Abstract
Accumulating evidence has shown that microRNA (miR) derived from M1 macrophage-derived exosomes can regulate the progression of hepatocellular carcinoma (HCC). However, the effect of miR-326 derived from M1 macrophage-derived exosomes on HCC has not been reported. Therefore, the objective of the present study was to explore the mechanism of exosomal miR-326 from M1 macrophages in regulating HCC cell progression. RT-qPCR detected miR-326 expression in HCC cell lines. miR-326 expression in HCC was altered by transfection, and the effect of miR-326 on CD206 and NF-κB expression, cell proliferation, colony formation, migration, apoptosis and invasion was detected. Subsequently, exosomes were isolated from M1 macrophages. RT-qPCR identified miR-326 expression in M1 macrophage-derived exosomes. miR-326 expression in M1 macrophage-derived exosomes was changed by transfection. M1 macrophage-derived exosomes were co-cultured with HCC cells to figure out their effects on the biological progress of HCC cells. Finally, in vivo experiments were performed to verify the in vitro results. MiR-326 was decreased in HCC cells and enriched in M1 macrophage-derived exosomes. Up-regulating miR-326 would inhibit HCC cell proliferation, colony formation, migration, invasion, and CD206 and NF-κB expression and promoted apoptosis, and inhibited the growth of HCC tumors in vivo, while down-regulating miR-326 showed opposite effects. M1 macrophage-derived exosomes inhibited HCC cell proliferation, colony formation, migration, invasion, and CD206 and NF-κB expression and enhanced apoptosis, while overexpression of miR-326 enhanced the effect of M1 macrophage-derived exosomes on HCC cells. It is revealed that M1 macrophages-derived exosomal miR-326 suppresses proliferation, migration and invasion as well as advances apoptosis of HCC through down-regulating NF-κB expression.
Keyword: M1 macrophage, Exosomes, MicroRNA-326, Hepatocellular carcinoma, Migration, Invasion
Introduction
Hepatocellular carcinoma (HCC) is the fifth commonest cancer globally and the commonest primary liver cancer [1]. Manifested by the data from the China National Cancer Registry, mortality rate of primary liver cancer is listed the third, while the incidence is the fourth among common malignancies [2]. The major risk factors for HCC are chronic infection with hepatitis C virus and hepatitis B virus, aflatoxin-contaminated foodstuffs, obesity, smoking, heavy alcohol intake and type 2 diabetes [3]. Transarterial chemoembolization is an established treatment for HCC in intermediate stage, which improves survival in the majority of HCC patients in intermediate or advanced stages [4]. Currently, the diagnosis of HCC majorly depends on serum biomarkers and imaging techniques [5]. HCC-related 5-year survival rate is only about 60%, and the incidence raises year by year in recent years [6]. Given that, researching for accurate therapeutic target is a priority in treatment of HCC.
Macrophages are effector cells and major regulators of the immune system, exert enormous functions on tissue remodeling and repair, and perform orchestrate metabolic functions in almost all tissues in vivo [7]. It is revealed that M1 macrophages exert tumor-promoting effects and enhance the motility of HCC cells [8]. Exosome is a discoid vesicle with a diameter of 40–150 nm [9]. According to Xu et al., exosomal microRNAs (miRNAs) have functions on proliferation, invasiveness, metastasis and drug resistance of HCC via modulating gene expression in the target cells [10]. A study has demonstrated that miR-326 containing exosomes could be a potential clinical target in multiple sclerosis [11]. MiRNAs could act as either oncogenes or tumor inhibitors through modulating a large number of protein-encoded genes expression by means of mRNA degradation and translation blockage in a specific sequence [12]. A study has discussed that miR-326 suppresses HCC cell growth through disturbing cell-cycle progression as well as enhancing apoptosis, in addition suppresses cell invasion via decreasing the epithelial–mesenchymal transition phenotype [13]. Another study has reported that miR-326 could be a potential therapeutic target for HCC patients treatment [14]. Therefore, this present study discussed the mechanism of exosomal miR-326 regulating the invasion and migration of HCC cells.
Materials and Methods
Ethics Statement
All animal experiments were in compliance with the Guide for the Care and Use of Laboratory Animal by International Committees. The protocol was approved by the Institutional Animal Care Use Committee of The Third Hospital of Jilin University.
Induction and Identification of Macrophages
Human monocytic cell line THP-1 (Kunming Institute of Zoology, CAS, Kunming, China) was cultured in RPMI 1640 medium (Gibco, CA, USA; Thermo Fisher Scientific, MA, USA) containing 10% heat-inactivated fetal bovine serum (FBS). THP-1 cells were reacted with 150 ng/mL phorbol 12-myristate 13-acetate (PMA; P8139, Sigma-Aldrich, SF, CA, USA) and incubated in RPMI medium for 24 h to obtain M0 macrophages. Then, the cell morphology before and after induction was observed by Wright staining. THP-1 cells and induced macrophages were resuspended in 5 μL PBS, dropped on the glass slide, stained by Wright's dye solution and mixed with the buffer solution at 1:2. Dyed for 10 min and rinsed with running water, the cells were observed under the microscope. In addition, M0 macrophage markers (CD68 and CD206) were measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR). Next, macrophages were induced into M1 macrophages through incubation with 20 ng/mL Interferon (IFN)-γ (#285-IF; R&D Systems, MN, USA) and 10 pg/mL LPS (#8630; Sigma-Aldrich) for 18 h. M1 macrophage markers (IDO1 and IL-12 p35) were examined by RT-qPCR [15].
Extraction of Exosomes
The exosomes were extracted by exosomes separation kit (ExoEasy Maxi Kit, Qiagen, Hilden, Germany). Macrophage supernatant was amassed to a 15-mL centrifuge tube under aseptic conditions and filtrated by 0.8-μM filtering film. Cell supernatant in each group was appended with XBP buffer (1: 1) and then centrifuged with exoEasy membrane affinity centrifugation column at 500g. Cells were supplemented with 10 mL XWP buffer and centrifuged with 3000–5000g. The exoEasy membrane affinity centrifugation column was hatched with 400 μL XE eluting buffer and centrifuged at 500g. The eluting buffer was transferred to exoEasy membrane affinity centrifugation column and centrifuged at 500g. The eluting buffer was stored for 24 h at 4 °C and then used for identification. The rest was stored at − 80 °C.
TEM Observation and Nanoparticle Tracking Analysis (NTA)
The above obtained exosomes were dropped in the carbon-supported membrane copper mesh and added with 2% phosphotungstic acid. The sample was observed under a TEM, and the optimal image was collected and analyzed.
The impurities and particles in PBS were removed by a 0.22-μM microporous filter. According to the particle density of the exosomes, the filtered PBS was diluted to the appropriate concentration and detected using a Nanosight NS300 nanoparticle detector (Malvern, Westborough, MA, USA).
After identification, macrophage-derived exosomes transfected with miR-326 inhibitor and miR-326 inhibitor negative control (NC), miR-326 mimic and miR-326 mimic NC were extracted by exosomes separation kit (Invitrogen).
RT-qPCR
The total RNA was extracted by Trizol Reagent (Thermo Fisher), and real-time PCR was performed using the SYBR-Green PCR Master Mix (Roche) and the ABI 7500 Real-Time PCR System (Life Technologies, Grand Island, NY, USA). The primer sequences are shown in Table 1. The quantitative analysis was carried out using the method of 2−△△Ct.
Table 1.
Primer sequence
| Gene | Sequence (5′–3′) |
|---|---|
| IDO1 | F: TATTTGTCTGGCTGGAAAGGC |
| R: GGAGGAACTGAGCAGCATGTC | |
| IL-12p35 | F: GATGGCCCTGTGCCTTAGTA |
| R: TCAAGGGAGGATTTTTGTGG | |
| miR-326 | F: ACTGTCCTTCCCTCTGGGC |
| R: AATGGTTGTTCTCCACTCTCTCTC | |
| CD206 | F: AAAACTGACTGGGCTTCCGT |
| R: CCTCTCGAGCACAGGTCATC | |
| CD68 | F: GCTTTGCAATCTCCCTGTTG |
| R: TTGATCCGGGGGTTCTTACCTG | |
| NF-κB | F: GAAATTCCTGATCCAGACAAAAAC |
| R: ATCACTTCAATGGCCTCTGTGTAG | |
| U6 | F: ATTGGAACGATACAGAGAAGATT |
| R: GGAACGCTTCACGAATTTG | |
| GAPDH | F: AGAACATCATCCCTGCCTCTACT |
| R: GATGTCATCATATTTGGCAGGT |
F, forward; R, reverse; IDO1, indoleamine 2,3-dioxygenase 1; IL-12p35, interleukin-12p35; miR-326, microRNA-326; NF-κB, nuclear factor κB; GAPDH, glyceraldehyde phosphate dehydrogenase
Western Blot Analysis
The total protein of cells and exosomes was extracted. The protein concentration was determined by bicinchoninic acid (BCA) kit (Boster Biological Technology Co. Ltd., Wuhan, Hubei, China). The protein was appended with sample buffer and boiled at 95 °C, and each well was loaded with 30 µg. The protein was separated with 10% polyacrylamide gel electrophoresis (Boster Biological Technology) and electroblotted onto a polyvinylidene fluoride membrane, which was followed by sealing in 5% bovine serum albumin (BSA). The membrane was hatched with primary antibody against CD63 (1: 1000, Developmental Studies Hybridoma Bank, University of Iowa, Ames, IA, USA), CD181 (1: 1000, R&D Systems), GAPDH (1: 2000, Jackson ImmunoResearch Laboratories, PA, USA) and with the secondary antibody (1: 500, Jackson ImmunoResearch Laboratories) labeled by horseradish peroxidase. The images were obtained by Odyssey dual color infrared fluorescence scanning imaging system, and the gray values of bands were measured by Quantity One image analysis software.
Cell Culture and Screening
Normal liver cell line HL-7702 and human HCC cell line BEL-7404, HepG2, SMMC-7721 and QGY-7703 were selected and cultured in Gibco RPMI Media 1640 with 10% fetal bovine serum (FBS), penicillin (100 U/mL) and streptomycin (100 mg/mL). MiR-326 expression was detected by RT-qPCR, and the suitable cell lines were screened.
Exosomes Labeling and Uptake of Exosomes
The exosomes were resuspended with 250 μL Diluent C and gently triturated avoiding making damage to the exosome’s membrane. The PKH67 dye (1 μL, Sigma-Aldrich) was added to 250 μL Diluent C to reach 500 μL and incubated. The solution was appended with 500 μL 1% BSA and incubated at 37 °C for 1 min. The exosomes were obtained by centrifugation at 120,000g 4 °C for 2 h. The exosomes labeled with PKH67 were obtained by centrifugation at 120,000g 4 °C for 2 h. The exosomes were re-suspended with 6 mL RPMI-1640 medium avoiding light. Then, the labeled exosomes were co-cultured with HCC cells for 12 h. After that, the culture medium was removed and washed with PBS for 3 times, 5 min/time, and the fluorescent-labeled exosomes which were not internally absorbed by HCC cells were thoroughly washed off. The exosomes were fastened with 4% paraformaldehyde and dyed with 4′-6-diamidino-2-phenylindole. After sealing, the fluorescence distribution was observed by a laser confocal microscope.
Cell Grouping and Treatment
HepG2 cells and SMMC-7721 cells were seeded in the 12-well plate at 0.5–1 × 106 cells/well. With 50–60% confluence, cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). HepG2 cells were distributed into miR-326-mimic group (transfected with miR-326 mimic) and NC-mimic group (transfected with miR-326 mimic NC). SMMC-7721 cells were assigned into miR-326-inhibitor group (transfected with miR-326 inhibitor) and NC-inhibitor group (transfected with miR-326 inhibitor NC). miR-326-mimic, miR-326-inhibitor and their NCs were mixed with Lipofectamine 2000 for transfection. HepG2 cells and SMMC-7721 cells without any treatment were set as the blank group. miR-326-mimic, miR-326-inhibitor and their NC were devised and composed by Guangzhou RibBio Co., Ltd. (Guangzhou, China) (Table 1).
Co-culture of M1 Macrophage-Derived Exosomes with HCC Cells
The protein concentration of M1 macrophage-derived exosomes suspension was detected by BCA method, and the volume of corresponding exosomes suspension with 50 μg protein was calculated. HepG2 cells and SMMC-7721 cells were seeded in 12-well plate at 1 × 105 cells/mL per well. HepG2 cells were distributed into 4 groups: control group (HepG2 cells not co-cultured with exosomes), exosomes (Exo) group (HepG2 cells co-cultured with M1 macrophages-derived exosomes), Exo-miR-326-mimic group (HepG2 cells co-cultured with M1 macrophage-derived exosomes which transfected with miR-326 mimic), Exo-NC-mimic group (HepG2 cells co-cultured with M1 macrophage-derived exosomes which transfected with miR-326 mimic NC). SMMC-7721 cells were also assigned into 4 groups: blank group (SMMC-7721 cells not co-cultured with exosomes), Exo group (SMMC-7721 cells co-cultured with M1 macrophages-derived exosomes), Exo-miR-326-inhibitor group (SMMC-7721 cells co-cultured with M1 macrophage-derived exosomes which transfected with miR-326 inhibitor), Exo-NC-inhibitor group (SMMC-7721 cells co-cultured with M1 macrophage-derived exosomes which transfected with miR-326 inhibitor NC).
3-(4, 5-Dimethylthiazol-2-yl)-2, 5-Diphenyltetrazolium Bromide (MTT) Assay
The cells were detached with trypsin and seeded on 96-well plate with the cell density of 4 × 104 cells per well. The culture medium was abandoned after culturing 12, 24, 36, 48, 60 h, respectively. Incubated with 500 μL 0.5 g/L MTT solution, the cells were appended with 200 μL dimethyl sulfoxide solution, triturated and hatched. Optical density (OD, 490 nm) values were measured by a microplate reader.
Colony Formation Assay
Cultured for 24 h and detached with trypsin, the cells were seeded in a 35-mm small dish with 300 cells per dish. The solution was replaced every 3 d. After 10 d of culture, the cells were fixed with 40 g/L−1 paraformaldehyde and dyed with 1 g/L−1 crystal violet solution and dried. Colony number (more than 50 cells) was computed under a microscope.
Transwell Assay
Cells (1 × 105) were suspended with 200 μL blank culture media. Experiments were conducted in conformity with the instruction of Transwell chamber (Corning Glass Works, Corning, N.Y., USA) (matrigel was needed for invasion experiment, but not for migration experiment). RPIM 1640 (10% FBS, 600 μL) was added into the lower chamber. The upper and lower chambers were separated by a Transwell membrane precoated by matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Cultured for 24 h, the chamber was fixed with 95% alcohol. After staining with crystal violet solution, cells were observed in five visual fields under the microscope.
Flow Cytometry
Cell cycle: cells were detached by trypsin. Cells (1 × 106) were suspended with 0.5 mL PBS and triturated into single suspension. Mixed with 4.5 mL pre-cooled 70% ethanol on ice, the cells were centrifuged at 3000g, rinsed with 5 mL PBS and centrifuged again at 3000g. Followed by that, the cells were suspended with 1 mL PI/Triton X-100 staining solution (20 μg PI/0.1% Triton X-100) containing 0.2 mg RnaseA. Cell cycle was detected by flow cytometry.
Cell apoptosis: Trypsinized cells (1 × 106) were suspended with 1 mL PBS, triturated and centrifuged at 3000g. Cells were rinsed in turn with incubation buffer (10 mmol/L Hepes/NaOH, pH 7.4, 140 mmol/L NaCl, 5 mmol/L CaCl2) and centrifuged at 3000g. Next, cells were incubated with 100 μL marking solution (FITC-Annexin V and PI were added to the incubation buffer to reach 1 μg/mL), centrifuged at 3000g, washed once with incubation buffer and hatched with fluorescent (SA-FLOUS) solution. Cell apoptosis was detected by flow cytometry. The wavelength of flow cytometry was 488 nm, and FITC fluorescence was detected by band filter at 515 nm, while PI with wavelength larger than 560 nm. The results were analyzed automatically by computer.
Tumor Xenograft in Nude Mice
Forty mice (College of Animal Science, Jilin University, Jilin, China) aged 4–6 w were randomly distributed into 8 group, with 5 mice in each group. The mice were fed in a specific pathogen-free grade animal laboratory for 1 w, and the feed, cushion and water bottle were replaced in time. The health status of mice should be observed every day. One week later, HCC cells were prepared into cell suspension and subcutaneously injected into the neck and back at 0.1 mL cell suspension (1 × 106). The growth of the tumor was observed after 3–5 d. The nude mice were weighed every 4 d, and the tumor volume was measured. The nude mice were euthanized 20 d after injection.
Statistical Analysis
All data were interpreted by SPSS 17.0 software (SPSS Statistics, Chicago, IL, USA). Measurement data were indicated as mean ± standard deviation. Comparisons between two groups were formulated by the t-test, while comparisons among multiple groups were assessed by one-way analysis of variance (ANOVA). P value < 0.05 was indicative of statistically significant difference.
Results
Identification of M1 Macrophage and Exosomes
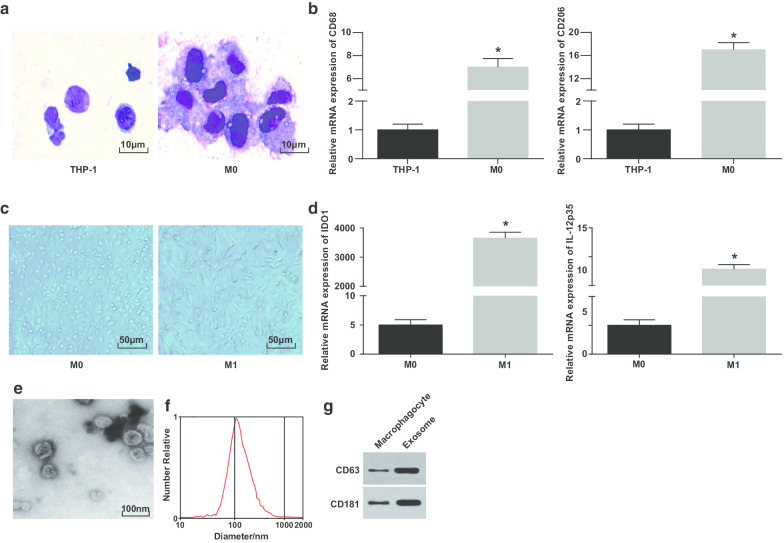
Wright's staining was utilized to observe the morphology of THP-1 cells induced by PMA. It was pictured that the volume of THP-1 cells before induction was small and the proportion of caryoplasm was higher; the morphology of cells after induction was irregular, the volume became larger, and the proportion of caryoplasm decreased; the cytoplasm was richer and performed light blue, with abundant particles and a few vacuoles; the nucleus was purplish red and often inclined to one side, showing that the cells had the typical morphological characteristics of macrophages (Fig. 1a).
Fig. 1.
Identification of M1 macrophage and exosomes. a Wright’s staining for observing the morphology of THP-1 cells before and after PMA treatment. b CD68 and CD206 expression in THP-1 cells and PMA-treated THP-1 cells detected by RT-qPCR; c morphology of macrophages and M1 macrophages. d Expression of IDO1 and IL-12 p35 in M0 macrophages and LPS and INF-γ-treated M0 macrophages detected by RT-qPCR. e TEM for exosomes observation. f Detection of particle size distribution of exosomes by NTA. g Protein bands of CD63 and CD181. In panel b, *P < 0.05 versus THP-1 cells; In panel d, *P < 0.05 versus M0 macrophages. Measurement data were depicted as mean ± standard deviation (N = 3), comparisons between two groups were conducted by t test
To further verify the successful induction of macrophages, CD68 and CD206 expression before and after induction was tested by RT-qPCR. It was manifested that CD68 and CD206 expression were elevated since PMA induction, indicating the PMA successfully induced THP-1 cells into M0 macrophages (Fig. 1b). Then, M0 macrophages were polarized into M1 macrophages by induction of LPS and INF-γ. The morphology, and M1 type macrophage surface marker IDO1 and IL-12 p35 expression of macrophages were observed and tested. M0 macrophages showed various and irregular adherent morphologies, showing round, elliptical or spindle shape. The morphology of macrophages stimulated by IFN-γ demonstrated with more pseudopods and protrusions and fusiformis (Fig. 1c). RT-qPCR outlined (Fig. 1d) that after LPS and INF-γ treatment, M1 macrophages showed an increase in their markers (IDO1 and IL-12p35).
Subsequently, the exosomes derived from macrophages were observed by the TEM. It was revealed that the exosomes derived from macrophages were rich and the shape was round or oval, with membranous structure, uniform size and less pollutants (Fig. 1e). NTA displayed that exosomes with a centralized peak of MODE curve and smooth linear had more concentrated diameter and less impurities (Fig. 1f). Western blot analysis reported that versus macrophages, the specific marker protein CD63 and CD181 expression heightened in exosomes derived from macrophages (Fig. 1g). These results indicate that we successfully induced monocytes to differentiate into macrophages and polarized them into M1 macrophages.
M1 Macrophage-Derived Exosomes Deliver miR-326 to HCC Cells and Affect miR-326 Expression in HCC Cells
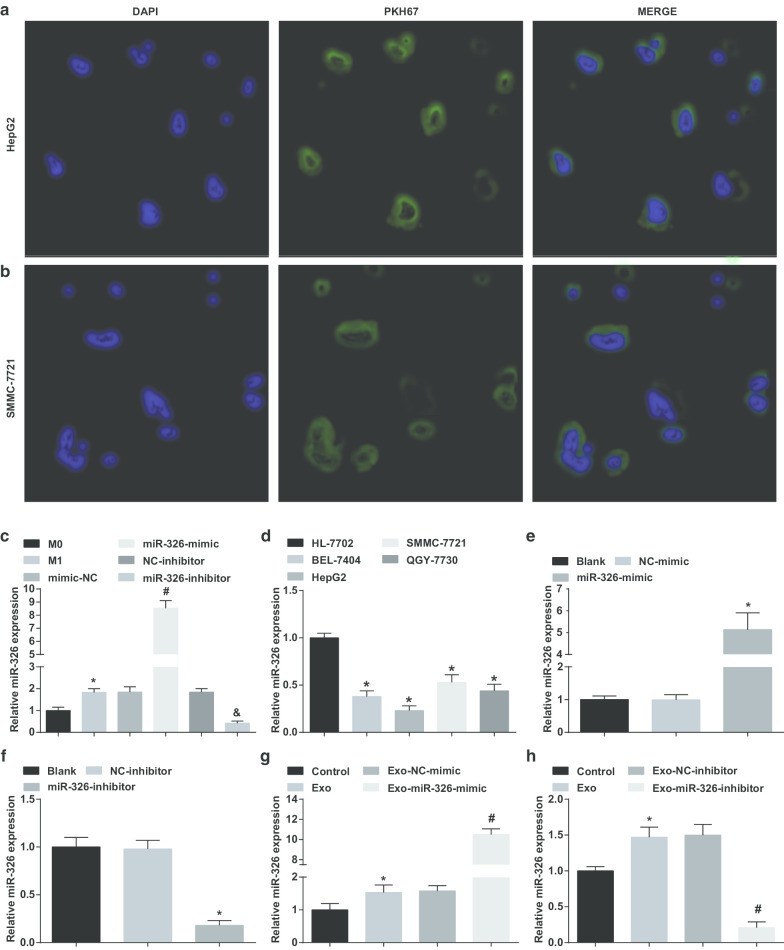
To verify whether exosomes derived from M1 macrophages transported miR-326 to HCC cells, HepG2 and SMMC-7721 cells were co-cultured with exosomes. It could be recognized that a large number of exosomes were assimilated by HepG2 and SMMC-7721 cells upon 4-h transfection under the fluorescence microscope (Fig. 2a, b).
Fig. 2.
M1 macrophage-derived exosomes deliver miR-326 to HCC cells and affect miR-326 expression in HCC cells. a Uptake of M1 macrophages derived exosomes by HepG2 cells. b Uptake of M1 macrophages-derived exosomes by SMMC-7721 cells. c Comparison of miR-326 expression of macrophage exosomes in each group before and after INF-γ and LPS induction. d miR-326 expression in HCC cell lines (BEL-7404, HepG2, SMMC-7721, QGY-7703) and human normal hepatocyte HL-7702 cell line detected by RT-qPCR. e RT-qPCR detected the effect of miR-326 mimic on miR-326 expression in HepG2 cells. f RT-qPCR detected the effect of miR-326 inhibitor on miR-326 expression in SMMC-7721 cells. g RT-qPCR detected the effects of miR-326 mimic-transfected M1 macrophage-derived exosomes on miR-326 expression in HepG2 cells. h RT-qPCR detected the effects of miR-326 inhibitor-transfected M1 macrophage-derived exosomes on miR-326 expression in SMMC-7721 cells. In panel c, *P < 0.05 versus the M0 macrophage; In panel d, *P < 0.05 versus HL-7702 cells; In panel e, *P < 0.05 versus the NC-mimic group; In panel f, *P < 0.05 versus the NC-inhibitor group; In panel g, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-mimic group; In panel h, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-inhibitor group. Measurement data were depicted as mean ± standard deviation (N = 3), comparisons among multiple groups were conducted by one-way analysis of variance
Subsequently, miR-326-mimic/inhibitor was transfected into macrophages and checked miR-326 expression in their exosomes before and after transfection. It was displayed that versus the exosomes from M0 macrophages, miR-326 expression was raised in M1 macrophages-derived exosomes. Elevated miR-326 expression was seen in the M1 macrophages-derived exosomes with miR-326-mimic treatment. MiR-326 expression reduced in M1 macrophages-derived exosomes with miR-326-inhibitor treatment (Fig. 2c).
Then, miR-326 expression in HCC cell lines was tested. As manifested, miR-326 expression was decreased in BEL-7404, HepG2, SMMC-7721 and QGY-7703 cells compared to HL-7702 cells, among which HepG2 cells manifested with the lowest expression, while SMMC-7721 cells with the highest expression (Fig. 2d).
Followed by that, miR-326-mimic and miR-326-inhibitor were transfected into HepG2 and SSMC-7721 cells, respectively, and examined their effects on miR-326 expression. MiR-326 mimic elevated miR-326 expression in HepG2 cells, while miR-326 inhibitor reduced miR-326 expression in SSMC-7721 cells (Fig. 2e, f).
Next, the exosomes from M1 macrophages transfected with miR-326-mimic and miR-326-inhibitor were co-cultured with HepG2 and SSMC-7721 cells, respectively. It was highlighted that co-culture with exosomes increased miR-326 expression in HCC cells, miR-326-mimic-transfected M1 macrophages-derived exosomes further raised miR-326 expression in HepG2 cells, while miR-326-inhibitor-transfected M1 macrophages-derived exosomes degraded miR-326 expression in SSMC-7721 cells (Fig. 2g, h). It is suggested that M1 macrophage-derived exosomes deliver miR-326 to HCC cells and affect miR-326 expression in HCC cells.
M1 Macrophage-Derived Exosomal miR-326 Reduces Cell Proliferation and Colony Formation Ability in HCC Cells
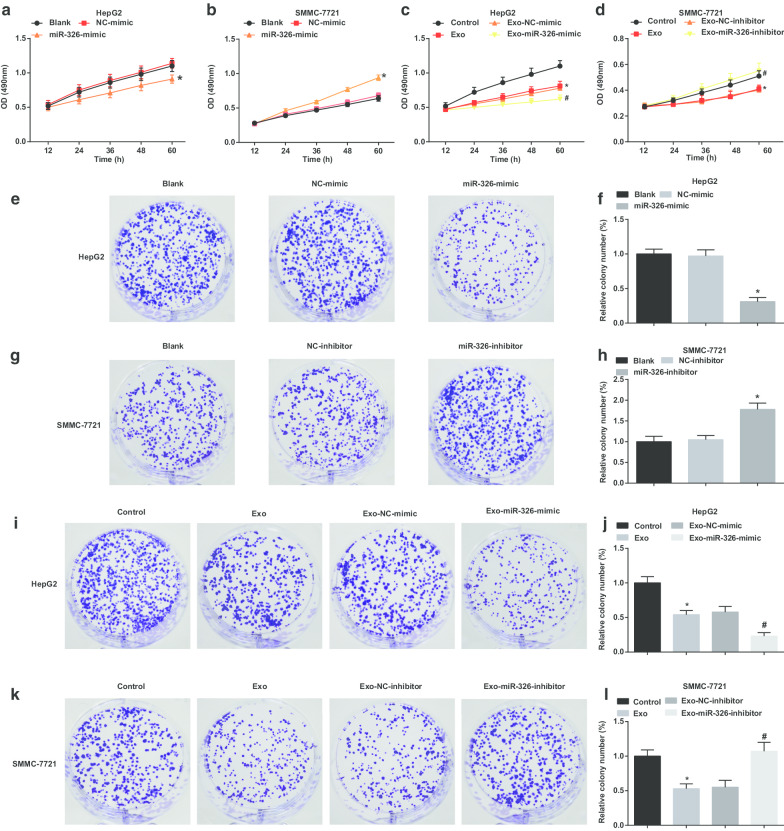
In exploring the effect of exosomal miR-326 on the proliferation of HCC cells, MTT and colony formation assays were conducted to examine HCC cell proliferation. It was suggested that in HepG2 cells, miR-326 restoration impaired cell proliferation and colony formation ability (Fig. 3a, e, f). M1 macrophages-derived exosomes obstructed HepG2 cells to proliferate and form colonies. MiR-326 mimic-transfected M1 macrophages-derived exosomes further impaired cell proliferation and colony formation ability (Fig. 3c, i, j).
Fig. 3.
M1 macrophage exosomal miR-326 inhibits cell proliferation and colony formation in HCC cells. a MTT assay detected the effect of transfection of miR-326 mimic on the proliferation of HepG2 cells. b MTT assay detected the effect of transfection of miR-326 inhibitor on the proliferation of SMMC-7721 cells. c MTT assay detected the effect of co-culture with miR-326 mimic-transfected M1 macrophage-derived exosomes on the proliferation of HepG2 cells. d MTT assay detected the effect of co-culture with miR-326 inhibitor-transfected M1 macrophage-derived exosomes on the proliferation of SMMC-7721 cells. e Colony formation assay detected the effect of transfection of miR-326 mimic on the colony formation ability of HepG2 cells. f Colony numbers of HepG2 cells. g Colony formation assay detected the effect of transfection of miR-326 inhibitor on the colony formation ability of SMMC-7721 cells. h Colony numbers of SMMC-7721 cells. i Colony formation assay detected the effect of co-culture with miR-326 mimic-transfected M1 macrophage-derived exosomes on the colony formation ability of HepG2 cells. j Colony numbers of HepG2 cells treated with exosomes. k Colony formation assay detected the effect of co-culture of miR-326 inhibitor-transfected M1 macrophage-derived exosomes on the colony formation ability of SMMC-7721 cells. l Colony numbers of SMMC-7721 cells treated with exosomes. In panel a and f, *P < 0.05 versus the NC-mimic group; In panel b and h, *P < 0.05 versus the NC-inhibitor group; In panel c and j, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-mimic group; In panel d and l, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-inhibitor group. Measurement data were depicted as mean ± standard deviation (N = 3), comparisons among multiple groups were conducted by one-way analysis of variance
In SMMC-7721 cells, miR-326 knockdown enhanced cell proliferation and colony formation ability (Fig. 3b, g, h). In SMMC-7721 cells treated with M1 macrophages-derived exosomes, cell proliferation and colony formation ability were decreased. MiR-326-inhibitor-transfected M1 macrophages-derived exosomes further encouraged cell proliferation and colony formation ability (Fig. 3d, k, l). It is hinted that miR-326 derived from M1 macrophage-derived exosomes impedes the proliferation of HCC cells.
M1 Macrophage-Derived Exosomal miR-326 Suppresses Migration and Invasion of HCC Cells
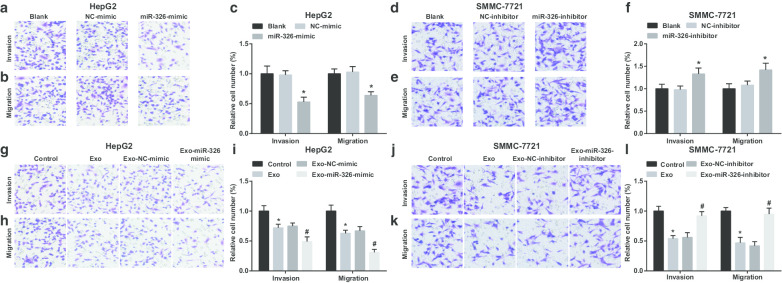
Then, the effect of exosomal miR-326 on the invasion and migration of HCC cells was examined. It was demonstrated that in HepG2 cells, restoration of miR-326 restricted invasion and migration (Fig. 4a–c). M1 macrophages-derived exosomes disturbed HepG2 cells to invade and migrate. Invasion and migration further degraded when HepG2 cells were co-cultured with miR-326-mimic-transfected M1 macrophages-derived exosomes (Fig. 4g–i).
Fig. 4.
M1 macrophage exosomal miR-326 inhibits migration and invasion of HCC cells. a Transwell assay detected the effect of transfection of miR-326 mimic on the invasion of HepG2 cells. b Transwell assay detected the effect of transfection of miR-326 mimic the migration of HepG2 cells. c The number of invasion and migration of HepG2 cells. d Transwell assay detected the effect of transfection of miR-326 inhibitor on the invasion of SMMC-7721 cells. e Transwell assay detected the effect of transfection of miR-326 inhibitor on the migration of SMMC-7721 cells. f The number of invasion and migration of SMMC-7721 cells. g Transwell assay detected the effect of co-culture with miR-326 mimic-transfected M1 macrophage-derived exosomes on the invasion of HepG2 cells. h Transwell assay detected the effect of co-culture with miR-326 mimic-transfected M1 macrophage-derived exosomes on the migration of HepG2 cells. i The number of invasion and migration of HepG2 cells co-cultured with M1 macrophage-derived exosomes. j Transwell assay detected the effect of co-culture with miR-326 inhibitor-transfected M1 macrophage-derived exosomes on the invasion of SMMC-7721 cells. k Transwell assay detected the effect of co-culture with miR-326 inhibitor-transfected M1 macrophage-derived exosomes on the migration of SMMC-7721 cells. l The number of invasion and migration of SMMC-7721 cells co-cultured with M1 macrophage-derived exosomes. In panel c, *P < 0.05 versus the NC-mimic group; In panel f, *P < 0.05 versus the NC-inhibitor group; In panel i, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-mimic group; In panel l, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-inhibitor group. Measurement data were depicted as mean ± standard deviation (N = 3), comparisons among multiple groups were conducted by one-way analysis of variance
MiR-326 knockdown resulted in enhancements in SMMC-7721 cell invasion and migration (Fig. 4d–f). When treated with M1 macrophages-derived exosomes, SMMC-7721 cells were exhibited with decreased invasion and migration. However, SMMC-7721 cell invasion and migration were boosted upon co-culture with miR-326-inhibitor-transfected M1 macrophages-derived exosomes (Fig. 4j–l). It is implied that miR-326 derived from M1 macrophage exosomes impedes the invasion and migration of HCC cells.
M1 Macrophage-Derived Exosomal miR-326 Promotes Apoptosis of HCC Cells
When examining the effect of exosomal miR-326 on the cell cycle and apoptosis of HCC cells, PI single staining and Annexin V-FITC/PI double staining were applied. It was illustrated that miR-326 overexpression increased cells arrested at G0/G1 phase, reduced cells arrested at S and G2/M phases and raised apoptosis in HepG2 cells (Fig. 5a–d). Co-culturing with M1 macrophages-derived exosomes increased cells arrested at G0/G1 phase, reduced cells arrested at S and G2/M phases and raised cell apoptosis of HepG2 cells. Co-cultivation with exosomes from M1 macrophages transfected with miR-326-mimic further enhanced these effects (Fig. 5i–l).
Fig. 5.
M1 macrophage exosomal miR-326 promotes apoptosis of HCC cells. a, b Flow cytometry detected the effect of transfection of miR-326 mimic on HepG2 cell cycle; c, d flow cytometry detected the effect of transfection of miR-326 mimic on HepG2 cell apoptosis; e, f flow cytometry detected the effect of transfection of miR-326 inhibitor on SMMC-7721 cell cycle; g, h flow cytometry detected the effect of transfection of miR-326 inhibitor on SMMC-7721 cell apoptosis; i, j flow cytometry detected the effects of co-culture of with miR-326 mimic-transfected M1 macrophage-derived exosomes on HepG2 cell cycle; k, l flow cytometry detected the effect of co-culture with miR-326 mimic-transfected M1 macrophage-derived exosomes on HepG2 cell apoptosis; m, n flow cytometry detected the effect of co-culture with miR-326 inhibitor-transfected M1 macrophage-derived exosomes on SMMC-7721 cell cycle; o, p, flow cytometry detected the effect of co-culture with miR-326 inhibitor-transfected M1 macrophage-derived exosomes on SMMC-7721 cell apoptosis. In panel b and d, *P < 0.05 versus the NC-mimic group; In panel f and h, *P < 0.05 versus the NC-inhibitor group; In panel j and l, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-mimic group; In panel n and p, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-inhibitor group . Measurement data were depicted as mean ± standard deviation (N = 3), comparisons among multiple groups were conducted by one-way analysis of variance
In SMMC-7721 cells, miR-326 down-regulation reduced cells arrested at G0/G1 phase, elevated cells arrested at S and G2/M phases, and declined cell apoptosis (Fig. 5e–h). Untransfected M1 macrophages-derived exosomes increased cells arrested at G0/G1 phase, reduced cells arrested at S and G2/M phases, and heightened cell apoptosis. MiR-326-inhibitor-transfected M1 macrophages-derived exosomes degraded cells arrested at G0/G1 phase, elevated cells arrested at S and G2/M phases, and decreased cell apoptosis (Fig. 5m–p). Briefly, it is summarized that miR-326 derived from M1 macrophage exosomes arrests cell cycle in G0/G1 phase and induces cell apoptosis in HCC.
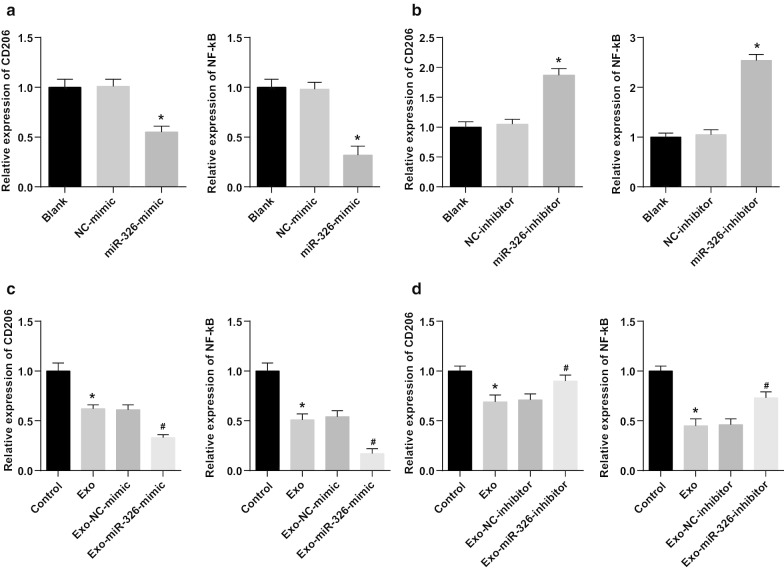
M1 Macrophage-Derived Exosomal miR-326 Declines CD206 and NF-κB Expression in HCC Cells
Next, the potential mechanism of miR-326 derived from M1 macrophage exosomes in the biological progress of HCC cells was explored. NF-κB is the key link between inflammation and cancer. Many regulatory proteins and miRNAs could inhibit the excessively activated NF-κB signaling to suppress cancer [16]. Such a beneficial effect may include the polarization of M2 macrophages into M1 macrophages. CD206 and NF-κB expression in HepG2 and SMMC-7721 cells was tested by RT-qPCR. It was suggested that miR-326 restoration decreased CD206 and NF-κB expression in HepG2 cells, while miR-326 knockdown enhanced CD206 and NF-κB expression in SMMC-7721 cells (Fig. 6a, b). Moreover, co-culture with M1 macrophage exosomes significantly reduced CD206 and NF-κB expression in HepG2 cells, while co-culture with M1 macrophage exosomes-overexpressing miR-326 further decreased CD206 and NF-κB expression. Treated with untransfected M1 macrophages-derived exosomes, CD206 and NF-κB expression was decreased in SMMC-7721 cells. Co-cultured with miR-326-inhibitor-transfected M1 macrophages-derived exosomes, SMMC-7721 cells were featured by heightened CD206 and NF-κB expression (Fig. 6c, d). It was concluded that miR-326 from M1 macrophage exosomes played a tumor suppressor by inhibiting NF-κB in HCC cells.
Fig. 6.
M1 macrophage exosomal miR-326 declines CD206 and NF-κB expression in HCC cells. a RT-qPCR detected the effect of transfection of miR-326 mimic on the expression of CD206 and NF-κB in HepG2 cells; b RT-qPCR detected the effect of transfection of miR-326 inhibitor on the expression of CD206 and NF-κB in SMMC-7721 cells. c RT-qPCR detected the effect of co-culture with miR-326 mimic-transfected M1 macrophage-derived exosomes on the expression of CD206 and NF-κB in HepG2 cells; d RT-qPCR detected the effect of co-culture with miR-326 inhibitor-transfected M1 macrophage-derived exosomes on the expression of CD206 and NF-κB in SMMC-7721 cells. In panel a, *P < 0.05 versus the NC-mimic group; In panel b, *P < 0.05 versus the NC-inhibitor group; In panel c, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-mimic group; In panel d, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-inhibitor group. Measurement data were depicted as mean ± standard deviation (N = 3), comparisons among multiple groups were conducted by one-way analysis of variance
miR-326 from M1 Macrophage Exosomes Inhibits HCC Tumor Growth In Vivo
Finally, the in vivo results were validated through tumor xenografts. As displayed, miR-326 overexpression decreased volume and weight of tumors in HepG2 cells (Fig. 7a–c). In mice transplanted with HepG2 cells co-cultured with exosomes, the treatment with M1 macrophage exosomes significantly reduced the tumor volume and weight of HepG2 cells, while treatment with M1 macrophages-overexpressing miR-326 further reduced the tumor volume and weight of HepG2 cells (Fig. 7g–i).
Fig. 7.
M1 macrophage exosomal miR-326 reduces the volume and weight of HCC tumor in vivo. a–c The effect of transfection of miR-326 mimic on the tumor volume and tumor of nude mice xenografted with HepG2 cells. d–f The effect of transfection of miR-326 inhibitor on the tumor volume and tumors of nude mice xenografted with SMMC-7721 cells. g–i The effect of co-culture with miR-326 mimic-transfected M1 macrophage-derived exosomes on the tumor volume and tumor of nude mice xenografted with HepG2 cells. j–l The effect of co-culture with miR-326 inhibitor-transfected M1 macrophage-derived exosomes on the tumor volume and tumor of nude mice xenografted with SMMC-7721 cells. In panel b and c, *P < 0.05 versus the NC-mimic group; In panel e and f, *P < 0.05 versus the NC-inhibitor group; In panel h and i, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-mimic group; In panel k and l, *P < 0.05 versus the control group, #P < 0.05 versus the Exo-NC-inhibitor group. Measurement data were depicted as mean ± standard deviation (n = 5), comparisons among multiple groups were conducted by one-way analysis of variance
In SMMC-7721 cells, miR-326 suppression increased volume and weight of tumors (Fig. 7d–f). Untransfected M1 macrophages-derived exosomes obstructed tumor growth in volume and weight of SMMC-7721 cells. Co-cultured with miR-326 inhibitor-transfected M1 macrophages-derived exosomes, SMMC-7721 cells were injected into mice and caused elevations in tumor volume and weight (Fig. 7j–l). Summarily, miR-326 derived from M1 macrophage exosomes depressed tumor growth of HCC in vivo.
Discussion
HCC is a common cancer that is characterized with high morbidity and mortality, difficult early diagnosis and treatment, poor prognosis and 5-year survival rate [17]. Recently, a study has highlighted that the lowly expressed lncRNA cox-2 declines the ability of M1 macrophages to suppress HCC cell growth, invasiveness, angiogenesis migration and promote apoptosis [18]. Hu et al. have discussed that miR-326 is obviously degraded in HCC tissues and cell lines, while down-regulated miR-326 is connected to the TNM stage, lymph node metastasis and differentiation of HCC patients [14]. It is customarily considered that HCC cells-derived exosomes can form a fertile environment to facilitate HCC cells growth, invasiveness and metastasis as well as development of drug resistance [19]. The current study was designed to explore the mechanism of exosomal miR-326 in regulating invasion and migration of HCC cells.
Our results indicated that miR-326 expression decreased in HCC cells but increased in exosomes. A recent study has pointed out that miR-326 expression is declined in HCC tissues [20]. Another study has presented miR-326 expression is notably reduced in HCC cell lines and tissues and its down-regulation predicts a poor prognosis in HCC [13]. It is reported that miR-326 acts a tumor-suppression role and is greatly depressed in HCC cells [21]. All these aforementioned evidences are in line with our findings. A study has purported that in comparison with the controls, miR-326 expression is raised in Tconv-derived exosomes which is observed in relapsing–remitting multiple sclerosis patients [11].
Other results emerge from our data that highly expressed exosomal miR-326 reduced cell proliferation, colony formation, migration and invasion as well as facilitated apoptosis of HCC cells in vitro and reduced the volume and weight of HCC tumor in vivo. It has been suggested previously that HCC cell growth can be suppressed via overexpression of miR-326, and HCC cell migration and invasion ability are markedly attenuated through elevating miR-326 [21]. It is reported that the up-regulated miR-326 expression suppresses HCC cell growth and invasiveness as well as stimulates cell apoptosis in vitro [14]. Besides that, a prior study has verified that overexpression of miR-326 declined tumor growth in vivo [13]. A study has revealed that ectopic expressed miR-326 markedly attenuates cell growth, and suppresses cellular migration and invasiveness in non-small cell lung cancer cell lines [22]. Moreover, it is found that miR-326 decreases profibrotic genes like MMP-9, implying its repressive function in cancer cell proliferation [23]. Also, it is presented that miR-326 represses Bcl-2 protein expression and elevates Bax expression so as to affect the apoptosis [24]. Similar to our findings, there are some miRNAs interacting with exosomes to play a role in HCC development. It is displayed that highly expressed exosomal miR-638 can repress the proliferation of HCC cells, involving the potential impact on carcinogenesis [25]. Another study also proves that HCC cells-derived exosomal miR-451a suppresses tumor angiogenesis via disrupting endothelial functions as apoptosis, tube formation, migration and permeability [26]. A prior research generally confirms that when treated with the overexpression of miR-744 exosomes, the proliferation of HCC cells is dramatically suppressed [27].
Conclusion
To briefly conclude, our study provides evidence that M1 macrophage-derived exosomal miR-326 suppresses proliferation, migration and invasion as well as advances apoptosis of HCC cells, supplying a new insight in a novel target therapy for HCC. Due to the limited sample size and limited known researches, the exact mechanism of miR-326 is not fully elucidated, and therefore, further large-scale studies are required to illustrate the underlying mechanism.
Acknowledgements
We would like to acknowledge the reviewers for their helpful comments on this paper.
Abbreviations
- HCC
Hepatocellular carcinoma
- miRNAs
MicroRNAs
- FBS
Fetal bovine serum
- M/CSF
Macrophage colony-stimulating factor
- PBS
Phosphate buffered saline
- DMEM
Dulbecco’s modified eagle medium
- PPAR
Peroxisome proliferator-activated receptor
- TEM
Transmission electron microscope
- NTA
Nanoparticle tracking analysis
- NC
Negative control
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- BCA
Bicinchoninic acid
- BSA
Bovine serum albumin
- Exo
Exosomes
- ANOVA
One-way analysis of variance
Authors’ contributions
XZ contributed to study design; ZB and HL contributed to manuscript editing; CL contributed to experimental studies; CS contributed to data analysis. All authors read and approved the final manuscript.
Funding
None.
Ethics approval and consent to participate
All animal experiments were in compliance with the Guide for the Care and Use of Laboratory Animal by International Committees. The protocol was approved by the Institutional Animal Care Use Committee of The Third Hospital of Jilin University.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Availability of data and materials
Not applicable.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: 10.1186/s11671-022-03763-8
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhen-zi Bai and Hong-yan Li co-first authors
Change history
12/13/2022
This article has been retracted. Please see the Retraction Notice for more detail: 10.1186/s11671-022-03763-8
References
- 1.Yoo SH, et al. Early development of de novo hepatocellular carcinoma after direct-acting agent therapy: comparison with pegylated interferon-based therapy in chronic hepatitis C patients. J Viral Hepat. 2018;25(10):1189–1196. doi: 10.1111/jvh.12918. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Elalfy H, et al. Monocyte/granulocyte to lymphocyte ratio and the MELD score as predictors for early recurrence of hepatocellular carcinoma after trans-arterial chemoembolization. Br J Biomed Sci. 2018;75(4):187–191. doi: 10.1080/09674845.2018.1494769. [DOI] [PubMed] [Google Scholar]
- 5.Ao L, et al. A qualitative signature for early diagnosis of hepatocellular carcinoma based on relative expression orderings. Liver Int. 2018;38(10):1812–1819. doi: 10.1111/liv.13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang D, et al. Long non-coding RNA SNHG1 functions as a competitive endogenous RNA to regulate PDCD4 expression by sponging miR-195-5p in hepatocellular carcinoma. Gene. 2019;714:143994. doi: 10.1016/j.gene.2019.143994. [DOI] [PubMed] [Google Scholar]
- 7.Muller J, et al. Differential S1P receptor profiles on M1- and M2-polarized macrophages affect macrophage cytokine production and migration. Biomed Res Int. 2017;2017:7584621. doi: 10.1155/2017/7584621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zong Z, et al. M1 macrophages induce PD-L1 expression in hepatocellular carcinoma cells through IL-1beta signaling. Front Immunol. 2019;10:1643. doi: 10.3389/fimmu.2019.01643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, et al. lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 2018;21(3):651–659. doi: 10.3233/CBM-170727. [DOI] [PubMed] [Google Scholar]
- 10.Xu X, et al. The role of microRNAs in hepatocellular carcinoma. J Cancer. 2018;9(19):3557–3569. doi: 10.7150/jca.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azimi M, et al. Altered expression of miR-326 in T cell-derived exosomes of patients with relapsing-remitting multiple sclerosis. Iran J Allergy Asthma Immunol. 2019;18(1):108–113. [PubMed] [Google Scholar]
- 12.Ghaemi Z, Soltani BM, Mowla SJ. MicroRNA-326 functions as a tumor suppressor in breast cancer by targeting ErbB/PI3K signaling pathway. Front Oncol. 2019;9:653. doi: 10.3389/fonc.2019.00653. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Mo Y, et al. Gold nano-particles (AuNPs) carrying miR-326 targets PDK1/AKT/c-myc axis in hepatocellular carcinoma. Artif Cells Nanomed Biotechnol. 2019;47(1):2830–2837. doi: 10.1080/21691401.2018.1489266. [DOI] [PubMed] [Google Scholar]
- 14.Hu S, et al. MicroRNA-326 inhibits cell proliferation and invasion, activating apoptosis in hepatocellular carcinoma by directly targeting LIM and SH3 protein 1. Oncol Rep. 2017;38(3):1569–1578. doi: 10.3892/or.2017.5810. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, et al. IL-16 regulates macrophage polarization as a target gene of mir-145-3p. Mol Immunol. 2019;107:1–9. doi: 10.1016/j.molimm.2018.12.027. [DOI] [PubMed] [Google Scholar]
- 16.Chaturvedi MM, et al. NF-kappaB addiction and its role in cancer: 'one size does not fit all'. Oncogene. 2011;30(14):1615–1630. doi: 10.1038/onc.2010.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu M, et al. Associated measurement of fucosylated levels of AFP, DCP, and GPC3 for early diagnosis in hepatocellular carcinoma. Int J Biol Mark. 2019;34(1):20–26. doi: 10.1177/1724600818812472. [DOI] [PubMed] [Google Scholar]
- 18.Ye Y, et al. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J Cell Biochem. 2018;119(3):2951–2963. doi: 10.1002/jcb.26509. [DOI] [PubMed] [Google Scholar]
- 19.Chen R, et al. Exosomes in hepatocellular carcinoma: a new horizon. Cell Commun Signal. 2019;17(1):1. doi: 10.1186/s12964-018-0315-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Q, et al. LncRNA SNHG3 promotes hepatocellular tumorigenesis by targeting miR-326. Tohoku J Exp Med. 2019;249(1):43–56. doi: 10.1620/tjem.249.43. [DOI] [PubMed] [Google Scholar]
- 21.Wei LQ, et al. Involvement of H19/miR-326 axis in hepatocellular carcinoma development through modulating TWIST1. J Cell Physiol. 2019;234(4):5153–5162. doi: 10.1002/jcp.27319. [DOI] [PubMed] [Google Scholar]
- 22.Sun C, et al. Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget. 2016;7(7):8341–8359. doi: 10.18632/oncotarget.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das S, et al. MicroRNA-326 regulates profibrotic functions of transforming growth factor-beta in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2014;50(5):882–892. doi: 10.1165/rcmb.2013-0195OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu S, et al. miR-326 targets antiapoptotic Bcl-xL and mediates apoptosis in human platelets. PLoS ONE. 2015;10(4):e0122784. doi: 10.1371/journal.pone.0122784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi M, et al. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem. 2018;119(6):4711–4716. doi: 10.1002/jcb.26650. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, et al. Exosomal miR-451a functions as a tumor suppressor in hepatocellular carcinoma by targeting LPIN1. Cell Physiol Biochem. 2019;53(1):19–35. doi: 10.33594/000000118. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, et al. Exosomal MiR-744 inhibits proliferation and sorafenib chemoresistance in hepatocellular carcinoma by targeting PAX2. Med Sci Monit. 2019;25:7209–7217. doi: 10.12659/MSM.919219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.