Abstract
Lung cancer is the leading worldwide cause of cancer mortality, as it is often detected at an advanced stage. Since 2011, low-dose CT scan-based screening has promised a 20% reduction in lung cancer mortality. However, effectiveness of screening has been limited by eligibility only for a high-risk population of heavy smokers and a large number of false positives generated by CT. Biomarkers have tremendous potential to improve early detection of lung cancer by refining lung cancer risk, stratifying positive CT scans, and categorizing intermediate-risk pulmonary nodules. Three biomarker tests (Early CDT-Lung, Nodify XL2, Percepta) have undergone extensive validation and are available to the clinician. The authors discuss these tests, with their clinical applicability and limitations, current ongoing evaluation, and future directions for biomarkers in lung cancer screening and detection.
Keywords: biomarkers, lung cancer screening, early detection, autoantibodies, lung cancer
Introduction
Lung cancer is the leading worldwide cause of cancer mortality.(1) A major contributor to the lethality of lung cancer is that most lung cancers are diagnosed at an already advanced stage.(2) Screening for lung cancer was generally not considered successful until 2011, when the National Lung Screening Trial (NLST) demonstrated a reduction in lung cancer mortality of 20% through low-dose CT screening of a selected pool of high-risk patients.(3) Although the trial was a success, the screening protocol produced a high number of false positive results, with 24% of scans read as positive and only 6% of these confirmed as true positive. Through the three years of screening in the trial, 39% of the CT-screened subjects had at least one false positive scan. In the United States, the accepted entry criteria (>55 years old, >30 pack-year history of smoking, currently smoking or <15 years since quit) are somewhat restrictive, with only 27% of lung cancer diagnoses estimated to occur in screening-eligible patients. The expense and logistics of implementing such a screening trial are thus complex, and the false positive rate is a tangible downside for patients considering screening.(4,5) Thus, in the United States, only 4% of eligible high-risk patients undergo screening yearly for lung cancer (Figure 1).(6) As a compounded limitation, while CT screening is recommended by the United States Preventive Services Task Force, adoption by other health services worldwide has been piecemeal at best.(7,8) Even with full implementation, only around 10% of US lung cancer deaths could be averted.(9)
Figure 1:
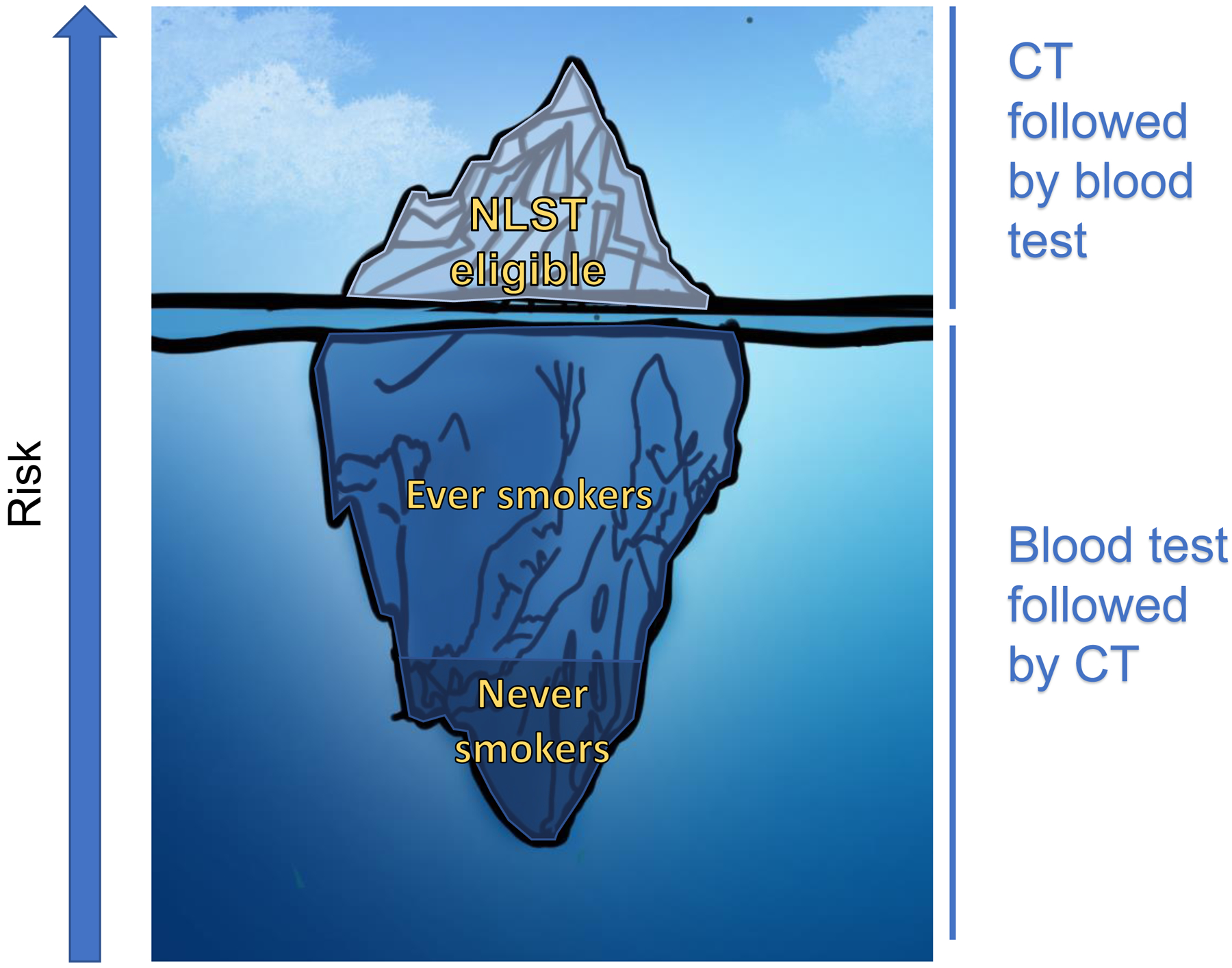
Like an iceberg, the bulk of cases of lung cancer are not only not detected by screening, but occur in patients not eligible for screening. Approximately 4% of NLST-eligible patients undergo screening. If all NLST-eligible patients underwent screening, only 27% of lung cancers would be detected, similar to the portion of an iceberg seen above water. The remaining 73% of lung cancers occur in those ineligible for screening, for instance those with only a light smoking history, those who have quit > 15 years ago, or those who have never smoked. Biomarkers in high-risk individuals can decrease the rate of false positives after CT-based screening. In lower-risk individuals, biomarkers can be used to identify patients at higher risk who may benefit from screening.
Outside of true cancer screening, approximately 2% of the US population undergoes CT of the chest yearly for various reasons.(10) Pulmonary nodules are incidentally found on 24–31% of these CTs. Although there has been work to better classify and stratify pulmonary nodules, mostly through risk calculators and radiographic characteristics, much of the diagnostic workflow involves watchful waiting, PET imaging, or invasive procedures.(11,12)
For both low-dose CT screening and incidentally discovered nodules, there is a substantial need for biomarkers that could accurately discriminate benign lesions from early cancers at the time of imaging. Several diagnostic biomarkers have come to market, with several other biomarkers in active development. Here, we will explore the uses and limitations of existing biomarkers and the path to development of additional, clinically useful biomarkers for early detection of lung cancer.
Discussion
Clinical utility
There are many potential roles for lung cancer biomarkers, both within the setting of low-dose CT screening and independent of combined testing. In patients being considered for screening, a biomarker may help to stratify risk, optimally performing to refine existing lung cancer risk models based on age, smoking, and family history.(13–15) Such a risk model would then help to identify subjects at a high risk for lung cancer who may benefit from CT screening but are excluded by standard NLST or National Comprehensive Cancer Network (NCCN) criteria. It also may encourage a subset of patients who meet the established criteria but who are reluctant to undergo screening. These patients may test negative with a biomarker and thus could be clinically followed with less frequent CT scans or serial biomarker testing.
After scanning, when nodules are detected, there is an obvious role for biomarkers in classifying nodule risk. Again, an effective biomarker should perform in combination with a clinical nodule risk score, for instance the Lung-RADS criteria or a model such as proposed by McWilliams et al or Swensen et al.(14,15) Here, a biomarker may be of utility both in the setting of CT screening and in the workup of incidentally discovered lung nodules. It is in this diagnostic setting that this field has seen most progress, with three approved tests available. These tests, two of which are blood-based and the other performed on airway epithelial brushings collected during bronchoscopy, are reviewed below.
Given the multiple roles of biomarkers and different approaches to screening, assessment of a biomarker’s performance clearly necessitates reporting its sensitivity, specificity, negative and positive predictive value, as well as likelihood ratio. For instance, the clinical utility of a biomarker intended for use in combination with a risk score or cutoff can be easily assessed using a diagnostic likelihood ratio. As an example, one can point to the Lung-RADS criteria used in CT-based screening.(12) In this population, the pre-test probability for lung cancer in the setting of a positive CT scan is 4%. With classification of these positive scans using the Lung-RADS guidelines, it is recommended that patients undergo more intensive follow-up if they are in category 4A or higher, whereby the pre-test probability of malignancy is >5%. In the Lung-RADS guidelines, nodules in class 2 have a pre-test probability of <1% and thus return to standard annual screening is advised. Lung-RADS category 4B has a pre-test probability of >15% and further workup by PET-CT or biopsy is indicated. These should then be our goalposts, such that if we could move the post-test probabilities to >15% or <1%, our clinical decision making would be enhanced. Given the pre-test probabilities, we can use Bayes’ theorem to calculate the needed positive and negative likelihood ratios (LR+ and LR-) to deliver these post-test probabilities. We can thus estimate that a diagnostic test, given a pre-test probability of 5%, should have a LR+ of 3.38 to deliver a post-test probability of 15%, and should have a LR- of 0.191 to deliver a post-test probability of <1%.
Another outlook is to consider a test’s negative or positive predictive value (NPV or PPV). This allows a rapid assessment of a value of a test: if negative, a test with an NPV of 95% would only miss 5% of cases. However, the NPV and PPV depend on disease prevalence, which varies both in the populations on which these tests were calibrated on and in their intended clinical use. For example, a test with 95% specificity would still yield a low PPV in most at-risk smoking populations for lung cancer bringing in hundreds of patients without disease for further assessment.
Blood-based biomarkers
Detection of biomarkers in the blood holds numerous advantages, including the relative noninvasive nature of blood draws and well-established laboratory pipelines for isolation and analyses of various assays from plasma, exosomes, circulating nucleic acids, and circulating cells. Two tests, mostly intended for classification of indeterminate pulmonary nodules, are currently available.
EarlyCDT-Lung (OncImmune), is a 7-autoantibody panel, first developed in 2010 and extensively validated in seven different cohorts.(16–20) This panel, consisting of autoantibodies against p53, CAGE, NY-ESO-1, SOX2, GBU4–5, HuD, and MAGE-A4, has shown good performance in classifying indeterminate pulmonary nodules, with important information produced both when the panel is positive or negative. Work to assemble this panel began in 2006, through several different assessments of lung cancer autoantibodies. These utilized forward discovery tools such as phage display and Serological Analysis of Tumor Antigens by Recombinant cDNA (SEREX), as well as work analyzing humoral response to known cancer antigens. From these, a six-autoantibody panel was selected, with clinical validation in three different matched cohorts of patients with newly diagnosed lung cancer, showing a specificity of approximately 90% at sensitivities of around 40%. The panel was technically validated in three separate cohorts and four additional post-validation cohorts of newly diagnosed lung cancer patients versus control.(21,22) Further development included testing and validation of a related seven-autoantibody panel (dropping annexin I and adding HuD and MAGE A4), which showed an improvement in performance, raising specificity from 82% to 90% in a prospective validation in an at-risk population from the previous six-autoantibody panel.(23) Based on this, the seven-marker panel has replaced the original six-marker panel as the commercially available EarlyCDT-Lung test, and in a post-marketing audit of over 1600 patients presenting with a nodule, showed a similar sensitivity of 41% at a specificity of 87%.(24) A cost effectiveness study indicated that the use of EarlyCDT-Lung in patients presenting with nodules of approximately 8–30 mm is around $24,000 per quality-of-life adjusted life year gained.(25)
This test, while extensively validated in newly diagnosed lung cancer and nodule cohorts, has not been evaluated extensively as a part of traditional low-dose CT based screening program. However, if one were to assume that it would perform similarly in a screening population after the detection of nodules, with a sensitivity of 40% at specificity of 90%, this would generate a positive diagnostic likelihood ratio (LR+) of 4, calculated as sensitivity/(1-specificity), which is enough to reclassify some low probability nodules into a category where they should have radiographic follow-up sooner. Interestingly, mathematical modeling of a high- and low-specificity version of EarlyCDT-Lung has shown the ability to both reclassify nodules into a higher and a lower risk group.(26) Recently, a double blinded randomized trial was published describing the use of EarlyCDT-Lung followed by CT scan in a population at higher risk for lung cancer.(27) Participants were randomized into an intervention arm that began with the EarlyCDT-Lung test then CT scan if the biomarker test was positive versus control, or standard of care which did not include CT scans and relied on symptomatic presentation. Over two years, EarlyCDT-Lung did not increase the frequency of detection of lung cancer, but lung cancers detected in the intervention arm were at earlier stage. They detected 56 cancers in the intervention arm, of which 23 were early stage (41.1%), and 71 in the control arm, of which 19 were early-stage (26.8%). Since most subjects did not under CT scanning, sensitivity and specificity were estimated using cancer registry data. Sensitivity was lower than in previous trials, at 32.1%, with a preservation of specificity at 90.3%. Of note, sensitivity was particularly low for stage III or IV cancers, at 18.2%. Continued longitudinal monitoring, to better measure the true cancer incidence in each arm, is warranted.
A second test, Nodify XL2 (Biodesix), is also available for the classification of indeterminate pulmonary nodules.(28) This test measures a panel of blood proteins using a mass-spectrometry based assay. This test originated in a 2013 study which reported a 13-protein proteomic classifier, with scoring by a logistic regression model that gave a 90% negative predictive value (NPV) for benign nodules.(29) The model was developed in a discovery set of patients with IPNs between 4 and 20 mm, and with a lung cancer incidence of 20%. Markers were measured using a mass spectrometry multiple reaction monitoring (MRM) technique. The panel was then validated in two independent sets, in which it gave similar performance. Later work by the same group used a 5-marker subset of the original 13 proteins plus 6 normalization markers, which they validated in a similar population.(30) A subsequent study claimed clinical utility based on the test’s negative predictive value potentially sparing invasive procedures for 31.8% of subjects.(31)
The panel underwent continued refinement on a cohort of 222 patients presenting with nodules 8–20 mm in size who had undergone invasive workup.(32) The panel was integrated with clinical risk factors, with a specific focus on the marker performance in this high prevalence population (81% diagnosed with lung cancer). Using decision tree analyses, an integrated model was found to have mildly improved performance versus the Mayo clinical risk model or the proteomic classifier individually (ROC from 58% for the clinical risk score, 60% for the proteomic classifier alone, to 63% for the integrated model). This integrated model, termed Xpresys Lung (XL2, Integrated Diagnostics), was then prospectively validated in the PANOPTIC study of 685 patients presenting with 6–30 mm nodules.(33) While XL2 was tuned for a high incidence population, PANOPTIC revealed its best performance in a subgroup with clinician-assessed pretest probability of cancer of <50%. In this subgroup of 178 patients, who had a lung cancer prevalence of 16%, the classifier showed a sensitivity of 97% and a specificity of 44%, with an NPV of 98% (and a LR- of 0.07). These findings supported the use of this panel for identifying low-risk pulmonary nodules, outperforming PET/CT, physician estimates, and lung nodule risk scores.
Airway gene expression classifiers
Cigarette smoking produces gene-expression alterations throughout the epithelial cells that line the respiratory tract, leading to an airway “field of injury.” (34–36) Further, cancer-associated gene-expression patterns are found in cytologically-normal epithelium collected from the bronchial airways of current and former smokers with lung cancer.(37) As an initial proof of concept in 2007, investigators identified an 80-gene expression set, measured from brushings of histologically normal bronchial airways that could distinguish smokers with and without lung cancer with 80% sensitivity and 84% specificity in a validation set of samples.(37) Accuracy was maintained in early stage cancers, with 90% sensitivity in stage I tumors. Interestingly, the accuracy of the panel was not affected by the location of the airway brushing relative to the cancer, implying a broad field of injury throughout the bronchial epithelium. This panel was additive to bronchoscopic cytopathology obtained during the same procedure. Cytopathology alone only yielded a diagnosis of cancer in 32 of 60 subjects with cancer and ruled out cancer in 5 of 69 subjects without cancer. Among the cytopathologically nondiagnostic bronchoscopies, the classifier performance was consistent (89% sensitivity, 83% specificity).
Based on these findings, two large prospective multicenter trials (AEGIS-1 and AEGIS-2) were conducted, enrolling patients undergoing bronchoscopy for suspicion of lung cancer.(38) During these bronchoscopies, airway brushings were performed on the normal-appearing mainstem bronchi and underwent RNA expression profiling by microarray. Both cohorts had high prevalence of lung cancer (74% and 78%), with the cancer subjects being older, heavier smokers. Bronchoscopy was non-diagnostic on 272 patients of the 639 in both cohorts. In the two trials, the classifier showed a similar sensitivity (88% and 89%) but lower specificity (47% in both). Combining the classifier with bronchoscopy increased sensitivity from 74–76% for bronchoscopy alone to 96–98%. In patients with non-diagnostic bronchoscopies, the sensitivity was maintained, at 86–92%, with no impact from size or location of the lesion, cancer stage, or involvement of lymph nodes. To define a clinical utility, the authors further examined the performance of the classifier based on physician-assessed pre-bronchoscopy probability of cancer. In patients with an intermediate pretest probability and a negative bronchoscopy (who had cancer prevalence of 41%), the classifier had a 91% NPV. In a combined group of low and intermediate probability patients with nodules < 3 cm, the reported sensitivity was 88% with an NPV of 94%. Combining bronchoscopy and the classifier produced a negative LR of 0.06, which can produce a posttest probability of < 10% in patients with pretest probabilities of up to 66%. A negative classifier in patients with a nondiagnostic bronchoscopy and an intermediate probability of cancer may allow physicians to avoid unnecessary invasive procedures. The bronchial genomic classifier, originally developed by Allegro Diagnostics Inc, was acquired by Veracyte Inc. who launched the Percepta™ test in 2015.
These findings spurred additional research on the classifier, including work defining possible clinical utility and cost-effectiveness within the AEGIS collection.(39,40) Use of Percepta was projected to reduce unnecessary invasive procedures by as much as 50%, with a false negative rate of 11%. Use of the classifier reduced costs for invasive procedures, with a small benefit in quality adjusted life years, giving a modest gain in incremental cost effectiveness ratio. The test has been successfully deployed and in clinical use since 2015, with continued post-marketing data collection that facilitated refinement of the test. In 2019, an updated test based on next-generation RNA transcriptome sequencing, termed Percepta Genomic Sequencing Classifier (GSC), was released which allows for both up-classification and down-classification among those at intermediate pre-test risk of lung cancer.
Future Directions
Given the high failure rate of cancer biomarkers, there needs to be substantial rigor in experimental design and extensive critical assessment of derived panels. There have been many guidelines published on best practices for validation of biomarkers, including the Institute of Medicine and REMARK guidelines.(41,42) However, we would add guidelines that mandate independent validation not only of the panel itself, but of the collection, the analysis, and the statistical techniques particularly when claims of clinical utility are made and the test is offered to patients. A comprehensive analysis of the missed costs of over- and under-diagnosis in a lung nodule cohort has not been performed. For instance, EarlyCDT-Lung is used to identify high-risk nodules, with false positive tests leading to overdiagnosis. Even small cumulative risks such as additional radiation from CT scans and PET-CTs, could overwhelm a small improvement in mortality from early detection of a subset of lung cancers. For Nodify XL2, a false negative test could lead to delayed diagnosis. However, it is difficult to estimate whether any potential delay in diagnosis would be outweighed by the benefits of fewer unnecessary invasive procedures performed. Some of the analyses on Percepta have addressed these questions, and continued post-marketing data analysis is necessary to see if these predictions correspond with real-world performance.(39) These are difficult questions to answer, and will require extensive, long-term investigation.
These biomarkers have demonstrated their performance through repeated validation. However, their best utility is in a limited clinical role, with EarlyCDT-Lung showing good specificity and Nodify XL2 showing good sensitivity in indeterminate pulmonary nodules, which thus precludes head-to-head comparison. However, modeling such a comparison, with both tests used in a potential “real world” scenario outside of their intended use, may still be valuable. Percepta a high-sensitivity test done using bronchoscopic samples, likely will find usage in a much higher prevalence population than a blood-based biomarker. However, a comparison of Nodify XL2 and Percepta in patients with nodules undergoing bronchoscopy would be enlightening.
There are still several unmet needs in the lung cancer biomarker field. As mentioned, even with a full implementation of lung cancer screening under current guidelines, only a small fraction of lung cancer deaths would be averted.(9) In a blinded validation study, a biomarker panel that combines a previously validated marker (43) with three additional markers has shown the ability to improve a clinical smoking-based risk model.(44) A low-cost, easily implementable test has a high potential to alter lung cancer screening and diagnostics in the future. More accurate risk stratification, through a combined clinical and biomarker risk score, has the potential to reduce non-effective screening (and thus false positives) and also identify higher risk subjects who do not meet NLST screening criteria yet may benefit from lung cancer screening. Future studies will need to assess the utility of this panel combined with screening.
An additional area for future exploration is the process of biomarker discovery. We believe that adopting rational evaluation of biomarker discovery would benefit the field, centering on biological plausibility of the marker panel. Biomarkers can be found in plasma, brushings and sputum. For example, promoter methylation has shown promise in both plasma DNA, sputum, and effusions.(45,46) In an initial study in plasma, patients were divided into two groups, ground glass opacity (n = 23) and cancerous tumors (n = 70). Plasma DNA from age-matched nodule-free individuals were used as controls (n = 80). 73% of patients with cancerous tumors showed methylation of at least one gene with a specificity of 71% and a methylation marker was found in only 22% of those with a ground glass opacity.(46) Promoter methylation of a 6 gene panel was detected in 92.2% (83/90) of a training cohort with a specificity of 72.0% (18/25) and in 93.0% (40/43) of an independent cohort of stage IA primary NSCLC.(47) Tissue and cell-free DNA-Based epigenomic approaches for cancer detection were recently reviewed elsewhere.(48)
Additionally, new biomarker panels should reflect the heterogeneity of the disease, including driver and passenger mutations, differences in immune response, and histological subtypes. For biomarker discovery, the collection, isolation, and analysis protocols should be robust and reproducible, and based in biology. The members of a multi-component panel need to be individually validated, preferably on a variety of platforms and in more than one center. Batch effect may be introduced by platform calibration, sample collection, sample preparation, and at numerous other steps. This could be addressed through reanalysis of replicates of the original biospecimens done at a later time. There should be a dose-response between the detected abundances and the size of a lesion, to help to establish biologic plausibility behind a correlation.
Conclusions
Biomarkers are playing an emerging role in the early detection of lung cancer. There are many potential roles for biomarkers, from risk stratification to classification of nodules detected incidentally or through low-dose screening programs. Several multi-analyte biomarker panels are available and have shown performance in classification of indeterminate pulmonary nodules. However, these panels must be used in an appropriate clinical context. Additionally, there is still much work to be done on unfulfilled needs within and outside of CT-based screening.
Acknowledgements
The authors would like to acknowledge Lila Ostrin for preparation of the iceberg illustration in Figure 1.
Relevant Financial support:
EJO American Lung Association Lung Cancer Discovery Award 619882
DS NIH/NCI R01CA206027,NIH/NCI R01CA208709
AS NIH/NCI 1U2CCA23323801S1, NIH/NCI 1U2CCA233238-01, NIH/NCI 1U2CCA233238-01
SMH NIH/NCI 1U01CA194733-01A1, NIH/NCI 1U01CA213285-01A1, NIH/NCI 1U19CA203654
Footnotes
Conflict of Interest Disclosure Statement:
A.S. is an employee of Johnson & Johnson. S.H. has IP filings related to lung cancer biomarkers with applications to cancer detection.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69(1):7–34 doi 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, et al. 2019. SEER Cancer Statistics Review, 1975–2016. National Cancer Institute <https://seer.cancer.gov/csr/1975_2016/>. [Google Scholar]
- 3.National Lung Screening Trial Research T, Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365(5):395–409 doi 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinsinger LS, Anderson C, Kim J, Larson M, Chan SH, King HA, et al. Implementation of Lung Cancer Screening in the Veterans Health Administration. JAMA Intern Med 2017;177(3):399–406 doi 10.1001/jamainternmed.2016.9022. [DOI] [PubMed] [Google Scholar]
- 5.Gesthalter YB, Koppelman E, Bolton R, Slatore CG, Yoon SH, Cain HC, et al. Evaluations of Implementation at Early-Adopting Lung Cancer Screening Programs: Lessons Learned. Chest 2017;152(1):70–80 doi 10.1016/j.chest.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Jemal A, Fedewa SA. Lung Cancer Screening With Low-Dose Computed Tomography in the United States-2010 to 2015. JAMA Oncol 2017;3(9):1278–81 doi 10.1001/jamaoncol.2016.6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moyer VA, Force USPST. Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2014;160(5):330–8 doi 10.7326/M13-2771. [DOI] [PubMed] [Google Scholar]
- 8.Pinsky PF. Lung cancer screening with low-dose CT: a world-wide view. Transl Lung Cancer Res 2018;7(3):234–42 doi 10.21037/tlcr.2018.05.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma J, Ward EM, Smith R, Jemal A. Annual number of lung cancer deaths potentially avertable by screening in the United States. Cancer 2013;119(7):1381–5 doi 10.1002/cncr.27813. [DOI] [PubMed] [Google Scholar]
- 10.Gould MK, Tang T, Liu IL, Lee J, Zheng C, Danforth KN, et al. Recent Trends in the Identification of Incidental Pulmonary Nodules. Am J Respir Crit Care Med 2015;192(10):1208–14 doi 10.1164/rccm.201505-0990OC. [DOI] [PubMed] [Google Scholar]
- 11.MacMahon H, Naidich DP, Goo JM, Lee KS, Leung ANC, Mayo JR, et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society 2017. Radiology 2017;284(1):228–43 doi 10.1148/radiol.2017161659. [DOI] [PubMed] [Google Scholar]
- 12.2014 American College of Radiology. Lung CT Screening Reporting and Data System (Lung-RADS). American College of Radiology <http://www.acr.org/Quality-Safety/Resources/LungRADS>.
- 13.Swensen SJ, Silverstein MD, Edell ES, Trastek VF, Aughenbaugh GL, Ilstrup DM, et al. Solitary pulmonary nodules: clinical prediction model versus physicians. Mayo Clin Proc 1999;74(4):319–29 doi 10.4065/74.4.319. [DOI] [PubMed] [Google Scholar]
- 14.Swensen SJ, Silverstein MD, Ilstrup DM, Schleck CD, Edell ES. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Arch Intern Med 1997;157(8):849–55. [PubMed] [Google Scholar]
- 15.McWilliams A, Tammemagi MC, Mayo JR, Roberts H, Liu G, Soghrati K, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med 2013;369(10):910–9 doi 10.1056/NEJMoa1214726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong L, Coe SP, Stromberg AJ, Khattar NH, Jett JR, Hirschowitz EA. Profiling tumor-associated antibodies for early detection of non-small cell lung cancer. J Thorac Oncol 2006;1(6):513–9. [PubMed] [Google Scholar]
- 17.Tan EM. Autoantibodies as reporters identifying aberrant cellular mechanisms in tumorigenesis. J Clin Invest 2001;108(10):1411–5 doi 10.1172/JCI14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houghton AN. Cancer antigens: immune recognition of self and altered self. The Journal of experimental medicine 1994;180(1):1–4 doi 10.1084/jem.180.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tureci O, Usener D, Schneider S, Sahin U. Identification of tumor-associated autoantigens with SEREX. Methods Mol Med 2005;109:137–54 doi 10.1385/1-59259-862-5:137. [DOI] [PubMed] [Google Scholar]
- 20.OncImmune. EarlyCDT Lung: For the early detection of lung cancer. <https://oncimmune.com/products/earlyctd-lung>.
- 21.Boyle P, Chapman CJ, Holdenrieder S, Murray A, Robertson C, Wood WC, et al. Clinical validation of an autoantibody test for lung cancer. Ann Oncol 2011;22(2):383–9 doi 10.1093/annonc/mdq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam S, Boyle P, Healey GF, Maddison P, Peek L, Murray A, et al. EarlyCDT-Lung: an immunobiomarker test as an aid to early detection of lung cancer. Cancer Prev Res (Phila) 2011;4(7):1126–34 doi 10.1158/1940-6207.CAPR-10-0328. [DOI] [PubMed] [Google Scholar]
- 23.Chapman CJ, Healey GF, Murray A, Boyle P, Robertson C, Peek LJ, et al. EarlyCDT(R)-Lung test: improved clinical utility through additional autoantibody assays. Tumour Biol 2012;33(5):1319–26 doi 10.1007/s13277-012-0379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jett JR, Peek LJ, Fredericks L, Jewell W, Pingleton WW, Robertson JF. Audit of the autoantibody test, EarlyCDT(R)-lung, in 1600 patients: an evaluation of its performance in routine clinical practice. Lung Cancer 2014;83(1):51–5 doi 10.1016/j.lungcan.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Edelsberg J, Weycker D, Atwood M, Hamilton-Fairley G, Jett JR. Cost-effectiveness of an autoantibody test (EarlyCDT-Lung) as an aid to early diagnosis of lung cancer in patients with incidentally detected pulmonary nodules. PloS one 2018;13(5):e0197826 doi 10.1371/journal.pone.0197826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Healey GF, Macdonald IK, Reynolds C, Allen J, Murray A. Tumor-Associated Autoantibodies: Re-Optimization of EarlyCDT-Lung Diagnostic Performance and Its Application to Indeterminate Pulmonary Nodules. Journal of Cancer Therapy 2017;8(5) doi 10.4236/jct.2017.85043. [DOI] [Google Scholar]
- 27.Sullivan FM, Mair FS, Anderson W, Armory P, Briggs A, Chew C, et al. Earlier diagnosis of lung cancer in a randomised trial of an autoantibody blood test followed by imaging. Eur Respir J 2020. doi 10.1183/13993003.00670-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biodesix. 2019 NODIFY XL2™ TEST. <https://www.biodesix.com/products/nodule-management/nodify-xl2>.
- 29.Li XJ, Hayward C, Fong PY, Dominguez M, Hunsucker SW, Lee LW, et al. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Science translational medicine 2013;5(207):207ra142 doi 10.1126/scitranslmed.3007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vachani A, Pass HI, Rom WN, Midthun DE, Edell ES, Laviolette M, et al. Validation of a multiprotein plasma classifier to identify benign lung nodules. J Thorac Oncol 2015;10(4):629–37 doi 10.1097/JTO.0000000000000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vachani A, Hammoud Z, Springmeyer S, Cohen N, Nguyen D, Williamson C, et al. Clinical Utility of a Plasma Protein Classifier for Indeterminate Lung Nodules. Lung 2015;193(6):1023–7 doi 10.1007/s00408-015-9800-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kearney P, Hunsucker SW, Li XJ, Porter A, Springmeyer S, Mazzone P. An integrated risk predictor for pulmonary nodules. PloS one 2017;12(5):e0177635 doi 10.1371/journal.pone.0177635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silvestri GA, Tanner NT, Kearney P, Vachani A, Massion PP, Porter A, et al. Assessment of Plasma Proteomics Biomarker’s Ability to Distinguish Benign From Malignant Lung Nodules: Results of the PANOPTIC (Pulmonary Nodule Plasma Proteomic Classifier) Trial. Chest 2018;154(3):491–500 doi 10.1016/j.chest.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadara H, Wistuba II. Field cancerization in non-small cell lung cancer: implications in disease pathogenesis. Proc Am Thorac Soc 2012;9(2):38–42 doi 10.1513/pats.201201-004MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steiling K, Ryan J, Brody JS, Spira A. The field of tissue injury in the lung and airway. Cancer Prev Res (Phila) 2008;1(6):396–403 doi 10.1158/1940-6207.CAPR-08-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proceedings of the National Academy of Sciences of the United States of America 2004;101(27):10143–8 doi 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nature medicine 2007;13(3):361–6 doi 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 38.Silvestri GA, Vachani A, Whitney D, Elashoff M, Porta Smith K, Ferguson JS, et al. A Bronchial Genomic Classifier for the Diagnostic Evaluation of Lung Cancer. N Engl J Med 2015;373(3):243–51 doi 10.1056/NEJMoa1504601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vachani A, Whitney DH, Parsons EC, Lenburg M, Ferguson JS, Silvestri GA, et al. Clinical Utility of a Bronchial Genomic Classifier in Patients With Suspected Lung Cancer. Chest 2016;150(1):210–8 doi 10.1016/j.chest.2016.02.636. [DOI] [PubMed] [Google Scholar]
- 40.Feller-Kopman D, Liu S, Geisler BP, DeCamp MM, Pietzsch JB. Cost-Effectiveness of a Bronchial Genomic Classifier for the Diagnostic Evaluation of Lung Cancer. J Thorac Oncol 2017;12(8):1223–32 doi 10.1016/j.jtho.2017.04.030. [DOI] [PubMed] [Google Scholar]
- 41.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015;351:h5527 doi 10.1136/bmj.h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Micheel CMNSJ, Omenn GS In: Micheel CM, Nass SJ, Omenn GS, editors. Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington (DC)2012. [PubMed] [Google Scholar]
- 43.Sin DD, Tammemagi CM, Lam S, Barnett MJ, Duan X, Tam A, et al. Pro-surfactant protein B as a biomarker for lung cancer prediction. J Clin Oncol 2013;31(36):4536–43 doi 10.1200/JCO.2013.50.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Integrative Analysis of Lung Cancer E, Risk Consortium for Early Detection of Lung C, Guida F, Sun N, Bantis LE, Muller DC, et al. Assessment of Lung Cancer Risk on the Basis of a Biomarker Panel of Circulating Proteins. JAMA Oncol 2018;4(10):e182078 doi 10.1001/jamaoncol.2018.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Begum S, Brait M, Dasgupta S, Ostrow KL, Zahurak M, Carvalho AL, et al. An epigenetic marker panel for detection of lung cancer using cell-free serum DNA. Clinical cancer research : an official journal of the American Association for Cancer Research 2011;17(13):4494–503 doi 10.1158/1078-0432.CCR-10-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ostrow KL, Hoque MO, Loyo M, Brait M, Greenberg A, Siegfried JM, et al. Molecular analysis of plasma DNA for the early detection of lung cancer by quantitative methylation-specific PCR. Clinical cancer research : an official journal of the American Association for Cancer Research 2010;16(13):3463–72 doi 10.1158/1078-0432.CCR-09-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ooki A, Maleki Z, Tsay JJ, Goparaju C, Brait M, Turaga N, et al. A Panel of Novel Detection and Prognostic Methylated DNA Markers in Primary Non-Small Cell Lung Cancer and Serum DNA. Clinical cancer research : an official journal of the American Association for Cancer Research 2017;23(22):7141–52 doi 10.1158/1078-0432.CCR-17-1222. [DOI] [PubMed] [Google Scholar]
- 48.Leal A, Sidransky D, Brait M. Tissue and Cell-Free DNA-Based Epigenomic Approaches for Cancer Detection. Clin Chem 2019. doi 10.1373/clinchem.2019.303594. [DOI] [PubMed] [Google Scholar]


