Abstract
Activating mutations in RAS GTPases drive many cancers, but limited understanding of less-studied RAS interactors, and of the specific roles of different RAS interactor paralogs, continues to limit target discovery. We developed a multistage discovery and screening process to systematically identify genes conferring RAS-related susceptibilities in lung adenocarcinoma. Using affinity purification mass spectrometry, we generated a protein-protein interaction map of RAS interactors and pathway components containing hundreds of interactions. From this network, we constructed a CRISPR dual knockout library targeting 119 RAS-related genes that we screened for KRAS-dependent genetic interactions (GIs). This approach identified new RAS effectors, including the adhesion controller RADIL and the endocytosis regulator RIN1, and >250 synthetic lethal GIs, including a potent KRAS-dependent interaction between RAP1GDS1 and RHOA. Many GIs link specific paralogs within and between gene families. These findings illustrate the power of multiomic approaches to uncover synthetic lethal combinations specific for hitherto untreatable cancer genotypes.
Keywords: KRAS, Interaction Mapping, Synthetic Lethality, RAP1GDS1, lung cancer
INTRODUCTION
RAS proteins are mutated in 20% of cancers, yet few targeted therapies are available in the clinic (1). Although RAS oncogenes were discovered over 35 years ago (2), the consequences of RAS activation remain only partly understood. K-, H-, and NRAS are small GTPases that cycle between GTP and GDP-bound states with the aid of exchange factor (GEF) and activating (GAP) proteins. While K-, H-, and NRAS are all known oncogenes, KRAS is the most frequently mutated, notably in pancreatic, colon and lung adenocarcinomas (LUAD) (1). Oncogenic mutants of RAS proteins have reduced GAP-mediated GTP hydrolysis (3) and strongly favor the GTP-bound active state in which they bind tumorigenic effectors. Some RAS effectors, like BRAF and PI(3) kinase, are themselves oncoproteins (3), and therefore are attractive therapeutic targets. RAS proteins have many other potentially important effectors and regulators with poorly understood function (3). Many of these factors are represented by multiple paralogs, which potentially compensate for one another in screening experiments, concealing their collective importance (4). The extent to which paralogous RAS effectors or regulators, such as the three RAF kinases, are functionally distinct remains incompletely understood. However, therapeutic inhibitors are often not paralog-selective and can modulate multiple paralogous targets through related binding sites. The ensemble of RAS protein effectors and regulators thus likely contains both new therapeutic targets and unexplored paralog-selective targets for unmet clinical needs (3).
The development of targeted therapies relies on the concept that genetic alterations in cancer cells cause pathway “rewiring,” making less-essential mutant and wild type genes more essential in neoplastic cells (5,6). Targeting such cancer-specific dependencies is a major strategy guiding efforts to treat RAS-driven cancers (3) and has been successful in other cancer types (7,8). While many cancer-specific susceptibilities are known, little systematic investigation of pathway rewiring has been performed. Genome-wide knockout screens (9–11) have identified new cancer pathway components, and patient sequencing initiatives (1) have identified candidate driver mutations. However, these searches for single-gene targets neither address pathway rewiring, nor the possibility of novel combination targets selective for the rewired cancer cell.
Here, we present the integrated results of two direct and orthogonal analyses of functional interactions in the RAS pathway (Figure 1A). First, we mapped detailed protein-protein interactions (PPIs) using highly-optimized affinity purification/mass spectrometry (AP/MS) (12). This approach enables unbiased detection of “prey” proteins co-purifying with chosen “bait” proteins. Beginning with KRAS itself, using GTP and GDP-locked (sometimes called apo) mutant forms as baits, we iteratively expanded the PPI network using preys discovered in earlier experiments as baits for later experiments, yielding a dense PPI network of RAS pathway interactions. Second, focusing on the genes identified in this PPI network and on known RAS pathway genes, we mapped genetic interactions (GIs) using CRISPR dual knockout (CDKO) technology (13). CDKO screens identify synergistic or buffering GIs through interference of knockout phenotypes of two genes. GIs can link pairs of redundant or interacting genes. GI screens can therefore reveal essential activities carried out by pairs of related genes that would be maintained if only one gene were lost, and so suggest new possible combination therapy strategies (13,14). From the identified pairwise links, we constructed networks that produce testable hypotheses regarding the biology of RAS-driven cancers. The complementary approaches of GI and PPI mapping clearly profile both known and new effectors and regulators of the major RAS oncoproteins. They also show distinctive patterns of PPIs and GIs between paralogous and non-paralogous proteins, identifying surprising new connections among RAS and other signaling pathways.
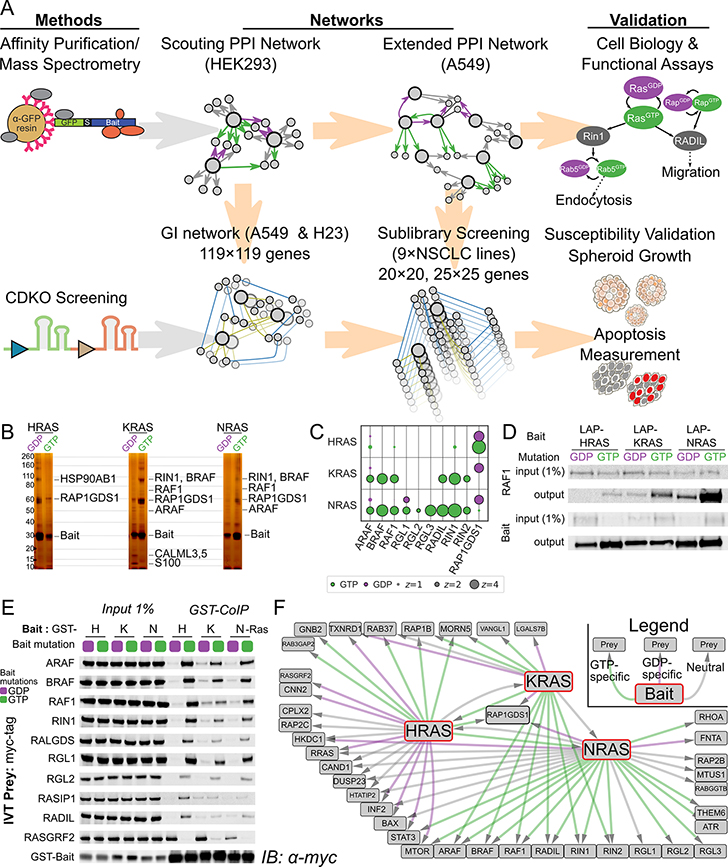
Figure 1. Multiomic Interaction Mapping Reveals Strong Selectivity of RAS Isoforms.
A Schematic outlining experimental approach. Top row: Proteins of known interest, expressed as GFP-tagged constructs, were used as bait for AP/MS to discover protein-protein interactions (PPIs). Proteins from the PPI network were selected for CRISPR Dual Knockout (CDKO) genetic interaction (GI) screening. sgRNA cassettes targeting these genes were transduced into Cas9-expressing KRAS-driven LUAD lines and assayed for enrichment or depletion after 21 days growth in culture. Relative depletion of single- and double-targeting cassettes was used to establish synergy or buffering between gene knockout phenotypes. GIs that could form the basis of new therapies were validated by both CDKO and AP/MS in other LUAD lines, as well as functional and growth assays. B Silver stain PAGE of tandem affinity purification of H-, K-, and NRAS from A549 cells shows distinct GTP-specific PPIs. Known effectors are marked. C Bubble plot of 6 AP/MS experiments with GTP- and GDP-locked mutant GTPases as baits (rows), showing the enrichment of selected preys (columns). Dark borders indicate FDR < 0.05. D Western blot of co-immunoprecipitation reactions from A549 cells expressing tagged H, K, or NRAS with mutations locking them in their GTP or GDP-bound forms, probing for endogenous RAF1. HRAS binds significantly less RAF1 than does either other RAS. E Cell-free co-immunoprecipitation of selected prey proteins by recombinant purified mutant H, K, and NRAS, imaged by immunoblot. Negligible RAS isoform specificity is observed. F Network diagram showing PPIs of RAS proteins and selected prey proteins. Green and purple lines indicate PPIs from GTP- and GDP-locked preys respectively; gray lines indicate that the PPI is not nucleotide-state specific. All diagrammed interactions have FDR < 0.05 (see Methods)
The combined GI and PPI networks connect genes important for KRAS-driven LUAD and together provide a physical and functional “wiring diagram” linking drivers of RAS cancer biology. This combination mapping approach provides a wealth of testable candidates for new combination therapies and demonstrates a generalizable method for mapping cell biological pathways altered by disease.
RESULTS
Mapping the RAS Pathway through Physical and Genetic Interactions: an Outline
Interaction maps were assembled by an iterative process. First, we performed AP/MS of KRAS using mutations that confine it to a GTP-locked (3) or GDP-locked (or apo) state (15)(Methods, Supplementary Table 1). Using high-efficiency tandem affinity purification (12,16) in HEK293 cells, we isolated each bait with co-purifying proteins, which were identified by mass spectrometry and quantified as described (17). We expanded the network by repeating the procedure using 5 other RAS pathway proteins (RALA, RALB, RALGDS, RAP1B, and RAP2B) as baits, creating a network of 451 proteins including both known and unknown RAS pathway members and incorporating public PPI data (Supplementary Table 2, Figure S1A, S1B).
We selected the most confidently identified proteins from this network and cross-referenced them with public data sets to select 119 genes for GI analysis by CDKO in two KRAS-mutant LUAD cell lines (A549 and H23). This analysis of 119 genes (7021 pairs) revealed hundreds of mostly novel GIs. We then repeated and expanded the PPI mapping experiments in A549 cells with 14 baits, including most members of the RAL and RAP GTPase families (KRAS, NRAS, HRAS, RALA/B, RAP1A/B, RAP2A/B, RALGDS, RGL1, RGL2, RADIL, and RASGRF2). This produced a network of 930 proteins, to which we added relevant published interaction data (Supplementary Tables 2 and 3, Figure S2A). Informed by the results of both PPI maps and the initial GI maps, we created two smaller 20×20- and 25×25-gene CDKO libraries to screen a set of 9 LUAD lines (see Methods). These cell lines were found to have a range of dependencies on KRAS, allowing us to stratify GIs into classes positively or negatively associated with KRAS activity. Here we describe new insights into KRAS-driven biology derived from this multi-omic strategy, including the identification of novel potential combinatorial susceptibilities (Figure 1A, S2B).
Affinity Purification/Mass Spectrometry Mapping Reveals Strong Selectivity of RAS Isoforms
Oncogenic forms of RAS proteins cause transformation in specific tumors through enhanced binding to downstream effectors (3). Different RAS isoforms can selectively bind distinct effectors, yet no systematic comparison of RAS PPIs with their regulators and effectors has been performed. From AP/MS experiments in HEK293 cells, we identified efficient interactors of KRAS, both well-known (RAF1, BRAF, ARAF) and less studied (RALGEFs RALGDS, RGL1, RGL2, RGL3, RAP1GDS1, RADIL) (Figure S1B).
As a comparison, we conducted copurifications with GTP- and GDP-locked mutants of H-, K-, and NRAS stably expressed in A549 cells, a KRAS-transformed LUAD line. RAS effector proteins with known preference for the GTP-locked RAS state bound specifically to that mutant form. We observed striking differences in co-purifying proteins between each RAS protein, and between their mutant forms, as apparent by silver stain (Figure 1B) and by mass spectrometry (Figure 1C, S2A,C). Surprisingly, there were notable differences among the specific effectors copurified by activated H-, K-, and NRAS (Figure 1C). The HRAS interactome differed most strongly from the other two. Most strikingly, RAF kinases (ARAF, BRAF, RAF1) were highly enriched by either GTP-locked K- or NRAS baits but not HRAS. We analyzed small-scale coimmunoprecipitation experiments in lung cancer cells by immunoblot (Figure 1D) and confirmed that RAF1 bound GTP-locked HRAS much less efficiently than GTP-locked K- or NRAS, even with similar expression of RAS bait proteins. The same was true for three other RAS effectors containing canonical RAS-association (RA) domains: RIN1, RIN2, and RADIL. In contrast, three members of the RALGEF family – RGL1, −2, and −3 – that also contain RA domains copurified efficiently with NRAS but poorly with H- or KRAS (Figure 1C).
We considered whether observed binding specificities reflected protein-intrinsic properties that could be recapitulated in cell-free assays. Using purified GTP- and GDP-locked H-, K-, and NRAS proteins, we reconstituted binding to in vitro-translated forms of the known and new RAS-interacting proteins identified (Figure 1E, S2D). Although all RAS-effector interactions were strongly GTP-dependent, we found negligible differences in binding efficiency between the three GTP-locked RAS proteins and these RAS effector proteins in vitro. This and experiments below indicate that other cellular processes (such as regulation of localization, protein signaling and modifications or effector availability) are comparably important to RAS nucleotide state in determining the efficiency of RAS-effector engagement in tumor cells.
A number of known RAS-binding proteins were not identified by our AP/MS experiments. PPIs between GTPases and their coordinate GEFs have been historically difficult to isolate by AP/MS (see (18)), so the absence of signal from GEFs like SOS in these purifications was expected. The absence of other RAS-binding proteins, like PIK(3)-CA or TIAM1, requires further study. These PPIs may only occur in specific cell lines or under stimuli not supplied by our culture method, or are simply inefficient without overexpression of preys.
It is worth noting that not all interactions efficiently observed by AP/MS are necessarily direct, since our large-scale purifications often identify complexes and secondary interactions (see (16)). Combining our AP/MS data with data from public PPI databases including BioGRID (19), we constructed a PPI network of RAS interactors (Figure S2C). A selective representation of this network is shown in Figure 1F.
AP/MS Identifies New KRAS Effectors Underlying RAS-Driven Macropinocytosis and Migration
Interactions between RAS family proteins and effectors are mediated by the effector binding domain of RAS proteins and RA domains in target proteins (20). In addition to the effectors of KRAS mentioned above, we found two proteins that co-purified efficiently with both GTP-locked K- and NRAS that contain canonical RA domains: RIN1 and RADIL (Figure 2A).
Figure 2. AP/MS Identifies New KRAS Effectors Underlying RAS-Driven Macropinocytosis and Migration.
A Network diagram showing PPIs between RAS proteins, RIN1 and RADIL. B Schematic of assay to assess macropinocytosis (MPC), derived from (25). C MPC was quantified by TMR-dextran (red) signal under fluorescence microscopy. RAS inhibition reduced dextran uptake in wild-type, but not RIN1 knockout cells. Error bars indicate 95% confidence intervals estimated from 76 fields from the wild type and 226 from knockouts (Supplementary Methods). * : p<0.05 by two-sided Student’s t-test D Left: Superposition of crystal structures of the RA domain of RAF1 (PDB ID: 4G0N, brown) and RADIL (PDB ID: 3EC8, gray). RADIL R99 superimposes near RAF1 R89, which is required for RAS-RAF binding (20). E Cell-free co-immunoprecipitations indicated myc-tagged constructs as prey, and indicated GST-RAS fusion proteins as bait. myc-RADIL binds RAS directly, and its RA domain (residues 61–164) is sufficient. R99 is required. F Quantitative Real-Time PCR of 3 indicated siRNAs against RADIL (see Supplementary Methods). Total RNA was isolated from transfected H23 cells at 24, 48, and 72hrs post-transfection and the relative levels of RADIL transcript were quantified. For experiments with RADIL knockdown, a 1:1:1 pool of all three siRNAs were used. n=4 for all samples. G Schematic of scratch-wound assay. Cells transfected with anti-RADIL siRNA or a negative control were replated into a confluent monolayer. A wound was introduced in the monolayer, and the closure of the wound was monitored over time. H Quantification of cellular velocity into the wound over 24 hours; RADIL knockdown increased migration speed, but not when KRAS was inhibited. 5–6 wounds were quantified per condition, imaged hourly (Supplementary Methods). * p<0.05 by two-sided Student’s t-test.
RIN1 has been shown to bind HRAS in vitro (21), and functions as a nucleotide exchange factor for RAB5 GTPases, which are central organizers of endocytosis (22). Recent work shows KRAS-transformed cancers use macropinocytosis, a specific form of endocytosis, to scavenge nutrients from their environment (23,24). The identification of RAS-RIN PPIs suggested that KRAS might directly regulate macropinocytosis through RIN1. We assayed macropinocytosis as described previously (25). Serum-starved H23 cells take up 70kDa fluorescent dextran by macropinocytosis, as visualized by fluorescence microscopy (Figure 2B). Three clonal CRISPR RIN1 knockout lines (Figure S3A) showed decreased macropinocytosis relative to cells transduced with a negative control sgRNA. Upon pretreatment with ARS853, a KRASG12C-specific inhibitor (26), macropinocytosis was substantially suppressed in the control cells but not in the RIN1 knockout cells. These cells appear insensitive to further inhibition of macropinocytosis by ARS853, consistent with the model that RAS-driven macropinocytosis is substantially coupled to RIN1 activity (Figure 2C).
RADIL, which has not previously been shown to bind KRAS, also strongly co-purified specifically with GTP-locked KRAS in both A549 and HEK293 lines. The N-terminal RA domain in RADIL resembles that of other RAS effectors (Figure S3B) and has been crystallized (27). Arginine 99 (R99) in RADIL occupies a position in the RA domain comparable to RAF1 R89 (Figure 2D), a site required for RAS interaction (20). We find that RADIL specifically and efficiently binds in vitro to GTP-locked H-, K-, and NRAS, that the RA domain is sufficient for this PPI and that the R99 mutant fails to bind KRAS-GTP (Figure 2E, S3C). Previous studies of RADIL focus on its role in RAP1A-driven cell adhesion (28,29), and our results suggest a direct link between KRAS and RADIL, at least in LUAD. To validate that RADIL also affects adhesion and motility in these cells, we conducted a scratch-wound assay (30) and found that RADIL knockdown increased the migration of these cells, in agreement with other studies (28). However, RADIL knockdown did not increase the migration rate of cells treated with ARS853, consistent with a mechanism where RADIL-regulated cell migration is enhanced by activated KRAS (Figure 2F-H).
Systematic Mapping of the RAS/RAP/RAL Signaling Pathway Shows a Network of Interconnected GTPases
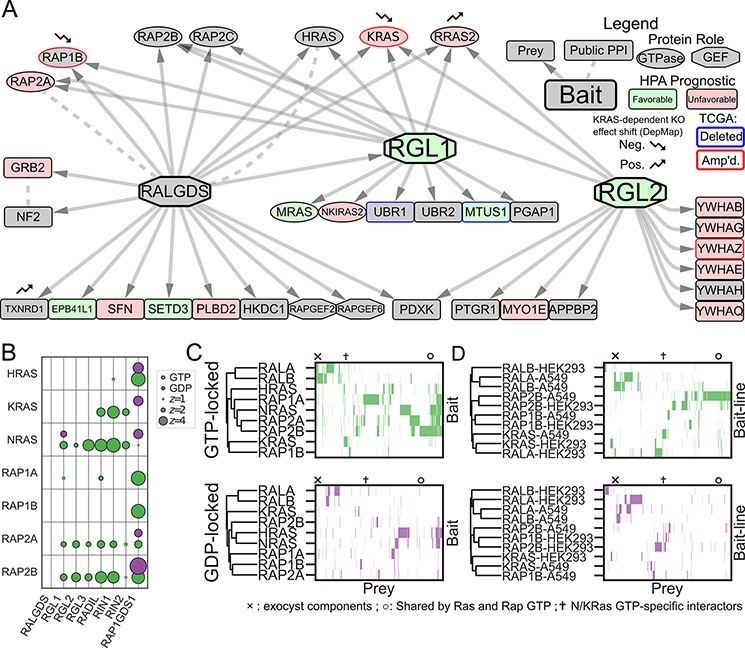
We identified strong GTP-dependent PPIs between KRAS and the RALGEFs RALGDS, RGL1, and RGL2 in HEK293 cells. While RALGEFs and their RAL GTPase substrates are linked to cell proliferation and carcinogenesis (31–33), they remain incompletely understood. RALGEFs also bind RAP GTPases and different family members have distinct but unknown RAL-independent activities (33,34). Reasoning that an unbiased survey of RALGEF PPIs would provide important clues to their function, we conducted AP/MS experiments with RALGDS, RGL1, and RGL2 as bait proteins. Each RALGEF co-purified a wide variety of upstream GTPases, including RAS and RAP family members. Up-regulation of many of these proteins is associated with unfavorable LUAD prognosis, and the genes that encode them include conditional dependencies of KRAS-dependent and independent cancers (Figure 3A; see Supplementary Methods). Here we identified other interactors unique to each RALGEF, implicating them in signaling pathways including the Hippo (NF2) and NF-κB pathways (NKIRAS2).
Figure 3. Systematic Mapping of the RAS/RAP/RAL Signaling Pathway Shows a Network of Interconnected GTPases.
A Network diagram of PPIs in A549 cells with RALGEF family members as baits. The RALGEFs differ in their PPIs, but collectively link numerous genes involved in signaling transduction and LUAD progression. Nodes are filled in green or red by their Human Protein Atlas (HPA) prognostic category, which is associated with the mRNA expression of the corresponding gene. Nodes are outlined in blue or red if their encoding genes are unusually up- or down-regulated by copy number alteration in LUAD (see Supplementary Methods). Nodes marked with small arrows show significant changes in knockout effect between KRAS-dependent and independent DepMap (11) cell lines. B Bubble plot of 14 AP/MS experiments with GTP- and GDP-locked mutant GTPases as baits (rows), showing the enrichment of selected preys (columns). RAP2A and RAP2B bind some canonical RAS effectors in their GTP-locked states. C Clustermap of prey proteins significantly enriched (FDR<0.05; see Methods) in each experiment with the indicated baits in A549 cells. Prey protein order is preserved across panels C and D, but prey proteins not significantly enriched in at least one experiment were removed for simplicity. Symbols indicate columns in the clustermap corresponding to proteins of interest: × Exocyst components; ✝N/KRAS-GTP specific interactors; ○ Common interactors of K- and NRAS and of RAP2. D Clustermaps as in C but comparing GTPases that were used as bait proteins for AP/MS experiments in both A549 and HEK293 cells.
To better understand RALGEF activity-specific interactions and effects, we conducted AP/MS experiments with the downstream RAL (RALA, RALB) and linked RAP (RAP1A, RAP1B, RAP2A, RAP2B) GTPases in their GTP- and GDP-locked forms. Like RAS proteins, RAP proteins engage effectors, including the RALGDS family, through effector RA domains. A comparison of common PPI partners of RAS and RAP family members in A549 cells is shown in Figure 3B, indicating that RADIL, RALGEF and RIN-family effectors are preferentially engaged by NRAS over H- or KRAS, and RAP2A and -B over RAP1 isoforms. Clustering analysis of RAS, RAP, and RAL GTPases showed that RAS and RAP GTPases bind surprisingly similar effectors including RALGEFS, RIN1/2 and RADIL (marked ○) (Figure 3C). In contrast, the RALA and -B GTPases show a distinct set of interactors including all components of the exocyst complex (marked ×). A subset of AP/MS experiments were conducted in both A549 and HEK293 cells. An analogous clustering analysis (Figure 3D) shows that, generally, each GTPase interacted with a similar set of proteins in both lines. Most strikingly, clustering analysis does not distinguish RAS from RAP proteins, indicating an underappreciated intersection between RAS and RAP interactions and signaling. Both cooperative and antagonistic relationships between oncogenic RAS GTPase and the families of RAP GTPases are possible. Our genetic interactions, presented below, shed some light on this phenomenon.
These PPI maps show the benefit of systematic mapping of paralogs and similar families. The many specific PPIs of RAP, RAL, and RALGEF family proteins suggest the existence of an elaborate network of dynamic interactions controlling signaling and trafficking at plasma and organellar membranes. Curated subsets of these PPIs are diagrammed in Figures S1B and S2A. Complete PPI data can be found in Supplementary Table 2, and online via NDEX. Importantly, these PPIs predict and rationalize many genetic interactions, as we validate below.
CRISPR Dual Knockout Screens of KRAS Interactome Components Reveal Functional Relations and Potential Susceptibilities
Having established that this high-confidence PPI network contains many uncharacterized but potentially important interactors, we next sought to map genetic interactions (GIs) between them to better understand the RAS pathway and to identify combination target pairs. To map GIs, we used CRISPR Dual Knockout (CDKO) screening (13), in which genes or pairs of genes are systematically knocked out by a CRISPR library transduction approach. Because the number of knockout combinations scales geometrically with the number of targets, CDKO screens are limited to a smaller target gene set than single-target screens.
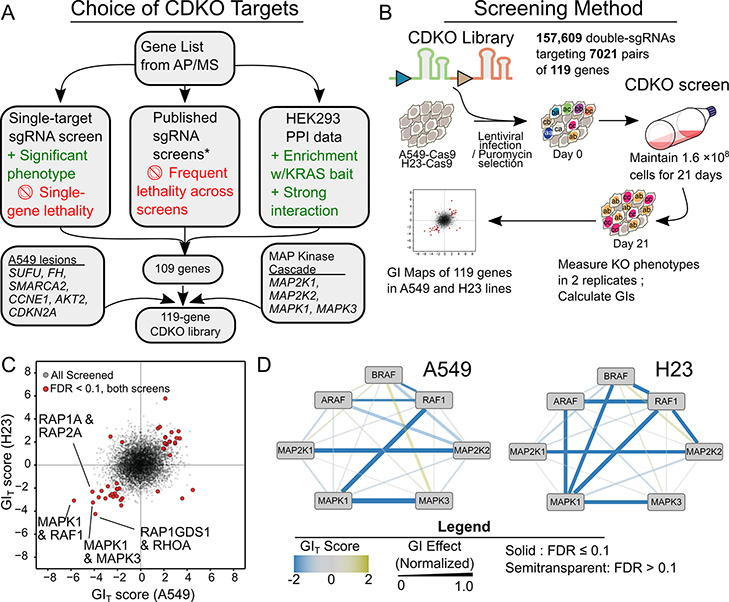
Beginning with the genes encoding proteins in our HEK293 PPI network, we sought to identify the ones most likely to show genetic interactions. We first constructed a single-sgRNA library targeting 223 genes based on their protein identification in AP/MS experiments (Figure S1). The library contained 10 sgRNAs per gene and was transduced into A549-Cas9 cells. Cells were grown for 21 days and DNA isolated and sequenced to identify depleted or enriched sgRNAs (Figure S4A, B, Supplementary Table 4). This screen identified some essential genes, which would be less likely to show GIs and thus were removed from further consideration. Similarly, we removed genes previously shown to be essential in a majority of cancer cell lines (9,10,35). Finally, we prioritized the remaining genes by the strength of their interactions in our AP/MS data, and whether that interaction involved KRAS. Of these, we selected 109 genes. We then added genes encoding MEK and ERK kinases, and others reflecting genetic lesions in A549 cells for a total of 119 genes (Figure 4A, Supplementary Table 5).
Figure 4. CRISPR Dual Knockout Screens of KRAS Interactome Components Reveal Functional Relations and Potential Susceptibilities.
A Schematic illustrating gene selection for the CDKO (CRISPR Dual Knockout) screen. Genes involved in strong PPIs, especially with KRAS, were prioritized, while genes producing weak or outright lethal phenotypes were discarded. *Studies used were (9,10,35). B Schematic showing CDKO screening. Cas9-expressing LUAD cells are transduced with pairs of sgRNAs. Cells are sequenced before and 21 days after transduction. Relative sgRNA abundances are used to calculate their effects. C Scatterplot showing GIs in the A549 and H23 lines. Negative, “synergistic,” phenotypes are more severe than would be expected from the two single-gene effects in combination. D GIs between components of the MAPK pathway in A549 (left) cells and H23 (right) cells differ. All GIs have FDR<0.1 by Mann-Whitney U test followed by Benjamini-Hochberg correction.
Next, we constructed a CDKO library including 3 sgRNAs per gene (13), enabling systematic deletion of 7,021 gene pairs. We included 60 “safe” sgRNAs targeting nonfunctional genomic loci, previously validated to have minimal effects on cell growth (13), to control for intrinsic effects of Cas9 activity and to allow measurement of single gene deletion phenotypes. The library was transduced into two Cas9-expressing KRAS-mutant LUAD lines, A549 and H23. The abundance of each guide pair was measured before and after the 21-day growth period (Figure 4B). We quantified GIs by comparing the abundance of targeting to safe guide pairs and double-targeting guide pairs (Figure S4C, D, Supplementary Table 5, Methods), as described previously (13).
We identified 548 GIs with FDR<0.1 in A549 cells, and 447 in H23 cells, of which 59 overlapped (Figure 4C, Methods). The consistently interacting pairs included paralogs such as RAP1A with RAP2A or genes in the same pathway, such as MAPK1 with RAF1. Many of the strongest synergistic GIs in either line were found between paralogous MAPK pathway components (e.g. MAPK1 with MAPK3, MAP2K2 with MAP2K4). The observed strong dependencies on this known central pathway confirms that the screening captured relevant GIs. By combining GI and PPI data, we found that roughly one in six GIs paralleled direct (first-degree) PPIs, while many more GI partner pairs are connected to second or third-degree PPIs (Figure S5A).
Representative GI/PPI subnetworks, showing GIs common to both cell lines tested, are diagrammed in Supplementary Figure S5B. These combined maps provide testable hypotheses about mechanisms underlying observed GIs. For example, a synergistic GI links the PI(3) kinase subunit PIK3R1 and RAF1. Both are constituents of RAS effector pathways, and this GI is supported by pharmacological combinations (36).
GI strength is quantified by GIT score, which describes the degree of synergy of a given GI, and accounts for assay noise (13). As one strong example, KRAS-mutant H23 cells infected with the safe/safe, RAP1GDS1/safe, and safe/RHOA sgRNA cassettes doubled once every 51, 53 and 64 hours, respectively. Those infected with a double-targeting guide doubled only every 120 hours (not shown). These changes in doubling time correspond to a GIT score of −4.2. Strong GIT scores may therefore indicate dramatic changes in fitness. The efficacy of this combination is confirmed in multiple assay systems, as we show below.
We included the MAPK pathway genes MAP2K1, MAP2K2, MAPK1, and MAPK3 in the CDKO library as positive controls because of known synthetic lethal GIs among these genes (37). As predicted, we observed strong synthetic lethal GIs between these paralogous pairs. Combined knockout of MAPK1 and RAF1 was also strongly synergistic, but surprisingly, knockout of RAF1 and MAPK3 was much less so. In this screen and screens below, the specific link from MAPK1 to RAF1 is consistently seen, suggesting that MAPK1 (also called ERK2) has specific link to RAF1 and therefore different regulatory properties from MAPK3 (or ERK1). A549 cells showed strong dependency on RAF1 in combination with other pathway members, but limited dependency on ARAF or BRAF. In H23 cells, however, GIs could be observed between all three RAF genes and other MAPK pathway components (Figure 4D). The seven MAPK genes showed numerous but highly varied GIs with other genes in these two cell lines (Figure S5C). This suggests that the MAPK pathway signals to other RAS effector pathways, but does so differently in different tumor lines. Therefore, paralog-selective inhibitors of ERK or RAF kinases might have more specific anti-tumor activity in certain patients.
Although the presence of synergistic GIs common to both lines showed that CDKO screening could uncover consistent combination susceptibilities, the preponderance of cell line-specific GIs suggested that screening additional cell lines would reveal additional tumor-selective patterns of susceptibility, especially with regard to KRAS dependence.
GI Mapping Identifies Exploitable Synergistic Interactions in RAS-Driven Lung Adenocarcinoma
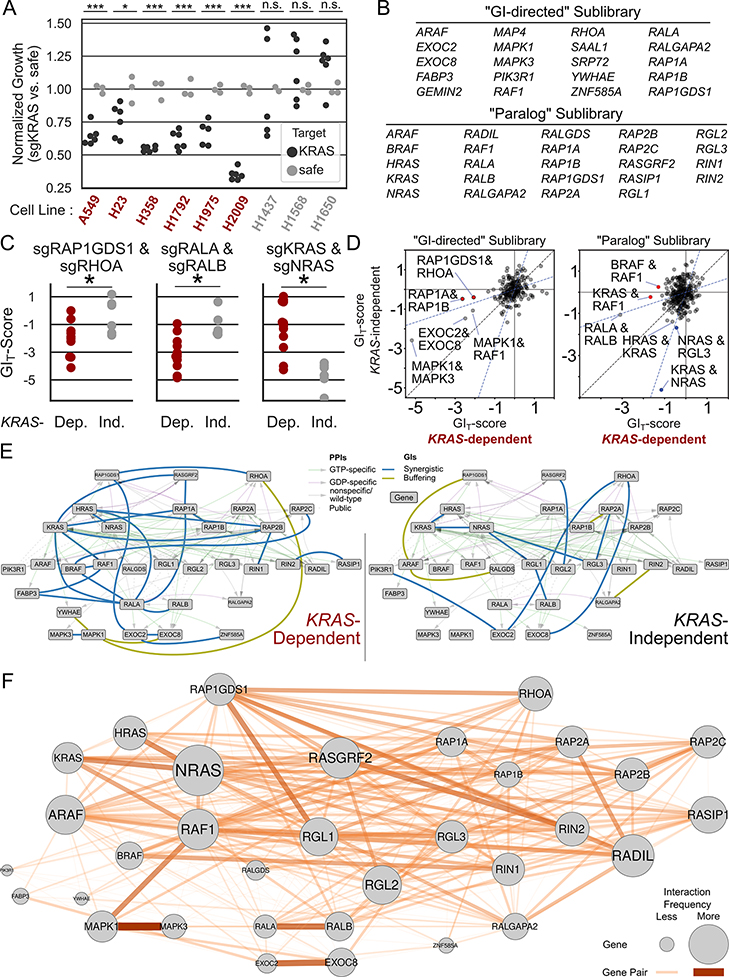
To identify GIs specific to KRAS-driven LUAD, we first assayed the KRAS dependence of 9 LUAD lines by measuring their growth with and without CRISPR/Cas9-mediated KRAS knockout. We found that six lines were highly KRAS-dependent, and three were KRAS-independent (Figure 5A, S6A). Of the six KRAS-dependent lines, five had codon 12 mutations in KRAS and the one that did not (H1975) had activating mutations in EGFR. None of the KRAS-independent lines had RAS mutations (Figure S6B, C).
Figure 5. GI Mapping Identifies Exploitable Synergistic Interactions in RAS-Driven Lung Adenocarcinoma.
A Nine engineered LUAD lines stably expressing Cas9 were assayed for KRAS dependence. Lines were stratified into KRAS-dependent (labeled red) and KRAS-independent (gray) cohorts. *: p<0.05 ***: p<0.001 by one-sided t test. B Two smaller “sublibraries” were screened for GIs in the lines identified in panel A. One sublibrary (“GI-Directed”) targeted the strongest interactors from the first screens, and the other (“paralog”) was targeted families of RAS-interacting proteins. C GIs were identified with different patterns of interaction in the KRAS-dependent (red; n=12) or –independent (gray; n=6) cohorts. The GIT scores of each GI across individual replicates are rendered as box plots. *: p<0.05 by Mann-Whitney U test. D Scatter plot of mean GIT score for each gene pair in KRAS-dependent (x axis) and –independent cohorts (y axis) in both sublibraries. Noteworthy gene pairs labeled. E Combined PPI/GI networks. GIs enriched to one cohort or another (determined by Mann-Whitney U test) are plotted in each network. F Frequency of GI observation by gene and gene pair. Larger genes (circles) had a greater frequency of observed interaction; wider and more opaque edges correspond to a greater frequency of observed interactions. Frequencies are calculated as (# observed interactions with FDR < 0.1)/(# of interactions possible to observe).
Therapeutically useful combination target pairs should show synthetic lethal synergistic GIs in KRAS-dependent, but not KRAS-independent lines. To identify such pairs, we constructed two smaller CDKO libraries and conducted focused screens across the cell line panel. The first, “GI-Directed” library, targeted 20 genes with strong interactions identified in the first CDKO screen described above (Figure 5B, Supplementary Tables 6 and 7). The second, “paralog” library, targeted paralogous families of RAS regulator and effector proteins, focusing on those found in the A549 PPI network. The screens were conducted with both libraries in all 9 lines following the protocol described above. In both screens we found multiple examples of gene pairs whose synergy was greater in the KRAS-dependent than -independent lines. Among the strongest hits in the “GI-Directed” library was RAP1GDS1 and RHOA (Figure 5C, left), which had shown a strong synthetic lethal GI in the two earlier larger screens. RAP1GDS1 is a GEF for RHOA and RHOC, and also regulates RAS and RAP GTPases (38,39). We will discuss this interaction extensively in the next section.
One of the strongest synthetic lethal pairs specific to KRAS-dependent lines in the “paralog” library was RALA/RALB (Figure 5C, middle). This pair previously showed synthetic lethality in a genetically engineered mouse model (37). As shown in Figure 3A, the immediate upstream regulators of RAL GTPases, the RALGEFs, link a network of proteins important for signal transduction in a variety of important pathways, as well as a variety of KRAS-related dependencies and prognostic markers.
In contrast to the RAP1GDS1/RHOA and RALA/B pairs, KRAS/NRAS double knockout strongly synergized specifically in KRAS-independent lines (Figure 5C, right). This observation suggests that while KRAS-dependent cells are “addicted” specifically to KRAS, KRAS-independent cells may rely jointly on normal K- and NRAS proteins to accomplish tumor-driving signaling functions.
Many gene pairs had a greater tendency toward genetic interaction in either KRAS-dependent or -independent lines. Some GIs common to both A549 and H23 cells in the original screen, such as MAPK1/MAPK3, and MAPK1/RAF1, were recapitulated in the sublibrary screens (Figure 5D).
GIs specific to either the KRAS-dependent or -independent cohort, and the PPI networks linking them, are plotted in Figure 5E. While few synthetic lethal pairs involving exactly one MAPK pathway component were consistently more severe across the KRAS-dependent lines, there were significantly more such GIs in the KRAS-dependent than -independent cohorts (Figure S6D). Pairs of paralogous genes were strongly enriched for GIs in both the KRAS-dependent and independent cohorts (Figure S6E). Certain GI pairs, involving many different pathway members, were associated with non-KRAS mutations (Figure S6F).
GI targets and interrogated gene pairs are diagrammed in Figure 5F, with nodes and edges scaled by the frequency of observed interactions across all cell lines. Interestingly, we found that RIN1 and −2, RADIL, and RALGEF genes, which encode central members of our PPI network, were among the most frequent interacting partners across all nine cell lines, regardless of their KRAS-dependence, pointing to their central importance in RAS and RAP signaling. All GIs with FDR < 0.1 observed in all 9 cell lines and two libraries are diagrammed in Supplementary Figure S7.
RAP1GDS1 and RHOA form a Synthetic Lethal Gene Pair in RAS-Driven Lung Adenocarcinoma
One of the strongest interactions we observed in KRAS-dependent cells was a synthetic lethal GI between RAP1GDS1 and RHOA (Figure 5C, left). RHOA encodes a small GTPase critical for actin filament assembly and membrane organization (40). RAP1GDS1 (SmgGDS) has been described as a GEF for RHOA and RHOC (38,39). RAP1GDS1 differs structurally from canonical RHO and RAS GEFs (41) and may also act as a chaperone for RHO, RAS, RAP and other families of GTPases (41–43), potentially facilitating their prenylation and delivery to the membrane. RAS-driven oncogenesis also depends on RHO proteins (11). The CDKO screens point to specific GIs among RAP1GDS1, RHOA and mutant KRAS not previously reported that raise the possibility of targeting RHOGEFs like RAP1GDS1 for cancer therapies.
To validate the specificity of this GI for KRAS-dependent cell lines, we used CRISPR/Cas9 to knockout RHOA, RAP1GDS1, or both genes together in a set of KRAS-dependent and –independent LUAD cell lines and in human bronchial epithelial cells (HBECs) (Figure S8A). Double knockout of RAP1GDS1 and RHOA selectively impaired proliferation in three KRAS-dependent LUAD lines but showed minimal synergy in two KRAS-independent lines (Figure 6A, S8B). Notably, the RAP1GDS1/RHOA double knockout is comparably deleterious to KRAS knockout in the KRAS-dependent lines. Neither single-gene knockout had a strong effect on proliferation, indicating that only the loss of both genes in combination confers a vulnerability in KRAS-dependent cells. Analysis of apoptosis by flow cytometry revealed that double knockout induced cell death only in KRAS-dependent cells (Figure 6B, C).
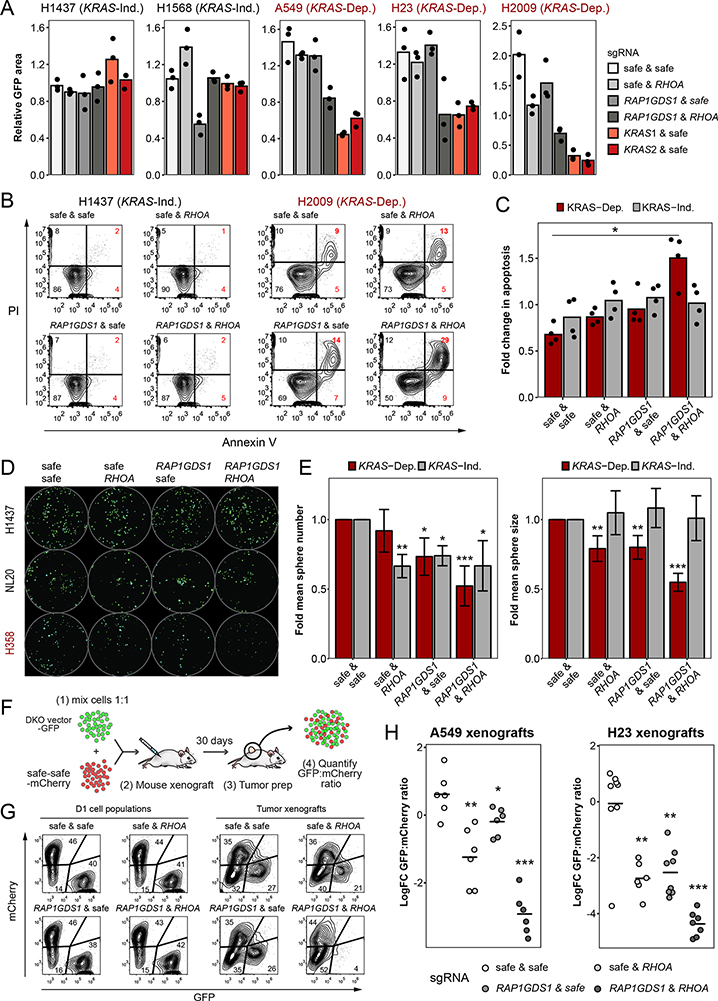
Figure 6. RAP1GDS1 and RHOA form a Synthetic Lethal Gene Pair in RAS-Driven Lung Adenocarcinoma.
A. Combined loss of RAP1GDS1 and RHOA leads to decreased proliferation in KRAS-dependent cell lines (H23, H2009, A549) but not KRAS-independent cell lines (H1437, H1568). Proliferation of GFP-positive cells was monitored using IncuCyte. Area of GFP-positive cells at the endpoint is shown relative to T0. B Flow cytometry of KRAS-dependent (H2009) and KRAS-independent (H1437) cells stained with Annexin V-APC and propidium iodide (PI). C Quantification of apoptosis in a series of cell lines. Red bars – KRAS-dependent cells (H2009, H358, H23), grey bars – KRAS-independent cells and human bronchial epithelial cells (HBECs) (H1568, H1437, NL20). Bars represent means, *: FDR<0.05 by two-sided t-test with Benjamini-Hochberg correction between the tested sample and safe-safe control. D Images of GFP-expressing spheroids in two LUAD cell lines (H1437 and H358) and one HBEC cell line (NL20) expressing Cas9 and the indicated sgRNA combinations. E Quantification of sphere number (left) and size (right) in KRAS-dependent (H23, H358, H2009) and KRAS-independent (H1568, H1437, NL20) cell lines. Graph represents means of 9 independent experiments ± standard deviation, *: FDR<0.05, **: <0.01, ***: <0.001 by two-sided t-test with Benjamini-Hochberg correction between the tested sample and the corresponding safe-safe control. F Schematic for the in vivo competitive-growth assay. G Flow cytometry of day 1 A549 cell populations (left) and representative endpoint tumor cell populations (right). H Log-fold change in GFP/mCherry ratios between A549 and H23 xenograft tumor cell populations at endpoint and day 1 cell populations. Lines represent means, *: FDR<0.05, **: <0.01, ***: <0.001 by two-sided t-test with Benjamini-Hochberg correction.
As we and others have shown previously, growth in spheroid culture reproduces important aspects of cancer biology not fully recapitulated by 2D cultures (44). To further validate the importance of RAP1GDS1/RHOA synthetic lethality in KRAS-dependent cells, we tested the susceptibility of LUAD cell lines to dual RAP1GDS1 and RHOA knockout in 3D culture. Individual knock-out of RAP1GDS1 or RHOA had a modest and variable effect on sphere number and size in KRAS-dependent and KRAS-independent cells (Figure 6D, E). Double knockout significantly decreased both sphere number and sphere size, but only in KRAS-dependent cells. Notably, the double knockout had a minimal effect on untransformed HBEC cells (Figure 6D, S8C).
Since RHO Kinases (ROCKs) are key effectors of RHOA function (45,46) and pharmacological inhibitors are available, we combined ROCK inhibition with RAP1GDS1 knockout to determine if this would phenocopy dual loss of RHOA and RAP1GDS1. Treatment of two LUAD cell lines with different doses of the ROCK inhibitor Fasudil (50, 100 and 150 μM) resulted in a dose-dependent decrease in sphere growth. Fasudil treatment of RAP1GDS1 knockout cells decreased sphere growth in KRAS-dependent H23 cells comparably to RHOA knockout alone (Figure S8D, E). However, we also observed decreased sphere growth from Fasudil treatment in RHOA knockout and RAP1GDS1/RHOA double-knockout cells, suggesting that ROCK is not a strong determinant of the synergy of RAP1GDS1/RHOA double knockout. Additionally, we cannot exclude the possibility that ROCK inhibitors may affect targets beyond ROCK1/2. In contrast to H23 cells, in KRAS-independent H1437 cells Fasudil treatment had no further effects on growth in any of the knock-out conditions.
Finally, to validate this synthetic lethal interaction in vivo, we performed competitive growth assays using xenografts of two KRAS-dependent LUAD cell lines (Figure 6F). We injected NSG mice subcutaneously with cells transduced with a safe/safe construct expressing mCherry mixed 1:1 with one of four DKO constructs expressing GFP: safe/safe, safe/RHOA, RAP1GDS1/safe, or RAP1GDS1/RHOA. Tumors were allowed to develop for 30 days, after which the mice were sacrificed and tumor cell suspensions analyzed by flow cytometry to quantify the ratio of GFP-positive knockout to mCherry-positive control cells. We found that in RAP1GDS1/RHOA tumors of both cell lines the GFP-positive population was barely detectable, confirming that the double knock-out of RAP1GDS1 and RHOA is highly deleterious in vivo (Figure 6G, H).
Taken together, these studies identify a susceptibility of KRAS mutant LUAD cells to combined suppression of both RAP1GDS1 and RHOA. Further evaluation of the mechanistic basis for this interaction may provide a new strategy for targeting KRAS-dependent cancer.
DISCUSSION
The Complex Drivers of KRAS-Mutant Cancers Motivate a Multiomic Mapping Approach
Genetic mutations cause pathway rewiring in cancer cells that both drives pathological behaviors and introduces susceptibilities that can be exploited in the clinic. Understanding this rewiring requires both knowledge of the molecules involved and the organization of interactions between them. Large-scale tumor profiling efforts (1) and single gene CRISPR and RNAi screening efforts (9–11) have both made valuable progress in assembling a “parts list” of cancer pathways. However, these methods have limited ability to identify the linkage of cancer-related genes in cellular pathways. Cancer pathways have complex features that make potential pharmacological targets invisible to these methods. Such features identified in the RAS pathway, like the RAF inhibitor paradox (47) and feedback inhibition (48) have important implications for cancer therapy development. The question of which pathway components are redundant is particularly important in RAS signaling, since many factors, including the RAS proteins themselves, are represented by multiple paralogs. Many agents in clinical use, like pan-RAF, pan-MEK, and pan-PI(3)K inhibitors, target families of proteins rather than individual paralogs. Single-gene knockouts cannot mimic the family-wide inactivation caused by these drugs. Rewiring events may also introduce combination susceptibilities where cancer cells become sensitive to treatment by multiple agents. These possibilities, too, are not addressed by conventional, single-target screening methods, which do not directly address complex, multigene dependencies.
We reasoned that we could address these shortcomings by systematic interaction mapping. By combining AP/MS and CRISPR-based CDKO genetic interaction (GI) mapping of RAS pathway components, we both expanded our knowledge of the RAS pathway and revealed new physical and genetic relationships between pairs of components. We have identified functional redundancies and differences between RAS, RAP, RAF, and RALGEF family members. The GIs we observe point to previously undescribed crosstalk between the RAF kinases and other RAS effector proteins. Finally, we have discovered new prospects for combination therapies involving both well-known targets like the RAF kinases or RAL GTPases and newer ones like RAP1GDS1. This combined PPI and GI map both aids our understanding of the RAS pathway and serves as a model for similar approaches to elucidate other disease pathways (Figure 7).
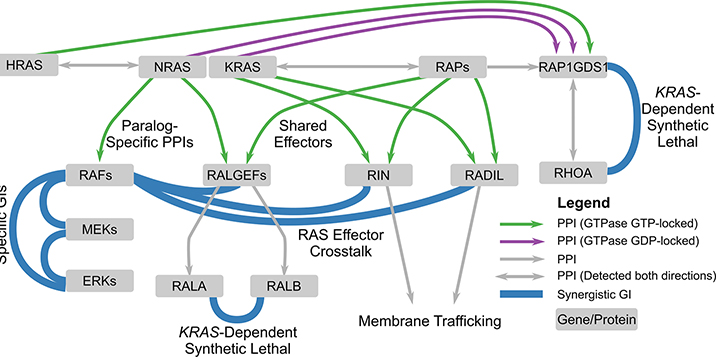
Figure 7. Genetic and Physical Interaction Mapping Reveal New Facets of RAS Pathway Biology.
Schematic of RAS pathway features identified in this study. Among them are new, paralog-specific RAS/effector GI and PPIs, heterogeneity in GIs between MAPK pathway components, new roles in membrane dynamics for the RAS effectors RIN and RADIL, a striking intersection between RAS and RAP signaling, and synthetic lethal GIs between RALA and RALB and between RAP1GDS1 and RHOA that are contingent on KRAS activity.
Deep PPI Networks Illuminate New Facets of RAS Pathway Biology
Protein-protein interaction mapping by AP/MS has several features ideal for the particular challenges of RAS pathway biology. Among these are the unbiased identification of interacting proteins, the ability to capture PPIs as they occur in situ, and the ability to modulate sets of observed PPIs by judicious mutation of bait proteins.
To exemplify the utility of these experiments, we focused on two proteins that interacted with GTP-locked mutant KRAS: RIN1 and RADIL. While a RIN1-HRAS PPI was previously observed (21), RIN proteins have received little attention in RAS signaling. We demonstrate that RIN1 is a significant regulator of macropinocytosis, a process which has recently been specifically associated with RAS activation (Figure 2B, C) (23,24). These observations provide a mechanistic link between RAS effectors and macropinocytosis and provide a context for more detailed screening (49).
We also demonstrate that KRAS binds and regulates RADIL via its RA domain similarly to known KRAS targets (Figure 2D-H). RADIL is a new RAS pathway member previously implicated in adhesion and cell migration (28,29), which we confirm and find linked to KRAS activity. Though important for cell migration, further work is required to determine how RADIL aids small GTPase activity.
By combining the results of several AP/MS experiments in networks, we and other groups can make powerful comparisons among proteins, especially paralogs (17,50–52). By comparing the interactomes of H-, K-, and NRAS in A549 cells, we determined that the PPIs of HRAS with canonical RAS effector proteins are not observed in these cells (Figure 1B-F). K- and NRAS, meanwhile, copurified many known RAS effector proteins, including the RAF kinase family and RALGEFs. Because the six RAS protein baits were expressed at comparable levels (Figure 1B), the strong differences likely reflect differences in biological regulation rather than artefacts of overexpression. We note that there are some other, “known” interactions, that were not recapitulated by these experiments. In some cases, such as interactions between GTPases and their upstream GEFs, we are aware that the interaction is difficult to observe with AP/MS-based methods (18). In other cases, such as the absence of TIAM1 or PIK3CA/p110α, we cannot differentiate between “false negatives” and interactions actually down-regulated by cellular mechanisms. In the comparisons we make in this study, we are careful to compare PPIs involving members of the same protein families (e.g. RAS proteins and RA-domain-containing effectors) in the same cell line to avoid these ambiguities as much as possible.
Virtually every RA-domain-containing protein observed interacting with any GTP-locked RAS protein in vivo co-purified with all GTP-locked RAS isoforms tested in cell-free systems. Judging by the PPI data, it appears that only a subset of these effectors are activated by a given RAS protein in a given cellular context. Therefore, some cellular regulation independent of the nucleotide state of each RAS protein modulates each RAS-effector PPI. Control of RAS and RAS effector localization is one likely mechanism by which different PPI patterns could be determined (53). Similarly, by mapping the different interactomes of three RALGEFs, we provide support for long-suspected differences in their cellular roles (31). By examining differences in PPIs between these different proteins, we can identify functional differences between similar proteins and more confidently map their links to different regulators, effectors, and cellular processes.
We were struck by the observation that proteins copurifying with RAS GTPases also copurify with RAP GTPases. Clustering analysis demonstrates that these two families share such strongly overlapping sets of interacting partners that they are not easily resolved (Figure 3C). While some links between these GTPases, and their sequence similarity, have been noted previously (34), the extent of PPI overlap suggests RAS-RAP crosstalk has hitherto been greatly underestimated. The regulation of RAP GTPase PPIs with RAS effectors, and their influences on RAS signaling, is an important problem. Our genetic interaction data (Figure S7) shows differing patterns of synergistic and buffering interactions between RAS and RAP family members, suggesting that their functional alignment may itself be context-dependent. Further research should characterize signaling upstream of RAP GTPases and to test whether RAS and RAP GTPase interactions with their common effectors are cooperative or antagonistic.
The Combination of GI and PPI Mapping Produces Wiring and Rewiring Diagrams of Lung Adenocarcinoma
Having identified direct physical links between pathway components by PPI mapping, we conducted GI mapping to identify indirect functional links between components. The GI networks recovered predicted synthetic lethal GIs between the two MEK kinases (MAP2K1 and MAP2K2) and the two ERK kinases (MAPK1 and MAPK3). However, we found that GIs within the MAPK pathway differed between cell lines (Figure 4D, S5C, S7). Pan-RAF, pan-MEK and Pan-ERK inhibitors, while approved for some indications, often meet either toxicity limitations or adaptive resistance in clinical use (3,54). The variable and specific GIs observed here support the investigation of isoform-specific RAF or ERK inhibition in RAS-driven cancers. Our subsequent comparison of GIs across multiple cell lines (Figure 5) showed that MAPK pathway genes have more synthetic lethal GIs with non-MAPK genes in KRAS-dependent than -independent cells (Figure S6D). However, these GIs involve different gene pairs in different cell lines (Figure S7). The existence of these GIs, though, indicates a surprising degree of crosstalk between the MAPK pathway and other branches of the RAS pathway.
The heterogeneous susceptibilities we observe among cell lines agrees with other work (14). However, the lines compared in that study (A549, HEK293, and HeLa) had very different primary sources, while the nine we compare are all LUAD. Though we might expect similar provenance of these lines to yield similar GI patterns, we find many important differences (Figure 5E). These apparently rewired GIs present a high-dimensional and somewhat daunting problem for the discovery of combination therapies; some may be modulated by other tumor suppressors or secondary oncogenes (Figure S6F). While PPIs help identify GIs, we observe most GIs between proteins distant in physical interaction space (Figure S5A), which could only be suggested by genetic or functional evidence. Notwithstanding, several GIs were sufficiently consistent to identify new predictions for combination therapies or the use of selective inhibitors.
For example, we found a strong synthetic lethal GI between RALA and RALB specific to KRAS-dependent lines (Figure 5C, middle). While RALs are well-known RAS pathway members, they have yet to gain much interest as a therapeutic target. Though a RAL inhibitor has been developed targeting RAL interactions with RALBP1 (55), our PPI and GI data suggest RAL binding of the exocyst complex may be a more relevant endpoint to assay.
Another noteworthy synthetic lethal GI between KRAS and NRAS was specific to cells not dependent on KRAS (Figure 5C, right). Given that the PPI results indicate that K- and NRAS both engage common effectors that HRAS does not (Figure 1C), we suggest that by knocking out these two paralogs we have impaired the critical aspects of RAS signaling. The fact that KRAS-dependent cells showed less synergy between K- and NRAS knockout suggests that these cells have indeed become specifically addicted to KRAS, rather than to RAS signaling more generally. While we have only examined a few KRAS-independent lines here, these results show that at least some nominally KRAS-independent lines are jointly dependent on multiple RAS genes.
We have classified GIs by their association with KRAS dependence, and not their KRAS mutation, though genetic alterations are easier to identify in the clinic. We expect aberrant KRAS activity, and not mutation per se, to modulate GIs. We note, however, that in both this work and others, KRAS-mutant lines are largely a subset of KRAS-dependent lines (11). Many KRAS-dependent lines, such as the H1975 line we examine, have upstream activating mutations, notably in EGFR (1). Therefore, we expect the combination susceptibilities of this work to apply both in KRAS-mutant LUAD as well as some undetermined subset of KRAS-wildtype LUAD.
Though we and other groups have established functional links between RAS and the effectors RIN1, RADIL, and RALGDS-family proteins, they were not involved in the most salient genetic interactions in this study. These genes do not generally show high mutation rates in patients (1), nor are they frequently crucial dependencies in cancers (11). However, they are among the most frequently-interacting genes screened in the two smaller GI libraries (Figure 5F). How can we resolve this apparent paradox?
As we have noted before, many genes in this pathway are represented by multiple paralogs. Though there are genes, like RAF1, that are uniquely critical in their gene family across a variety of cell lines, this seems to be an exception rather than the rule. Many GIs we observed link the same gene families in either line (Figure S7), but involve different members of those families in different lines. These may hint at gene family/gene family synthetic lethal interactions that would require several knockouts to observe genetically but could potentially be phenocopied by treatment with two drugs that inhibited all members of one protein family each. The numerous genetic interactions involving RADIL, RIN1 and 2, and RALGEFs, motivate further investigation of these pathways.
RAP1GDS1 and RHOA as a Synthetic Lethal Susceptibility in RAS-Driven Lung Adenocarcinoma
The GI maps pointed to a KRAS-associated susceptibility of combined loss of RAP1GDS1 and RHOA. Mutant RAS has previously been shown to require intact RHO signaling for transformation (56), which may be required for cell cycle progression in KRAS mutant cancers (57). DepMap reveals RHOA and RHOC dependency in a number of KRAS-dependent lines, including LUAD, supporting its activity in these cancers.
Previous studies have established that RAP1GDS1 acts as a non-canonical GEF for RHOA and RHOC (38). It also promotes prenylation and membrane trafficking of RHO and RAS proteins, which is critical for their signaling (42,43). We identified RAP1GDS1 in the PPI data with all of the RAS and RAP GTPases as baits, making it a highly central member of this signaling network (Figure 1F, 3B, S1B, S2A, C). The importance of these binding partners in cancer signaling make RAP1GDS1 a noteworthy therapeutic target.
In vitro validation in 2D and 3D cultures of LUAD cell lines confirmed that dual RAP1GDS1 and RHOA knockout selectively impaired growth of KRAS-dependent cells (Figure 6, S8), an effect we find tightly linked to induction of apoptosis. The mechanism behind this GI is unclear. One parsimonious explanation is that the two knockouts are a multi-hit insult to RHOA signaling, compromising both RHOA and its RAP1GDS1-mediated prenylation and/or activation. RAP1GDS1 also serves as a GEF for RHOC, and it may be that the RAP1GDS1/RHOA double knockout effectively impairs two branches of RHO signaling (38). A RHO-focused GI network including RAP1GDS1 would greatly clarify this point. Two recent studies also point to a link between RAP1GDS1 and KRAS prenylation and trafficking to the plasma membrane, suggesting that RAP1GDS1 loss may modulate KRAS activity more directly (39,58). Finally, disruption of RAP1GDS1 has been shown to alter global prenylation of many GTPases from the RAP, RAB, RHO and RAS families (59). Therefore, it could be that these other GTPases synergize with RHO and KRAS signaling and that the simultaneous disruption of all of these pathways by RAP1GDS1 deletion results in the synthetic lethality we observe.
Although identifying the precise mechanism of this genetic interaction will require further study, our results establish that this GI manifests specifically in KRAS-dependent cells. In vivo analysis using LUAD xenografts further suggests that combined RAP1GDS1 and RHOA inhibition may provide a promising strategy to treat KRAS-driven tumors.
Conclusion
The concept of cancer pathway rewiring – a change of the linkage of cell biological pathways – underlies the core theories of targeted therapy development. In this work, we provide a systematic survey of RAS pathway interactions in RAS-transformed LUAD. Combining protein interaction mapping with genetic screens allows us to map pathway wiring directly. In doing so, we have gained new insights into RAS-driven tumor biology and discovered new susceptibilities of RAS-driven tumors and provided a resource for other investigators to do the same.
METHODS
Plasmids
pMCB306, pKHH30 and p293 Cas9-BFP were previously described (13). pCMV-VSV-G and pCMV-dR8.2 dvpr were gifts from Prof. Bob Weinberg (Addgene plasmid #8454 and #8455) (60). pWPXLd LAP plasmids were also previously described (61). Cassettes were introduced into GST-fusion, myc-fusion, and LAP-fusion plasmids by Gateway cloning (Supplementary Methods). Point mutations in bait proteins were introduced by QuikChange mutagenesis using primers in Supplementary Table 1.
Cell Lines
HEK293, A549, H23, H358, H1437, H1568, H1650, H1975, H2009, and NL20 cells were obtained from American Type Culture Collection (ATCC). HEK293-LAP cell lines were generated using HEK293-FlpIn cells (61). Culture methods are described in Supplementary Methods. Cas9- or inducible Cas9-expressing strains of all cell lines were generated using lentiviral vectors (Addgene #52962 and # 83481, respectively) and tested for Cas9 cutting efficiency as described previously (13). Cell lines were screened for mycoplasma contamination with Cell Check 9 Plus Test (IDEXX) and kept below 15 passages.
Affinity Purification / Mass Spectrometry
Affinity purification/mass spectrometry was carried out as described in (61), and quantified as in (17), with changes described in Supplementary Methods.
Co-Immunoprecipitation Experiments
4×107 cells of each line expressing a LAP-RAS mutant construct were lysed in 300 μL of 150mM KCl, 50mM HEPES pH 7.4, 1mM MgCl2 10% glycerol, and 0.3% NP-40 (lysis buffer) supplemented with 3μL phosphatase inhibitor cocktail 2 (Sigma-Aldrich) and 1μg/mL each of leupeptin, pepstatin, and chymostatin (Sigma-Aldrich) on ice for 10 minutes. Cells were centrifuged for 15min at 4C. Supernatant concentration was measured by Bradford assay and diluted to 9mg/mL protein in 300μL lysate. For a 5% input sample, 9 μL were withheld and mixed with 4× LDS sample buffer +2.5% β-mercaptoethanol. 4.83 μL protein A beads conjugated to Rabbit αGFP antibody (Thermo Fisher) were added to the remaining supernatant. Samples were incubated 16h at 4C. Beads were washed 5× with 200μL 200mM KCl, 50mM HEPES pH 7.4, 1mM MgCl2 10% glycerol, and 0.3% NP-40 (lysis buffer), then immunoblotted as described (61), probed 16h at 4°C with rabbit αRAF1 (Abcam) and mouse αGFP (Invitrogen).
Recombinant Protein Purification
Purifications of recombinant GST-tagged HRAS, KRAS, and NRAS with the indicated mutations performed as described in (61). Briefly, proteins were purified from Rosetta 2 E. Coli cells transformed with plasmids encoding the indicated constructs.
Cell-Free Co-Immunoprecipitation Experiments
Cell free co-immunoprecipitation experiments were performed as described in (61). Primary antibodies used were rabbit αGST (Sigma-Aldrich) and mouse α-myc clone 9E10 (EMD Millipore).
Fluorescence Microscopy
Images were acquired on a Marianas spinning disk confocal (SDC) inverted microscope (Intelligent Imaging Innovations) using a Zeiss Plan-NEO 40×/1.3 Oil Ph3 objective, a Yokogawa csu22 confocal scanning unit and a Photometrics Cascade 1K camera.
sgRNA Vectors
Small guide RNAs against target genes or control safe-targeting sgRNAs were cloned into the MCB306 or KH30 (MCB306 with an hU6 promoter) vectors. For double sgRNA vectors the U6-sgRNA cassette was excised from the KH30 vector and cloned into the pMCB306 vector. All sgRNA sequences are listed in Supplementary Table 8. Lentivirus was produced in 293T cells as previously described (61), filtered, and applied directly to cells for infection at a MOI<1. Clonal RIN1 knockout generation is described in Supplementary Methods.
Macropinocytosis Assay
Macropinocytosis assays were conducted as described in (25), and are elaborated in Supplementary Methods.
Scratch-Wound Assay
Scratch-wound assays were performed as described (30), and are elaborated in Supplementary Methods.
Quantitative Real-Time PCR
Quantitative Real-Time PCR for the RADIL knockdown experiments began with reverse transfection (Supplementary Methods), except that siRNAs were pooled. Cells were harvested and frozen 24, 48, and 72 hours post-transfection. Relative RNA levels were quantified using the TaqMan RADIL Assay (Thermo) with a compatible GAPDH Assay (Life) as a control probe using the TaqMan manufacturer’s protocol in 96-well MicroAmp Optical reaction plates (Applied Biosystems, N8010560)
Clustering Analysis
Clustering analysis was conducted by the UPGMA (unweighted pair group method with arithmetic mean) method using the cosine distance metric, using the “linkage” method provided by the “scipy” module (62). Bait-prey interactions were binarized based on an FDR of < 0.05, and this adjacency matrix was used as input.
CRISPR/CDKO screening
CDKO screening was conducted as previously described (13), and is elaborated in Supplementary Methods.
Proliferation assays
Short-term cell viability assays were performed using the IncuCyte S3 (Essen Bioscience). Five days post infection the cells were trypsinized, counted and 500–1000 viable cells were seeded in 200μl of puromycin-containing medium into each well of a 96 well plate. Each condition was seeded in triplicate. Cells were imaged every 12h for the duration of the experiment. Cell area mask was calculated on the green channel to include only GFP-positive cells.
Sphere formation assay
For anchorage-independent sphere growth, cells were seeded into 24-well ultra-low attachment (Corning, #3473) plates (20,000 viable cells per well) in 2ml of complete medium supplemented with 0.5% methylcellulose. The spheres were allowed to form for 9–20 days (depending on the cell line). GFP-positive spheres were imaged using the Leica DMi8 fluorescence microscope. Sphere size and number were quantified using ImageJ (NIH, Bethesda, Maryland, USA). For sphere growth assays in the IncuCyte S3 (Essen Bioscience) cells were seeded into complete medium supplemented with 1.5% methylcellulose (500–1000 cells/well) in 96-well ultra-low attachment plates (Corning, #3474). Each condition was seeded in triplicate.
Competition assay in Tumor Xenografts
A549-iCas9 cells were infected with the indicated double sgRNA vectors and puromycin selection was applied for 7 days. Stably infected cell populations were trypsinized and transferred to 15cm dishes to expand. Three days prior to implantation the cells were treated with doxycycline (1μg/ml). On the day of the implantation (D1) cells were trypsinized, counted and analyzed by flow cytometry. Cells expressing mCherry (safe-safe sgRNA vector) and GFP (gene-targeting sgRNAs) were pooled 1:1, centrifuged and re-suspended in a mixture of serum-free DMEM (Corning, #15–017-CV) and Matrigel (Corning, #356237). Cells were injected subcutaneously into both flanks of NSG mice (2×106 cells per flank, 3 mice per condition for A549 cells and 5×106 cells per flank, 4 mice per condition for H23 cells). All the procedures involving mice were are approved by the Institutional Animal Care and Use Committee (IACUC) at UCSF (protocol #AN15761). Thirty days after implantation (D30), tumors were dissected, chopped and dissociated into a single-cell suspension using PBS supplemented with Collagenase/Dispase (Roche, #11097113001) and DNAse I (Worthington, #LS002006) at 37°C for 30 min with agitation. Digested samples were passed through a 40μm filter, resuspended in complete DMEM medium and centrifuged. Cell pellets were incubated for 1 min in RBC buffer (155mM NH4Cl, 12mM NaHCO3, 0.1 mM EDTA) to remove red blood cells, resuspended in complete DMEM and centrifuged. Resulting cell pellets were resuspended in PBS with 1% serum and analyzed by flow cytometry on Accuri C6 (BD Biosciences). The ratio of GFP to mCherry fluorescence in tumor samples at D30 was normalized to D1 cell populations and represented as log fold-change between these two time points.
Statistical significance was calculated using unpaired Student’s t-test between the tested sample and the corresponding safe-safe control. Resulting p-values were corrected for multiple-hypothesis testing using the Benjamini–Hochberg procedure.
Statistical Analysis
Analyses of statistical significance were performed using the tests described in the texts, using the “scipy” library in Python. Where shown, 95% confidence intervals were calculated using the “statsmodels” library in Python (www.statsmodels.org).
Analysis of mutation rates in Figure S7 relied on the provisional TCGA LUAD data from cbioportal.org (1). Using the EllipticalEnvelope method provided by scikit-learn (63), the mahalanobis distance of each gene from a common centroid based on relative mutation rates and CNA frequencies was calculated.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
We establish a deep network of protein-protein and genetic interactions in the RAS pathway. Many interactions validated here demonstrate important specificities and redundancies among paralogous RAS regulators and effectors. By comparing synthetic lethal interactions across KRAS-dependent and -independent cell lines, we identify several new combination therapy targets for RAS-driven cancers.
ACKNOWLEDGEMENTS
We thank Alex Loktev, PhD and Kevin Wright, PhD and Christopher Adams, PhD and Ryan Leib, PhD for their help with mass spectrometry. We thank Dr. Ji Luo for advice on growth of lung cancer cell lines in spheroid culture.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
REFERENCES
- 1.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddy EP, Reynolds RK, Santos E, Barbacid M. A point mutation is responsible for the acquisition of transforming properties by the T24 human bladder carcinoma oncogene. Nature. 1982;300:149–52. [DOI] [PubMed] [Google Scholar]
- 3.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging Ras Back in the Ring. Cancer Cell. 2014;25:272–81. [DOI] [PubMed] [Google Scholar]
- 4.Ewen-Campen B, Mohr SE, Hu Y, Perrimon N. Accessing the Phenotype Gap: Enabling Systematic Investigation of Paralog Functional Complexity with CRISPR. Dev Cell. 2017;43:6–9. [DOI] [PubMed] [Google Scholar]
- 5.Weinstein IB. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–4. [DOI] [PubMed] [Google Scholar]
- 6.Kaelin WG Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–98. [DOI] [PubMed] [Google Scholar]
- 7.Tangutoori S, Baldwin P, Sridhar S. PARP inhibitors: A new era of targeted therapy. Maturitas. 2015;81:5–9. [DOI] [PubMed] [Google Scholar]
- 8.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. [DOI] [PubMed] [Google Scholar]
- 9.Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell. 2017;168:890–903.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–26. [DOI] [PubMed] [Google Scholar]
- 11.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–76.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torres JZ, Miller JJ, Jackson PK. High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics. 2009;9:2888–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol. 2017;35:463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat Methods. 2017;14:573–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nassar N, Singh K, Garcia-Diaz M. Structure of the dominant negative S17N mutant of Ras. Biochemistry. 2010;49:1970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding S, Mooney N, Li B, Kelly MR, Feng N, Loktev AV, et al. Comparative Proteomics Reveals Strain-Specific β-TrCP Degradation via Rotavirus NSP1 Hijacking a Host Cullin-3-Rbx1 Complex. PLoS Pathog. 2016;12:e1005929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, Kwong M, et al. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011;25:2347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabian JR, Vojtek AB, Cooper JA, Morrison DK. A single amino acid change in Raf-1 inhibits Ras binding and alters Raf-1 function. Proc Natl Acad Sci U S A. 1994;91:5982–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han L, Colicelli J. A human protein selected for interference with Ras function interacts directly with Ras and competes with Raf1. Mol Cell Biol. 1995;15:1318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell. 2001;1:73–82. [DOI] [PubMed] [Google Scholar]
- 23.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fennell M, Commisso C, Ramirez C, Garippa R, Bar-Sagi D. High-content, full genome siRNA screen for regulators of oncogenic HRAS-driven macropinocytosis. Assay Drug Dev Technol. 2015;13:347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Commisso C, Flinn RJ, Bar-Sagi D. Determining the macropinocytic index of cells through a quantitative image-based assay. Nat Protoc 2014;9:182–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patricelli MP, Janes MR, Li L-S, Hansen R, Peters U, Kessler LV, et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov. 2016;6:316–29. [DOI] [PubMed] [Google Scholar]
- 27.Wisniewska M The crystal structure of the RA domain of FLJ10324 (RADIL). 2008. [Google Scholar]
- 28.Liu L, Aerbajinai W, Ahmed SM, Rodgers GP, Angers S, Parent CA. Radil controls neutrophil adhesion and motility through β2-integrin activation. Mol Biol Cell. 2012;23:4751–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed SM, Thériault BL, Uppalapati M, Chiu CWN, Gallie BL, Sidhu SS, et al. KIF14 negatively regulates Rap1a-Radil signaling during breast cancer progression. J Cell Biol. 2012;199:951–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cory G Scratch-wound assay. Methods Mol Biol. 2011;769:25–30. [DOI] [PubMed] [Google Scholar]
- 31.Cascone I, Selimoglu R, Ozdemir C, Del Nery E, Yeaman C, White M, et al. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 2008;27:2375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.González-García A, Pritchard CA, Paterson HF, Mavria G, Stamp G, Marshall CJ. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell. 2005;7:219–26. [DOI] [PubMed] [Google Scholar]
- 33.Neel NF, Martin TD, Stratford JK, Zand TP, Reiner DJ, Der CJ. The RalGEF-Ral Effector Signaling Network: The Road Less Traveled for Anti-Ras Drug Discovery. Genes Cancer. 2011;2:275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bos JL. All in the family? New insights and questions regarding interconnectivity of Ras, Rap1 and Ral. EMBO J. 1998;17:6776–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34:634–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin N, Jiang T, Rosen DM, Nelkin BD, Ball DW. Synergistic action of a RAF inhibitor and a dual PI3K/mTOR inhibitor in thyroid cancer. Clin Cancer Res. 2011;17:6482–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drosten M, Barbacid M. Modeling K-Ras-driven lung adenocarcinoma in mice: preclinical validation of therapeutic targets. J Mol Med. 2016;94:121–35. [DOI] [PubMed] [Google Scholar]
- 38.Hamel B, Monaghan-Benson E, Rojas RJ, Temple BRS, Marston DJ, Burridge K, et al. SmgGDS Is a Guanine Nucleotide Exchange Factor That Specifically Activates RhoA and RhoC. J Biol Chem. 2011;286:12141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.García-Torres D, Fierke CA. The chaperone SmgGDS-607 has a dual role, both activating and inhibiting farnesylation of small GTPases. J Biol Chem. 2019;294:11793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–35. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu H, Toma-Fukai S, Kontani K, Katada T, Shimizu T. GEF mechanism revealed by the structure of SmgGDS-558 and farnesylated RhoA complex and its implication for a chaperone mechanism. Proc Natl Acad Sci U S A. 2018;115:9563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berg TJ, Gastonguay AJ, Lorimer EL, Kuhnmuench JR, Li R, Fields AP, et al. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J Biol Chem. 2010;285:35255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuld NJ, Vervacke JS, Lorimer EL, Simon NC, Hauser AD, Barbieri JT, et al. The chaperone protein SmgGDS interacts with small GTPases entering the prenylation pathway by recognizing the last amino acid in the CAAX motif. J Biol Chem. 2014;289:6862–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y, de la Cruz CC, Sayles LC, Alleyne-Chin C, Vaka D, Knaak TD, et al. A rare population of CD24(+)ITGB4(+)Notch(hi) cells drives tumor propagation in NSCLC and requires Notch3 for self-renewal. Cancer Cell. 2013;24:59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amano M, Nakayama M, Kaibuchi K. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton. 2010;67:545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Julian L, Olson MF. Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases. 2014;5:e29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cox AD, Der CJ. The RAF inhibitor paradox revisited. Cancer Cell. 2012;21:147–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol. 2018;20:1064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redelman-Sidi G, Binyamin A, Gaeta I, Palm W, Thompson CB, Romesser PB, et al. The Canonical Wnt Pathway Drives Macropinocytosis in Cancer. Cancer Res. 2018;78:4658–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eckhardt M, Zhang W, Gross AM, Von Dollen J, Johnson JR, Franks-Skiba KE, et al. Multiple Routes to Oncogenesis Are Promoted by the Human Papillomavirus-Host Protein Network. Cancer Discov. 2018;8:1474–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kovalski JR, Shanderson RL, Khavari PA. Ras functional proximity proteomics establishes mTORC2 as new direct ras effector. Oncotarget. 2019;10:5126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castellano E, Santos E. Functional specificity of ras isoforms: so similar but so different. Genes Cancer. 2011;2:216–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer. 2015;15:577–92. [DOI] [PubMed] [Google Scholar]
- 55.Yan C, Liu D, Li L, Wempe MF, Guin S, Khanna M, et al. Discovery and characterization of small molecules that target the GTPase Ral. Nature. 2014;515:443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zohn IM, Campbell SL, Khosravi-Far R, Rossman KL, Der CJ. Rho family proteins and Ras transformation: the RHOad less traveled gets congested. Oncogene. 1998;17:1415–38. [DOI] [PubMed] [Google Scholar]
- 57.Olson MF, Paterson HF, Marshall CJ. Signals from Ras and Rho GTPases interact to regulate expression of p21Waf1/Cip1. Nature. 1998;394:295–9. [DOI] [PubMed] [Google Scholar]
- 58.Nissim S, Leshchiner I, Mancias JD, Greenblatt MB, Maertens O, Cassa CA, et al. Mutations in RABL3 alter KRAS prenylation and are associated with hereditary pancreatic cancer. Nat Genet. 2019;51:1308–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brandt AC, McNally L, Lorimer EL, Unger B, Koehn OJ, Suazo KF, et al. Splice switching an oncogenic ratio of SmgGDS isoforms as a strategy to diminish malignancy. Proc Natl Acad Sci U S A. 2020;117:3627–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanie T, Abbott KL, Mooney NA, Plowey ED, Demeter J, Jackson PK. The CEP19-RABL2 GTPase Complex Binds IFT-B to Initiate Intraflagellar Transport at the Ciliary Base. Dev Cell. 2017;42:22–36.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.