Abstract
Background:
Autoantibody responses in cancer are of great interest, as they may be concordant with T-cell responses to cancer antigens or predictive of response to cancer immunotherapies. Thus, we sought to characterize the antibody landscape of metastatic castration-resistant prostate cancer (mCRPC).
Methods:
Serum antibody-epitope repertoire analysis (SERA) was performed on patient serum to identify tumor-specific neoepitopes. Somatic mutation-specific neoepitopes were investigated by associating serum epitope enrichment scores with whole-genome sequencing results from paired solid-tumor metastasis biopsies and germline blood samples. A protein-based immunome-wide association study (PIWAS) was performed to identify significantly enriched epitopes, and candidate serum antibodies enriched in select patients were validated by ELISA profiling. A distinct cohort of patients with melanoma was evaluated to validate the top cancer-specific epitopes.
Results:
SERA was performed on 1,229 serum samples obtained from 72 men with mCRPC and 1,157 healthy control patients. 29 of 6,636 somatic mutations (0.44%) were associated with an antibody response specific to the mutated peptide. PIWAS analyses identified motifs in eleven proteins including NY-ESO-1 and HERVK-113 as immunogenic in mCRPC, and ELISA confirmed serum antibody enrichment in candidate patients. Confirmatory PIWAS, IMUNE, and ELISA analyses performed on serum samples from 106 patients with melanoma similarly revealed enriched cancer-specific antibody responses to NY-ESO-1.
Conclusions:
We present the first large-scale profiling of autoantibodies in advanced prostate cancer, utilizing a new antibody profiling approach to reveal novel cancer-specific antigens and epitopes. Our study recovers antigens of known importance and identifies novel tumor-specific epitopes of translational interest.
Background
The role of adaptive immunity in cancer is of great translational interest given the recent development of novel, clinically effective immunotherapies that focus on generating T cell responses to tumor antigens. While the T-cell landscape of numerous cancer types has been explored in some depth, the role of humoral immunity in cancer is much less well-characterized. Several studies have demonstrated that a distinct antibody signature may be detectable in the serum of breast1, prostate2, and lung3 cancer patients and may thus be useful for cancer detection. Additionally, studies have demonstrated that B-cell infiltration into the tumor microenvironment is associated with prolonged patient survival and enhanced response to immunotherapy in melanomas, renal cell carcinomas, and sarcomas4–9, with several studies suggesting that B-cell autoantibodies may play a direct role in mounting an anti-tumor response10,11. In the setting of cancer vaccines, preclinical data indicate that IgG anti-tumor antibody responses to neoantigens in a mouse model of breast cancer can predict corresponding T cell responses to the same epitopes12. Furthermore, in a completed phase III trial that led to approval of the autologous cellular vaccine sipuleucel-T for mCRPC, which was one of the first immunotherapies approved by the FDA for solid tumors, productive antibody responses to the immunogen were correlated with longer overall survival in retrospective analysis13. Finally, anti-tumor immune responses can also be stimulated by proteins ectopically expressed outside of immune-privileged sites in somatic tumor tissues, the prototype of which is cancer-testis antigen NY-ESO-1. The prevalence of autoantibodies to the NY-ESO-1 peptide and putative conservation of B- and T-cell epitopes has led to over 30 NY-ESO-1 T-cell receptor immunotherapy clinical trials, at various stages of completion, in diverse cancer types14,15. Altogether, these findings support the notion that a patient’s antibody repertoire may reflect a specific immune response to the patient’s cancer and may have potential diagnostic and therapeutic implications.
Tumor-associated antibodies detectable in patient serum are traditionally profiled using microarray-based methods16–18, phage-display approaches19–21, or techniques incorporating principles of the two22–24. One key limitation of candidate protein-based approaches is the throughput and subsequently limited number of antigens that can be profiled and the inability to detect patient- or tumor-specific sequence variants generated by mutation. The serum epitope repertoire analysis (SERA) tool leverages a randomized bacterial-display library paired with next generation sequencing (NGS) to identify peptides binding to serum antibodies25. By leveraging the randomized library, SERA is able to examine both wild type and mutant sequences without any modification to the experimental protocols. Protein-based Immunome Wide Association Study (PIWAS) builds on top of the SERA assay to identify proteome-constrained antigenic signals from the SERA assay. PIWAS calculates, for each sample and protein, a smoothed log-enrichment value across a window of overlapping kmers to identify a protein (gene)-level enrichment score while retaining epitope-level resolution for the signal source. By comparing PIWAS values between cohorts using the outlier sum, PIWAS is able to identify autoantigens against the human proteome26.
While autoantibody enrichment has previously been demonstrated in prostate cancer, these studies were limited by smaller discovery cohorts27,28 or relatively restrictive peptide libraries29,30. It also appears that autoantibody enrichment may be context-specific. For example, one large study that leveraged a phage-display approach developed a signature for prostate cancer screening but found that this signature could be found only in a minority of patients with castration-resistant disease2. Thus, the autoantibody landscape for patients with metastatic castration-resistant prostate cancer (mCRPC) has yet to be elucidated.
Given that metastatic castration-resistant prostate cancer (mCRPC) represents one of the leading causes of cancer-associated death in men, we sought to characterize the autoantibody landscape of this disease. Utilizing SERA, PIWAS, and IMUNE25,31, we performed an unbiased analysis of autoantibodies enriched in the serum of mCRPC patients compared to healthy controls. Specifically, we leveraged DNA-sequence level information from the assay to identify not only the proteins but also the specific epitopes (sub-peptides) within the full-length proteins that were putatively antigenic in mCRPC. We also integrated the serum antibody-profiling results with whole-genome sequencing performed on metastatic tumor biopsies and peripheral blood (germline) specimens from the same patients to assess the immunogenicity of antigens resulting from somatic mutations. We validated our top candidate antigen in NY-ESO-1, a known immunogenic tumor marker across cancer types, using an independent cohort of melanoma patients. We further validated the PIWAS-based seropositive results of our top motifs using ELISA experiments performed on the same serum specimens. In total, our study both recovered previously identified cancer antigens and identified novel, putative cancer-specific antigens in mCRPC.
Materials and Methods
Data acquisition and sample processing
A prospective IRB-approved study (NCT02432001) was conducted by a multi-institutional consortium that obtained serum, peripheral blood, and fresh-frozen, image-guided biopsy samples of metastases from mCRPC patients. Serum samples for each patient were prospectively obtained at time of study enrollment, at three-month follow-up, and at time(s) of cancer progression, if applicable. Blood was drawn at start of therapy, 3 months into therapy, and at clinically determined disease progression in serum separator tubes of 6mL (BD #367815) or 10mL (BD #367820). Tubes were spun within 90 minutes of collection (1500rcf for 10min), aliquoted into 2mL cryovials, and frozen on dry ice and shipped to a central lab at UCSF. Vials were stored upon arrival at −80°C until batch shipping on dry ice for processing at Serimmune. Solid-tumor metastases biopsies were sequenced using whole-genome sequencing and RNA-seq as previously described32,33. Serum samples from a control group consisting of 1,157 individuals without known history of cancer or other predicate disease were obtained from the Serimmune database of samples. A cohort of 106 melanoma patients was used for validation of specific antigens. The prospective IRB-approved study (11–003254) of these patients was conducted at University of California Los Angeles (UCLA) that obtained peripheral blood and biopsy samples for various analysis from patients treated for advanced melanoma malignancies. Plasma samples for each patient were prospectively obtained at time of study enrollment, at approximately three-month follow-up and at further follow-up time(s) as prescribed. At baseline and after approximately 3 months of treatment, blood was collected in K3-EDTA lavender tubes of 9mL (Greiner Bio-One# 455036) and so forth. Tubes were spun within 24-hours after collection (1200rcf for 10 min, brake off), aliquoted at 500uL into 2mL cryovials for long term storage at −80°C. A total of 106-subject aliquots were prepared as 120uL and overnight shipped on dry ice for processing at Serimmune. The study was performed after approval by an institutional review board (IRB) and was conducted in accordance with the ethical principles of the Declaration of Helsinki. All patients provided written informed consent.
Serum antibody-epitope profiling
An E. coli bacterial-display library consisting of plasmids encoding random 12-mer peptides at a diversity of 8×109 was constructed and prepared as previously described25. Serum samples were screened on this library as previously described26. Briefly, serum samples, at a 1:25 dilution, were added to each well of a 96 well deep well plate containing 8×1010 (10-fold over-sampling) induced library cells and incubated with orbital shaking at 4°C for 1 hour. Cells were washed once with PBS containing 0.05% Tween-20 (PBST) and then incubated with Protein A/G Sera-Mag SpeedBeads (GE Life Sciences, 17152104010350) for 1 hour at 4°C with orbital shaking. Cells displaying peptides bound to serum IgG antibodies were captured by magnetic separation and washed five times with PBST. Selected cells were grown overnight in LB supplemented with 34 μg/mL chloramphenicol and 0.2% wt/vol glucose at 37°C with shaking at 250 rpm.
Amplicon preparation and NGS sequencing were performed as previously described26. Briefly, plasmids were isolated from selected library cells using the Montage Plasmid MiniprepHTS Kit (MilliPore, LSKP09604) on a MultiscreenHTS Vacuum Manifold (MilliPore, MSVMHTS00) following the manufacturer’s instructions. Next, DNA encoding the 12mer variable regions was amplified and barcoded by two rounds of PCR. Finally, after normalizing DNA concentrations, pooled samples were sequenced using a NextSeq 500 (Illumina) and a High Output v2, 75 cycle kit (Illumina, FC-404-2005) with PhiX Run Control (Illumina, FC-110-3001) at 40% of the final pool concentration.
Identifying mutation-specific epitopes
Previously-published results of whole-genome sequencing performed on fresh-frozen metastasis biopsies and paired peripheral blood samples of the same patients32,34 was analyzed to identify somatic protein-coding point and frameshift mutations present in each patient’s tumor. Data from the SERA platform were broken into 5mers and 6mers for every sample and enrichments were calculated25. Using the same approach as PIWAS, these enrichments were tiled against both the wild type and mutated protein sequences26. The enrichment values for the wild type sequence were subtracted from the mutant sequence to identify differential signal. The maximum differential value was calculated for every mutated protein and the associated patient sample. The data were fit to an exponential distribution and the probability density function was used to estimate P-values for every protein. P-values were corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure35.
PIWAS approach
Using the prostate cancer patients as cases and the individuals without known cancer as controls, we ran a PIWAS analysis against the human proteome26. PIWAS was parameterized to have a window size of 5, the number of standard deviation approach, and the maximum peak signal. The outlier sum false discovery rate as defined previously was used to prioritize antigens26. The reference human proteome was dogwnloaded from Uniprot on February 28, 2019.
The validation PIWAS was run using the same parameterizations with the melanoma cohort as cases and the individuals without known cancers as controls.
The PIWAS-IMUNE approach
For top antigens NY-ESO-1 and HERV-K, additional steps were taken to develop a motif panel for these antigens. In both cases, the PIWAS-IMUNE algorithm was used to identify linear mapping motifs. In the initial PIWAS stage, prostate cancer samples with an antigen PIWAS score >6 were identified. The positive prostate cancer samples and 30 random healthy controls were used as input to the second stage IMUNE algorithm, which was parameterized with 20% sensitivity and 100% specificity25. Motifs that mapped linearly to the target antigen were retained. For each retained motif, the mean and standard deviation (SD) of enrichment scores was calculated using the 1,157 control specimens as a reference group. z-scores were calculated for every cancer specimen. Then, for each cancer specimen, enrichment scores for each motif were z-scored (based on the enrichment score mean and SD of the control group) and summed to generate a composite score for each specimen. Thus, the final composite PIWAS-IMUNE “panel score” was defined as the sum of motif z-scores for each specimen. Thresholds for positivity on the panel were set at a 99% specificity.
ELISA
Briefly, NY-ESO-1 recombinant protein (Origene) at 0.5 ug/ml, or control recombinant protein CENPA (Origene) at 0.5 ug/ml, or HERVK-5 recombinant protein (MyBiosource) at 1 ug/ml or control protein Bovine Serum Albumin (Sigma) at 1 ug/ml in PBS were coated onto flat bottom, 96 well plates (Nunc MaxiSorp), 50 ul per well at 4°C overnight. Plates were washed with PBS containing 0.1% Tween 20 and blocked with 5% non-fat milk in PBS for 2 hours at room temperature. Plates were then incubated with 100 ul of patient serum diluted 1/200 or 1/2000 in 5% non-fat milk in PBS for 2 hours at room temperature. Following washing, plates were incubated with peroxidase conjugated goat anti-human IgG secondary (1/10,000 in 5% non-fat milk in PBS; Jackson ImmunoResearch) for 1 hour at room temperature. After a last wash step, the reaction was developed with 3,3’,5,5’-tetramethylbenzidine substrate solution (ThermoFisher) for 1–10 minutes and stopped with 1M hydrochloric acid. Absorbance at 450 nm was measured on a plate reader. ELISA values were calculated as the mean difference between the testing recombinant protein and the control protein. Due to reagent availability constraints, sera reactivity to HERVK-5 was assessed in lieu of reactivity of HERVK-113 given the high sequence similarity of the HERVK-5 and HERVK-113 proteins (95.2% per BLAST analysis).
Statistical methods and survival analysis
Overall survival was measured from time of mCRPC diagnosis. Survival analyses were conducted using the Kaplan-Meier method with log-rank testing for significance. The χ2 test was performed to assess the relationship between ELISA and PIWAS antibody enrichment results. All independence and hypothesis tests were performed using a two-sided significance level of 0.05. Multiple hypothesis testing correction was performed using the Benjamini-Hochberg procedure.
Results
Serum specimens were obtained from a cohort of 72 mCRPC patients with a mean age of 72 years at time of mCRPC diagnosis (Table 1). The cohort was predominantly Caucasian (87%) with high-grade primary tumors in 54%. Visceral metastases were observed in 15 of 72 (21%) patients. Sera obtained at more than one timepoint were available for 79% of patients (Table S1).
Table 1:
Patient clinicopathologic characteristics
| Characteristic | Num. patients (N=72) |
|---|---|
| Age – years (SD) | 72 (8.4) |
| Race | |
| Asian | 4 (5.9) |
| Black or African American | 5 (7.4) |
| White | 59 (86.8) |
| Missing | 4 (5.9) |
| Gleason score at diagnosis | |
| 8+ | 35 (53.8) |
| < 8 | 30 (46.2) |
| Missing | 7 (10.8) |
| Metastatic sites at time of biopsy | |
| Liver | 7 (9.7) |
| Visceral metastases (non-liver) | 8 (11.1) |
| Bone +/− lymph node | 51 (70.8) |
| Lymph node only | 6 (8.3) |
Note: all clinicopathologic variables were measured at time of first solid-tumor biopsy and are presented as “Number (%).”
Integrating serum antibody profiling results with whole-genome sequencing results, we first sought to assess whether somatic, protein-coding mutations were associated with an antibody response specific to the mutant peptide in mCRPC. 29 of the 6,636 protein-coding somatic mutations observed in our cohort were associated with a significant enrichment (exponential FDR < 0.05) in antibodies specific to the mutated peptide (Figure 1A, Table S2). These 29 mutations were approximately evenly distributed between frameshift and missense mutations (Figure S1). These events constituted the minority of mutations (0.44%), consistent with literature that suggests that most protein-coding mutations do not elicit an immune response36. Each of the mutation-specific antibodies was enriched in only one patient. However, the somatic mutations that coded the epitopes were also private to individual patients. This suggested that the observed antibody response was specific to the individual in which the mutant antigen was available. In 11 of 20 mutant epitopes derived from patients with multiple serum specimens available, multiple independent serum samples obtained from the same individual at different timepoints confirmed the mutation-specific antibody enrichments (Table S2). Across all patients with multiple serum specimens obtained at different timepoints, there did not appear to be a consistent trend in favor of either increasing or decreasing enrichment scores over time. Nine of 20 mutations were associated with a progressively increasing and eleven of 20 mutations were associated with progressively decreasing enrichment values over time. We highlight an example of a patient with a point mutation and a patient with a frameshift mutation that demonstrated an enriched autoantibody response to the corresponding mutant epitope across multiple timepoints (Figures 1B, 1C).
Figure 1.
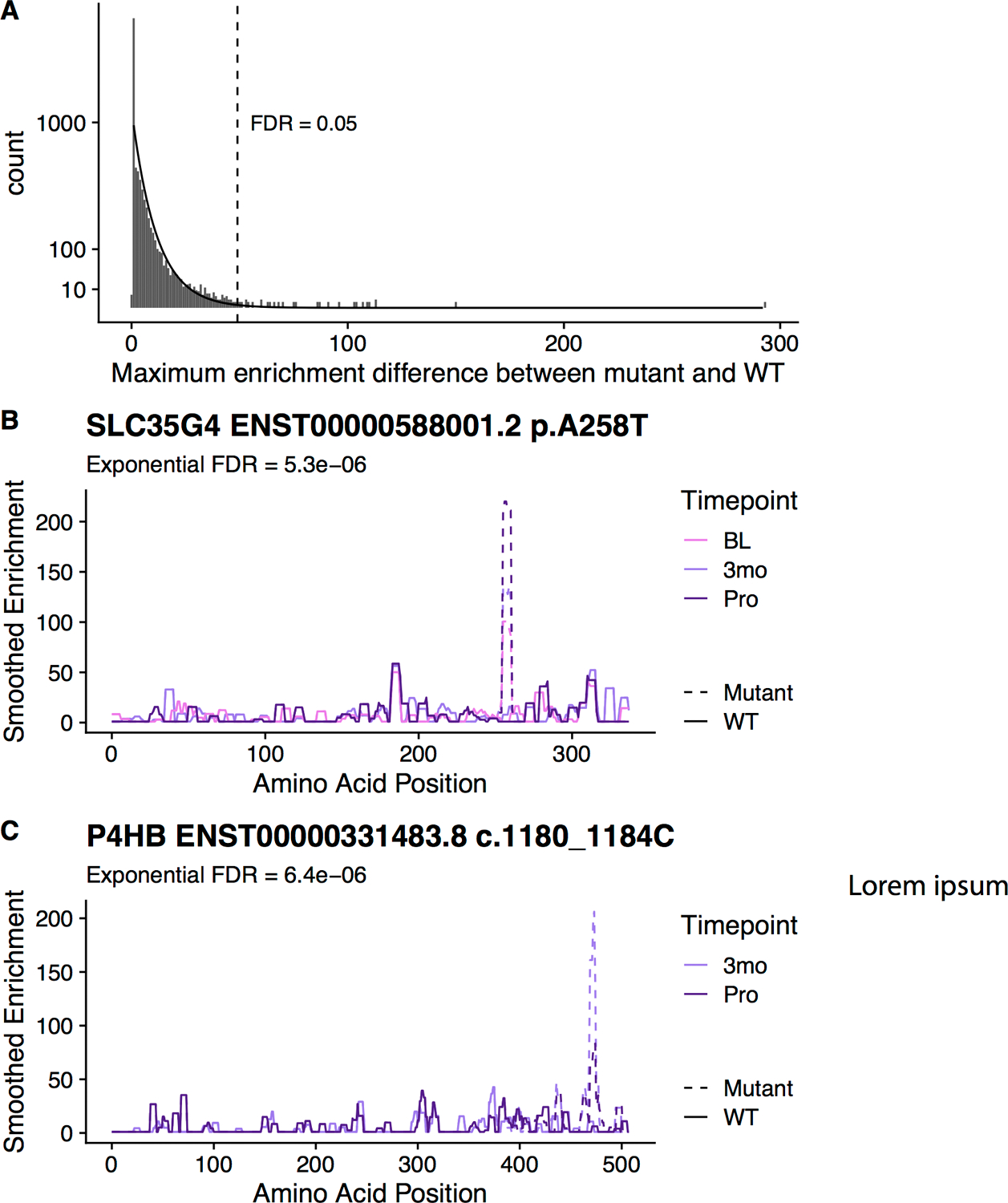
Analysis of mutation-specific epitopes in the prostate cancer patients. (A) Enrichments were calculated for every mutated protein in the affected patients (n mutations = 6,636; n patients = 72). An exponential distribution (indicated by the line) was fit to the data to calculate significance of each mutation. Difference between enrichment values in the mutant sequence and the wild type sequence in that patient are shown along the x-axis. (B) A high scoring point mutation in SLC35G4 is shown for patient DTB-129. (C) A high scoring frameshift mutation in P4HB is shown for patient DTB-102. (BL, baseline; 3mo, 3months after enrollment in study; Pro, disease progression). Note that no BL serum specimen was available for patient DTB-102 at time of study.
Next, to investigate cancer-specific autoantibodies resulting from non-mutant proteins, we performed a protein-based immunome-wide association study (PIWAS). We found 11 proteins to be significantly enriched for antibodies in mCRPC patients compared to healthy controls (Figure 2, Table 2/S3). The top two candidates were cancer-testis antigens NY-ESO-1 and NY-ESO-2, with the dominant epitope occurring in a conserved region between the proteins. Eight of 72 (11%) patients demonstrated PIWAS values > 6, all of which mapped to amino acids 11–30 of NY-ESO-1 (Figures 3A, S3A). PIWAS values for seven of these eight patients remained above the threshold at all timepoints (Figure S3B). Of note, this dominant B-cell epitope had been described in a previous study using a peptide approach and was found to be present in prostate cancer at a similar frequency37. In order to identify additional NY-ESO-1 antigenic regions, we applied the previously described IMUNE algorithm to identify peptide motifs that were significantly enriched in prostate cancer patients relative to healthy controls25. For this analysis, eight NY-ESO-1 PIWAS positive samples were analyzed by IMUNE using 30 healthy patient samples as controls. A total of nine cancer-specific motifs were identified that mapped to NY-ESO-1 (Figure 3B). While seven of the nine motifs aligned to the same portion of NY-ESO-1 identified by PIWAS, two of the motifs align to a new epitope around that 100th amino acid that is additionally present in samples without the PIWAS epitope. Samples with composite panel scores greater than 6.6 (based on a pre-defined 99% specificity threshold) were designated positive. Using this panel, nine of 72 patients (12.5%) including one patient without enrichment of the dominant epitope were positive for NY-ESO-1 at a specificity of 99% (Figures 3C, S3C).
Figure 2.

Manhattan plot of protein-based immunome wide association study (PIWAS) results highlighting antigens significantly enriched in prostate cancer compared to healthy control patients. Outlier sum FDRs are shown for every protein in the human proteome. Labels are shown for all proteins with an FDR < 0.05. Co-positivity of proteins with FDR < 0.05 are shown in Figure S2.
Table 2.
Top epitopes from prostate cancer PIWAS.
| Protein | Outlier Sum FDR | Top epitopes |
|---|---|---|
| Cancer/testis antigen 2 (NY-ESO-2) | 2.3E-12 | GIPDGPGGNAG, PDGPGGNAGGP, SPMEAELVRRI |
| Cancer/testis antigen 1 (NY-ESO-1) | 5.7E-11 | GIPDGPGGNAG, PDGPGGNAGGP, PPSGQRR |
| Immunoglobulin lambda variable 1–47 (IGLV1–47) | 4.1E-05 | YWYQQLPGTAP |
| Endogenous retrovirus group K member 113 Gag polyprotein (HERVK-113) | 0.0033 | NDWAIIKAALE, VIYPETLKLEG, IQPFVPQGFQG, QGFQGQQPPLS, GFQGQQPPLSQ, PLSQVFQGISQ |
| Uncharacterized protein | 0.0043 | YDPKEYDPFYM, FYMSKKDPNFL, SKKDPNFLKVT, ISNSRHFITPN |
| Solute carrier family 2, facilitated glucose transporter member 5 (SLC2A5) | 0.0074 | DQSMKEGRLTL, PLVNKFGRKGA, FFPESPRYLLI, VAEIRQEDEAE, AIYYYADQIYL, YYADQIYLSAG, IEINQIFTKMN |
| Receptor-interacting serine/threonine-protein kinase 3 (RIPK3) | 0.02 | HPPPVGSQEGP |
| Alpha-2,8-sialyltransferase 8E (ST8SIA5) | 0.02 | GPFEYNSTRCL, QEIFRMFPKDM |
| T cell receptor beta variable 25–1 (TRBV25–1) | 0.03 | YQQDPGMELHL |
| Squamous cell carcinoma antigen recognized by T-cells 3 (SART3) | 0.03 | MGPAWDQQEEG, DVEPPSKQKEK, MDGMTIKENII |
| Endogenous retrovirus group K member 24 Gag polyprotein (HERVK-24) | 0.033 | PEQGTLDLKDW, NDWAIIKAALE, VIYPETLKLEG, QGFQGQQPPLS, GFQGQQPPLSQ, PLSQVFQGISQ |
Figure 3.
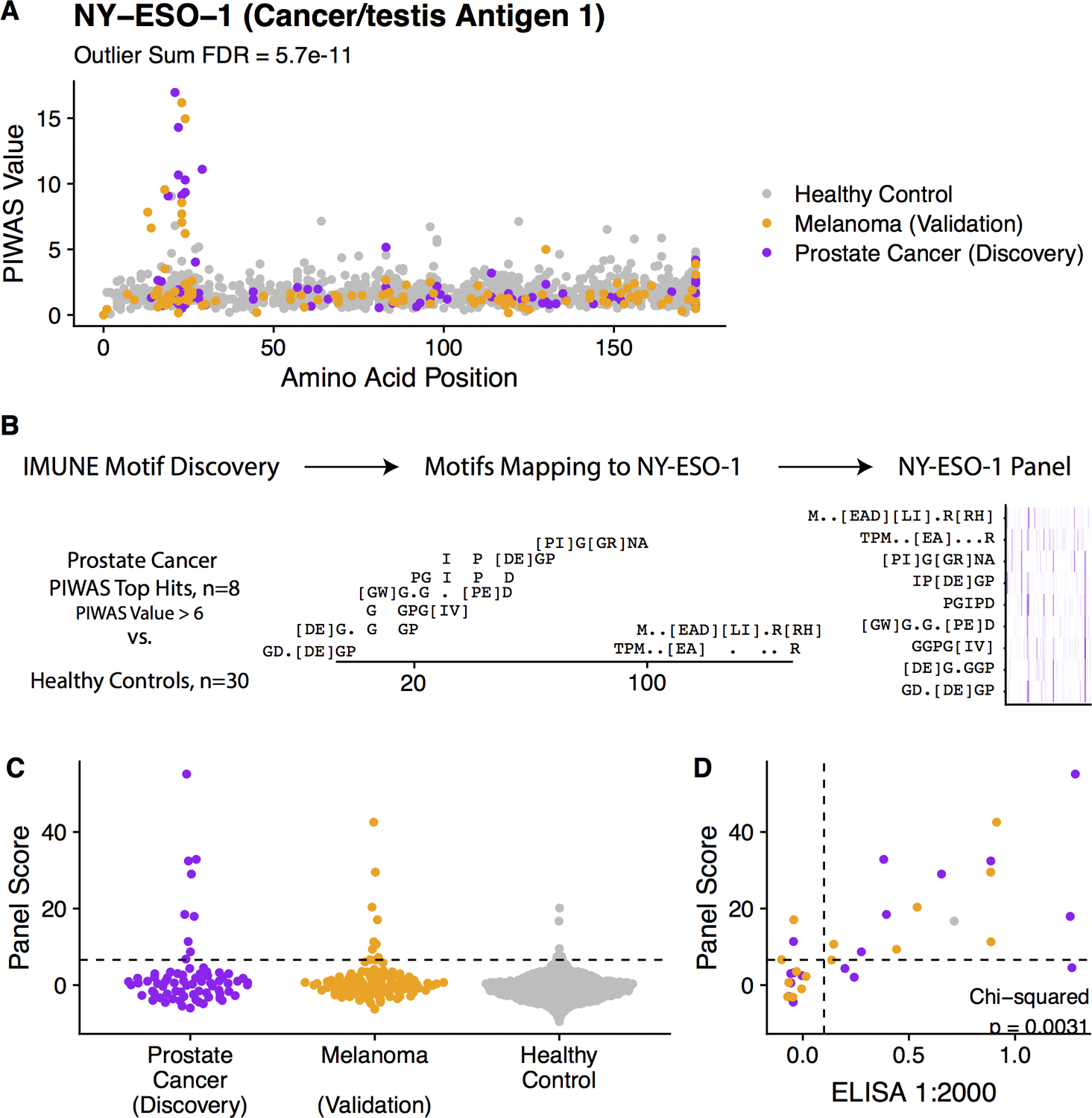
Discovery and validation of NY-ESO-1 antigenic signal. (A) Manhattan plot visualizing PIWAS values for NY-ESO-1, with one point per patient being shown and colored by patient subgroup (purple = prostate cancer (discovery, n=72), orange = melanoma (validation, n =218), (grey = controls without known disease (n=1,157)). (B) Prostate cancer samples that are positive by PIWAS are compared to healthy controls using IMUNE motif discovery algorithm. Motifs which map linearly to NY-ESO-1 are included. A panel score is calculated by summing enrichment z-scores across all motifs. (C) Dot plot of panel score for serum specimens stratified by patient subgroup. (D) Scatterplot demonstrating concordance between NY-ESO-1 panel score and ELISA results, assessed using the Chi-square test of independence. Points are colored based on patient subgroup. Additional results provided in Figure S3.
To validate this finding with an orthogonal serum profiling approach, the composite panel score results were benchmarked against a NY-ESO-1 ELISA experiment performed on the same prostate cancer serum samples. We found that the panel score and ELISA results were strongly associated (Cohen’s kappa = 0.57, Figure 3D). RNA-seq expression data revealed that NY-ESO-1 was expressed in the metastases of six of nine patients demonstrating NY-ESO-1 antibody enrichment at time of initial metastatic tumor biopsy, confirming antigenic availability in these patients (Table S4). Altogether, these findings demonstrated the ability of the joint PIWAS-IMUNE (PIWAS-I) approach to identify disease-specific epitopes in prostate cancer.
While cancer-specific, the dominant NY-ESO-1 epitope was previously known to be a tumor marker in not only advanced prostate cancer but also other cancers including melanoma. Specifically, a prior study found autoantibodies to the dominant NY-ESO-1 epitope to be enriched in 12.5% of melanoma samples37. To assess whether a similar finding would be observed using the PIWAS-I approach, we profiled the serum antibody repertoires of an independent cohort consisting of 106 melanoma patients. We observed the dominant NY-ESO-1 epitope enriched in 8 of 106 (7.5%) samples (Figures 3A, 3C), consistent with the prior report. This finding further validated the robustness of the PIWAS-I approach.
In addition to NY-ESO-1, the HERV (HML-2) family of proteins were also found by PIWAS to be significantly enriched for autoantibodies in mCRPC. 9 of 72 (12.5%) patients demonstrated significant enrichment of autoantibodies to HERVK-113, with recurrent epitopes identified near the 155th amino acid and C-terminus of the protein (Figures 4A, S4A). The presence of autoantibody enrichment to HERVK-113 was consistent across different timepoints in patients with multiple independently sampled serum specimens (Figure S4B). A motif panel for HERVK-113 was generated as described above using 9 PIWAS positive samples for IMUNE-based motif discovery (Figure 4B). The panel scores were enriched in 16 of 72 (22.2%) patients (Figures 4C, S4C). The panel scores were highly concordant with confirmatory ELISA testing (Cohen’s kappa = 0.91, Figure 4D). Additionally, RNA-seq expression of HERVK-113 was observed in all patients panel-score positive for HERVK-113, suggesting antigenic availability (Table S4).
Figure 4.
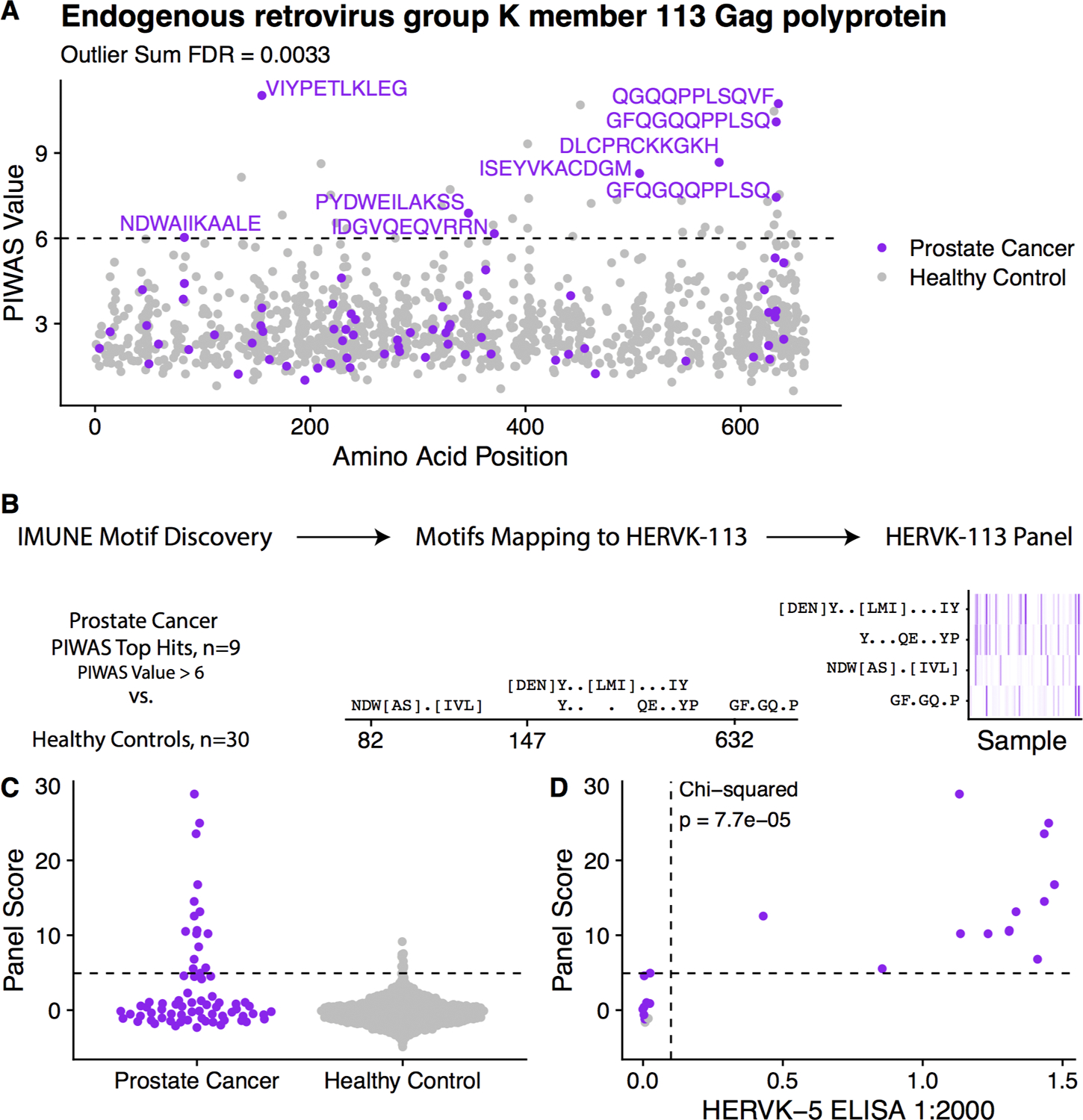
Discovery and validation of HERVK-113 antigenic signal. (A) Manhattan plot visualizing antibody-enrichment scores for HERVK-113. Epitopes associated with samples with a PIWAS value greater than 6 are labeled. (B) Prostate cancer samples that are positive by PIWAS are compared to healthy controls using IMUNE motif discovery algorithm. Motifs which map linearly to HERVK-113 are included. Panel score is calculated by summing enrichment z-scores across all motifs. (C) Dot plot of panel scores for prostate cancer patients compared to healthy controls. (D) Scatterplot of HERVK-113 panel score vs. HERVK-5 ELISA titer score. Additional results provided in Figure S4.
Additional mCRPC-specific epitopes were identified using the PIWAS approach. These included epitopes in the protein products of SART3, RIPK3, ST8SIA5, IGLV1–47, TRBV25–1, and SLC2A5 (Figure 2, Table S3). Seven of 72 (9.7%) patients demonstrated significant enrichment of autoantibodies to SART3, 6 of 72 (8.3%) demonstrated enrichment of autoantibodies to RIPK3, 5 of 72 (6.9%) demonstrated enrichment of autoantibodies to ST8SIA5, and 4 of 72 (5.6%) demonstrated enrichment of autoantibodies to IGLV1–47 (Figure S2). To assess autoantibody co-enrichment patterns, we assessed pairwise correlation between autoantibody enrichment scores of the eleven putative mCRPC-specific epitopes using our cohort of 72 patients (Figure S2A–B). Enrichment of autoantibodies to NY-ESO-1 and HERVK-113 was mutually exclusive on PIWAS analysis, as no patients demonstrated significant antibody enrichment to both NY-ESO-1 and HERVK-113 (Figure S2A). Presence of autoantibody enrichment to epitopes in NY-ESO-1 or HERVK-113 were not prognostic of overall survival (Figures S3D, S4D). We also examined the association between individual autoantibody enrichments and clinical variables including Gleason Grade Group, site of metastasis, prior abiraterone therapy, and prior enzalutamide therapy (Figure S5-S8). None of these variables were significantly associated with observed autoantibody enrichments, at least partially due to limited statistical power given relatively low enrichment prevalences and modest cohort size.
PSA and PSMA are of particular interest given their utility as prostate cancer biomarkers and potential therapeutic targets. Thus, although our unbiased PIWAS analysis did not identify autoantibodies to PSA or PSMA as being significantly enriched in mCRPC, we performed a tiled enrichment analysis to directly assess the potential antigenicity of these proteins. We confirmed that no autoantibody enrichment to sub-peptides of PSA or PSMA was observed (Figure S9).
Discussion
Herein, we have characterized the autoantibody landscape of metastatic castration-resistant prostate cancer. We observed cancer-specific enrichment of antibodies to mutant peptides in select genes and to non-mutant peptides in the NY-ESO-1 and HERVK-113 proteins among others.
Previous reports demonstrated that disease-specific neoepitopes may include defective gene products resulting from somatic alterations such as mutations38 and errors in protein translation39. The extension of this principle to cancer is supported by prior studies in lung and colorectal cancers, which found that tumors with missense mutations in TP53 and frameshift mutations in select genes were associated with autoantibodies to the mutant protein products40,41. We performed the first comprehensive assessment of cancer-specific B cell neoantigens to date and observed several examples of this phenomenon in genes such as SLC35G4 and P4HB. However, the majority (99.6%) of somatic mutations did not result in antibody-specific epitopes in our cohort. This finding is consistent with prior T-cell studies, which found that only a minority of mutations stimulate a specific T-cell response36,42. In the present study, mutations generating the strongest detected responses were approximately equally distributed between missense and frameshift mutations.
The mutations that were observed to be associated with an epitope-specific humoral immune response tended to be private to individual patients rather than shared among multiple individuals. This observation too is consistent with a prior report in colorectal cancer42 and may be due to the fact that somatic mutations themselves (and hence the resulting aberrant mutant protein) tend not to be recurrent across mCRPC patients. Nevertheless, we observed that the autoantibodies to mutant peptides were often present in multiple serum specimens collected independently from the same patient. These data validate the specificity of the epitope profiling and PIWAS approaches and support the notion that select mutations may induce a humoral immune response in mCRPC.
We observed enrichment of autoantibodies to not only mutant but also wild-type epitopes. This is supported by prior studies which suggest that cancer-specific overexpression of non-mutant antigens may comprise the majority of tumor-associated antigens43–46. NY-ESO-1, or cancer testis antigen 1B (CTGAG1B), has been well-characterized as an antigen that elicits humoral immune responses in various cancers including melanoma and breast, lung, bladder, ovarian, and prostate cancers47–49. Additionally, given its cancer-specific expression pattern outside of the testes49–52, NY-ESO-1 has shown great promise as a potential target for T-cell immunotherapies in various cancers47. In our unbiased approach to identifying immunogenic antigens, we found that NY-ESO-1 was the top candidate. Moreover, by experimental design, we were able to identify the specific recurrent motif that has been previously demonstrated to be immunogenic in multiple cancer types37,53. These findings, along with empiric validation via the ELISA approach, support the notion that PIWAS-I can be used to reliably recover immunogenic motifs in cancer.
The PIWAS-I approach also identified epitopes in HERVK-113. Human endogenous retroviruses (HERVs) comprise a family of retroviruses whose genetic material has previously been integrated into the human germline and whose gene products have been implicated in cancer pathogenesis54,55. HERVs have been previously described as being transcriptionally activated and potentially antigenic in the context of cancer: prior studies of renal cancers and seminomas identified a cancer-specific IgG response to ERVK-1022,56,57. Humoral responses to HERVs have similarly been reported in melanomas58 and ovarian59, breast60,61, and prostate62 cancers. Additionally, the gene product of HERV-K may be not only a biomarker of disease but also a therapeutic target, as a preclinical model demonstrated that monoclonal antibodies against the HERV-K env protein was associated with inhibition of tumor growth in breast cancer63. In prostate cancer particularly, detection of autoantibodies to the HERV-K gag protein has been shown to be enriched in advanced prostate cancer relative to early prostate cancer (21% vs. 1.4%) and associated with poor survival outcomes64. We observed a similar prevalence of 22% for HERV-K antibody enrichment in our cohort of advanced prostate cancer patients. However, we observed no significant difference in overall survival (OS) between mCRPC patients with and without HERV-K antibody enrichment. In contrast to the previously studied cohort, our cohort was comprised exclusively of advanced prostate cancer patients. Thus, our findings suggest that autoantibodies to the HERV-K protein may be associated with disease burden but may not be prognostic of OS amongst patients with advanced disease. We also found that autoantibody enrichments to HERV-K and NY-ESO-1 were mutually exclusive on PIWAS analysis. This observation may reflect a difference in underlying tumor biology driving different patients’ cancers, although the finding should be interpreted with caution due to the relatively modest size of our cohort. Additional prospective studies are needed to explore the prognostic value of HERV-K antibody enrichment in greater detail.
Additional antigens identified through the PIWAS-I approach included IGLV1–47, TRBV25–1, SART3 and RIPK3. Autoantibodies to IGLV1–47 and TRBV25–1 have been described as elevated in various inflammatory states, including in patients with systemic lupus erythematosus and rheumatoid arthritis65–68. Thus, they are more likely to be nonspecific markers of chronic inflammation rather than cancer-specific biomarkers. On the other hand, SART3 and RIPK3 have been previously implicated as biomarkers or potential regulators of cancer progression. SART3 is a cancer testis antigen that is expressed specifically in various cancer tissues (excluding normal testis)69,70. SART3 has been shown to induce both a humoral and cellular adaptive immune response in a vaccination study of patients with advanced colorectal cancer71. RIPK3 is a tumor suppressor whose downregulation has been associated with tumorigenesis, immunomodulation, and poor clinical prognosis in colorectal cancer72,73, although its exact role in the adaptive immunity is still under investigation. While confirmatory studies are needed, the present study in conjunction with supporting studies in other cancer types nominates potential immune biomarkers and therapeutic targets in mCRPC.
In addition to recovering known and novel epitopes of interest in mCRPC, the findings of the present study highlight potentially conserved B- and T-cell cancer-specific epitopes and a combined B- and T-cell response to cancer. NY-ESO-1 has been found to elicit a high-titer IgG humoral response as well as a cellular immune response in patients with melanoma47,74. SART3, or “Squamous cell carcinoma antigen recognized by T-cells 3,” was initially discovered as a T-cell epitope and was later found to also stimulate a correlated IgG response69,71. More generally, cancer vaccination studies have demonstrated how the humoral immune response to cancer-associated antigens may provide insights into targets of the endogenous cellular immune system12,71. Future work profiling paired patient serum and T-cells may more definitively assess the overlap between B- and T-cell epitopes. Additionally, profiling patients on clinical trials such as KEYNOTE-199 would be instrumental in identifying potential immune markers of exceptional response to immunotherapy75.
The present study is not without limitations. First, we acknowledge that our mCRPC cohort is of relatively modest size. Also, while the PIWAS-IMUNE approach has been successfully applied to immunologic26 and now oncologic diseases to recover validated B cell epitopes, ELISA-based validation experiments were performed on only the top two candidate epitopes (NY-ESO-1 and HERVK-113) in our mCRPC cohort. The patients described in this study should be considered a discovery cohort, and independent cohorts ideally of larger size are needed to validate our findings. Finally, the present study is limited by a lack of available paired peripheral blood mononuclear cell specimens and inability to investigate concomitant B- and T-cell responses. Additional studies are needed to further elucidate the frequency of epitopes on shared antigens among the humoral and cellular immune systems in mCRPC and the extent to which each contributes to antitumor activity.
In summary, we leveraged recently published epitope profiling techniques to characterize the autoantibody landscape of mCRPC and identify cancer-specific antigens and epitopes. By pairing patient serum profiling with whole-genome sequencing results of paired solid-tumor biopsies, we identified 29 novel epitopes to mutant peptides generated by patient-specific somatic mutations. We also identified 11 conserved protein antigens, with several supported by prior reports in other cancer cohorts. Our findings and the presented next generation sequencing-based approach to autoantibody profiling provide insight into immune biomarkers and potential therapeutic targets in advanced prostate cancer.
Supplementary Material
Statement of significance:
Autoantibodies have been shown to inform treatment response and candidate drug targets in various cancers. We present the first large-scale profiling of autoantibodies in advanced prostate cancer, utilizing a new next-generation sequencing-based approach to antibody profiling to reveal novel cancer-specific antigens and epitopes.
Acknowledgements
This study was funded in part by a Stand Up To Cancer-Prostate Cancer Foundation-Prostate Dream Team Translational Cancer Research Grant. This research grant is made possible by the generous support of the Movember Foundation. Stand Up To Cancer is a division of the Entertainment Industry Foundation. The research grant is administered by the American Association for Cancer Research. MS was supported by the Swedish Research Council (Vetenskapsrådet) with grant number 2018–00382 and the Swedish Society of Medicine (Svenska Läkaresällskapet). A.R. was funded by the Parker Institute for Cancer Immunotherapy, the Ressler Family Fund, support from Ken and Donna Schultz, and NIH grants R35 CA197633 and P01 CA244118. DYO was supported by a Young Investigator Award from the Prostate Cancer Foundation and the following NIH grant: NIH/NIAID 5K08AI139375-02. FYF was funded by Prostate Cancer Foundation Challenge Awards and the following NIH grants: NIH/ NCI 1R01CA230516–01, NIH / NCI 1R01CA227025–01A1, NIH 2U10CA180868–06, NIH P50CA186786. We thank the Serimmune team for supporting the processing and analysis of samples, including: Jack Reifert, Joel Bozekowski, Brian Martinez, Gregory Jordan, Timothy Johnston, Cameron Gable, Steve Kujawa, Elisabeth Baum-Jones. Special thanks to Elizabeth Stewart for editing this manuscript.
Footnotes
Disclosure of Potential Conflicts of Interest
W.A. Haynes reports a patent titled immunome wide association studies to identify condition-specific antigens for PCT/US20/38856 pending, owned by Serimmune, not licensed yet, and employment with and shareholder at Serimmune. R. Waitz reports personal fees from Serimmune (employment and ownership of company shares) during the conduct of the study and outside the submitted work. K. Kamath reports other from Serimmune Inc (receives salary and stock options) outside the submitted work, as well as has an unlicensed patent for PCT/US20/38856 methods and compositions for assessing antibody specificities pending and a patent for pub. no.: US 2018/0267056 A1 issued to Serimmune Inc. M. Zhang reports other from Serimmune Inc (employee who receives equity compensation) outside the submitted work. S.G. Zhao reports patent applications for biomarkers unrelated to this work in prostate cancer with Decipher Biosciences, Celgene, and in breast cancer with PFS Genomics, and that a family member holds a leadership role in PFS Genomics. J. Chou reports grants from A.P. Giannini Foundation during the conduct of the study. T.M. Beer reports grants from Stand Up To Cancer—Prostate Cancer Foundation Dream Team during the conduct of the study, Alliance Foundation Trials, Corcept Therapeutics, Endocyte Inc, Harpoon Therapeutics, Janssen Research & Development, Medivation, Inc., Sotio, Theraclone Sciences/OncoResponse, and Zenith Epigenetics, personal fees and other from Arvinas (consulting and stock ownership), personal fees from AstraZeneca, Bayer, Bristol-Myers Squib, Clovis Oncology, GlaxoSmithKline, Janssen Biotech, Merck, Novartis, Pfizer, Tolero, and Sanofi, grants and personal fees from Astellas Pharma and Boehringer Ingelheim, and other from Salarius Pharmaceuticals (stock ownership) outside the submitted work. M. Rettig reports grants from Novartis, personal fees from Johnson & Johnson (speakers’ bureau), Bayer (speakers’ bureau), Ambrx (consultant), and Amgen (consultant), other from Constellation (consultant), nonfinancial support from Astellas (drug for clinical trial), Pfizer (drug for clinical trial), and Merck (drug for clinical trial) during the conduct of the study, as well as has a patent for N-terminal androgen receptor antagonist for treatment of prostate cancer pending to no licensee yet, owned by UCLA. C.P. Evans reports grants from Stand Up To Cancer during the conduct of the study. K.N. Chi reports grants from Stand Up to Cancer during the conduct of the study, grants and personal fees from Amgen, Astellas, Janssen, Astra Zeneca, Sanofi, and Novartis, and personal fees from Merck, Daiichi Sankyo, and Point Biopharma. J.J. Alumkal reports grants from Stand Up to Cancer Foundation during the conduct of the study, Zenith Epigenetics (research support to institution), Gilead Sciences (research support to institution), and Aragon Pharmaceuticals (research support to institution), grants and personal fees from Astellas (consulting and research support to institution) and Janssen Biotech (consulting and research support to institution), and personal fees from Merck (consulting) and Dendreon (consulting) outside the submitted work. A. Ashworth reports other from Tango Therapeutics (cofounder), Ovibio Corporation (cofounder), Azkarra Therapeutics (cofounder), SPARC (consultant), Bluestar (consultant), ProLynx (consultant), Earli (consultant), Cura (consultant), GenVivo (consultant), GlaxoSmithKline (consultant), Genentech (SAB), Circle (SAB), and Gladiator (SAB), grants from SPARC (research support) and AstraZeneca (research support), and other from Cambridge Science Corporation (board) outside the submitted work, as well as has patents on the use of PARP inhibitors held jointly with AstraZeneca from which benefitted financially (and may do so in the future). R. Aggarwal reports grants from AstraZeneca, Janssen, Novartis, Amgen, Zenith Epigenetics, and Xynomic Pharmaceuticals, personal fees from Dendreon and Clovis, and grants and personal fees from Merck outside the submitted work. P.S. Daugherty reports a patent 8361933 issued, licensed, and with royalties paid from Serimmune and a patent 8293685 issued, licensed, and with royalties paid from Serimmune, both owned by the University of California and licensed to Serimmune, both on the topic of display of peptides on the outer surface of E. coli bacteria (i.e., E. coli surface display). This technology is part of the method Serimmune uses to generate datasets, as well as is an employee stockholder, and director of Serimmune Inc. A. Ribas reports personal fees from Amgen, AstraZeneca, Checkmate, Merck, Novartis, Sanofi, Vedanta (honoraria from consulting), Advaxis, CytomX, Five Prime, RAPT, Isoplexis, Kite-Gilead (stock owner), 4C Biomed, Apricity, Arcus, Highlight, Compugen, ImaginAb, MapKure, Merus, Rgenix, Lutris, PACT Pharma, and Tango (scientific advisory board and stock owner) and grants from Agilent, Bristol-Myers Squibb (research support) outside the submitted work. D.Y. Oh reports grants from Prostate Cancer Foundation (Young Investigator Award) during the conduct of the study, other from Roche/Genentech (research support) and Merck (research support), and personal fees from Maze Therapeutics (consulting) outside the submitted work. J.C. Shon reports personal fees from Serimmune (Employee) during the conduct of the study, as well as a patent for PIWAS method pending. F.Y. Feng reports other from SerImmune (scientific advisor and stock options) during the conduct of the study, as well as personal fees from Astellas (Consultant), Genentech (Consultant), Celgene (Consultant), Roivant (Consultant), Myovant (Consultant), Janssen (Consultant), Blue Earth Diagnostics (Consultant), PFS Genomics (Co-Founder), and Sanofi (Consultant) outside the submitted work. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Chapman C et al. Autoantibodies in breast cancer: their use as an aid to early diagnosis. Annals of Oncology 18, 868–873 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Wang X et al. Autoantibody Signatures in Prostate Cancer. N Engl J Med 353, 1224–1235 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Zhong L et al. Antibodies to HSP70 and HSP90 in serum in non-small cell lung cancer patients. Cancer Detection and Prevention 27, 285–290 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Hennequin A et al. Tumor infiltration by Tbet+ effector T cells and CD20+ B cells is associated with survival in gastric cancer patients. OncoImmunology 5, e1054598 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berntsson J, Nodin B, Eberhard J, Micke P & Jirström K Prognostic impact of tumour-infiltrating B cells and plasma cells in colorectal cancer: 2.1.5 Tumor Immunology and Microenvironment. Int. J. Cancer 139, 1129–1139 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Garg K et al. Tumor-associated B cells in cutaneous primary melanoma and improved clinical outcome. Human Pathology 54, 157–164 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Knief J et al. High Density of Tumor-infiltrating B-Lymphocytes and Plasma Cells Signifies Prolonged Overall Survival in Adenocarcinoma of the Esophagogastric Junction. AR 36, 5339–5346 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Helmink BA et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577, 549–555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petitprez F et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 577, 556–560 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Montgomery RB, Makary E, Schiffman K, Goodell V & Disis ML Endogenous anti-HER2 antibodies block HER2 phosphorylation and signaling through extracellular signal-regulated kinase. Cancer Res 65, 650–656 (2005). [PubMed] [Google Scholar]
- 11.Tabuchi Y et al. Protective effect of naturally occurring anti-HER2 autoantibodies on breast cancer. Breast Cancer Res Treat 157, 55–63 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Hulett TW et al. Coordinated responses to individual tumor antigens by IgG antibody and CD8+ T cells following cancer vaccination. j. immunotherapy cancer 6, 27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheikh NA et al. Sipuleucel-T immune parameters correlate with survival: an analysis of the randomized phase 3 clinical trials in men with castration-resistant prostate cancer. Cancer Immunol Immunother 62, 137–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarin DA, Fain KM, Dobbins HD, Tse T & Williams RJ 10-Year Update on Study Results Submitted to ClinicalTrials.gov. N. Engl. J. Med 381, 1966–1974 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowicki TS et al. A Pilot Trial of the Combination of Transgenic NY-ESO-1-reactive Adoptive Cellular Therapy with Dendritic Cell Vaccination with or without Ipilimumab. Clin Cancer Res 25, 2096–2108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson WH, Steinman L & Utz PJ Protein arrays for autoantibody profiling and fine-specificity mapping. Proteomics 3, 2077–2084 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Kijanka G & Murphy D Protein arrays as tools for serum autoantibody marker discovery in cancer. J Proteomics 72, 936–944 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Bouwman K et al. Microarrays of tumor cell derived proteins uncover a distinct pattern of prostate cancer serum immunoreactivity. Proteomics 3, 2200–2207 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Mintz PJ et al. Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat Biotechnol 21, 57–63 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Smith GP Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985). [DOI] [PubMed] [Google Scholar]
- 21.Giordano RJ, Cardó-Vila M, Lahdenranta J, Pasqualini R & Arap W Biopanning and rapid analysis of selective interactive ligands. Nat. Med 7, 1249–1253 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Sahin U et al. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc. Natl. Acad. Sci. U.S.A 92, 11810–11813 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klade CS et al. Identification of tumor antigens in renal cell carcinoma by serological proteome analysis. Proteomics 1, 890–898 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Zaenker P & Ziman MR Serologic autoantibodies as diagnostic cancer biomarkers--a review. Cancer Epidemiol. Biomarkers Prev 22, 2161–2181 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Pantazes RJ et al. Identification of disease-specific motifs in the antibody specificity repertoire via next-generation sequencing. Sci Rep 6, 30312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haynes WA, Kamath K, Daugherty PS & Shon JC Protein-based Immunome Wide Association Studies (PIWAS) for the discovery of significant disease-associated antigens http://biorxiv.org/lookup/doi/10.1101/2020.03.18.997759 (2020) 10.1101/2020.03.18.997759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mintz PJ et al. Discovery and horizontal follow-up of an autoantibody signature in human prostate cancer. Proc Natl Acad Sci USA 112, 2515–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graff JN, Puri S, Bifulco CB, Fox BA & Beer TM Sustained Complete Response to CTLA-4 Blockade in a Patient with Metastatic, Castration-Resistant Prostate Cancer. Cancer Immunology Research 2, 399–403 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie C et al. A novel multiplex assay combining autoantibodies plus PSA has potential implications for classification of prostate cancer from non-malignant cases. J Transl Med 9, 43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwek SS et al. Diversity of antigen-specific responses induced in vivo with CTLA-4 blockade in prostate cancer patients. J. Immunol 189, 3759–3766 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamath K et al. Antibody epitope repertoire analysis enables rapid antigen discovery and multiplex serology. Sci Rep 10, 5294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quigley DA et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 174, 758–769.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen WS et al. Genomic Drivers of Poor Prognosis and Enzalutamide Resistance in Metastatic Castration-resistant Prostate Cancer. European Urology S0302283819302064 (2019) 10.1016/j.eururo.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen WS et al. Genomic Drivers of Poor Prognosis and Enzalutamide Resistance in Metastatic Castration-resistant Prostate Cancer. European Urology 76, 562–571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 57, 289–300 (1995). [Google Scholar]
- 36.Efremova M, Finotello F, Rieder D & Trajanoski Z Neoantigens Generated by Individual Mutations and Their Role in Cancer Immunity and Immunotherapy. Front. Immunol 8, 1679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng G et al. Dominant B cell epitope from NY-ESO-1 recognized by sera from a wide spectrum of cancer patients: Implications as a potential biomarker. Int. J. Cancer 114, 268–273 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Bassani-Sternberg M et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 7, 13404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kracht MJL et al. Autoimmunity against a defective ribosomal insulin gene product in type 1 diabetes. Nat Med 23, 501–507 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Winter SF et al. Development of antibodies against p53 in lung cancer patients appears to be dependent on the type of p53 mutation. Cancer Res 52, 4168–4174 (1992). [PubMed] [Google Scholar]
- 41.Schwitalle Y et al. Immune Response Against Frameshift-Induced Neopeptides in HNPCC Patients and Healthy HNPCC Mutation Carriers. Gastroenterology 134, 988–997 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Angelova M et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol 16, 64 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe N, Arase H, Onodera M, Ohashi PS & Saito T The quantity of TCR signal determines positive selection and lineage commitment of T cells. J. Immunol 165, 6252–6261 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Hong Y Autoantibodies against tumor-associated antigens for detection of hepatocellular carcinoma. WJH 7, 1581 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodell V et al. Level of HER-2/neu protein expression in breast cancer may affect the development of endogenous HER-2/neu-specific immunity. Molecular Cancer Therapeutics 7, 449–454 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Zaenker P, Gray ES & Ziman MR Autoantibody Production in Cancer—The Humoral Immune Response toward Autologous Antigens in Cancer Patients. Autoimmunity Reviews 15, 477–483 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Thomas R et al. NY-ESO-1 Based Immunotherapy of Cancer: Current Perspectives. Front. Immunol 9, 947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugita Y et al. NY-ESO-1 Expression and Immunogenicity in Malignant and Benign Breast Tumors. Cancer Res 64, 2199–2204 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Gati A et al. NY-ESO-1 expression and immunogenicity in prostate cancer patients. Tunis Med 89, 779–783 (2011). [PubMed] [Google Scholar]
- 50.Old LJ & Chen Y-T New Paths in Human Cancer Serology. J Exp Med 187, 1163–1167 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boon T, Coulie PG & Van den Eynde B Tumor antigens recognized by T cells. Immunol. Today 18, 267–268 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Fosså A et al. NY-ESO-1 protein expression and humoral immune responses in prostate cancer. Prostate 59, 440–447 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Robbins PF et al. Tumor Regression in Patients With Metastatic Synovial Cell Sarcoma and Melanoma Using Genetically Engineered Lymphocytes Reactive With NY-ESO-1. JCO 29, 917–924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson PN Demystified … Human endogenous retroviruses. Molecular Pathology 56, 11–18 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schulte AM et al. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc. Natl. Acad. Sci. U.S.A 93, 14759–14764 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauter M et al. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol 69, 414–421 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauter M et al. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res 56, 4362–4365 (1996). [PubMed] [Google Scholar]
- 58.Humer J et al. Identification of a melanoma marker derived from melanoma-associated endogenous retroviruses. Cancer Res 66, 1658–1663 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Wang-Johanning F et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int. J. Cancer 120, 81–90 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Golan M et al. Human Endogenous Retrovirus (HERV-K) Reverse Transcriptase as a Breast Cancer Prognostic Marker. Neoplasia 10, 521–IN2 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang-Johanning F et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res 68, 5869–5877 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishida T et al. Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun 8, 15 (2008). [PMC free article] [PubMed] [Google Scholar]
- 63.Wang-Johanning F et al. Immunotherapeutic Potential of Anti-Human Endogenous Retrovirus-K Envelope Protein Antibodies in Targeting Breast Tumors. JNCI: Journal of the National Cancer Institute 104, 189–210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reis BS et al. Prostate Cancer Progression Correlates with Increased Humoral Immune Response to a Human Endogenous Retrovirus GAG Protein. Clinical Cancer Research 19, 6112–6125 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Marchalonis JJ, Kaymaz H, Schluter SF & Yocum DE Naturally Occurring Human Autoantibodies to Defined T-Cell Receptor and Light Chain Peptides in Immunobiology of Proteins and Peptides VII (ed. Atassi MZ) vol. 347 135–145 (Springer US, 1994). [DOI] [PubMed] [Google Scholar]
- 66.Schluter SF, Wang E, Winfield JB, Yocum DE & Marchalonis JJ Autoregulation of TCR V Region Epitopes in Autoimmune Disease in Immunobiology of Proteins and Peptides VIII (eds. Atassi MZ & Bixler GS) vol. 383 231–236 (Springer US, 1995). [DOI] [PubMed] [Google Scholar]
- 67.Adelman MK, Schluter SF, Robey IF & Marchalonis JJ PART I. Peptide ImmunotherapyNatural and Autoantibodies to Human T-Cell Receptor Vβ Segments: Potential Roles in Immunomodulation. Crit Rev Immunol 27, 221–232 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Lobo PI Role of Natural Autoantibodies and Natural IgM Anti-Leucocyte Autoantibodies in Health and Disease. Front. Immunol 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang D et al. Identification of a gene coding for a protein possessing shared tumor epitopes capable of inducing HLA-A24-restricted cytotoxic T lymphocytes in cancer patients. Cancer Res 59, 4056–4063 (1999). [PubMed] [Google Scholar]
- 70.Ito M et al. Identification of SART3-derived peptides capable of inducing HLA-A2-restricted and tumor-specific CTLs in cancer patients with different HLA-A2 subtypes. Int. J. Cancer 88, 633–639 (2000). [DOI] [PubMed] [Google Scholar]
- 71.Miyagi Y et al. Induction of cellular immune responses to tumor cells and peptides in colorectal cancer patients by vaccination with SART3 peptides. Clin. Cancer Res 7, 3950–3962 (2001). [PubMed] [Google Scholar]
- 72.Yan G et al. A RIPK3-PGE 2 Circuit Mediates Myeloid-Derived Suppressor Cell–Potentiated Colorectal Carcinogenesis. Cancer Res 78, 5586–5599 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Conev NV et al. RIPK3 expression as a potential predictive and prognostic marker in metastatic colon cancer. CIM 42, E31–E38 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Jäger E et al. Monitoring CD8 T cell responses to NY-ESO-1: correlation of humoral and cellular immune responses. Proc. Natl. Acad. Sci. U.S.A 97, 4760–4765 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antonarakis ES et al. Pembrolizumab for Treatment-Refractory Metastatic Castration-Resistant Prostate Cancer: Multicohort, Open-Label Phase II KEYNOTE-199 Study. JCO 38, 395–405 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


