AR 2013. Inflammation produces catecholamine resistance in obesity via activation of PDE3B by the protein kinases IKKε and TBK1. Elife, 2, e01119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OBSTFELD AE, SUGARU E, THEARLE M, FRANCISCO AM, GAYET C, GINSBERG HN, ABLES EV & FERRANTE AW 2010. C-C chemokine receptor 2 (CCR2) regulates the hepatic recruitment of myeloid cells that promote obesity-induced hepatic steatosis. Diabetes, 59, 916–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OLEFSKY JM & GLASS CK 2010. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol, 72, 219–46. [DOI] [PubMed] [Google Scholar]
- ORAL EA, REILLY SM, GOMEZ AV, MERAL R, BUTZ L, AJLUNI N, CHENEVERT TL, KORYTNAYA E, NEIDERT AH, HENCH R, RUS D, HOROWITZ JF, POIRIER B, ZHAO P, LEHMANN K, JAIN M, YU R, LIDDLE C, AHMADIAN M, DOWNES M, EVANS RM & SALTIEL AR 2017. Inhibition of IKKɛ and TBK1 Improves Glucose Control in a Subset of Patients with Type 2 Diabetes. Cell Metab, 26, 157–170.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAIS R, BARRITT AS, CALMUS Y, SCATTON O, RUNGE T, LEBRAY P, POYNARD T, RATZIU V & CONTI F 2016. NAFLD and liver transplantation: Current burden and expected challenges. J Hepatol, 65, 1245–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASQUA T, MAHATA S, BANDYOPADHYAY GK, BISWAS A, PERKINS GA, SINHA-HIKIM AP, GOLDSTEIN DS, EIDEN LE & MAHATA SK 2016. Impact of Chromogranin A deficiency on catecholamine storage, catecholamine granule morphology and chromaffin cell energy metabolism in vivo. Cell Tissue Res, 363, 693–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QUIROS PM, GOYAL A, JHA P & AUWERX J 2017. Analysis of mtDNA/nDNA Ratio in Mice. Curr Protoc Mouse Biol, 7, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REILLY SM, AHMADIAN M, ZAMARRON BF, CHANG L, UHM M, POIRIER B, PENG X, KRAUSE DM, KORYTNAYA E, NEIDERT A, LIDDLE C, YU RT, LUMENG CN, ORAL EA, DOWNES M, EVANS RM & SALTIEL AR 2015. A subcutaneous adipose tissue-liver signalling axis controls hepatic gluconeogenesis. Nat Commun, 6, 6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REILLY SM, CHIANG SH, DECKER SJ, CHANG L, UHM M, LARSEN MJ, RUBIN JR, MOWERS J, WHITE NM, HOCHBERG I, DOWNES M, YU RT, LIDDLE C, EVANS RM, OH D, LI P, OLEFSKY JM & SALTIEL AR 2013. An inhibitor of the protein kinases TBK1 and IKK-ɛ improves obesity-related metabolic dysfunctions in mice. Nat Med, 19, 313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROGERS GW, BRAND MD, PETROSYAN S, ASHOK D, ELORZA AA, FERRICK DA & MURPHY AN 2011. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One, 6, e21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALTIEL AR & OLEFSKY JM 2017. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest, 127, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHENK S, SABERI M & OLEFSKY JM 2008. Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest, 118, 2992–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHENKMAN JB & CINTI DL 1978. Preparation of microsomes with calcium. Methods Enzymol, 52, 83–9. [DOI] [PubMed] [Google Scholar]
- SCHONFELD G 2003. Familial hypobetalipoproteinemia: a review. J Lipid Res, 44, 878–83. [DOI] [PubMed] [Google Scholar]
- SCHUSTER S, CABRERA D, ARRESE M & FELDSTEIN AE 2018. Triggering and resolution of inflammation in NASH. Nat Rev Gastroenterol Hepatol, 15, 349–364. [DOI] [PubMed] [Google Scholar]
- SCOTT RA, SCOTT LJ, MÄGI R, MARULLO L, GAULTON KJ, KAAKINEN M, PERVJAKOVA N, PERS TH, JOHNSON AD, EICHER JD, JACKSON AU, FERREIRA T, LEE Y, MA C, STEINTHORSDOTTIR V, THORLEIFSSON G, QI L, VAN ZUYDAM NR, MAHAJAN A, CHEN H, ALMGREN P, VOIGHT BF, GRALLERT H, MÜLLER-NURASYID M, RIED JS, RAYNER NW, ROBERTSON N, KARSSEN LC, VAN LEEUWEN EM, WILLEMS SM, FUCHSBERGER C, KWAN P, TESLOVICH TM, CHANDA P, LI M, LU Y, DINA C, THUILLIER D, YENGO L, JIANG L, SPARSO T, KESTLER HA, CHHEDA H, EISELE L, GUSTAFSSON S, FRÅNBERG M, STRAWBRIDGE RJ, BENEDIKTSSON R, HREIDARSSON AB, KONG A, SIGURÐSSON G, KERRISON ND, LUAN J, LIANG L, MEITINGER T, RODEN M, THORAND B, ESKO T, MIHAILOV E, FOX C, LIU CT, RYBIN D, ISOMAA B, LYSSENKO V, TUOMI T, COUPER DJ, PANKOW JS, GRARUP N, HAVE CT, JØRGENSEN ME, JØRGENSEN T, LINNEBERG A, CORNELIS MC, VAN DAM RM, HUNTER DJ, KRAFT P, SUN Q, EDKINS S, OWEN KR, PERRY JRB, WOOD AR, ZEGGINI E, TAJES-FERNANDES J, ABECASIS GR, BONNYCASTLE LL, CHINES PS, STRINGHAM HM, KOISTINEN HA, KINNUNEN L, SENNBLAD B, MÜHLEISEN TW, NÖTHEN MM, PECHLIVANIS S, BALDASSARRE D, GERTOW K, HUMPHRIES SE, TREMOLI E, KLOPP N, MEYER J, STEINBACH G, et al. 2017. An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes, 66, 2888–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHU C, SANKARAN B, CHATON CT, HERR AB, MISHRA A, PENG J & LI P 2013. Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure, 21, 1137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SVENSSON RU, PARKER SJ, EICHNER LJ, KOLAR MJ, WALLACE M, BRUN SN, LOMBARDO PS, VAN NOSTRAND JL, HUTCHINS A, VERA L, GERKEN L, GREENWOOD J, BHAT S, HARRIMAN G, WESTLIN WF, HARWOOD HJ, SAGHATELIAN A, KAPELLER R, METALLO CM & SHAW RJ 2016. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med, 22, 1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANG Y, ZHOU J, HOOI SC, JIANG YM & LU GD 2018. Fatty acid activation in carcinogenesis and cancer development: Essential roles of long-chain acyl-CoA synthetases. Oncol Lett, 16, 1390–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINER S, CHAN Y, PALTSER G, TRUONG D, TSUI H, BAHRAMI J, DORFMAN R, WANG Y, ZIELENSKI J, MASTRONARDI F, MAEZAWA Y, DRUCKER DJ, ENGLEMAN E, WINER D & DOSCH HM 2009. Normalization of obesity-associated insulin resistance through immunotherapy. Nat Med, 15, 921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YKI-JÄRVINEN H 2014. Non-alcoholic fatty liver disease as a cause and a consequence of metabolic syndrome. Lancet Diabetes Endocrinol, 2, 901–10. [DOI] [PubMed] [Google Scholar]
- ZHAO P, WONG KI, SUN X, REILLY SM, UHM M, LIAO Z, SKOROBOGATKO Y & SALTIEL AR 2018. TBK1 at the Crossroads of Inflammation and Energy Homeostasis in Adipose Tissue. Cell, 172, 731–743.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO W 2013. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett, 587, 542–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze dataset.