Abstract
FBW7 functions as a tumor suppressor by targeting oncoproteins for degradation. Our previous study found FBW7 was low expressed in pancreatic cancer due to sustained activation of Ras-Raf-MEK-ERK pathway, which destabilized FBW7 by phosphorylating at Thr205. MicroPET/CT imaging results revealed that FBW7 substantially decreased 18F-fluorodeoxyglucose uptake in xenograft tumors. Mechanistically, FBW7 inhibited glucose metabolism via c-Myc/TXNIP axis. But in these studies, we observed FBW7 down-regulated genes were widely involved in redox reaction and lipid metabolism. Here we reanalyzed previous gene expression profiling and conducted targeted cell metabolites analysis. Results revealed that FBW7 regulated lipid peroxidation and promoted ferroptosis, a non-apoptotic form of cell death. Mechanistically, we found FBW7 inhibited the expression of stearoyl-CoA desaturase (SCD1) via inhibiting nuclear receptor subfamily 4 group A member 1 (NR4A1). SCD1 was reported to inhibit both ferroptosis and apoptosis, which was consistent with the function of FBW7 and NR4A1, another FBW7 down-regulated gene in the gene expression profiling. Moreover, FBW7 potentiated cytotoxic effect of gemcitabine via activating ferroptosis and apoptosis. Combination ferroptosis inducers and apoptosis activators could also significantly potentiated cytotoxic effect of gemcitabine in pancreatic cancer. Therefore, our findings might provide new strategies for the comprehensive treatment of pancreatic cancer.
Keywords: Pancreatic cancer, FBW7, Apoptosis, Ferroptosis
Graphical abstract
Highlights
-
•
Ferroptosis possesses great potential in pancreatic cancer therapy.
-
•
FBW7 synchronously induces apoptosis and ferroptosis.
-
•
Activation of apoptosis and ferroptosis potentiates cytotoxic effect of gemcitabine.
1. Introduction
Pancreatic cancer ranks the fourth leading cause of cancer death in the USA [1]. Surgical resection is the only chance to cure pancreatic cancer, but due to later diagnosis, most patients lost the chance of surgical excision. Moreover, pancreatic cancer responds poorly to most chemotherapeutic agents [2]. Hence, it is urgent to understand the biological mechanisms and molecular mechanisms to find new strategies for the comprehensive treatment of pancreatic cancer.
FBW7 (F-box and WD repeat domain-containing 7) is the substrate recognition component of the Skp1-Cul1-F-box (SCF) ubiquitin ligase complex and targets various oncoproteins for degradation, including MYC, cyclin E, Notch and JUN. Furthermore, loss of FBW7 function leads to chromosomal instability and tumorigenesis [3]. FBW7 gene is one of the most frequently mutated genes in human cancers [4]. However, in our previous studies, sequencing analysis revealed that fewer than 2% of pancreatic cancer samples harbored FBW7 mutations. Nonetheless, we found FBW7 was low expression in most pancreatic cancer samples. Mechanistically, we found that ERK kinase phosphorylated FBW7 at the T205 site, which caused destabilization of FBW7 in pancreatic cancer [5]. Functionally, we detected that FBW7 negatively regulated glucose metabolism in pancreatic cancer. With gene expression profiling, we further confirmed that FBW7 regulated glucose metabolism via c-Myc/TXNIP axis [6]. However, we noticed that in gene expression profiling, FBW7 down-regulated genes were widely involved in redox reaction and lipid metabolism. Hence, FBW7 might regulate lipid peroxidation.
Ferroptosis is an iron-dependent form of non-apoptotic cell death, which is characterized by lipid peroxidation [7]. Emerging evidence shows ferroptosis is the nexus between metabolism, redox biology, and human health. Fortunately, triggering ferroptosis exhibited great potential for cancer therapy, particularly for eradicating aggressive malignancies that responded poorly to traditional therapies [8,9]. Ferroptosis has a long-standing relationship with RAS mutation. Originally, Brent R. Stockwell screened small molecules that could specifically kill RAS mutant cancer cells. They detected that the death triggered by some small molecules, such as erastin and RAS-selective lethal 3 (RSL3), was an iron-dependent, non-apoptotic form of cell death [10,11]. Coincidentally, approximately 95% of pancreatic tumors possess KRAS gene mutation [12]. However, the study concerning ferroptosis in pancreatic cancer is insufficient. We previously reported FBW7 was inhibited by Ras–Raf–MEK–ERK pathway [6]. Furthermore, gene expression profiling indicated FBW7 might involve in lipid peroxidation. Hence, we speculated FBW7 could regulate ferroptosis in pancreatic cancer.
Previously, FBW7 was reported to promote apoptosis in various cells [13,14]. Furthermore, FBW7 had potential in regulating the sensitivity to chemotherapeutics [15,16]. Gemcitabine is the cornerstone for the comprehensive therapy of pancreatic cancer. But due to pancreatic cancer cells readily acquired the ability to mitigate gemcitabine-induced apoptosis, gemcitabine resistance developed within weeks of chemotherapy initiation [17]. Ferroptosis is a non-apoptotic cell death. Emerging evidence shows ferroptosis has enormous potential in cancer-acquired drug resistance [8,18,19]. Hence, it is reasonable to speculate that the combination of ferroptosis and apoptosis enhances the lethality to cancer cells.
In this study, we investigated the impact of FBW7 on ferroptosis in pancreatic cancer. We found FBW7 could activate both ferroptosis and apoptosis. Mechanistically, FBW7 exerted these effects by inhibiting the expression of NR4A1 and SCD1, which could also induce both ferroptosis and apoptosis. Eventually, FBW7 powerfully potentiated cytotoxic effect of gemcitabine in pancreatic cancer. Since some small molecules that specifically target FBW7-NR4A1-SCD1 axis, ferroptosis and apoptosis has exhibited potential clinical application value, our findings might provide new strategies for the comprehensive treatment of pancreatic cancer.
2. Materials and methods
2.1. Cell culture
The human pancreatic cancer cell lines PANC-1 and SW1990 were obtained from the American Type Culture Collection (ATCC), and were authenticated by DNA fingerprinting in 2015 and passaged in our laboratory fewer than 6 months after their receipt. PANC-1 cells were cultured in dulbecco's modified eagle medium (DMEM) (Hyclone) supplemented with 10% FBS (Wisent). SW1990 cells were cultured in L-15 medium supplemented with 10% FBS. All of the cell culture media contained 100 U/mL penicillin and 100 mg/mL streptomycin.
2.2. Chemicals
Ferroptosis inducer, RSL3 and erastin, and ferroptosis inhibitor, ferrostatin-1 (Fer-1), were purchased from selleckchem. Ferroptosis inducer, FIN56, was purchased from MedChemExpress. Other reagents used in this article: CAY10566, Z-VAD-FMK, oleic acid, palmitoleic acid, DIM-C-pPhOH, gemcitabine were purchased from MedChemExpress.
2.3. Cell viability and cell death
Cells (3 × 103) were seeded in 96 well plates and treated with corresponding chemicals. After incubation for 48–72 h, cell viability was assessed using calcein-AM (beyotime) and detected by synergy H4 (BioTek). Furthermore, cell death was assessed using propidium iodide (beyotime) analysis by FACS (Beckman Coulter).
2.4. Plasmids
The coding sequences of human FBW7, the relevant T205A or R465H mutant FBW7, or coding sequences of human SCD1 were cloned into the lentiviral vector pCDH-CMV-MCS-EF1-puro (SBI, USA) to generate FBW7 or SCD1 expression plasmids.
2.5. siRNA treatments
siRNA duplexes against FBW7 or NR4A1 were transfected into pancreatic cancer cells using Lipofectamine 2000 (Invitrogen). The siRNA duplex sense sequences were as follows:
si-FBW7-1: 5’-ACCTTCTCTGGAGAGAGAAATGC-3’;
si-FBW7-2: 5’-GTGTGGAATGCAGAGACTGGAGA-3’;
si-NR4A1-1: 5’-CAGTGGCTCTGACTACTAT-3’;
si-NR4A1-2: 5’-GAAGGAAGTTGTCCGAACA-3’;
2.6. Western blot
Western blotting was carried out as previously described [5]. Briefly, whole-cell protein lysates were extracted and then were separated by SDS-PAGE and blotted onto polyvinylidene fluoride membranes (Bio-Rad). After blocking, the membranes were incubated with corresponding antibodies: FBW7 antibody (Bethyl), SCD1 antibody (Cell Signaling Technology), NR4A1 antibody (Abcam), NR4A2 antibody (Abcam), β-actin (Proteintech). Next, the membranes were probed with secondary antibodies conjugated to HRP (Proteintech). Finally, immunoblots were incubated with an enhanced chemiluminescence detection kit (Millipore) and visualized by imaging systems (CLiNX).
2.7. RNA isolation and quantitative real-time PCR
Briefly, total RNA was extracted using TRIzol reagent (Invitrogen, USA). cDNA was obtained by reverse transcription using TaKaRa PrimeScript RT reagent Kit. Quantitative real-time PCR was conducted by an ABI 7900HT Real-Time PCR system (Applied Biosystems, USA). Primers used as follows, human FBW7: 5’-CCA CTG GGC TTG TAC CAT GTT-3’ (forward), 5’-CAG ATG TAA TTC GGC GTC GTT-3’ (reverse); human SCD1: 5’-AAA CCT GGC TTG CTG ATG-3’(forward), 5’- GGG GGC TAA TGT TCT TGT CA-3’ (reverse); human NR4A1: 5’- CTG AGT CAA GAA ACC AAG GCT-3’(forward), 5’-TCT GCA GCA TTT CTC CCA TCC-3’ (reverse); human NR4A2: 5’-GGT CCC TTT TGC CTG TCC A-3’ (forward), 5’- TGG CTT CAG CCG AGT TAC AG-3’ (reverse); human β-actin: 5’-CTA CGT CGC CCT GGA CTT CGA GC-3’ (forward), 5’-GAT GGA GCC GCC GAT CCA CAC GG-3’ (forward).
2.8. Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were conducted using the EZ-ChIP Kit (Millipore) according to the manufacturer's protocol. Primers to detect SCD1 promoter occupancy were listed as follows, Primer 1: 5’-GGT GGC TCA TGC CTG TAA TCC-3’ (F), 5’-TCT CGC TTC ACT GCA ACC TCT-3’ (R); Primer 2: 5’-AGG ACT GGA ATG CAG TCT CCT-3’ (F), 5’-CCA GAA ACA CCA TAT GAG AGC-3’ (R); Primer 3: 5’-TTC TTG TGA ATT GGC TTG CAGT-3’ (F), 5’- CCA CTA ACA TCT CCG TCC CG-3’ (R).
2.9. Promoter activity assessment by a dual-luciferase assay
The SCD1 promoter region, spanning from -2000 to +250 of the transcription start
site or relevant mutant sequence, was cloned into the pGL3-Basic vector. A dual-luciferase system (Promega, USA) was used to measure firefly and renilla luciferase activities according to the manufacturer's protocol.
2.10. C11-BODIPY staining
Cells were plated in 6-well cell culture plate (Corning) and then pretreated with ferroptosis inducers for 24 h. Before flow cytometry, cells were dissociated, resuspended, washed and then stained with 2 μmol/L C11-bodipy for 30 min. Fluorescence intensity was detected by flow cytometry (Beckman). For confocal imaging, cells were plated in round coverslip. Before detection, cells were incubated with 2 μmol/L C11-bodipy for 30 min. Next, cells were washed, fixed with 4% paraformaldehyde (Thermo Fisher Scientific) and images were acquired using inverted microscopy.
2.11. MDA and GSH/GSSG assay
Cells were plated in 6-well cell culture plate (Corning). Processing factors were consistent with C11-BODIPY assay. After cell homogenization, protein concentrations were measured using BCA protein assay kit (beyotime) and then detected MDA using lipid peroxidation MDA assay kit (beyotime). After obtaining the content of MDA, the ratio of MDA to protein concentration was calculated. The ratio of GSH/GSSG was measured according to the instruction (ab138881, Abcam) and protein concentrations of the cells lysates were used for normalization.
2.12. Metabolites analysis
Targeted cell metabolites analysis was conducted by Shanghai Bellas Biotech Co., Ltd. Briefly, amino acids were determined by Ultimate3000 DGLC (ThermoFisher) and an ACCQ-Tag TMULTRA C18 (100*2.1mm, 1.8um) liquid chromatography column was used. Medium and long chain fatty acids were determined by 7820A-5977B gas chromatograph-mass spectrometer (Agilent Technologies Inc. CA, UAS).
2.13. CoQ10 detection
CoQ10 was detected by an ELISA Kit. Briefly, Microporous plates were precoated with purified human CoQ10 antibody. Next, the solution to be detected containing CoQ10 was added to the microplate and then combined with HRP- labeled CoQ10 antibody to develop color in an acidic environment. The absorbance was measured at 450nm wavelength with a microplate reader. Protein concentrations of the cells lysates were used for normalization.
2.14. H2O2 measurement
Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen, Catalog no. A22188) was used to measure H2O2 released from cells. Cells were suspended in Krebs–Ringer phosphate (KRPG) buffer to keep active. All the procedures were conducted according to the protocol and the fluorescence was measured after incubation for selecting time points.
2.15. Transmission electron microscope (TEM) imaging
PANC-1 cells were plated in 10cm cell dish (Thermo) and then pretreated with corresponding factors for 24 h. Next, cells were collected and fixed with 2.5% glutaraldehyde. TEM imaging was conducted by Servicebio, wuhan, China.
2.16. Tissue immunofluorescence
Tissue immunofluorescence was conducted using paraffin sections. In brief, paraffin sections underwent the process of deparaffinating, antigen retrieval, blocking, antibody incubation, confocal imaging and scanning by Servicebio, wuhan, China.
2.17. Tissue specimens and immunohistochemical (IHC) staining
The clinical tissue samples used in this study was same as our previous study, which were histopathologically and clinically diagnosed at Fudan University Shanghai Cancer Center from 2010 to 2011, and prior patient consent and approval from the Institutional Research Ethics Committee were obtained [5]. IHC staining was performed as described previously [20]. Anti-FBW7 (1:100, Bethyl), anti-NR4A1 (1:50, Abcam), anti-SCD1 (1:50, Cell Signaling Technology) were used to detect protein expression.
2.18. Statistical analysis
Experiments were repeated at least three times. All data were analyzed by SPSS version 19.0 software (IBM) or GraphPad Prism 8. Two-tailed unpaired Student t tests were used to compare the differences between any two groups. The χ2 test was used to analyze the relationship between NR4A1 or SCD1 expression and corresponding clinicopathological characteristics. The survival curve was plotted using the Kaplan-Meier method and compared by the log-rank test. Differences were considered significant at *, P < 0.05; **, P < 0.01. N.S. means there was no significant difference.
3. Results
3.1. FBW7 promotes lipid peroxidation in pancreatic cancer cells
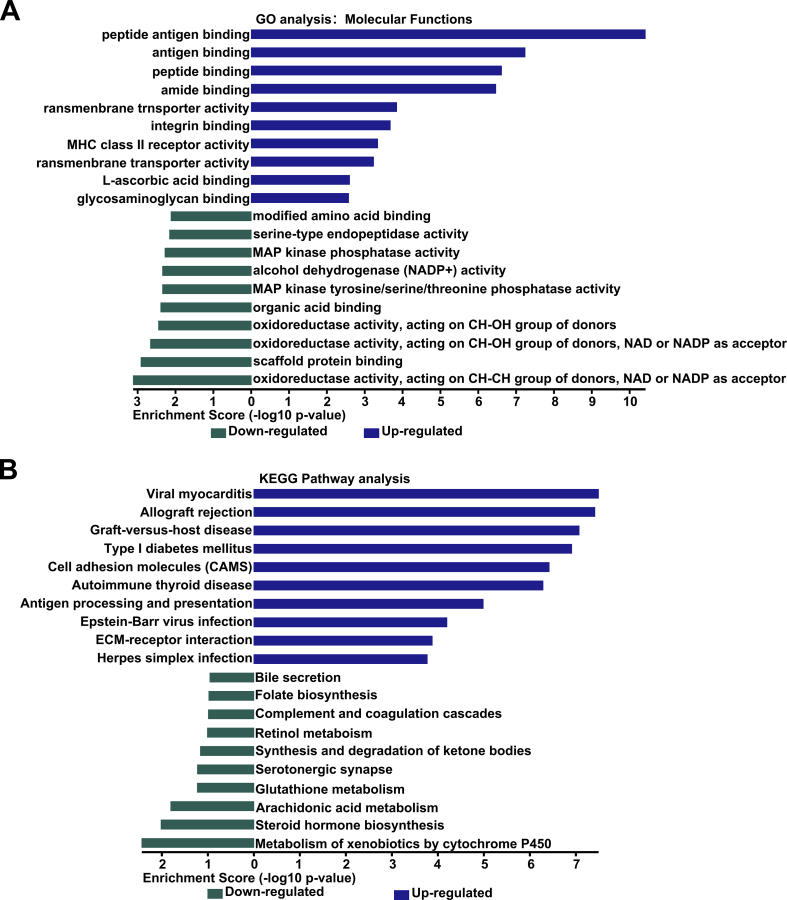
To further explore the function of FBW7 in pancreatic cancer, we reanalyzed our previous high-throughput gene expression profiling array that was conducted in SW1990 transfected with pCMV-FBW7 or empty vector (GEO: accession numbers GSE76443). Gene Ontology (GO) enrichment analysis showed that the significantly down-regulated genes could be categorized into oxidoreductases (Fig. S1A). KEGG Pathway analysis revealed that these down-regulated genes involved in arachidonic acid metabolism and glutathione metabolism (Fig. S1B). Next, we analyzed the effect of wild type FBW7 (FBW7WT), dominant-negative FBW7 mutant (FBW7R465H) or phospho-defcient T205A FBW7 mutant (FBW7T205A), the latter was resistant to ERK activation and enhanced the stability of FBW7 protein in our previous study [6], on cellular metabolites in PANC-1 and SW1990 cells. We focused on amino acids that were associated with glutathione metabolism and fatty acid metabolism. Our results revealed that serine, methionine, glycine and cystine were down-regulated by FBW7WT and FBW7T205A, while FBW7R465H had no effect on them (Fig. 1A). These amino acids could be catalyzed to form GSH (Fig. 1B). Thus GSH level can be speculated to be reduced. To confirm this conjecture, we tested GSH/GSSG ratio in PANC-1 and SW1990 stable cell lines ectopically expressing empty vector, FBW7WT or FBW7T205A. As expected, FBW7WT and FBW7T205A resulted in an increase in GSH/GSSG ratio (Fig. 1C). As for fatty acid metabolism, metabolites analysis revealed FBW7WT and FBW7T205A reduced arachidonic acid, palmitoleic acid and oleic acid level, and FBW7R465H had no effect on them (Figs. S2A and B). Palmitoleic acid and oleic acid are monounsaturated fatty acid. Further analysis found that FBW7WT and FBW7T205A also reduced the ratio of palmitoleic acid or linoleic acid compared to their corresponding saturated fatty acids (Fig. S2C). GSH is a major antioxidant, which scavenges free radicals and reduces ROS (reactive oxygen species). Thus FBW7 might affect ROS level. We used DCFH-DA probe to detect ROS and found FBW7 increased ROS level (Figs. S2D and E). Since FBW7 also affected fatty acid metabolism, we further surmised ROS occurred as lipid peroxidation. We used BODIPY 581/591C11 probe to detect oxidized lipid and further analyzed by flow cytometry and confocal microscopy. Fluorescence would change from red to green when lipid peroxidation occurred and our results revealed FBW7WT and FBW7T205A caused an increase in lipid peroxidation level (Fig. 1D and E). This result was further confirmed by the phenomenon that FBW7 also increased the level of MDA (malondialdehyde), the end product of lipid peroxidation (Fig. S2F). FBW7WT and FBW7T205A could also promote the H2O2 release in PANC-1 and SW1990 (Fig. S2G). In conclusion, FBW7WT and FBW7T205A promoted the ROS and lipid peroxidation in pancreatic cancer cells.
Fig. 1.
FBW7 promotes lipid peroxidation in pancreatic cancer cells. (A) Amino acids were detected in PANC-1 and SW1990 stable cell lines ectopically expressing empty vector, FBW7WT, FBW7R465H or FBW7T205A. (B) Schematic diagram demonstrated amino acids that were associated to glutathione metabolism were reduced. Amino acids on a white background were reduced. (C) GSH/GSSG ratio were detected in cells that stably expressed empty vector, FBW7WT or FBW7T205A. (D) Confocal imaging revealed the effect of FBW7 on lipid peroxidation. (E) BODIPY 581/591C11 was used to detected lipid peroxidation level.
3.2. FBW7 enhances ferroptosis in pancreatic cancer cells
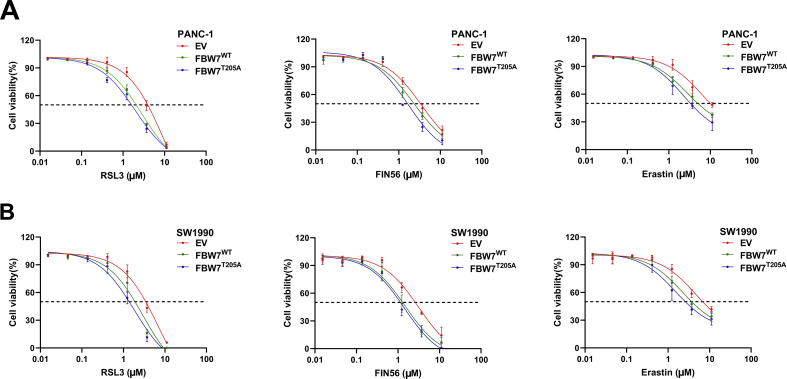
Lipid peroxidation is the most important hallmark of ferroptosis. FBW7 promotes lipid peroxidation. Thus FBW7 might enhance ferroptosis. To verify this, we tested the killing effect of ferroptosis inducers (RSL3, FIN56, erastin) in PANC-1 and SW1990 stable cell lines ectopically expressing FBW7WT, FBW7T205A or empty vector. Both FBW7WT and FBW7T205A enhanced the killing effect of ferroptosis inducers in PANC-1 and SW1990 cells, especially RSL3 (Fig. 2A and B, Figs. S3A and B). Moreover, ferroptosis inhibitor, ferrostatin-1(fer), could reverse lipid peroxidation caused by FBW7 overexpression (Fig. 2C and D). Ferrostatin-1 also partly rescued cell viability which was inhibited by FBW7 (Fig. 2E). These indicated that FBW7 enhanced ferroptosis in pancreatic cancer cells.
Fig. 2.
FBW7 enhances ferroptosis in pancreatic cancer cells. (A, B) Heatmap demonstrated FBW7 enhanced the sensitivity to ferroptosis inducers (RSL3, FIN56 and Erastin). (C) Flow cytometry analyzed the fluorescence of BODIPY 581/591C11 (left) and the relative content was calculated (right). (D) Concentrations of MDA were detected in FBW7 overexpressed PANC-1 and SW1990 cell lines in the presence or absence of 2 μmol/L Fer-1. (E) Cell viability was detected in FBW7 overexpressed PANC-1 and SW1990 cell lines in the presence or absence of 2 μmol/L Fer-1.
3.3. FBW7 promotes ferroptosis and apoptosis by down regulating SCD1
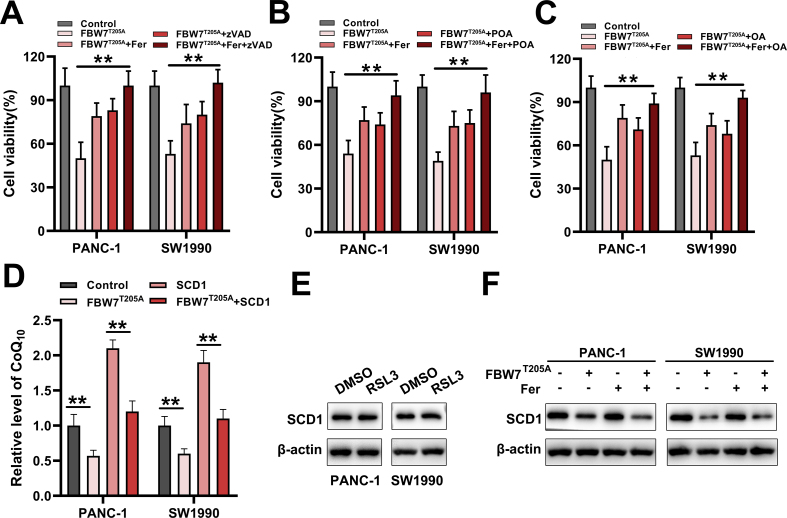
To find how FBW7 regulated ferroptosis, we reviewed our previous high-throughput gene expression profiling array. In these down-regulated genes, we noted that SCD1 was reported to inhibit both ferroptosis and apoptosis. Inhibition of SCD1 could induce ferroptosis [21,22]. We then speculated that FBW7 might promote ferroptosis by inhibiting the expression of SCD1. We conducted qRT-PCR and western blot to verify that FBW7 reduced SCD1 mRNA and protein level (Fig. 3A). Our preceding results demonstrated T205A mutant FBW7 had a more prominent effect in regulating ferroptosis, while T205A mutant FBW7 only enhanced the stability of FBW7 protein, thus in the next experiments, we mainly used T205A mutant FBW7 as the subject. To confirm FBW7 promotes ferroptosis via SCD1. We constructed pCMV-SCD1 plasmid to over express SCD1 in PANC-1 and SW1990 stable cell lines ectopically expressing T205A mutant FBW7 (Fig. 3B). Then we found overexpression of SCD1 could mostly reverse the lipid peroxidation caused by FBW7T205A (Fig. 3C and D). Moreover, we used small interfering RNA to silence FBW7 expression and detected the level of SCD1. FBW7 silencing leaded to high level SCD1 protein (Fig. 3E). More importantly, FBW7 silencing could partly reverse the cell viability reduced by RSL3 or SCD1 inhibitor (CAY 10566) (Fig. 3F). Both FBW7 and SCD1 were reported to regulate apoptosis in other cancer cells [13], thus we further validated whether FBW7 regulated apoptosis through inhibiting SCD1 in pancreatic cancer cells. The results revealed that overexpression of SCD1 could mostly reduce apoptosis caused by FBW7T205A overexpression (Fig. 3G). In order to observe the effect of FBW7 on cells more intuitively, we conducted TEM imaging in PANC-1 cells. Overexpression of FBW7T205A led to cells shrink, mitochondria shrink, and lipid droplets form, which was similar to the structure caused by combination of RSL3 and CAY 10566 (Fig. 3H). Further study demonstrated single ferroptosis inhibitor (Fer-1), apoptosis inhibitor (zVAD), or endproducts of SCD1 activity (POA, palmitoleic acid; OA, oleic acid) only partly restored cell viability reduced by FBW7T205A overexpression. But combination Fer-1 with zVAD, POA or OA entirely restored cell viability reduced by FBW7 T205A overexpression (Figs. S4A–C). SCD1 depletion was also reported to triggered ferroptosis by depleting CoQ10. We then tested the level of CoQ10 in PANC-1and SW1990 cells. Results showed that FBW7T205A leaded to a decrease in CoQ10 and overexpression of SCD1 could reverse this consequence (Fig. S4D). To eliminate the potential that FBW7 might inhibite the expression of SCD1 by promoting ferroptosis. We then detected whether ferroptosis inducer, RSL3, could affected the level of SCD1. Results showed that RSL3 had no significant influence on the protein level of SCD1 (Fig. S4E). Moreover, ferroptosis inhibitor, Fer-1, could not reverse the decreased SCD1 caused by FBW7T205A overexpression (Fig. S4F). These indicated FBW7 promotes both ferroptosis and apoptosis by down regulating SCD1.
Fig. 3.
FBW7 promotes ferroptosis and apoptosis by down regulating SCD1. (A) qRT-PCR (upper) and western blot (lower) to analyze the change of SCD1 caused by FBW7 overexpression. (B) FBW7 and SCD1 were detected in PANC-1 and SW1990 cells that overexpressed with FBW7T205A, SCD1 or both. (C, D) MDA and BODIPY 581/591C11 were detected in the presence of SCD1 or FBW7 or both in PANC-1 and SW1990 cells. (E) PANC-1 and SW1990 cells were pretreated with small interfering RNA of FBW7 for 48–72 h and then detected protein level of SCD1. (F) FBW7 silencing PANC-1 and SW1990 cells were treated with RSL3 (2 μmol/L) or CAY10566 (5 μmol/L) for 48–72 h and then detected cell viability. (G) Apoptosis was analyzed in the presence of SCD1 or FBW7 or both in PANC-1 and SW1990 cells. (H) TEM imaging was conducted in FBW7T205A overexpressed PANC-1 cells. RSL3 (2 μmol/L) and CAY10566 (5 μmol/L) treated PANC-1 cells functioned as the positive control. Red arrows showed the shrunken mitochondria. Blue arrows showed lipid droplets. The values below the western blot band represent the relative mean gray values of three independent experiments. Gray values were calculated by Adobe Photoshop CS6. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. FBW7 inhibits SCD1 transcription via NR4A1
To clarify how FBW7 regulate SCD1, we noted that NR4A family of nuclear receptors (NR4A1, NR4A2) were FBW7 down-regulated genes in our gene expression profiling array. Both NR4A1 and NR4A2 were reported to regulate apoptosis. Generally, NR4A2 functions as apoptosis suppressor genes, while NR4A1 has a proapoptotic role [23]. However, in pancreatic cancer, inhibition of NR4A1 could also cause apoptosis by reducing transcription of NR4A1-dependent antiapoptotic genes [24]. Thus we speculated FBW7 might inhibit SCD1 transcription via NR4A1 or NR4A2. To confirm our gene expression profiling, we conducted qRT-PCR and western blot to test the level of NR4A1 and NR4A2 in FBW7T205A overexpressed PANC-1 and SW1990 cells. FBW7T205A reduced both NR4A1 and NR4A2 in mRNA and protein level. Besides, we generated a dominant-negative FBW7 mutant, which lost its E3 ligase activity and was designated as FBW7R465H. Compared with FBW7T205A, FBW7R465H only marginally reduced NR4A1, NR4A2 and SCD1 expression (Fig. 4A and B). To probe NR4A1 or NR4A2 could regulate SCD1, we used small interfering RNA to silence NR4A1 or NR4A2 and detected the change of SCD1. Only NR4A1 silencing decreased SCD1 expression (Fig. 4C and D), NR4A2 silencing did not show significant difference (data not shown). Thus, FBW7 might regulated SCD1 trough NR4A1. We found NR4A1 had several binding sites in SCD1 promoter region and we then conducted CHIP assay used NR4A1 antibody to verify NR4A1 bound to SCD1 promoter. Our results showed NR4A1 bound to SCD1 promoter in the site corresponding to primer 2 (Fig. 4E). To further confirm the binding specificity and tested the effect of NR4A1 on SCD1 promoter, we conducted a luciferase reporter gene assay used a pGL3-SCD1 promoter plasmid that possessed mutations in NR4A1 binding site (MUT) and a wild type pGL3-SCD1 promoter plasmid (WT) as the control. NR4A1 silencing significantly reduced the luciferase activity in WT pGL3-SCD1 promoter but not MUT pGL3-SCD1 promoter (Fig. 4F). These indicated NR4A1 bound to SCD1 promoter and promoted SCD1 transcription. We further explored how FBW7 affected the relationship between NR4A1 and SCD1. CHIP assay with NR4A1 antibody demonstrated that overexpression of FBW7T205A reduced the binding of NR4A1 to SCD1 promoter, but overexpression of FBW7R465H did not show significant difference (Fig. 4G). The same trend was also obtained in luciferase assay and the group of MUT pGL3-SCD1 promoter did not show significant difference (Fig. 4H). Moreover, we found NR4A1 overexpression could counteract the effect of FBW7 T205A on SCD1 (Fig. 4I). Summing up, FBW7 inhibited SCD1 transcription by reducing the binding of NR4A1 to SCD1 promoter.
Fig. 4.
FBW7 inhibits SCD1 transcription via NR4A1. (A) qRT-PCR detected NR4A1 and NR4A2 mRNA in FBW7T205A or FBW7R465H overexpressed PANC-1 and SW1990 cells. (B) Western bolt detected the changes of NR4A1, NR4A2 and SCD1 in FBW7T205A or FBW7R465H overexpressed PANC-1 and SW1990 cells. (C, D) SCD1 protein and mRNA change were detected in NR4A1 silencing PANC-1 and SW1990 cells. (E) CHIP assay was conducted with NR4A1 antibody. Schematic diagram (left) demonstrated the potential binding site of NR4A1 on SCD1 promoter. Agarose gel electrophoresis (right) was used to confirm the binding site. (F) Schematic diagram (left) demonstrated the wild type and mutant binding site. Dual-luciferase reporter system (right) was used to confirm the effect of NR4A1 on SCD1 promoter activity. (G) Agarose gel electrophoresis demonstrated the effect of FBW7T205A or FBW7R465H on the binding of NR4A1 to SCD1 promoter. (H) Dual-luciferase reporter system was used to explore the effect of FBW7T205A or FBW7R465H on SCD1 promoter activity. (I) SCD1 protein was detected in PANC-1 and SW1990 cells that was overexpressed with NR4A1 in the presence or absence of FBW7T205A. The values below the western blot band represent the relative mean gray values of three independent experiments.
3.5. Pharmacological inhibition of NR4A1 promotes ferroptosis in pancreatic cancer cells
NR4A1 antagonist DIM-C-pPhOH was reported to induce apoptosis by inhibiting NR4A1 transactivation activity [24]. We then probed whether DIM-C-pPhOH influenced the transactivation activity of NR4A1 to SCD1. Luciferase reporter gene assay demonstrated DIM-C-pPhOH reduced the luciferase activity, but in the group possessed mutations in NR4A1 binding site (MUT), the luciferase activity did not show significant difference (Fig. 5A). This indicted DIM-C-pPhOH inhibited SCD1 transcription via NR4A1. Moreover, DIM-C-pPhOH could also reduce SCD1 protein in PANC-1 and SW1990 cells (Fig. 5B). We further explored whether DIM-C-pPhOH regulated ferroptosis in pancreatic cancer cells. DIM-C-pPhOH significantly reduced the IC50 (half maximal inhibitory concentration) of RSL3 in PANC-1 and SW1990 cells (Fig. 5C). DIM-C-pPhOH could also increase the level lipid peroxidation, and ferroptosis inhibitor, Fer-1, could largely reverse lipid peroxidation caused by DIM-C-pPhOH (Fig. 5D and E). Moreover, ferroptosis inhibitor (Fer-1), apoptosis inhibitor (zVAD), or endproducts of SCD1 activity (POA, OA) could partly rescue the cytotoxic effect caused by DIM-C-pPhOH (Fig. 5F). The same trends were also obtained in NR4A1 silencing cells (Fig. 5G).
Fig. 5.
Pharmacological inhibition of NR4A1 promotes ferroptosis in pancreatic cancer cells. (A) Dual-luciferase reporter system was used to explore the effect of DIM-C-pPhOH on SCD1 promoter activity. (B) SCD1 protein was detected in PANC-1 and SW1990 cells pretreated with DIM-C-pPhOH for 48–72 h. The values below the western blot band represent the relative mean gray values of three independent experiments. (C) IC50 was tested in PANC-1 and SW1990 cells in the presence or absence of DIM-C-pPhOH. (D, E) Lipid peroxidation was detected using MDA or BODIPY 581/591C11 in PANC-1 and SW1990 cells pretreated with DMSO, DIM-C-pPhOH (15 μmol/L), DIM-C-pPhOH (15 μmol/L) and Fer-1 (2 μmol/L) for 48 h. (F, G) DIM-C-pPhOH or small interfering RNA was used to inhibit or silence NR4A1 in PANC-1 and SW1990 cells. Then these cells were treated Fer-1 (2 μmol/L), zVAD (10 μmol/L), POA (80 μmol/L), OA (80 μmol/L) for 24 h. Cell viability was detected by calcein-AM.
3.6. FBW7 potentiates cytotoxic effect of gemcitabine via ferroptosis and apoptosis
Ferroptosis inducer, erastin, was reported to potentiate cytotoxic effect of gemcitabine and cisplatin in pancreatic cancer cells [25]. We further confirmed ferroptosis inducer, RSL3, could also increase the lethality of the gemcitabine in pancreatic cancer cells and ferroptosis inhibitor, Fer-1, had the opposite effect (Fig. 6A and B). These indicated activations of ferroptosis could mitigate gemcitabine resistance in pancreatic cancer. We then tested the effect of FBW7 on the gemcitabine sensitivity. Overexpression of FBW7T205A distinctly enhanced cytotoxic effect of gemcitabine, while overexpression of FBW7R465H did not show significant changes (Fig. 6C and D). Activation of ferroptosis or apoptosis could potentiate cytotoxic effect of gemcitabine, while FBW7 promoted both ferroptosis and apoptosis. Thus, we conjectured that FBW7 might potentiate cytotoxic effect of gemcitabine via both ferroptosis and apoptosis. To probe this, we pretreated FBW7T205A over-expressed cells with fer-1 or zVAD or both. Results revealed that both fer-1 and zVAD could partly rescue the cell viability caused by FBW7T205A overexpression, but combination of fer-1 and zVAD could entirely recuse it (Fig. 6E and F). These indicated that FBW7 potentiated cytotoxic effect of gemcitabine via both ferroptosis and apoptosis.
Fig. 6.
FBW7 potentiates cytotoxic effect of gemcitabine via ferroptosis and apoptosis. (A, B) PANC-1 and SW1990 cells were seeded with gemcitabine (0, 2.5, 5 or 10 μmol/L) or a combination of gemcitabine with either RSL3 (2 μmol/L) or Fer-1(2 μmol/L). After incubation for 48 h, cell viability was detected by calcein-AM. (C, D) PANC-1 and SW1990 cells that stably expressed empty vector, FBW7T205A or FBW7R465H were seeded with gemcitabine (0, 2.5, 5 or 10 μmol/L) for 48 h. Then cell viability was detected by calcein-AM. (E, F) PANC-1 and SW1990 cells that stably expressed empty vector or FBW7T205A were seeded with Fer-1(2 μmol/L), zVAD (10 μmol/L), or combination Fer-1 and zVAD in the presence or absence of gemcitabine (5 μmol/L). After incubation for 48 h, cell viability was detected by calcein-AM.
3.7. FBW7 is correlated with NR4A1 and SCD1 in pancreatic cancer tissues
In our previous study, we had confirmed FBW7 was lower expressed in most pancreatic cancer tissues and lower expression of FBW7 predicted a poor prognosis [5]. In this study, we examined the relationship between FBW7, NR4A1 and SCD1 expression in tissues from PADC patients. We conducted tissue immunofluorescence assay and FBW7, NR4A1, SCD1 were labeled with different fluorescent signals. Results showed that when FBW7 was low, NR4A1 and SCD1 were highly expressed. The opposite phenomenon was also found (Fig. 7A). These results were further confirmed by IHC staining (Fig. 7B). In IHC staining, we ranked the expression strength as negative, weak, moderate and strong, and defined strong as high expression, moderate as medium expression, negative and weak as low expression. The results revealed that FBW7 was negatively correlated with NR4A1 and SCD1 (Fig. 7C, and Fig. S5A). Both NR4A1 and SCD1 were high expression in pancreatic cancer tissues (Fig. S5B). We further confirmed the correlation between NR4A1 and SCD1 expression. IHC scores demonstrated NR4A1 was positively correlated with SCD1 in pancreatic cancer tissues (Fig. 7D, and Fig. S5C). Correspondingly, TCGA and GTEx data also demonstrated that NR4A1 and SCD1 were high expressed in pancreatic cancer and NR4A1 was positively correlated with SCD1 pancreatic cancer tissues (Fig. 7E and F). Next, we explored the relationship between NR4A1 expression and the clinicopathological features of PDAC. In our clinical data, we found that the high level of NR4A1 correlated with bigger tumor size and poor tumor differentiation (Table S1). The Kaplan-Meier survival curves and log rank test showed that high NR4A1 expression significantly correlated with poor overall survival (OS) in PDAC (Fig. 7G). Intriguingly, high level of SCD1 also correlated with bigger tumor size, poor tumor differentiation and poor overall survival in PDAC (Fig. 7H and Table S2).
Fig. 7.
FBW7 is correlated with NR4A1 and SCD1 in pancreatic cancer tissues. (A, B) Representative tissue immunofluorescence and IHC staining of FBW7, NR4A1, SCD1 in patients’ tissues. (C) Statistical analysis of the correlation between levels of FBW7 and NR4A1, or FBW7 and SCD1 (P value was obtained using a Pearson χ2 test). (D) The method to calculate IHC scores was shown in Fig. S5C. The correlation between levels of NR4A1 and SCD1 was analyzed (n = 86, spearman r = 0.8048, p < 0.001). (E) NR4A1 or SCD1 transcription was significantly higher in pancreatic cancer tissues than in normal tissues in the GEPIA dataset. Mean and SD are shown (num (T) = 179, num (P) = 171, P < 0.05 as determined by t-test). (F) NR4A1 was positively correlated with SCD1 in the GEPIA dataset. (G, H) Kaplan-Meier analysis of overall survival rate of pancreatic cancer patients according to the expression of NR4A1 (n = 86, P = 0.0022, log-rank test) or SCD1 (n = 86, P = 0.0004, log-rank test).
4. Discussion
Pancreatic cancer is a highly aggressive tumor. The overall 5-year survival for pancreatic cancer has changed little over the past few decades (less than 10%) [26]. Furthermore, therapeutic strategies for pancreatic cancer have not progressed significantly in recent years. The recommended chemotherapy regimens are still gemcitabine, FOLFIRINOX, and albumin-bound paclitaxel. But due to the rapid and common development of chemoresistance, the prognosis is still poor [27]. For instance, pancreatic cancer cells can quickly acquire the ability to mitigate gemcitabine-induced apoptosis within weeks. Although evidence revealed that inducing apoptosis could sensitize pancreatic cells to gemcitabine in vitro, it is still a long way from clinical application [28]. Hence, it is imperative to seek other methods to efficiently kill pancreatic cancer cells.
Ferroptosis is a non-apoptotic form of cell death. Emerging evidence shows that ferroptosis can be harnessed for cancer therapy, particularly for eradicating aggressive malignancies that are resistant to traditional therapies. Furthermore, pathological differences between cancerous and normal cells, such as the intracellular levels of iron, GSH, H2O2, etc. can be harnessed for targeting ferroptosis therapy [8,29]. Notably, nanotechnology provides additional options in ferroptosis‐based cancer therapy [30]. Ferroptosis also has potential in the treatment of pancreatic ductal adenocarcinoma (PDAC). Kenneth P. Olive et al. confirmed that exogenous cystine was needed for pancreatic cancer cells to avert ferroptosis and deletion of Slc7a11, a subunit system xc–, in KPC mice induced tumor ferroptosis and extended survival [31]. It was also verified that pharmacologic inhibition of system xc– potentiated the cytotoxic effects of both gemcitabine and cisplatin in PDAC cell lines [25]. Hence, in-depth exploration of the role of ferroptosis in pancreatic cancer is conducive to the treatment of pancreatic cancer.
In this study, we found FBW7 could reduce GSH/GSSG ratio and increase lipid peroxidation. Both reduced GSH/GSSG ratio and increased lipid peroxidation could induce ferroptosis and apoptosis [32,33]. Further experiments confirmed FBW7 activated both ferroptosis and apoptosis. To elucidate the underlying mechanism, we screened our previous gene expression profiling. In FBW7 down-regulated genes, we noticed that SCD1 was reported to inhibit both apoptosis and ferroptosis, which was opposed to the function of FBW7. SCD1 is a lipid-modifying enzyme that is up-regulated in numerous malignancies. It was reported that FBW7 could reduce SCD1 and regulate lipid metabolism in the mouse liver [34]. SCD1 depletion inhibited the synthesis of oleic acid and altering the ratio of monounsaturated to saturated fatty acids, which was corresponding to the change of cellular metabolites caused by FBW7 overexpression (Figs. S2A–C). Oleic acid could protect melanoma cells from ferroptosis in an Acsl3-dependent manner [35]. Moreover, oleic acid can be nitrated to form nitro-oleic acid. Nitro-oleic acid could induce multiple pleiotropic signaling responses, including anti-inflammatory and antioxidant effects, such as inhibiting inflammatory cell function, inducing HO-1 expression, inhibiting of NF-κB, promoting Keap1/Nrf2 activation and so on [36,37]. SCD1 depletion could also reduce the level of CoQ10 and then triggered ferroptosis [21]. We verified that FBW7 inhibited the transcription of SCD1, and overexpression of SCD1 or combination using endproducts of SCD1 activity could significantly alleviate FBW7-induced apoptosis and ferroptosis. Correspondingly, ferroptosis inhibitors or apoptosis inhibitors could partly rescue the cell viability reduced by FBW7 overexpression. But combination using ferroptosis inhibitors and apoptosis inhibitors could entirely rescue the cell viability caused by FBW7 overexpression.
To elucidate how FBW7 inhibit the transcription of SCD1. We screened transcription factors in previous gene expression profiling. Furthermore, this transcription factor should involve in apoptosis or ferroptosis. Intriguingly, NR4A1 and NR4A2, numbers of nuclear receptor subfamily 4 group A (NR4A), were inhibited by FBW7. Generally, NR4A2 inhibits apoptosis, while NR4A1 plays a proapoptotic role. However, inhibition of NR4A1 could also cause apoptosis in pancreatic cancer by reducing transcription of NR4A1-dependent antiapoptotic genes [23]. More coincidentally, NR4A could regulate fatty acid oxidation and glucose metabolism, which conformed to the function of FBW7 [38]. Further study confirmed that FBW7 regulated SCD1 via NR4A1. However, the limitation of this study was that we did not elucidate how FBW7 regulates NR4A1. We speculated that FBW7 might mediate ubiquitination of a transcription factor, which could bind to NR4A1 promotor to facilitate NR4A1 transcription. Because dominant-negative FBW7 mutant, FBW7R465H did not inhibit the expression of NR4A1 (Fig. 4B).
Intriguingly, another study on FBW7 reported that FBW7 deficient macrophages would inhibit the pentose phosphate pathway and aggravate the generation of intracellular ROS [39]. This seems contrary to our conclusion, because we confirmed that FBW7 would promote ROS, especially lipid peroxidation in pancreatic cancer. Indeed, these two conclusions are not drawn in the same context. In this research, the ubiquitin substrate of FBW7 is PKM2. In the case that cellular glucose uptake is not affected, reduced PKM2 would inhibit glycolysis and increase substrate flux through the pentose phosphate pathway to produce more NADPH and GSH. However, in our previous study, the downstream of FBW7 was TXNIP [6]. TXNIP functioned as a potent negative regulator of glucose uptake and aerobic glycolysis. Thereby, reduced TXNIP would inhibit aerobic glycolysis and decrease substrate flux through the pentose phosphate pathway to produce less NADPH and GSH. Moreover, the former occurs in the immune system, while ours conducted in pancreatic cancer cells. Contrary conclusions are possible in these two systems. However, we will further study the mechanism of this difference.
As for the clinical significance of this study, we confirmed that FBW7-NR4A1-SCD1 significantly potentiated cytotoxic effect of gemcitabine, which provided new targets for chemotherapeutic intervention. For instance, targeting FBW7 can overcome resistance to targeted therapy [40]. Novel agents targeting NR4A have been developed, due to NR4A modulating response to conventional chemotherapy [23]. Numerous chemical compounds that inhibit SCD1 have been developed and preclinically tested [41,42]. Furthermore, we also demonstrated that activating ferroptosis and apoptosis immensely increased gemcitabine sensitivity, which might provide strategies for the combination therapy for pancreatic cancer.
Declaration of competing interest
The authors declare no potential conflicts of interest.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (No., 81871950, 81702871 and 81972725); Clinical and Scientific Innovation Project of Shanghai Hospital Development Center (SHDC12018109); Scientific Innovation Project of Shanghai Education Committee (2019-01-07-00-07-E00057); Shanghai Municipal Commission of Health and Family Planning (No. 2018YQ06); Shanghai Municipal Science and Technology Commission (19QA1402100).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101807.
Contributor Information
Yi Qin, Email: yuxianjun@fudanpci.org.
Xianjun Yu, Email: qinyi@fudanpci.org.
Shunrong Ji, Email: jishunrong@fudanpci.org.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Fig. S2.
Fig. S3.
Fig. S4.
Fig. S5.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. Ca - Cancer J. Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. 2019. [DOI] [PubMed] [Google Scholar]
- 2.Vincent A., Herman J., Schulick R., Hruban R.H., Goggins M. Pancreatic cancer. Lancet. 2011;378(9791):607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welcker M., Clurman B.E. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Canc. 2008;8(2):83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 4.Davis R.J., Welcker M., Clurman B.E. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Canc. Cell. 2014;26(4):455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji S., Qin Y., Shi S., Liu X., Hu H., Zhou H. ERK kinase phosphorylates and destabilizes the tumor suppressor FBW7 in pancreatic cancer. Cell Res. 2015;25(5):561–573. doi: 10.1038/cr.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji S., Qin Y., Liang C., Huang R., Shi S., Liu J. FBW7 (F-box and WD repeat domain-containing 7) negatively regulates glucose metabolism by targeting the c-myc/TXNIP (Thioredoxin-Binding protein) Axis in pancreatic cancer. Clin. Canc. Res. 2016;22(15):3950–3960. doi: 10.1158/1078-0432.CCR-15-2380. [DOI] [PubMed] [Google Scholar]
- 7.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang C., Zhang X., Yang M., Dong X. Recent progress in ferroptosis inducers for cancer therapy. Adv. Mater. 2019;31(51) doi: 10.1002/adma.201904197. [DOI] [PubMed] [Google Scholar]
- 9.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolma S., Lessnick S.L., Hahn W.C., Stockwell B.R. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Canc. Cell. 2003;3(3):285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 11.Yang W.S., Stockwell B.R. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem. Biol. 2008;15(3):234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makohon-Moore A., Iacobuzio-Donahue C.A. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat. Rev. Canc. 2016;16(9):553–565. doi: 10.1038/nrc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inuzuka H., Shaik S., Onoyama I., Gao D., Tseng A., Maser R.S. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471(7336):104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeck J.D., Jandke A., Blake S.M., Nye E., Spencer-Dene B., Brandner S. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat. Neurosci. 2010;13(11):1365–1372. doi: 10.1038/nn.2644. [DOI] [PubMed] [Google Scholar]
- 15.Wertz I.E., Kusam S., Lam C., Okamoto T., Sandoval W., Anderson D.J. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471(7336):110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 16.Hu Q., Qin Y., Zhang B., Liang C., Ji S., Shi S. FBW7 increases the chemosensitivity of pancreatic cancer cells to gemcitabine through upregulation of ENT1. Oncol. Rep. 2017;38(4):2069–2077. doi: 10.3892/or.2017.5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Binenbaum Y., Na'ara S., Gil Z. Gemcitabine resistance in pancreatic ductal adenocarcinoma. Drug Resist. Updates. 2015;23:55–68. doi: 10.1016/j.drup.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Friedmann Angeli J.P., Krysko D.V., Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Canc. 2019;19(7):405–414. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 19.Wang W., Green M., Choi J.E., Gijon M., Kennedy P.D., Johnson J.K. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Q., Qin Y., Xiang J., Liu W., Xu W., Sun Q. dCK negatively regulates the NRF2/ARE axis and ROS production in pancreatic cancer. Cell Prolif. 2018;51(4) doi: 10.1111/cpr.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tesfay L., Paul B.T., Konstorum A., Deng Z., Cox A.O., Lee J. Stearoyl-CoA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Canc. Res. 2019;79(20):5355–5366. doi: 10.1158/0008-5472.CAN-19-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinha R.A., Singh B.K., Zhou J., Xie S., Farah B.L., Lesmana R. Loss of ULK1 increases RPS6KB1-NCOR1 repression of NR1H/LXR-mediated Scd1 transcription and augments lipotoxicity in hepatic cells. Autophagy. 2017;13(1):169–186. doi: 10.1080/15548627.2016.1235123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohan H.M., Aherne C.M., Rogers A.C., Baird A.W., Winter D.C., Murphy E.P. Molecular pathways: the role of NR4A orphan nuclear receptors in cancer. Clin. Canc. Res. 2012;18(12):3223–3228. doi: 10.1158/1078-0432.CCR-11-2953. [DOI] [PubMed] [Google Scholar]
- 24.Lee S.O., Abdelrahim M., Yoon K., Chintharlapalli S., Papineni S., Kim K. Inactivation of the orphan nuclear receptor TR3/Nur77 inhibits pancreatic cancer cell and tumor growth. Canc. Res. 2010;70(17):6824–6836. doi: 10.1158/0008-5472.CAN-10-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daher B., Parks S.K., Durivault J., Cormerais Y., Baidarjad H., Tambutte E. Genetic ablation of the cystine transporter xCT in PDAC cells inhibits mTORC1, growth, survival, and tumor formation via nutrient and oxidative stresses. Canc. Res. 2019;79(15):3877–3890. doi: 10.1158/0008-5472.CAN-18-3855. [DOI] [PubMed] [Google Scholar]
- 26.Balachandran V.P., Beatty G.L., Dougan S.K. Broadening the impact of immunotherapy to pancreatic cancer: challenges and opportunities. Gastroenterology. 2019;156(7):2056–2072. doi: 10.1053/j.gastro.2018.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M.P., Gallick G.E. Gemcitabine resistance in pancreatic cancer: picking the key players. Clin. Canc. Res. 2008;14(5):1284–1285. doi: 10.1158/1078-0432.CCR-07-2247. [DOI] [PubMed] [Google Scholar]
- 28.Edderkaoui M., Chheda C., Soufi B., Zayou F., Hu R.W., Ramanujan V.K. An inhibitor of GSK3B and HDACs kills pancreatic cancer cells and slows pancreatic tumor growth and metastasis in mice. Gastroenterology. 2018;155(6):1985–1998. doi: 10.1053/j.gastro.2018.08.028. e1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassannia B., Vandenabeele P., Vanden Berghe T. Targeting ferroptosis to iron out cancer. Canc. Cell. 2019;35(6):830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Shen Z., Song J., Yung B.C., Zhou Z., Wu A., Chen X. Emerging strategies of cancer therapy based on ferroptosis. Adv. Mater. 2018;30(12) doi: 10.1002/adma.201704007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badgley M.A., Kremer D.M., Maurer H.C., DelGiorno K.E., Lee H.J., Purohit V. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science. 2020;368(6486):85–89. doi: 10.1126/science.aaw9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malla J.A., Umesh R.M., Yousf S., Mane S., Sharma S., Lahiri M. A glutathione activatable ion channel induces apoptosis in cancer cells by depleting intracellular glutathione levels. Angew Chem. Int. Ed. Engl. 2020;59(20):7944–7952. doi: 10.1002/anie.202000961. [DOI] [PubMed] [Google Scholar]
- 33.Busciglio J., Yankner B.A. Apoptosis and increased generation of reactive oxygen species in Down's syndrome neurons in vitro. Nature. 1995;378(6559):776–779. doi: 10.1038/378776a0. [DOI] [PubMed] [Google Scholar]
- 34.Onoyama I., Suzuki A., Matsumoto A., Tomita K., Katagiri H., Oike Y. Fbxw7 regulates lipid metabolism and cell fate decisions in the mouse liver. J. Clin. Invest. 2011;121(1):342–354. doi: 10.1172/JCI40725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ubellacker J.M., Tasdogan A., Ramesh V., Shen B., Mitchell E.C., Martin-Sandoval M.S. Lymph protects metastasizing melanoma cells from ferroptosis. Nature. 2020;585(7823):113–118. doi: 10.1038/s41586-020-2623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freeman B.A., Baker P.R., Schopfer F.J., Woodcock S.R., Napolitano A., d'Ischia M. Nitro-fatty acid formation and signaling. J. Biol. Chem. 2008;283(23):15515–15519. doi: 10.1074/jbc.R800004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zatloukalova M., Mojovic M., Pavicevic A., Kabelac M., Freeman B.A., Pekarova M. Redox properties and human serum albumin binding of nitro-oleic acid. Redox Biol. 2019;24:101213. doi: 10.1016/j.redox.2019.101213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearen M.A., Muscat G.E. Minireview: nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol. Endocrinol. 2010;24(10):1891–1903. doi: 10.1210/me.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C., Chao Y., Xu W., Liu Z., Wang H., Huang K. Myeloid FBW7 deficiency disrupts redox homeostasis and aggravates dietary-induced insulin resistance. Redox Biol. 2020;37:101688. doi: 10.1016/j.redox.2020.101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye M., Zhang Y., Zhang X., Zhang J., Jing P., Cao L. Targeting FBW7 as a strategy to overcome resistance to targeted therapy in non-small cell lung cancer. Canc. Res. 2017;77(13):3527–3539. doi: 10.1158/0008-5472.CAN-16-3470. [DOI] [PubMed] [Google Scholar]
- 41.Tracz-Gaszewska Z., Dobrzyn P. Stearoyl-CoA desaturase 1 as a therapeutic target for the treatment of cancer. Cancers. 2019;11(7) doi: 10.3390/cancers11070948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Southam A.D., Khanim F.L., Hayden R.E., Constantinou J.K., Koczula K.M., Michell R.H. Drug redeployment to kill leukemia and lymphoma cells by disrupting SCD1-mediated synthesis of monounsaturated fatty acids. Canc. Res. 2015;75(12):2530–2540. doi: 10.1158/0008-5472.CAN-15-0202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.