Abstract
Plants undergo several important developmental transitions including flowering and senescence during their life cycle. Timing these transitions according to the environmental conditions increases plant fitness and productivity. The circadian clock senses various environmental cycles, including photoperiod, and synchronizes plant physiological processes to maximize plant fitness. Here, we propose that the cellular localization of GIGANTEA (GI), a key clock component, regulates leaf senescence and flowering in Arabidopsis thaliana. We show that GI, which connects the circadian clock with photoperiod-regulated flowering, induces leaf senescence depending on its subcellular localization. Overexpression of GI in the gi mutant rescued its delayed senescence phenotype but only when the GI protein was targeted to the nucleus, not when it was targeted to the cytosol. In the nucleus, EARLY FLOWERING 4 (ELF4) inhibited the binding of GI to ORESARA 1 (ORE1) promoter to regulate leaf senescence. GI also positively regulated the day-peak of ORE1 expression. These results indicate that like flowering, leaf senescence is also controlled by the location of GI in the cell. Taken together, our results suggest that ELF4 and GI act together to control flowering and senescence in Arabidopsis.
Keywords: circadian clock, leaf senescence, GIGANTEA, EARLY FLOWERING 4, subcellular localization, ORESARA 1, Arabidopsis
Introduction
Plants undergo important developmental events, such as flowering and senescence, during their life cycle (Huijser and Schmid, 2011). Leaves are the main photosynthetic organs of a plant. During the growth period, photosynthetic products are stored in young leaves; however, at the senescence stage, these products are transferred to newly developing leaves or seeds (Woo et al., 2019). Thus, proper timing of developmental events is critical for maximizing plant fitness, and this timing is determined by plants through sensing the surrounding environmental conditions.
The endogenous biological clock, also known as the circadian clock, regulates almost all developmental processes in plants, ranging from germination to plant maturation, by sensing the environmental conditions. For example, the circadian clock regulates flowering by sensing the photoperiod (de Montaigu et al., 2010). One of the circadian clock components, GIGANTEA (GI), regulates photoperiod-dependent flowering in Arabidopsis thaliana. GI directly binds to the promoter of CONSTANS (CO), a key flowering regulator, and activates CO expression. However, GI promotes CO expression, enabling it to reach peak expression levels, only under long-day (LD) photoperiod, not under short-day (SD) photoperiod. This coincidence between the environmental condition (LDs) and internal rhythm (circadian rhythm) promotes flowering in Arabidopsis, and is known as the external coincidence model (Sawa et al., 2008). Interestingly, the control of GI on flowering time depends on the location of GI in the cell. The GI protein localizes to both the nucleus and the cytoplasm; however, cytosolic GI does not affect flowering time. GI must be transferred to the nucleus for activating CO expression (Kim et al., 2013a). In the nucleus, another clock component, EARLY FLOWERING 4 (ELF4), physically interacts with and sequesters the GI protein, which inhibits GI from binding to the CO promoter (Kim et al., 2013b). Thus, the subcellular localization of GI is important for regulating flowering time in Arabidopsis.
Recently, several studies showed that circadian clock components control leaf senescence in Arabidopsis. Leaf senescence is a critical developmental process involving the transition from nutrient accumulation to nutrient transfer to newly developing organs. ORESARA 1 (ORE1), a key positive regulator of senescence, is under the control of the circadian clock and is regulated by several clock components, such as Circadian Clock Associated 1 (CCA1; Song et al., 2018), Pseudo Response Regulator 9 (PRR9; Kim et al., 2018), and the evening complex [EC; composed of ELF3, ELF4, and LUX ARRYTHMO (LUX); Zhang et al., 2018]. GI, a key component of the circadian clock that is important for flowering, is also involved in the response to oxidative stress. gi mutants exhibit tolerance to hydrogen peroxide and extended plant longevity (Kurepa et al., 1998). These studies suggest that the perception of environmental conditions by the circadian clock is important for regulating the timing of leaf senescence. Previously, we reported that almost all clock mutants of Arabidopsis exhibit an early or a delayed senescence phenotype. Additionally, clock mutants show a strong correlation between flowering and leaf senescence phenotypes (Kim et al., 2018).
Here, we show that GI controls leaf senescence, similar to flowering time, in a location-dependent manner in Arabidopsis. Unlike the cytosol-localized GI, the nuclear-localized GI induces leaf senescence, suggesting that plants utilize the same regulatory mechanism for regulating two major developmental events: flowering and senescence. In the nucleus, ELF4 directly interacts with GI and inhibits its binding to the ORE1 promoter. Taken together, these results suggest that GI acts as a mediator to couple flowering with leaf senescence for enhancing plant fitness and productivity.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild-type in all experiments. Arabidopsis thaliana mutants, gi-2 (Park et al., 1999), gi-ko (Martin-Tryon et al., 2007), elf4-209 (Kolmos et al., 2009), and gi-2 elf4-209 (Kim et al., 2012), and overexpression lines, CsV:GI-GFP, elf4 CsV:GI-GFP, and VGE:GI-GFP (Kim et al., 2013b), used in this study have been described previously. The CsV:GI-GFP-NLS and CsV:GI-GFP-NES vectors were constructed using the nuclear localization signal (NLS) (Kalderon et al., 1984) from SV4012 and nuclear export signal (NES) (Henderson and Eleftheriou, 2000) from MAPKII in the pPZP221 backbone (Hajdukiewicz et al., 1994). The CsV:GI-GFP-NLS and CsV:GI-GFP-NES constructs were introduced into the Col-0 line, and the gi-2 mutation was then introduced via a genetic cross.
Plants were grown in an environmentally controlled growth room at 22°C under a LD (16 h light/8 h dark) or SD (8 h light/16 h dark) photoperiod and 100 μmol ⋅ m–2 ⋅ s–1 light intensity using fluorescent lamps. All physiological experiments were carried out using the third and fourth rosette leaves of plants. Leaf samples were obtained by cutting the leaves at approximately the middle of the petiole using a sharp scalpel to minimize wounding stress.
Leaf Senescence Assay
Chlorophyll was extracted from individual leaves by heating in 95% ethanol at 85°C for 10 min. The chlorophyll concentration per fresh weight of leaf tissue was calculated as described previously (Lichtenthaler, 1987). The ratio of variable fluorescence to maximum fluorescence (Fv/Fm) was used as a measure of the photochemical efficiency of PSII (Oh et al., 1997), which was measured using an Imaging-PAM chlorophyll fluorometer (Heinz Walz GmbH, Effeltrich, Germany). Six biological replicates were performed.
Gene Expression Analysis
Total RNA was extracted from leaves using WelPrep (Welgene, Daegu, Korea) and then treated with DNase I (Ambion, Austin, TX, United States) to remove any traces of contaminating DNA. Subsequently, 0.75 μg of DNase I-treated total RNA was reverse-transcribed using the ImProm II reverse transcriptase (Promega, Madison, WI, United States). The quantity of transcript in each sample was measured using real-time PCR on a CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States) using universal SYBR Supermix (Bio-rad, Hercules, CA, United States). Fold changes in gene expression were calculated using the comparative threshold (CT) method and then normalized relative to the expression of Actin2 (ACT2; At3g18780). Three biological replicates were performed. Primers used in this study are listed in Supplementary Table 1.
Immunoblot Analysis
Immunoblot analyses of GI, HSP90, and H3 were performed as previously reported (Kim et al., 2003, 2007). Polyclonal anti-HSP90 antibodies were generated in rabbits and were diluted 1:5,000 for detection. Nuclear or cytosolic fractions were generated with/without the NP40 detergent in protein extraction buffer or using the CelLytic PN-Plant Nuclei Isolation Kit (Sigma-Aldrich).
Flowering Time Measurement
To measure flowering time, seeds were sown on soil following stratification (4°C for 2 days). Plants were grown under LD condition (16L/8D). Flowering time was measured by counting the number of rosette leaves when the first flower opened.
Chromatin Immunoprecipitation and Quantitative PCR (ChIP-qPCR)
Approximately 2 g of the third and fourth leaf tissue harvested from 5 weeks old plants was fixed in 1% formaldehyde and crosslinked under vacuum for 15 min. A final concentration of 0.25 M glycine was used to quench the formaldehyde for 5 min under vacuum. After washing twice with cold deionized water, the tissue was ground in liquid N2, and chromatin was extracted as described previously (Zhu et al., 2013). ChIP assays were performed using the ChIP Kit-Plants (ab117137, Abcam). Nuclei extraction and chromatin disruption were performed using a sonicator with 10 cycles on ice (10 s each at amplitude 25%/50 s cooling). Chromatin was immunoprecipitated using anti-GFP antibody (ab290, Abcam)-bound assay plate for 90 min. The resulting immunoprecipitated DNA was subjected to qPCR to examine the enrichment of target genes using primers listed in Supplementary Table 1. Three biological replicates were performed.
Chemical Induction of GI
To perform methoxyfenozide (MOF) treatments, inducible GI-GFP (VGE:GI-GFP) plants were grown in soil under LD conditions. After 3 weeks, VGE:GI-GFP plants were soaked in dimethyl sulfoxide (DMSO; mock control) or 50 μM MOF for 6 h. After 2 weeks, samples were collected for analyzing leaf senescence. Six biological replicates were performed.
Results
Leaf Senescence Is Altered by Day Length
In Arabidopsis, a facultative LD plant, flowering is induced by LDs (Corbesier et al., 1996). The circadian clock regulates various developmental processes including seedling growth and flowering time, depending on the photoperiod (de Montaigu et al., 2010). In Aspen tree, SD photoperiod acts as the main trigger of autumn senescence (Fracheboud et al., 2009). Therefore, we first examined whether leaf senescence in Arabidopsis is also photoperiod dependent, similar to flowering. We used the third and fourth rosette leaves for all experiments because both leaves represent a different age status (Zentgraf et al., 2004). Three-weeks-old wild-type plants grown under the LD condition were transferred to either LD or SD photoperiod to avoid any developmental issues, and leaf yellowing was used as a visual indicator of leaf senescence. Leaves of wild-type plants started yellowing at 28 days after emergence (DAE) under the LD photoperiod but did not start yellowing until 40 DAE under the SD photoperiod (Supplementary Figure 1A). Other senescence indicators, including Fv/Fm and chlorophyll content, were consistent with the leaf yellowing phenotype (Supplementary Figures 1B,C). These results suggest that leaf senescence is induced by LD photoperiod, similar to flowering, in Arabidopsis.
GI Positively Regulates Leaf Senescence
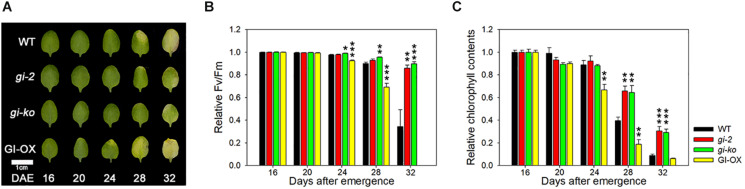
GI is known as a mediator between the circadian clock and photoperiod-dependent flowering (Park et al., 1999). The circadian clock senses the photoperiod and relays it to GI to regulate flowering (Sawa et al., 2007). While gi mutants exhibit late flowering, overexpression of GI rescues the late flowering phenotype of gi mutants (Sawa and Kay, 2011). Previously, gi-2 mutant showed the delayed leaf senescence phenotype in an age-dependent manner (Kim et al., 2018). In this study, we tested the leaf senescence phenotype of another gi mutant, gi-ko (gi-201), and examined the effect of GI overexpression in the gi-2 mutant. Under the LD photoperiod, the gi-ko mutant maintained green leaves, similar to the gi-2 mutant, until 32 DAE compared with the wild type. However, gi-2 mutant plants overexpressing GI showed yellowing of leaf tips at 24 DAE, with greater yellowing at 28 DAE (Figure 1A). The values of Fv/Fm and chlorophyll contents were consistent with the leaf yellowing phenotype (Figures 1B,C). This result indicates that GI positively regulates leaf senescence, similar to flowering.
FIGURE 1.
GI positively regulates leaf senescence in Arabidopsis. (A) Photographs showing yellowing of wild-type (Col-0), gi-2, gi-ko, and CsV:GI-GFP leaves at the indicated days after emergence (DAE). The third and fourth rosette leaves were used in this experiment. Scale bar: 1 cm. (B,C) Photochemical efficiency (Fv/Fm) (B) and chlorophyll content (C) of leaves of the indicated genotypes. Data are presented as mean ± standard error of mean (SEM). Six biological replicates were performed. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.005; Student’s t-test).
To further test the direct influence of GI on leaf senescence, we used MOF-inducible VGE:GI-GFP transgenic plants. After 3 weeks of growth under LD photoperiod, we soaked the transgenic plants in DMSO (control) or 50 μM MOF for 6 h. Leaf yellowing was significantly greater in the leaves of MOF-treated plants than in the leaves of control plants (Supplementary Figure 2A). The Fv/Fm values and chlorophyll contents were consistent with the leaf yellowing phenotype (Supplementary Figures 2B,C). These results suggest that GI directly affects leaf senescence in Arabidopsis.
GI Controls Leaf Senescence in a Location-Dependent Manner
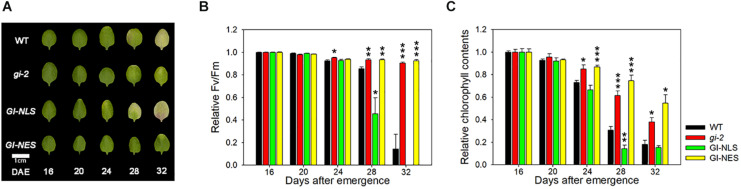
GI differentially controls the circadian clock and flowering, depending on its subcellular localization. While both nuclear and cytosolic GI regulate the circadian rhythm, only nuclear GI promotes flowering by binding to CO or FT promoter (Kim et al., 2013a). In the nucleus, ELF4 directly interacts with GI, thus inhibiting its binding to the target gene promoter (Kim et al., 2013b). Here, we hypothesized that GI controls leaf senescence in a manner similar to how it controls flowering time. To test this hypothesis, we first tested the effect of the location of GI in the cell on leaf senescence. We used transgenic lines expressing CsV:GI-GFP-NLS or CsV:GI-GFP-NES in the gi-2 mutant background, in which the GI-GFP fusion protein is preferentially localized to the nucleus or cytosol, respectively (Supplementary Figure 3A). We then examined alterations of circadian physiology, such as flowering time in these transgenic plants. CsV:GI-GFP complemented the flowering physiological lesions altered by the gi-2 mutation (Supplementary Figure 3B). CsV:GI-GFP-NLS and CsV:GI-GFP-NES differentially complemented the circadian alterations: the late flowering phenotype of gi-2 mutants was complemented by CsV:GI-GFP-NLS but not by CsV:GI-GFP-NES (Supplementary Figure 3B).
In the LD condition, CsV:GI-GFP-NLS plants showed an early leaf yellowing phenotype compared with wild-type plants, whereas CsV:GI-GFP-NES plants showed no leaf yellowing, similar to the gi-2 mutant (Figure 2A). Compared with the wild type, the Fv/Fm value of CsV:GI-GFP-NLS leaves was clearly reduced at 28 DAE, whereas that of CsV:GI-GFP-NES leaves was maintained until 32 DAE, similar to gi-2 mutant leaves (Figure 2B). Additionally, the chlorophyll content of CsV:GI-GFP-NLS leaves rescued the gi-2 mutant phenotype, whereas that of CsV:GI-GFP-NES leaves did not (Figure 2C), indicating that only nuclear-localized GI rescues the delayed senescence phenotype of the gi mutant. Taken together, these results suggest that the subcellular localization of GI determines leaf senescence, similar to flowering.
FIGURE 2.
Subcellular localization of GI determines its effect on leaf senescence. (A) Photographs showing yellowing of wild-type (Col-0), gi-2, CsV:GI-GFP-NLS, and CsV:GI-GFP-NES leaves. The third and fourth rosette leaves were used for this experiment. DAE, days after emergence. Scale bar: 0.5 cm. (B,C) Fv/Fm ratio (B) and chlorophyll content (C) of leaves of the indicated genotypes at the indicated time points. Data are presented as the mean ± SEM. Six biological replicates were performed. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.005; Student’s t-test).
ELF4 Negatively Regulates the Role of GI in Leaf Senescence
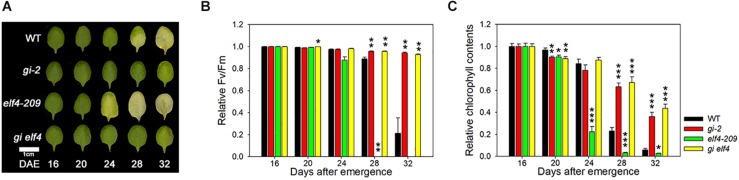
In the nucleus, ELF4 physically interacts with and inhibits the binding of GI to the CO promoter to induce flowering (Kim et al., 2013b). The gi mutant allele is epistatic to elf4 in photoperiod-dependent flowering time regulation (Kim et al., 2012). Thus, we first tested whether gi and elf4 share a similar hierarchy in the leaf senescence pathway. The elf4-209 single mutant showed early senescence, whereas the gi-2 mutant showed delayed senescence (Kim et al., 2018). The timing of leaf yellowing in the gi elf4 double mutant was similar to that of the gi single mutant under LDs (Figure 3A). We further quantitatively evaluated the senescence time of all mutant combinations by measuring Fv/Fm values and chlorophyll contents with leaf aging. The timing of leaf senescence in the gi elf4 double mutant was similar to that in the gi single mutant but not to that in the elf4 single mutant (Figures 3B,C). These results indicate that like the flowering phenotype, the early senescence phenotype of the elf4 single mutant is masked by the gi mutant.
FIGURE 3.
The gi-2 mutant allele is epistatic to elf4-209 in the leaf senescence pathway. (A) Photographs showing yellowing of leaves of wild-type (Col), gi-2, elf4-209, and gi elf4 plants at the indicated time points. DAE, days after emergence. The third and fourth rosette leaves were used for this experiment. Scale bar: 1 cm. (B,C) Fv/Fm ratio (B) and chlorophyll content (C) in leaves of the indicated genotypes at the indicated time points. Data are presented as mean ± SEM. Six biological replicates were performed. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.005; Student’s t-test).
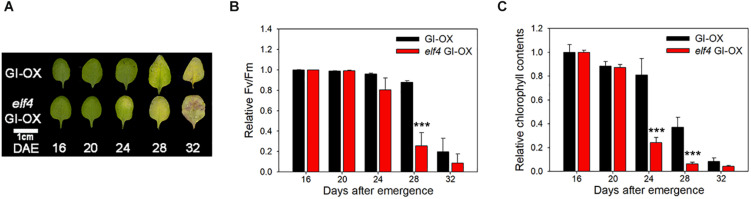
Next, we tested whether ELF4 negatively regulates the effect of GI on leaf senescence, similar to flowering (Kim et al., 2013b). We compared the leaf senescence phenotype of wild-type and elf4 mutant plants overexpressing GI. We found that early senescence of GI-overexpressing plants was more accelerated in the elf4 mutant. Leaf yellowing progressed at a faster rate in GI-OX/elf4 plants than in GI-OX/Col-0 plants (Figure 4A). The Fv/Fm values and chlorophyll contents of GI-OX/elf4 and GI-OX/Col-0 plants were consistent with their leaf yellowing phenotypes (Figures 4B,C). This result indicates that ELF4-free GI controls leaf senescence in a similar manner as it controls flowering.
FIGURE 4.
ELF4 alters the leaf senescence phenotype of GI overexpression (GI-OX) plants. (A) Photographs showing yellowing of CsV:GI-GFP (GI-OX) and elf4 CsV:GI-GFP (elf4 GI-OX) leaves at the indicated time points. DAE, days after emergence. The third and fourth rosette leaves were used for this experiment. Scale bar: 1 cm. (B,C) Fv/Fm ratio (B) and chlorophyll content (C) of leaves of the indicated genotypes at the indicated time points. Data are presented as mean ± SEM. Six biological replicates were performed. Asterisks indicate significant differences (***p < 0.005; Student’s t-test).
GI Positively Regulates Leaf Senescence by Activating ORE1
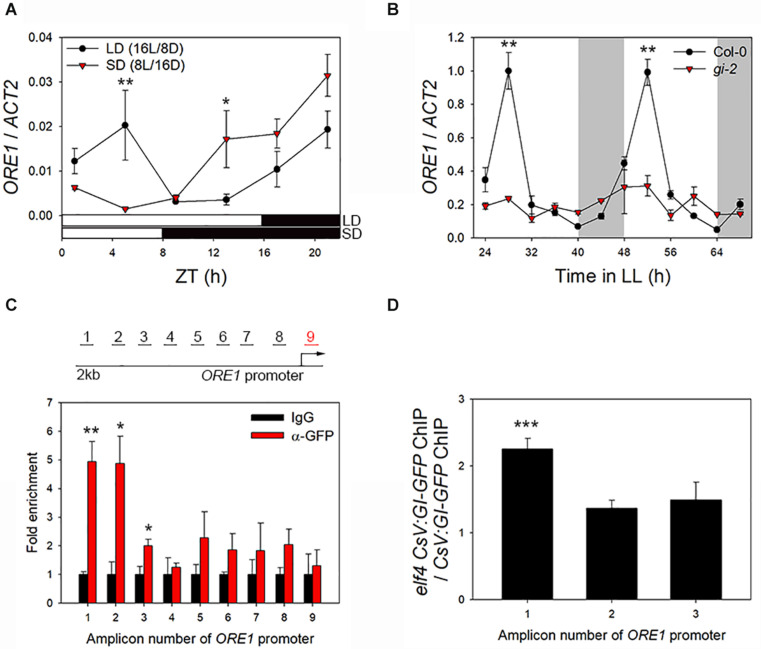
ORE1 promotes senescence in Arabidopsis (Kim et al., 2009). We previously reported that ORE1 is under the control of the circadian clock, and the clock component, PRR9, binds to the ORE1 promoter to induce leaf senescence (Kim et al., 2018). Additionally, we suggested that GI acts upstream of ORE1 (Kim et al., 2018). Here, we hypothesized that GI controls leaf senescence by inducing ORE1 expression. To test this hypothesis, we first examined the effect of the photoperiod on ORE1 expression. To avoid the difference in plant growth between SD and LD conditions, we transferred wild-type plants to either SD or LD photoperiod after 3 weeks of growth under LDs. ORE1 expression showed two peaks under the LD condition, morning peak (ZT4) and evening peak (ZT20) (Figure 5A). However, the morning peak of ORE1 was absent under the SD condition, despite higher expression level of ORE1 during the dark phase under the SD photoperiod than under the LD photoperiod (Figure 5A). We previously showed that in plants entrained under the LD condition and then moved to continuous light, ORE1 exhibits one peak in the morning, which is controlled by the circadian clock (Kim et al., 2018). These results suggest that the morning peak of ORE1 is important for the circadian clock-mediated regulation of leaf senescence. Thus, we tested expression of ORE1 in wild-type and gi-2 mutant plants under continuous light. The expression of ORE1 was significantly reduced in the gi-2 mutant at 28 h and 52 h in LL compared with the wild-type (Figure 5B). To further confirm the effect of GI on the expression of ORE1, we used VGE:GI-GFP transgenic plants. Ten-days-old VGE:GI-GFP seedlings grown in 1/2 B5 media were transferred to DMSO (control) or MOF-containing media at ZT10 (GI peak time). After 24 h, MOF treatment showed ∼2-fold increase in ORE1 transcript level compared with the control (Supplementary Figure 4). These results suggest that GI acts as a direct transcriptional activator of ORE1.
FIGURE 5.
GI binds to ORE1 promoter and activates its expression. (A) Diurnal expression of ORE1 under long-day (LD; 16 h light/8 h dark) and short-day (SD; 8 h light/16 h dark) photoperiod. (B) Abundance of ORE1 mRNA in wild-type and gi-2 mutant leaves under continuous light (LL). White bars indicate subjective day, and gray shading indicates subjective night. (C) ORE1 promoter-binding affinity of GI in CsV:GI-GFP transgenic plants. Schematic representation of amplicons 1–9 of the ORE1 promoter amplified in the chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assay are indicated. Amplicon 9 was used as an internal control. Leaves were harvested at 12 h after light on. Fold enrichment indicates the ratio of chromatin immunoprecipitated using anti-GFP antibody to that immunoprecipitated using anti-IgG antibody. Asterisks indicate statistically significant difference compared with the IgG control. (D) ORE1 promoter-binding activity of GI in the elf4 mutant relative to that in the wild type at amplicons 1, 2, and 3. Three biological replicates were performed. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.005; Student’s t-test).
In yeast (Saccharomyces cerevisiae), GI binds to the ORE1 promoter (Kim et al., 2018). Thus, we tested the direct binding of GI to the ORE1 promoter in Arabidopsis leaves by ChIP-qPCR. Leaves of GI-OX plants at ZT12 (GI protein peak time) were used for ChIP, and the binding of GI to eight different test regions (amplicons 1–8) or to an internal control region (amplicon 9) of the ORE1 promoter (∼2 kb upstream of the transcription start site) was tested by qPCR to screen the GI binding region on ORE1 promoter (Figure 5C, upper diagram). GI showed significantly greater binding affinity toward amplicons 1, 2, and 3 of the ORE1 promoter than the negative control (anti-IgG antibody) (Figure 5C). Enrichment of the internal control region (amplicon 9) showed no significant difference between the anti-IgG antibody (control) and anti-GFP antibody samples. These results suggest that GI regulates leaf senescence by directly binding to the ORE1 promoter and activating its expression. Previously, we showed that ELF4 inhibits the binding of GI to the CO promoter (Kim et al., 2013b). Therefore, we tested whether ELF4 affects the activity of GI as a transcriptional activator of ORE1 by conducting ChIP assays using CsV:GI-GFP and elf4 CsV:GI-GFP leaves. GI-GFP was enriched > twofold in the elf4 mutant background compared with the wild type at amplicon 1, but no significant difference was detected at the other amplicons (Figure 5D). These results suggest that nuclear-localized ELF4 and GI proteins together regulate leaf senescence. Thus, the regulation of leaf senescence in Arabidopsis is similar to that of flowering.
Discussion
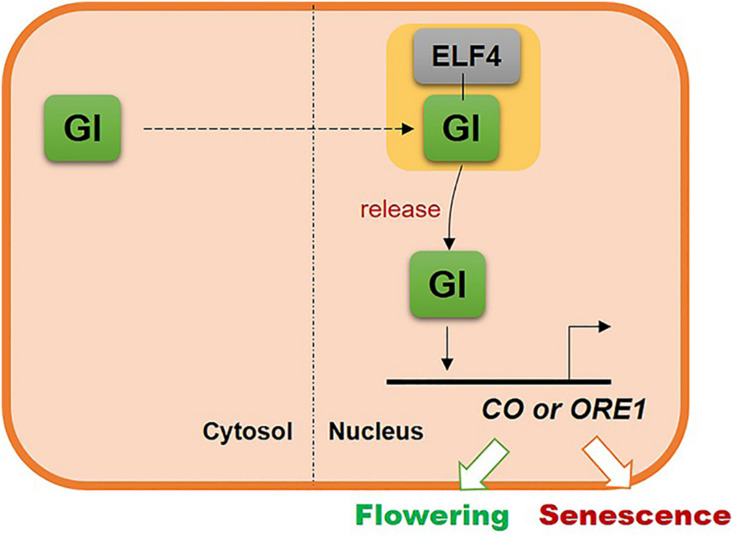
The results of this study suggest that GI, one of the clock components, controls leaf senescence in Arabidopsis via the same regulatory mechanism as that used to control flowering. The location of GI in the cell is critical for regulating the timing of leaf senescence, similar to flowering. In the nucleus, GI is sequestered by ELF4 and then released at the proper developmental stage to induce CO or ORE1 expression and consequently flowering or senescence, respectively. These results suggest that plants use a common regulatory mechanism to couple flowering with senescence (Figure 6).
FIGURE 6.
Schematic illustrating the common molecular mechanism underlying the regulation of flowering and leaf senescence by GI.
Depending on the subcellular localization, a single protein can regulate several signaling pathways and developmental processes such as flowering and senescence. These two developmental stages have a common purpose in the plant life cycle; flowering switches the transition from the vegetative growth phase to the reproductive phase, while senescence remobilizes the nutrients to newly developing leaves or seeds. This implies that flowering and senescence are closely connected and need similar regulatory process to maximize fitness. GI, a key clock regulator, connects both these developmental processes successfully.
GI is involved in multiple processes including redox signaling in Arabidopsis (Mishra and Panigrahi, 2015). Deficiency of GI results in increased tolerance to redox signals. Chlorophyll accumulation is higher in gi mutants than in the wild-type under oxidative stress conditions (Kurepa et al., 1998). However, the molecular mechanism that links the cellular localization of GI with redox signaling for plant developments including flowering and senescence is unknown. Further research is required to elucidate this molecular mechanism.
In this paper, we suggest that GI protein directly binds to ORE1 promoter to induce ORE1 expression. Additional explanation is required for this process since there is a significant temporal distance between the expression timing of these two molecules. The expression of GI protein peaks around ZT12, while ORE1 gene expression peaks around ZT4. We searched for a similar case in Arabidopsis circadian clock, and found that RVE8, which peaks in the morning, is known to directly activate evening genes, including PRR5 and TOC1 (Rawat et al., 2011). However, we cannot provide any results in this paper to suggest an underlying mechanism for the temporal gap between GI abundance and ORE1 expression. It would be an interesting topic for further study.
ORE1, a key senescence regulator, is controlled by the circadian clock. The role of ORE1 in senescence is similar to that of CO in flowering. Like CO, ORE1 shows two peaks under the LD photoperiod, and its peak during the subjective day disappears under the SD photoperiod. This indicates that day-peaks of ORE1 and CO expression are equally important for inducing senescence and flowering, respectively. These results suggest that GI regulates flowering and senescence in Arabidopsis using different partner but via the same mechanism. It thus appears that GI was co-opted during evolution to coordinate the reproductive phase and senescence process, which provides important insight into the concerted evolution of reproduction and senescence.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Author Contributions
HK and HGN designed all the experiments, prepared the figures, and wrote the manuscript. HK, SJP, and YK performed all the experiments. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank K. H. Suh and H. H. Jo for technical assistance.
Footnotes
Funding. This work was supported by the Institute for Basic Science (IBS-R013-D1).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.589707/full#supplementary-material
References
- Corbesier L., Gadisseur I., Silvestre G., Jacqmard A., Bernier G. (1996). Design in Arabidopsis thaliana of a synchronous system of floral induction by one long day. Plant J. 9 947–952. 10.1046/j.1365-313x.1996.9060947.x [DOI] [PubMed] [Google Scholar]
- de Montaigu A., Toth R., Coupland G. (2010). Plant development goes like clockwork. Trends Genet. 26 296–306. 10.1016/j.tig.2010.04.003 [DOI] [PubMed] [Google Scholar]
- Fracheboud Y., Luquez V., Bjorken L., Sjodin A., Tuominen H., Jansson S. (2009). The control of autumn senescence in European aspen. Plant Physiol. 149 1982–1991. 10.1104/pp.108.133249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. 10.1007/BF00014672 [DOI] [PubMed] [Google Scholar]
- Henderson B. R., Eleftheriou A. (2000). A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256 213–224. 10.1006/excr.2000.4825 [DOI] [PubMed] [Google Scholar]
- Huijser P., Schmid M. (2011). The control of developmental phase transitions in plants. Development 138 4117–4129. 10.1242/dev.063511 [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. (1984). A short amino acid sequence able to specify nuclear location. Cell 39(3 Pt 2), 499–509. 10.1016/0092-8674(84)90457-4 [DOI] [PubMed] [Google Scholar]
- Kim H., Kim H. J., Vu Q. T., Jung S., McClung C. R., Hong S., et al. (2018). Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115 8448–8453. 10.1073/pnas.1722407115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Woo H. R., Kim J., Lim P. O., Lee I. C., Choi S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323 1053–1057. 10.1126/science.1166386 [DOI] [PubMed] [Google Scholar]
- Kim W. Y., Fujiwara S., Suh S. S., Kim J., Kim Y., Han L., et al. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449 356–360. 10.1038/nature06132 [DOI] [PubMed] [Google Scholar]
- Kim W. Y., Geng R., Somers D. E. (2003). Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. U.S.A. 100 4933–4938. 10.1073/pnas.0736949100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Han S., Yeom M., Kim H., Lim J., Cha J. Y., et al. (2013a). Balanced nucleocytosolic partitioning defines a spatial network to coordinate circadian physiology in plants. Dev. Cell 26 73–85. 10.1016/j.devcel.2013.06.006 [DOI] [PubMed] [Google Scholar]
- Kim Y., Lim J., Yeom M., Kim H., Kim J., Wang L., et al. (2013b). ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Rep. 3 671–677. 10.1016/j.celrep.2013.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Yeom M., Kim H., Lim J., Koo H. J., Hwang D., et al. (2012). GIGANTEA and EARLY FLOWERING 4 in Arabidopsis exhibit differential phase-specific genetic influences over a diurnal cycle. Mol. Plant 5 678–687. 10.1093/mp/sss005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E., Nowak M., Werner M., Fischer K., Schwarz G., Mathews S., et al. (2009). Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 3 350–366. 10.2976/1.3218766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J., Smalle J., Van Montagu M., Inze D. (1998). Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J. 14 759–764. 10.1046/j.1365-313x.1998.00168.x [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H. K. (1987). Chlorophylls and Carotenoids - Pigments of Photosynthetic Biomembranes. Methods Enzymol. 148 350–382. 10.1016/0076-6879(87)48036-1 [DOI] [Google Scholar]
- Martin-Tryon E. L., Kreps J. A., Harmer S. L. (2007). GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 143 473–486. 10.1104/pp.106.088757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., Panigrahi K. C. (2015). GIGANTEA - an emerging story. Front. Plant Sci. 6:8. 10.3389/fpls.2015.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. A., Park J. H., Lee G. I., Paek K. H., Park S. K., Nam H. G. (1997). Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 12 527–535. 10.1046/j.1365-313x.1997.00489.x [DOI] [PubMed] [Google Scholar]
- Park D. H., Somers D. E., Kim Y. S., Choy Y. H., Lim H. K., Soh M. S., et al. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582. 10.1126/science.285.5433.1579 [DOI] [PubMed] [Google Scholar]
- Rawat R., Takahashi N., Hsu P. Y., Jones M. A., Schwartz J., Salemi M. R., et al. (2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7:e1001350. 10.1371/journal.pgen.1001350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Kay S. A. (2011). GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108 11698–11703. 10.1073/pnas.1106771108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Kay S. A., Imaizumi T. (2008). Photoperiodic flowering occurs under internal and external coincidence. Plant Signal. Behav. 3 269–271. 10.4161/psb.3.4.5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Nusinow D. A., Kay S. A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318 261–265. 10.1126/science.1146994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Jiang Y., Kuai B., Li L. (2018). CIRCADIAN CLOCK-ASSOCIATED 1 Inhibits Leaf Senescence in Arabidopsis. Front. Plant Sci. 9:280. 10.3389/fpls.2018.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H. R., Kim H. J., Lim P. O., Nam H. G. (2019). Leaf Senescence: systems and dynamics aspects. Annu. Rev. Plant Biol. 70 347–376. 10.1146/annurev-arplant-050718-095859 [DOI] [PubMed] [Google Scholar]
- Zentgraf U., Jobst J., Kolb D., Rentsch D. (2004). Senescence-related gene expression profiles of rosette leaves of Arabidopsis thaliana: leaf age versus plant age. Plant Biol. 6 178–183. 10.1055/s-2004-815735 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Wei H., Li N., Tian W., Chong K., et al. (2018). Circadian Evening Complex Represses Jasmonate-Induced Leaf Senescence in Arabidopsis. Mol. Plant 11 326–337. 10.1016/j.molp.2017.12.017 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Dong A., Shen W. H. (2013). Histone variants and chromatin assembly in plant abiotic stress responses. Biochim. Biophys. Acta 1819 343–348. 10.1016/j.bbagrm.2011.07.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.