Abstract
Accumulating evidence suggests that human genome can fold into non-B DNA structures, when appropriate sequence and favourable conditions are present. Among these, G-quadruplexes (G4-DNA) are associated with gene regulation, chromosome fragility and telomere maintenance. Although several techniques are used in detecting such structures in vitro, understanding their intracellular existence has been challenging. Recently, an antibody, BG4, was described to study G4 structures within cells. Here, we characterize BG4 for its affinity towards G4-DNA, using several biochemical and biophysical tools. BG4 bound to G-rich DNA derived from multiple genes that form G-quadruplexes, unlike complementary C-rich or random sequences. BLI studies revealed robust binding affinity (Kd = 17.4 nM). Gel shift assays show BG4 binds to inter- and intramolecular G4-DNA, when it is in parallel orientation. Mere presence of G4-motif in duplex DNA is insufficient for antibody recognition. Importantly, BG4 can bind to G4-DNA within telomere sequence in a supercoiled plasmid. Finally, we show that BG4 binds to form efficient foci in four cell lines, irrespective of their lineage, demonstrating presence of G4-DNA in genome. Importantly, number of BG4 foci within the cells can be modulated, upon knockdown of G4-resolvase, WRN. Thus, we establish specificity of BG4 towards G4-DNA and discuss its potential applications.
Keywords: non-B DNA, G4-DNA, G-quartets, fragile region, genomic instability
1. Introduction
Since its discovery, deoxyribo nucleic acid (DNA) was popularly thought to be in canonical B-form within cells and has been attributed to influence majority of functions of DNA such as replication, transcription and repair.1In silico analyses of eukaryotic genome have revealed the existence of non-B DNA motifs including cruciform, triplexes and G-quadruplexes.2,3Genome-wide studies revealed that G-quadruplex motifs are conserved across different eukaryotic organisms and are enriched in promoters of genes.4–6 Telomeric DNA repeats were also shown to adopt triplex and G-quadruplex structures.7–9 Z-DNA motifs have been abundantly observed in 5′ ends of human genes.10 Some eukaryotic intergenic regions and 3′ regions of genes have been reported to harbour cruciform-forming sequences.11 Such non-B DNA structures have been suggested to impart chromosomal fragility, leading to translocations amongst other outcomes, and eventually predisposing a cell to an oncogenic state.12–20
G-quadruplexes (G4) are helical structures of stacked G-quartets, resulting in four stranded DNA structures. Guanines in the G-quartets are connected through Hoogsteen hydrogen bonding in a square planar arrangement, with extruded loops of 1–7 nucleotides.21 These loops in turn determine the parallel, antiparallel or mixed orientation. Various cations, particularly K+, influence the topology and stability of G-quadruplexes.22 Quadruplexes can be formed by one (intramolecular G4), two or four separate strands (bi-/tri-/tetra-molecular G4) of DNA (or RNA).21–23. Most G-quadruplexes follow an empirical formula G≥3X G≥3X G≥3X G≥3, where X is loop length, ranging from one to seven.24 Recent studies suggest that even the interrupted G stretches, and loops longer than seven nucleotides can be accommodated in G-quadruplex structures, if they fold into duplex hairpins.25,26 Almost all bimolecular (dimeric) G4 are formed by association of two identical sequences, whereas tetramolecular G4 may be formed by four G-rich strands associating together.24
Over 375,000 putative classical G4 motifs have been reported to exist in human genome through computational analyses.3,27 In genomic context, retention of G4 motifs is observed in several functional regions and is conserved across species.28 Recent studies have demonstrated many non-conformities in G-quadruplex formation21,29–31 by virtue of GNG motifs within guanine stretches increasing the number of potential G4 motifs in vivo. G-quadruplex formation could be promoted by superhelical stress, molecular crowding32 or by specific G-quadruplex binding proteins.33
Biological relevance of G4 structures is an active area of research. Maximum number of G4 structures are expected to be at the telomeric regions, given the presence of 5 to 10,000 bp of tandemly arranged T2AG3 repeats, wherein G4 structures have been suggested to play a critical role in telomere stability.34,35 G4 motifs are abundantly present in gene promoters, borders between introns and exons, and in several human DNA replication origins, indicating their potential role in gene regulation.36
G-quadruplex DNA structures have been extensively studied in vitro using various biophysical techniques like nuclear magnetic resonance (NMR) spectroscopy, circular dichroism (CD) spectroscopy, X-ray crystallography25,26,37 and small molecule probes.38,39 However, in vivo existence and function of G4 structures have been controversial. In an important observation, high-affinity single-chain antibodies, Sty3 (for parallel G4) and Sty49 (for antiparallel and parallel G4), were generated by ribosome display, to visualize telomeric G4 of Stylonychia lemnae macronuclei.40 These were followed up by identification of several G4 structure-specific antibodies such as HF1, BG4, hf2, and 1H6 against G4 for imaging in cells and tissues.3,41–43 Among these, BG4, a monoclonal single chain antibody generated by phage display with high affinity and specificity for G43 is being used by several groups to investigate presence of G4 structures in the genome.19,29,44,45 BG4 has been raised for a broader range of G4 structures with an impressive Kd (∼2.0 nM).3 Immunostaining with BG4 in human cells showed the presence of G-quadruplex structures in all cell cycle phases, with maximum foci in S phase, which corroborates the replication-dependent formation of G4.3 Treatment of live cells with G4-intercalator pyridostatin (PDS), before immunostaining, increased the number of BG4 foci. The same group reported an impressive study where G4 ChIP-seq with BG4 led to detection of over 10,000 G4 in regulatory, nucleosome-depleted regions of human chromatin.46 Recently, a real-time detection of G4 in live cells using G4-specific fluorescent probe (SiR-PyPDS) was also shown.47 Collectively, such studies have provided insights into G4 formation in the nuclear context.
Although ELISA has been used to study BG4 specificity towards G-quadruplexes previously, several aspects of its specificity are less understood. In the present study, we establish that BG4 specifically binds to both inter- and intramolecular G4 DNA, particularly when it is in parallel orientation. Double-stranded DNA did not show any BG4 binding despite the presence of G4 motif, confirming structural specificity. Also consistent with robust binding observed for G4 DNA when present in oligomeric DNA (Kd = 17.4 nM), BG4 exhibited specific binding to G4 structure, when formed in a plasmid or in genome.
2. Materials and methods
2.1 Enzymes, chemicals and reagents
Chemical reagents were obtained from Sigma Chemical Co. (USA), Amresco (USA) and SRL (India). DNA modifying enzymes were from New England Biolabs (USA) and Fermentas (USA). Radioisotope-labeled nucleotides were from BRIT (India) and American Radiolabelled Chemicals (ARC) (USA). FBS and PenStrep were from Gibco BRL (USA).
2.2 Plasmids
Plasmid expressing BG4, pSANG10-3F-BG4 was a gift from Shankar Balasubramanian (Addgene plasmid # 55756). Plasmids harbouring the human telomere repeat sequences were from Addgene. pSXneo 135(T2AG3) (Addgene plasmid # 12402) with a 0.8-kb TTAGGG repeat sequence and pSXneo 270(T2AG3) (Addgene plasmid # 12403), with a 1.6-kb repeat were gifts from Titia de Lange (USA).54 To generate mutant plasmid lacking the telomere sequence with same plasmid backbone, pSXneo270 was double digested using BglII and XbaI and used for cloning a 1.19-kb fragment containing random sequence to generate pSV4. shRNA against WRN was used in the study and was purchased from shRNA Resource Center at Division of Biological Science, IISc, Bangalore (funded by DBT: BT/PR4982/AGR/36/718/2012), Indian Institute of Science (India).
2.3 Cell culture
HeLa and HEK 293 T (human kidney) cell lines were purchased from National Centre for Cell Science, Pune, India. Nalm6 and REH (B‐cell leukemia) were from Dr. M.R. Lieber (USA). REH and Nalm6 were cultured in RPMI1640 (Lonza) containing 10% FBS (Gibco BRL, USA), 100 µg/ml penicillin and 100 µg/ml streptomycin (Sigma-Aldrich, USA). HEK 293 T cells were cultured in DMEM medium (Gibco) with l‐glutamine, supplemented, while HeLa in MEM with 10% FBS and PenStrep. Cells were grown at 37°C in a humidified atmosphere containing 5% CO2.
2.4 Antibodies
Anti-AID (SC14680), anti-GFP (SC9996), anti-PAX5 (SC55515), anti-MBP (SC13912), anti-GAPDH (SC137179) and IgG (SC2027) were used for EMSA studies.
2.5 Preparation of BG4
BG4 was purified as described earlier.29 BL21(DE3) E. coli cells were transformed with pSANG10-3F-BG4 at 37°C using LB medium (30 μg/ml kanamycin). BG4 expression was induced at OD600 (0.4) in presence of IPTG (0.5 mM) and by incubation at 16°C (180 rpm, overnight). Cells were spun down at 4°C (7000 rpm for 10 min), resuspended in lysis buffer [20 mM Tris-Cl (pH 8.0), 400 mM NaCl, 5% glycerol, 1 mM PMSF, 2 mM MgCl2 and 1 mM β-mercaptoethanol] and incubated on ice for 10 min. Cells were sonicated, supernatant was collected by centrifugation at 4°C (14,000 rpm for 20 min) and loaded onto Ni-NTA column (Novagen). Column was washed in 10 mM imidazole, followed by 20 mM imidazole in phosphate-buffered saline (PBS) (pH 8.0). Purified BG4 antibody was eluted using an imidazole gradient (100–1,000 mM imidazole). Fractions of interest were dialysed against dialysis buffer [25 mM Tris (pH 8.0), 100 mM NaCl, 0.1 mM EDTA, 2 mM DTT, 10% glycerol] after pooling the pure ones. Aliquots of BG4 were stored at 4°C for immediate use and the stock fractions at −20°C.
2.6 Oligomeric DNA
All oligomers were gel purified as described earlier.66 The oligomers used in the present study are listed below. RT17, 5′-GCGCGGGGAGGGGAGAGGGGGCGGGAGCGGC-3′; MN89, 5′-GCCGCTCCCGCCCCCTCTCCCCTCCCCGCGC-3′; BTM6, 5′-GAAGGACCA-CAGGATATCTCTATATAAATCTGT-3′; KD50, 5′CTCCGCCCCGCCGGGACCCCGCCCCCGGCCCG 3′; KD49, 5′ CGGGCCGGGGGCGGGGTCCCGGCGGGGCGGAG 3′; KD51, 5′CGGGCCGGGGGCGGGGTCCCGGCGTTTCGGAG 3′; KD53, 5′-GCTCCGCCCCGCGCCCCCGGAAACCTCC-3′; KD52, 5′-GGAGGTTTCCGGGGGCGCGGGGCGGAGC-3′; KD16, 5′-ACTGGGGCTGGGGTGGGGGGTAATCCAGAACTGGATCGGGGT-3′; KD17, 5′-ACCCCG-ATCCAGTTCTGGATTACCCCCCACCCCAGCCCCAGT-3′; KD80, 5′GCCCCGGCCCCTAGCGCGCGACTCCT 3′; KD81, 5′ AGGAGTCGCGCGCTAGGGGCCGGGGC 3′; KD83, 5′AGGCCGGAGGGTGGGCGGCGCGCGG 3′; KD85, 5′ CGGGGCGGGGCTGCGGTTGCGGT 3′; SMJ36, 5′-GTCCCAATTC-CCATATTGAAATCCCCAATCCCAA-3′; SMJ37, 5′-CAGGGTTAAGGGTATAACTTTA-GGGGTTAGGGTT-3′; SMJ50, 5′TCGGGTTGCGGGCGCAGGGCACGGGCG 3′; SMJ51, 5′GTGGGGTTAGGGGTTAGGGGTTAGGGG 3′; SMJ52, 5′TTGGGGCCGGGGCCGGGGCCGGGGTT 3′; SMJ53, 5′ GTAGGGTTAGGGTTAGGGTTAGGG 3′; SMJ56, 5′ TGGGTTTGGGTTTGGGTTTGGGT 3′; SMJ58, 5′GGGGAGTAAAAGGGAGCGGGGTGCTGGG 3′; SMJ59, 5′GGGCCTTGTGGGCCTTGTGGGCCTTGTGGG 3′; SMJ61, 5′ GGGGTTTTGGGGTTTTGGGGTTTTGGGG 3′; DG1, 5′-GATCCCTCTAGACCGGTACTACTCGAGCTTGGTTGCAGATTAAGAGAGGCTCCTG-3′; SV23, 5′ GGGTTAGGGTTAGGGTTAGGG 3′; SV24, 5′ CCCTAACCCTAACCCTAACCC 3′; SV25, 5′ GGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGG 3′; SV26, 5′ CCCTAACCCTAACCCTAACCCTAACCCTAACCC 3′; SV27, 5′ GGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGG 3′; SV28, 5′ CCCTAACCCTAACCCTAACCCTAACCCTAACCCTAACCCTAACCC 3′; AKN11, 5′ GACCTGAGGGCGAGCTTTTTTCGAGTAACTTAACAG 3′; AKN18, 5′ CTGTTAAGTTACTCGAAAAAAGCTCGCCCTCAGGTC 3′.
2.7 5′ End labelling of oligomers
5′ end labelling of oligomeric DNA was done using T4 polynucleotide kinase and [γ-32P] ATP as described earlier.67 The labelled substrates were purified using Sephadex G25 (Sigma, USA) columns and stored at −20°C until further use.
2.8 Preparation of G4 DNA and other DNA substrates
For preparation of single-stranded substrates containing G-quadruplex DNA, radiolabelled oligomer (20 nM) was heat denatured in Tris-EDTA (TE) buffer (pH 8.0) containing 100 mM potassium chloride (KCl) at 95°C for 10 min and allowed to cool down to room temperature overnight. In order to prepare double-stranded DNA, radiolabelled oligomeric DNA (20 nM) was annealed with unlabelled complementary strand (5-fold excess) in 100 mM NaCl and 1 mM EDTA in boiling water for 10 min followed by gradual cooling as described earlier.68 For preparation of single-stranded substrates containing i-motifs, radiolabelled oligomer (20 nM) was heat denatured in 100 mM potassium acetate (pH 5.2) at 95°C for 10 min and allowed to cool down to room temperature for 2 h. These substrates were then utilized for gel mobility shift assays.
2.9 Gel mobility shift assay
Radiolabelled oligomers (20 nM) forming G-quadruplexes were incubated either in the presence or absence of 100 mM potassium chloride in Tris–EDTA (TE) buffer (pH 8.0) at 37°C for 1 h. In case of i-motifs, radiolabelled substrates (20 nM) were annealed either in the presence or absence of 100 mM potassium acetate (pH 5.2) at 95°C for 1 h. Reactions were incubated with BG4 or antibodies at 4°C for 1 h and the products were resolved on 6% native polyacrylamide gels containing 25 mM KCl at 100 V, at 4°C. The gels were dried, exposed to a screen, and the signal was detected using PhosphorImager Typhoon (GE, USA) as described previously.29,69
For competition assays, radiolabelled oligomers (20 nM) were incubated in presence of 100 mM KCl in Tris–EDTA (TE) buffer (pH 8.0) at 37°C for 1 h, followed by incubation with BG4 (1.28 µg) at 4°C for 1 h. Increasing concentrations (0, 05, 10, 20, 40, 80, 100, 150, 200 nM) of unlabelled (cold) oligomeric DNA substrates (RT17 or DG1) were incubated simultaneously at 4°C for 1 h. Reaction products were resolved on 6% native polyacrylamide gels containing 25 mM KCl (100 V, at 4°C). The gels were dried, exposed to a screen and the signal was detected using PhosphorImager Typhoon (GE, USA).
For quantification of BG4-bound complexes on native PAGE, area corresponding to the band of interest was selected from each lane. An identical box was selected in an area with no band from respective lane of the gel and was subtracted. Intensity measured in each lane was indicated as photostimulated luminescence (PSL) units (PSLU) and plotted using GraphPad Prism (ver 5.01, San Diego, CA, USA). For statistical analyses, intensities of positive control and competitor oligomer added samples have been compared by unpaired Student’s t-test. Values were considered statistically significant, if the P ≤ 0.05.
2.10 DMS protection assay
Radiolabelled oligomers (RT17, KD49, KD83) were incubated in TE in presence of 100 mM KCl (37°C for 1 h), followed by incubation with BG4 (1.5 µg) at 4°C for 1 h. Dimethyl sulphate (DMS) was added to the reaction (1/200 dilution) and incubated for 15 min at room temperature as described previously.18 An equal volume of piperidine (10%) was added to each tube, and the reaction was incubated at 90°C for 30 min. Reaction was diluted to double the volume and dried under vacuum. Resultant pellet was washed thrice with water and dried under vacuum. Reaction products were resolved on an 18% denaturing polyacrylamide gel, which was further dried and the signal was detected using PhosphorImager Typhoon (GE, USA).
2.11 Bio-layer interferometry (BLI)
ForteBio Octet RED 96 (Forte Bio, USA) and streptavidin-coated high-precision (SAX) sensors (Forte Bio, USA) were used to study the interaction between Hif1α G4-DNA and BG4 as described earlier.48–50 PBS (1×) was used as assay buffer for BLI studies and the study was conducted at 30°C. SAX sensor tips were hydrated in buffer for 30 min prior to use. Wells in 96-microwell plates were filled with 200 μl buffer or sample as the case and agitated at 1000 rpm. Hydrated sensors were immobilized and saturated with biotinylated DNA. A reference sensor without biotinylated DNA subjected to the same protocol as the DNA-loaded sensors served as a background control. Following immobilization with biotinylated DNA, binding interaction with different concentrations of BG4 was carried out, which consisted of baseline (60 s), association (150 s), dissociation (150 s), baseline (60 s). Kd value was calculated using global curve fit (1:1) model, using Octet data analysis (v 8.0).
2.12 Circular dichroism studies
G4-forming oligomers were annealed in TE in presence or absence of potassium chloride or lithium chloride (100 mM) in TE at 37°C for 1 h. For double-stranded random sequences, oligomers were annealed in TE in presence of sodium chloride (100 mM) followed by gradual cooling at room temperature. Sequences forming i-motif were annealed in presence or absence of potassium acetate (pH 5.2) (100 mM) for 10 min at 90°C and then cooled down to room temperature. Plasmids, pSXneo135, pSXneo270 and pSV4 were incubated in presence of potassium chloride (100 mM) in TE at 37°C for 1 h. All CD spectra were recorded at room temperature at wavelength range of 200–300 nm (10 cycles) for every sample, using a spectropolarimeter (JASCO J-810) at a scan speed of 50 nm/min. Separate spectrum was measured for buffer alone (15 cycles) and was subtracted from all the experimental spectra. Ellipticity was calculated using Spectra Manager and plotted as a function of wavelength.
2.13 Gel mobility shift assay on plasmids
Plasmids pSXNeo135, pSXNeo270 and pSV4 were incubated in presence of 100 mM KCl in TE (37°C for 1 h), followed by BG4 (0, 1.5, 2.3, 3 µg) at 4°C for 1 h. Products were resolved on 1.2% agarose gels containing 25 mM KCl at 100 V, at room temperature. The gels were stained with ethidium bromide (0.5 µg/ml) and photographed under transilluminator (UVItec, Cambridge).
2.14 Immunoblotting
For immunoblotting analysis, ∼30 μg of protein was resolved on 8–10% SDS-PAGE as described earlier.70 Briefly, following gel electrophoresis, proteins were transferred to PVDF membrane (Millipore), blocked with 5% skimmed milk powder, and probed with appropriate primary antibodies against BG4, WRN and Actin; and secondary antibodies. The blots were developed using chemiluminescent detection solution (ImmobilonTM Western, Millipore) and scanned using a gel documentation system (LAS 3000, Fuji). For quantifying the bands of interest, the area was selected in each lane, and identical rectangle was selected in an area with no band from each lane of the membrane for subtracting the background. Intensity measured in each lane was indicated as PSLU. Normalization of the protein of interest was done with loading control, β-actin and plotted using GraphPad Prism (ver 5.01).
2.15 WRN knockdown in human cells
HeLa cells (4 × 105) were transfected with shRNA cocktail against WRN using a stable cationic polymer, linear polyethylenimine (PEI) (10 µg/µl). Cells were harvested after 48 h, washed with PBS, resuspended in RIPA buffer [25 mM Tris, pH 7.6, 150 mM NaCl, 1% NP-40 (SRL, India), 1% sodium deoxycholate (Sigma-Aldrich, USA) and 0.1% SDS] containing protease inhibitors and incubated on ice. The cell suspension was sonicated and the supernatant containing proteins were collected by centrifugation. The protein concentration was determined by Bradford method.71 Efficiency of knockdown was confirmed using immunoblotting and cells were further processed for immunofluorescence.
2.16 Immunofluorescence
HeLa cells grown on glass coverslips were fixed in 2% paraformaldehyde and permeabilized using 0.1% Triton-X100 in PBS. Blocking was performed using 0.1% bovine serum albumin and 10% FBS. Immunofluorescence was performed using standard method with BG4, anti-FLAG (PRB-132C, Biolegend), anti-RAG1 (SC363) and anti-rabbit (62111028001 A, Millipore), followed by development using Streptavidin-FITC.72 Coverslips were mounted with DABCO/DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen). Digital images were recorded using Zeiss Apotome and analyzed with ImageJ software. Data were analyzed and plotted with standard mean of error using GraphPad Prism (ver 5.01, GraphPad Software, Inc.).
HeLa cells were seeded in presence and absence of 10% fetal bovine serum in culture medium. Cells were harvested after 48 h, processed for immunofluorescence and BG4 foci were analyzed.
3. Results
3.1 Purified fractions of BG4 antibody show efficient binding to single-stranded DNA containing G4 motif
BG4 was overexpressed and purified using affinity column chromatography. Imidazole fractions were analyzed on a denaturing PAGE, all fractions showed enrichment for BG4 (Supplementary Fig. S1A). Among that, selected fractions (800 mM to 1 M imidazole) were pooled, dialysed and used for experiments. Identity of the protein was confirmed by western blotting (Supplementary Fig. S1B). In order to test whether the fractions collected after purification from different batches of proteins were active, purified proteins (∼500 ng) were incubated with single-stranded oligomeric DNA, RT17, derived from Hif1α gene, possessing G-quadruplex motif (Supplementary Fig. S1C). Results showed specific binding of BG4 to RT17, when different batches of proteins were analyzed (Supplementary Fig. S1C, lanes 5–10 and data not shown). However, binding was absent when complementary C-rich sequence (MN89) derived from Hif1α was used (Supplementary Fig. S1C, lanes 1, 2). Thus, our results reveal that BG4 overexpressed and purified from bacteria was pure and functional.
3.2 DNA substrate titrations and competition assays reveal that binding of BG4 to G4 DNA is specific
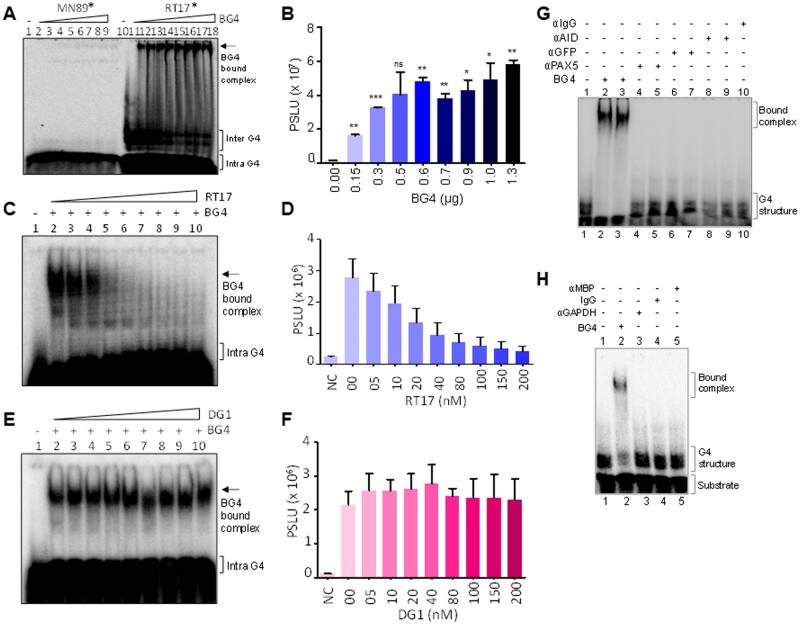
Titration of BG4 with G4 containing single-stranded oligomeric DNA derived from Hif1α (RT17) and its complementary C-rich sequence (MN89) showed that the antibody binds to the G4-forming DNA from lowest concentration (150 ng) onwards (Fig. 1A, lanes 10–18). Efficiency of binding improved with increase in BG4 concentration (Fig. 1A and B). Unlike RT17, the complementary C-rich oligomer, did not show BG4 binding even at the highest concentration tested (1.3 µg) (Fig. 1A, lanes 1–9). We also observed an increase in efficiency of BG4 binding, when concentration of RT17 was increased, further confirming the preference of BG4 to G-quadruplex forming DNA (Supplementary Fig. S2).
Figure 1.
Evaluation of specificity of BG4 antibody towards G-quadruplex DNA. (A) Gel image showing BG4 binding to G4 DNA. Radiolabelled Hif1α oligomeric substrate, RT17* (lanes 10–18), was incubated in KCl at 37°C, followed by incubation with no or increasing concentrations of BG4 (0.15, 0.3, 0.5, 0.6, 0.7, 0.9, 1.0 and 1.3 µg). Bound DNA was then electrophoresed on a 6% native PAGE and indicated using an arrow. Complementary C-rich sequence, MN89*, fails to show any binding (lanes 1–9). Asterisk represents radiolabelled substrate. (B) Bar diagram showing increase in formation of bound complex for RT17 with an increase in BG4 concentration (P < 0.05). (C) Competition assay using radiolabelled RT17, in presence of increasing concentration of unlabelled (cold) RT17, as the specific competitor. Briefly, radiolabelled RT17* was allowed to form G4 structure at 37°C, followed by incubation with BG4. Simultaneously, unlabelled RT17 (0, 05, 10, 20, 40, 80, 100, 150, 200 nM) was added to the reaction mixture, incubated and the products were resolved on a 6% native PAGE. Bound complex is indicated by an arrow. (D) Bar diagram showing reduction in bound complex when incubated with increasing concentration of unlabelled oligomer RT17 (P < 0.05), indicating specificity of antibody towards G4 DNA. (E) Competition assay to evaluate BG4 binding to radiolabelled RT17* in presence of cold random oligomeric DNA, DG1. Radiolabelled RT17* was allowed to form G4 at 37°C, followed by BG4 incubation. Unlabelled DG1 (0, 05, 10, 20, 40, 80, 100, 150, 200 nM) was added to reactions, and the products were resolved on a 6% native PAGE. (F) Bar diagram showing BG4 bound complex (P < 0.05), when cold random DNA substrate was added to the mixture confirming the specificity of antibody towards G4 DNA. (G, H) Gel images showing specificity of BG4. Various antibodies (200 ng) were incubated with RT17 in the same conditions as that of BG4, and the products were resolved on 6% native PAGE. Bound complex and substrate have been indicated in the gel images.
To evaluate the specificity of antibody interaction with the substrate, competition experiments were conducted wherein fixed concentration of BG4 antibody (1.3 µg) was incubated with radiolabelled RT17 (20 nM). For competition studies, increasing concentration (5, 10, 20, 40, 80, 100, 150, 200 nM) of either unlabelled RT17 or random unlabelled oligomer, DG1 was added and incubated to the mix (Fig. 1C and E). Results showed significant reduction in BG4 binding when unlabelled RT17 was used for competition studies (Fig. 1C and D). The extent of decrease in bound complex with increasing concentrations of cold RT17 demonstrated that antibody binding is indeed specific towards G4-forming sequence (Fig. 1D). However, upon addition of random unlabelled sequence, the band formed due to RT17-BG4 bound complexes stayed unchanged (Fig. 1E and F), reiterating the fact that BG4 recognizes G4-forming motif in a highly specific manner. To further test the specificity of BG4 to G4 DNA, we incubated RT17 with various antibodies against AID, GFP, PAX5, MBP, GAPDH, and IgG (Fig. 1G and H). It was observed that none of the antibodies tested could bind to RT17, thus underlining the specificity of BG4 towards G4 motif (Fig. 1G, lanes 4–10; Fig. 1H, lanes 3–5).
To quantitate BG4-binding specificity with G4 DNA, BLI experiments were performed.48–50 5′-biotinylated oligomeric DNA derived from Hif1α DNA was synthesized, immobilized to high precision streptavidin sensor surface, and allowed to interact with different concentrations (50, 80, 160, 320 nM) of BG4. Upon binding of antibody to sensors, the thickness of the sensor tip layer increases, and this difference was measured by the BLI machine as interference in wavelength shift. Sensors immobilized with 5′ biotinylated DNA were transferred to wells containing assay buffer and then to the wells with both buffer and BG4 to measure interaction. Reference probe with no antibody served as background signal. Results showed a significant shift in association curve upon BG4 binding in a concentration-dependent manner (Supplementary Fig. S1D) and confirm the ability of BG4 to interact with G4 DNA. Binding curves were fit globally to a 1:1 binding model, and equilibrium dissociation constant (Kd) was calculated. Importantly, robust binding of BG4 to G4 DNA was observed with a Kd value of 17.4 ± 0.588 nM (mean ± SD), further demonstrating the BG4-binding specificity to G4 structures.
3.3 BG4 binds to both inter- and intramolecular G4
We have compared binding efficiency of BG4 towards different DNA substrates (Fig. 2A). Previous studies have reported that G-quadruplex structures can be either inter- or intramolecular in nature.28 We observed that oligomeric DNA derived from Hif1α (RT17) can fold into both inter- and intramolecular G-quadruplexes (Fig. 2B, lane 1).15,18,29 Incubation with increasing concentrations of BG4 revealed a concentration-dependent binding of BG4 to inter- and intramolecular G-quadruplexes (Fig. 2B, lanes 2–4) suggesting that BG4 can bind to G4 DNA when formed within a strand of DNA molecule or between different strands of DNA (Supplementary Fig. S3). However, complementary C-rich strand (MN89) of RT17, or a random sequence of comparable length, but devoid of any non-B DNA motifs (BTM6) did not show any BG4 binding (Fig. 2B, lanes 5–12). Importantly, incubation of increasing concentration of heat denatured BG4 with RT17 or of BG4 with MN89, did not show any binding (Fig. 2C, lanes 5–12).
Figure 2.
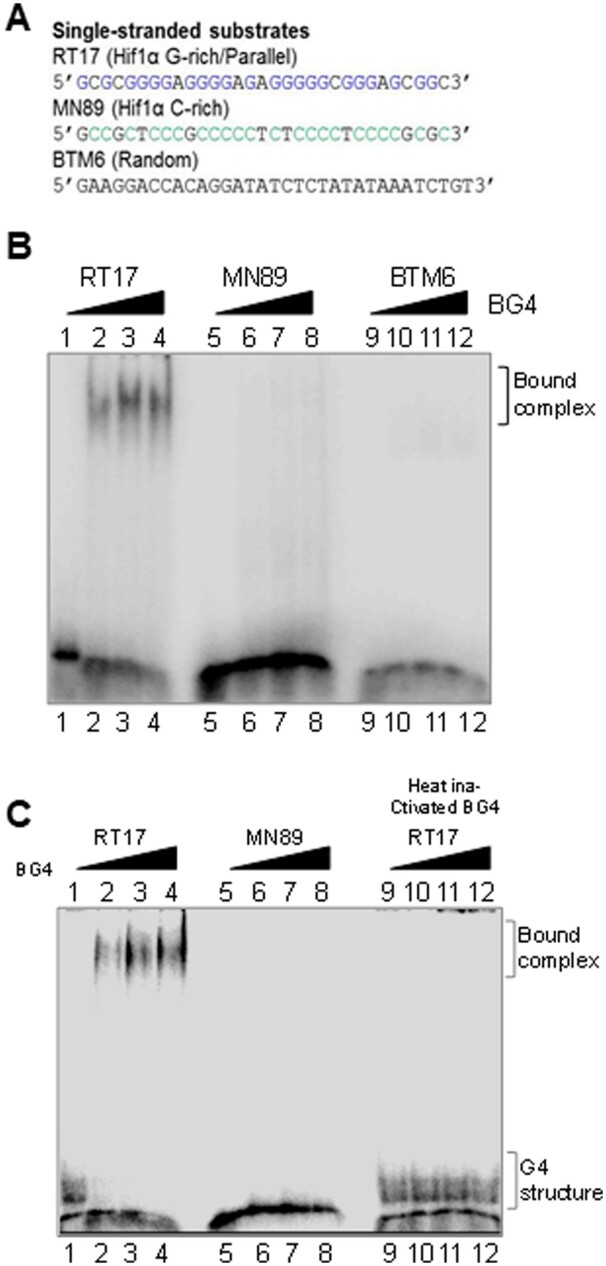
Assessment of BG4 binding to different single-stranded oligomeric DNA containing G-quadruplex motifs, random sequences. (A) Sequences of DNA substrates used for binding studies to test specificity of BG4. (B) Comparison of BG4 binding to oligomer containing G4 motif (RT17; lanes 1–4), its complementary sequence (MN89; lanes 5–8), and a random sequence (BTM6; lanes 9–12). Three different antibody concentrations (0.4, 0.6, 0.8 µg) were used in each case. (C) Evaluation of binding efficiency of active BG4 and heat-inactivated (95°C) BG4 to RT17 (lanes 1–4, 9–12) and MN89 (lanes 5–8). In all panels, oligomeric DNA was incubated at 37°C with BG4 or heat-inactivated BG4 and products were then electrophoresed on a 6% native PAGE. Bound complex is indicated in the images.
3.4 Mere presence of G4 motif in double-stranded DNA is not sufficient for BG4 binding
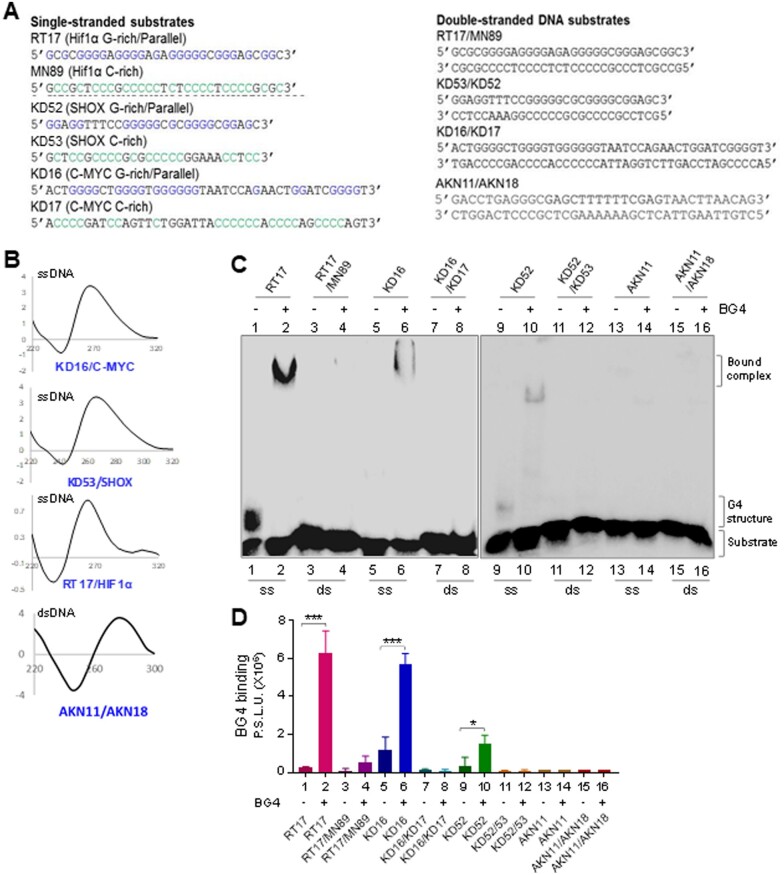
Further, BG4 binding to oligomeric DNA containing G-quadruplex motifs was compared with that of corresponding double-stranded DNA (Fig. 3A). For this, CD studies were performed using single-stranded DNA containing G4 motif (RT17, KD16 and KD52) to ensure G4 structure formation in the DNA sequence (Fig. 3B). In contrast, an oligomeric DNA containing random sequence (AKN11/AKN18) resulted in a spectra characteristic of B-DNA (Fig. 3B). To compare BG4 binding to G4 DNA and corresponding sequence when present as double-stranded B-DNA, G4 motifs containing oligomers were annealed with their C-rich complementary sequences and used for BG4 binding assay. Results showed that unlike single-stranded DNA derived from Hif1α, c-MYC and SHOX containing G4 structures, double-stranded DNA from corresponding sequences did not show any binding to BG4 (Fig. 3C and D). These results suggest that mere presence of G-quadruplex motif on double-stranded DNA is not sufficient for BG4 binding, in all the three regions tested; rather, it requires formation of G4 structures. A random single-stranded DNA (AKN11) and duplex B-DNA (AKN11/18) were also analyzed for BG4 binding (Fig. 3C, lanes 13–16, 3 D). BG4 showed no binding to either random single-stranded DNA or B-DNA duplex, further underscoring its specificity towards G4 motifs.
Figure 3.
Assessment of BG4 binding to different single-stranded and double-stranded DNA. (A) Sequences of DNA substrates (single-stranded, double-stranded) used for binding studies. (B) CD spectra were recorded for G4-forming sequences in presence of 100 mM KCl, and for B-DNA duplex, at room temperature, within wavelength range of 220–300 nm (10 cycles), using a spectropolarimeter (JASCO J-810) at scan speed of 50 nm/min. Absorption spectrum measured for buffer alone (15 cycles) was subtracted from each sample and resulting spectra were plotted. KD16 (c-MYC), KD53 (SHOX) and RT17 (HIF1α) showed a parallel orientation with a negative dip at 240 nm and a positive peak at ∼270 nm. In all cases ‘X’-axis denotes wavelength (nm) and ‘Y’-axis denotes molar ellipticity (mdeg). (C) Comparison of BG4-binding efficiency between single-stranded DNA containing G4 structures, RT17 (Hif1α; lanes 1, 2), KD16 (c-MYC; lanes 5, 6), KD52 (SHOX; lanes 9, 10), random DNA (AKN11) and double-stranded DNA containing G4 motifs, RT17/MN89 (Hif1α; lanes 3, 4), KD16/KD17 (c-MYC; lanes 7, 8), KD52/53 (SHOX; lanes 11, 12) and B-DNA duplex (AKN11/18). In all cases, BG4 (1.3 µg) was incubated with ssDNA or dsDNA and bound complexes were resolved on a 6% native PAGE. (D). Bar diagram showing BG4 binding across single-stranded and double-stranded DNA substrates derived from Hif1α, c-MYC and SHOX. Y-axis denotes radioactivity intensity depicting the BG4-bound complexes.
3.5 BG4 binds to parallel G4 structures
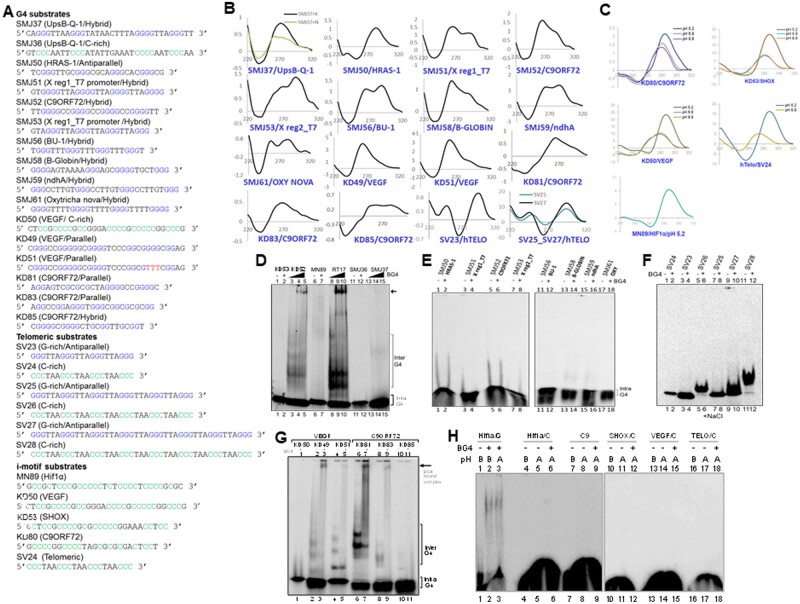
Previous studies have shown that based on orientation of the DNA strand involved, G-quadruplexes can be either parallel or antiparallel in nature.24 To test the BG4 binding preference with respect to orientation of G4 DNA, oligomeric DNA were designed from different regions of human genome that is known to form G-quadruplex DNA structures (Figs 3A and 4A). CD studies confirmed formation of parallel G4 DNA in case of oligomers derived from SHOX gene (KD52), C-MYC (KD16) and Hif1α (RT17). In contrast, oligomers derived from UpsB-Q-151 (SMJ37), SMJ50 (HRAS-1), SV23 (TELO), SV25 (TELO), and SV27 (TELO) showed G4 formation in antiparallel orientation (Fig. 4B). Oligomers SMJ51 (X region 1/T7 promoter), SMJ52 (C9ORF72), SMJ53 (X region 2/T7 promoter), SMJ56 (BU-1), SMJ58 (B-GLOBIN), SMJ59 (ndhA) and SMJ61 (OXY) showed G4 DNA in hybrid orientations, with prominent antiparallel peak (Fig. 4B).
Figure 4.
Evaluation of BG4 binding to different orientations of G-quadruplex DNA structures. (A) Sequences of oligomeric DNA substrates used for the study. (B) CD spectra showing G4 formation in parallel, antiparallel and hybrid orientation. DNA sequences used for studying G4 in parallel orientation were, SMJ50 (HRAS-1), KD49 (VEGF), KD51 (VEGF), KD81 (C9ORF72) and KD83 (C9ORF72). G4 structures studied in antiparallel orientation (in presence of 100 mM NaCl) were, oligomeric DNA-containing telomeric sequences SV23, SV25 and SV27. Oligomers SMJ37 (UpsB-Q-1), SMJ51 (X Reg1/T7 promoter), SMJ52 (C9ORF72), SMJ53 (X Reg2/T7 promoter), SMJ56 (BU-1), SMJ58 (B-GLOBIN), SMJ59 (ndhA), SMJ61 (OXY) showed more of hybrid G4 structures with dominant antiparallel orientation (in presence of 100 mM NaCl). For other details, refer Fig. 3B. (C) CD spectra showing i-motif formation in presence of 100 mM potassium acetate (pH 5.2), Tris buffer (pH 6.8) and Tris buffer (pH 8.8), at room temperature. Reading was taken from wavelength range of 220–320 nm (10 cycles), using a spectropolarimeter (JASCO J-810) at scan speed of 50 nm/min. Absorption spectrum measured for buffer alone (15 cycles) was subtracted from each sample and resulting spectra were plotted. In case of all the sequences (KD80, KD50, SV24, KD53, MN89), acidic pH of 5.2 promoted i-motif formation, as seen from positive peak at ∼285 nm and dip at ∼255 nm. (D) Comparison of BG4-binding efficiency between oligomeric DNA that can form parallel and antiparallel G-quadruplexes. Parallel G-quadruplex forming oligomers RT17 (Hif1α; lanes 8-10), KD52 (SHOX; lanes 3-5) and antiparallel G-quadruplex-forming oligomers SMJ37 (UpsB-Q-1; lanes 13-15) were incubated with increasing concentrations of BG4 (0.0, 0.4, 0.8 µg) and resolved on PAGE. In every case complementary DNA (KD53, lanes 1, 2; MN89, lanes 6, 7; SMJ36, lanes 11, 12) was used as control for BG4 binding. (E) Comparison of BG4-binding efficiency between oligomeric DNA that can form antiparallel G-quadruplexes. Antiparallel G-quadruplex forming oligomers SMJ50 (HRAS-1; lanes 1–2), SMJ51 (X Reg1/T7 promoter, lanes 3–4), SMJ52 (C9ORF72, lanes 5–6), SMJ53 (X Reg2/T7 promoter, lanes 7–8), SMJ56 (BU-1, lanes 11–12), SMJ58 (B-GLOBIN, lanes 13–14), SMJ59 (ndhA, lanes 15–16), SMJ61 (OXY, lanes 17–18) were incubated with BG4 (1.0 µg) and resolved on 6% native PAGE. (F) Assessment of BG4 binding to oligomers containing antiparallel G4 structures (SV23, lanes 3–4; SV25, lanes 7–8; SV27, lanes 9–10) and their complementary sequences (SV24, lanes 1–2; SV26, lanes 5–6; SV28, lanes 11–12). Substrates were incubated at 37°C in presence of 100 mM sodium chloride, followed by incubation with 1.5 µg of BG4 at 4°C, and the resulting complexes were resolved on a 6% native PAGE containing 100 mM NaCl. (G) Evaluation of binding efficiency of G4 DNA derived from VEGF and C9ORF72 regions, containing GNG motifs. Oligomers were incubated at 37°C in presence of 100 mM KCl, followed by incubation with 1.0 µg of BG4 at 4°C, and products were resolved on a 6% native PAGE. (H) Comparison of BG4-binding efficiency between DNA substrates containing G4 structure, RT17 (HIF1Α; lanes 1-3) and DNA containing i-motifs, MN89 (HIF1α; lanes 4–6), KD80 (C9ORF72; lanes 7–9), KD52 (SHOX; lanes 10-12), KD50 (VEGF; lanes 13–15) and SV24 (telomeric region; lanes 16–18). RT17 was incubated at 37°C, in presence of 100 mM KCl, whereas i-motif substrates were incubated with 100 mM potassium acetate (pH 5.2) at 95°C for 2 h and cooled to room temperature.
Incubation of increasing concentrations of BG4 with single-stranded substrates, KD52, RT17, SMJ37, and their respective complementary single-stranded C-rich oligomers revealed a concentration-dependent binding of BG4 to KD52 and RT17 unlike SMJ37. These results suggest that while BG4 efficiently binds to parallel G-quadruplexes but failed to do so with antiparallel structures (Fig. 4D, lanes 3–5, 8–10, 13–15). Consistent with above observations, BG4 failed to bind complementary C-rich sequences KD53, MN89, and SMJ36 (Fig. 4D, lanes 1–2, 6–7, 11–12). Further BG4 binding was also tested on oligomeric DNA that contain G4 motifs with GNG sequences KD49, KD81, KD83 and KD85 (Fig. 4A). CD studies confirmed formation of parallel G4 DNA confirmation on these oligomers (Fig. 4B). EMSA studies using BG4 antibody on these oligomers revealed that all G4 structures exhibited detectable binding to these oligomeric DNA except KD85 (Fig. 4G). Surprisingly, oligomers assuming hybrid and antiparallel orientations (SMJ50-61; Fig. 4A and B) consistently did not show any BG4 binding (Fig. 4A, B and E). Antiparallel G4 structures derived from the human telomeric regions (SV23, SV25, SV27) also did not show BG4 binding (Fig. 4A, B and F) Thus, it appears that BG4 recognizes primarily parallel G-quadruplexes and the collective observations suggest a possible preference of the antibody for parallel over antiparallel conformations.
3.6 BG4 does not bind to i-motifs
Since i-motif is a multi-stranded DNA structure formed by cytosines at acidic pH (intercalated motifs),52,53 we tested whether BG4 can bind to these DNA structures. We examined propensity of multiple telomeric and non-telomeric C-rich sequences to form intercalated motifs at acidic pH of 5.2 and compared these with structures assumed at pH 6.8 and 8.8 (Fig. 4A). CD analyses of these DNA revealed robust i-motif formation across all the sequences tested, at acidic pH (Fig. 4C). BG4-binding assays revealed no significant binding when DNA substrates containing i-motif structures were tested except in the case of C-strand derived from Hif1α (Fig. 4H). BG4 binding to G4 structure present in RT17 served as positive control (Fig. 4H, lanes 1–3).
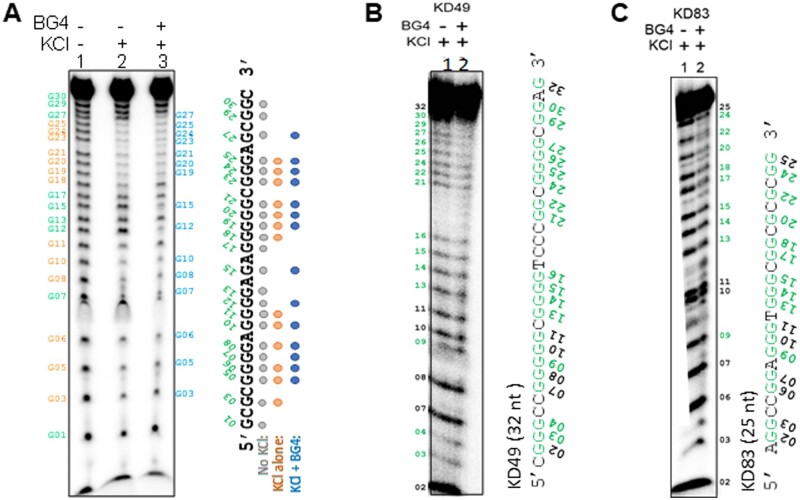
3.7 DMS protection assay reveal formation of G4 structures on oligomeric DNA and interference on DMS reactivity due to BG4 binding
DMS probing is generally used to determine formation of Hoogsteen hydrogen bonding within G-quadruplex structures as they cannot get methylated by DMS and hence are resistant to piperidine cleavage.18 First, formation of G4 structure was confirmed using DMS probing on an oligomeric DNA that was derived from Hif1α containing the G4 motif (RT17) by incubating in presence of KCl (Fig. 5A). Gel profile revealed DMS protection in guanine residues following incubation with KCl suggesting formation of G4 structures (Fig. 5A). Interestingly, incubation of BG4 with RT17 in KCl containing buffer, further shielded the structures from DMS reactivity and resulted in a pattern of differential protection in the guanine residues of G4 that are free of and bound by BG4 (Fig. 5A). Two other sequences, VEGF (KD49) (Fig. 5B) and C9ORF72 (KD83) (Fig. 5C) were also probed following G4 structure formation in presence of KCl. Both revealed a similar pattern of protection upon antibody binding (Fig. 5B and C). This result further suggests an interaction of the antibody with stacked G4 structure across the three plates.
Figure 5.
DMS probing of BG4-bound G-quadruplex DNA substrates. (A) DMS protection assay of RT17 (HIF1α) in presence or absence of KCl and BG4 antibody. Since G-quadruplex structure does not form in the absence of KCl on RT17, all guanines react to DMS resulting in specific bands (lane 1, guanine residues marked in grey). Addition of potassium stabilizes the G-quadruplex DNA, and those G residues are protected and indicated in brown (lane 2). BG4 binding shields the sequence from DMS leading to additional protection and footprint (lane 3, protected guanine residues are indicated in blue). RT17 was incubated at 37°C in presence of KCl, and then incubated with BG4. DMS probing was performed at 37°C on both samples, followed by piperidine cleavage and purification. The reaction products were resolved on 18% denaturing PAGE. Brown ovals indicate the guanine residues protected in presence of potassium and in the absence of BG4. Blue ovals indicate the protected residues in presence of both potassium and BG4. (B, C) DMS probing of KD49 (VEGF) and KD83 (C9ORF72) in presence of KCl and BG4 antibody. BG4 antibody was added (lane 2) to potassium-stabilized G4 (lane 1) and subjected to DMS protection assay. Guanine residues are marked and those protected are indicated in green.
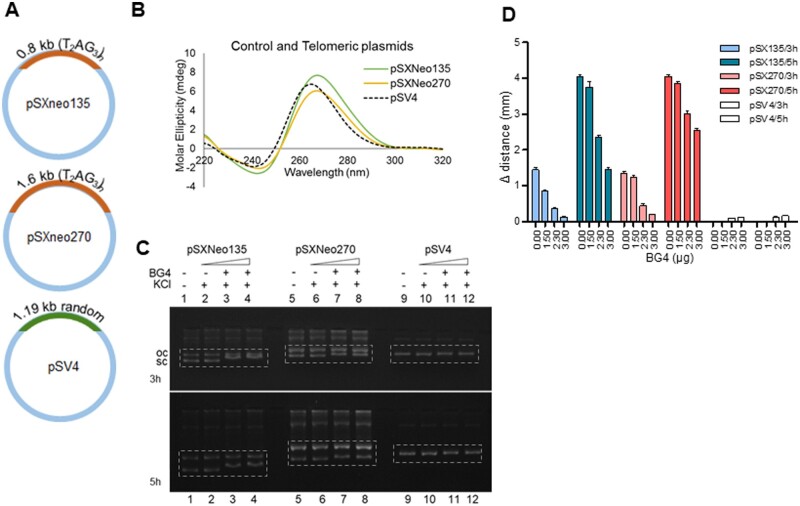
3.8 BG4 can bind to G4 structures when present on a plasmid DNA
It has been shown that telomeric repeats, when present on a plasmid DNA can fold into G-quadruplex DNA structures.54 In order to test BG4 affinity to plasmid DNA-bearing G4 structures, telomeric plasmids, pSXneo135 and pSXneo270, containing 0.8 kb and 1.6 kb long telomeric repeats (T2AG3)n (Fig. 6A) were first tested for their ability to form G4 structures by CD (Fig. 6B). Results showed a positive peak at 265 nm and negative trough at 240 nm when pSXneo135 and pSXneo270 plasmids were incubated in KCl suggesting the formation of parallel G4 structure (Fig. 6B), which was consistent with previous studies.54 Further, pSXneo135 or pSXneo270 was incubated with increasing concentrations (1.5, 2.3, 3 µg) of BG4 antibody and resolved on an agarose gel to evaluate BG4 binding to G4 structures (Fig. 6C). Results showed a shift in the mobility of supercoiled forms of both the plasmid DNA (pSXneo135 and pSXNeo270), when incubated with BG4 antibody (Fig. 6C and D). The open circular form, which does not have G4 structures, remained unbound by the antibody (Fig. 6C, Supplementary Fig. S4). It is also important to note that control plasmid, pSV4 (Fig. 6A), with random sequence did not exhibit such a distinct shift even at the highest antibody concentration (Fig. 6C, Supplementary Fig. S4). Interestingly, although the shift was marginal when the plasmids were electrophoresed for a period of 3 h, the mobility shift across various concentrations of BG4 was more prominent when electrophoresed for 5 h (Fig. 6D). Thus, our results suggest that BG4 is able to bind to G4 structures when present even on plasmid DNA.
Figure 6.
Evaluation of BG4 binding on a plasmid containing telomeric DNA sequences that have the ability to fold into G-quadruplexes. (A) pSXneo135(T2AG3) and pSXneo270(T2AG3) contain 0.8 and 1.6 kb long telomeric repeats [(T2AG3)n], respectively and is shown to have the ability to form G-quadruplexes.54 pSV4 contains 1.19 kb long region containing random sequence, devoid of any non-B DNA motifs. (B) CD spectra recorded for pSXneo135, pSXneo270 and pSV4 in presence and absence of 100 mM KCl, at room temperature, within wavelength range of 200–300 nm (10 cycles), using a spectropolarimeter (JASCO J-810) at a scan speed of 50 nm/min. Absorption measured for buffer alone (15 cycles) was subtracted from each sample and the resulting spectra was plotted. (C) Comparison of BG4-binding efficiency between pSXneo135 (lanes 1–4), pSXneo270 (lanes 5–8) and pSV4 (lanes 9–12) as determined by shift in mobility of supercoiled form of the plasmid, indicated using dotted box. In each case, plasmid DNA was incubated with 100 mM KCl at 37°C for 1 h, followed by BG4 (0, 1.5, 2.3, 3 µg) at 4°C for 1 h, and resolved on 1.2% agarose gel containing 25 mM KCl at 100 V, at room temperature. BG4-bound supercoiled forms of pSXneo135 shift substantially (lanes 1–4) as do those of pSXneo270 (lanes 5–8), unlike the control plasmid pSV4 (lanes 9–12), when electrophoresed for longer time periods (5 h compared to 3 h). (D) Bar diagram showing quantification of shift in the mobility of supercoiled plasmids upon addition of BG4 (0.00, 1.5, 2.3, 3 µg). Y-axis denotes the mobility shift of the supercoiled form.
3.9 BG4 binding within the cells reveal that G4 structures across different human cell types are comparable
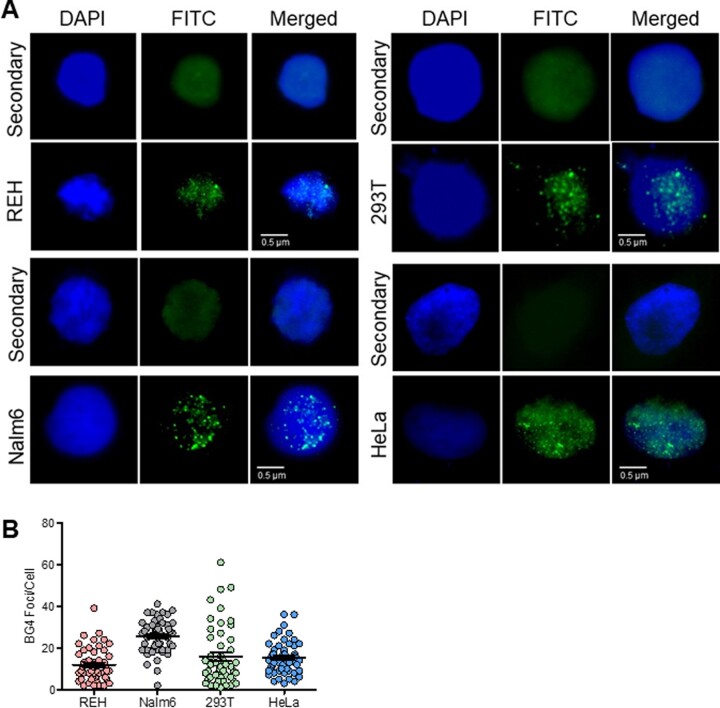
Formation of endogenous G4 structures in nuclei was examined by fluorescence microscopy using BG4 antibody.19 Cell lines of different origins, REH and Nalm6 (B cells), HEK293T (human embryonic kidney epithelial cells), and HeLa (cervical cells) were incubated with BG4, which was further detected using FLAG antibody, using biotinylated anti-rabbit secondary antibody tagged with FITC. Nuclei were counterstained with DAPI.19 Visualization of cells under Apotome fluorescence microscope revealed that BG4 foci were primarily restricted to the nuclear region. The foci pattern was of distinct punctate nature (Fig. 7). Cells treated with secondary antibody (FLAG antibody followed by anti-rabbit antibody) alone, did not result in any foci formation. RAG1 is known to be expressed in pre-B cells and is localized in the nucleus. Therefore, we used anti-RAG1 antibody in Nalm6 cells as a control to check for foci formation. It was observed that the foci formation is specific to BG4,19 whereas RAG1 showed complete fluorescence throughout the nucleus (Supplementary Fig. S5). Foci were specific to BG4 antibody addition in all cell types tested (Fig. 7). In case of each cell line, ∼50 cells were counted, and results showed that maximum number of foci was seen in Nalm6 cells, which was 26 foci per cell (Fig. 7B). In case of REH cells, average BG4 foci per cell were 12. Number of foci per HEK293T cell was 16, while it was 15 in case of HeLa cells (Fig. 7B). The difference in the number of foci observed in different cell lines could be due to multiple factors. Possibilities include the transcription and replication status of the cell, level of supercoiling of the genome, higher order chromatin structures, and the difference in the origin of the cell type. Although the number of foci observed can be an under-representation, our results reveal that BG4 antibody can be used in determining G4 structures within the cells, which was consistent with previous reports3,19 and they are mostly comparable among different cell types.
Figure 7.
Evaluation of BG4 foci in various cell lines to determine occurrence of G-quadruplex DNA structures within cells. (A) Evaluation of ability of BG4 binding to G4 structures within different human cell lines, REH, Nalm6, 293 T and HeLa. Cells were fixed in 2% paraformaldehyde and permeabilized with 0.1% Triton-X100, followed by blocking. Immunofluorescence was performed by incubating cells with BG4 at 4°C, followed by anti-FLAG, and anti-rabbit secondary antibody. Following streptavidin-FITC, coverslips were mounted with DABCO/DAPI and images were recorded using Zeiss Apotome, and analyzed with ImageJ. Data were analyzed and plotted as scatter plots using GraphPad Prism. BG4 was tagged with FLAG antibody and observed in FITC channel (green). Nuclei were counterstained with DAPI (blue). First row of every panel is secondary control (only anti-FLAG, anti-rabbit). Second row depicts the representative BG4-stained image. BG4 foci can be observed within cells in the nuclear region, in punctate form. (B) Scatter plots depict the quantification of BG4 foci for REH, Nalm6, HEK293T and HeLa cells.
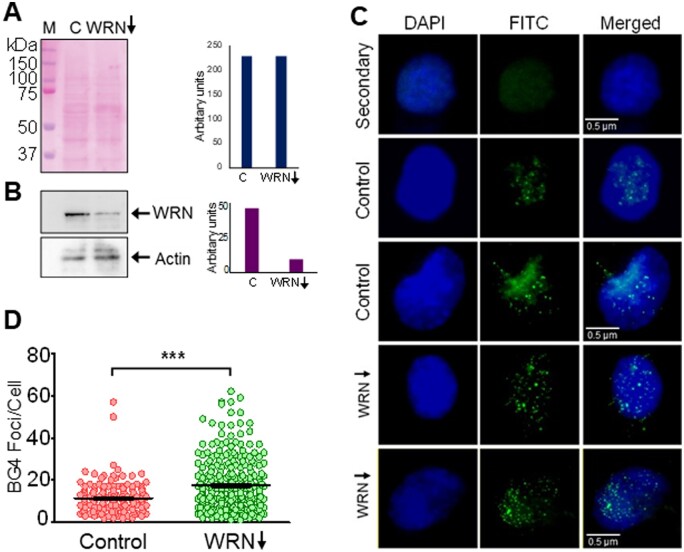
3.10 Levels of WRN helicase can modulate G-quadruplex formation within cell
It is known that Werner’s helicase (WRN) acts as a G-quadruplex resolvase.55 Thus, we wondered whether knock-down of WRN expression in cells will have an impact on BG4 foci formation. In order to address this, HeLa cells were transfected with WRN-shRNA and the knockdown was assessed by immunoblotting after equalization of the protein (Fig. 8A and B). Scrambled shRNA treated cells served as the control. Immunofluorescence studies following incubation with BG4 antibody, as described above revealed a significantly higher number of BG4 foci per cell following knockdown of WRN in the cellular population (Fig. 8C and D). The average number of foci seen per cell was ∼20 following WRN knock down, while it was only ∼10 in scrambled shRNA-treated cells (Fig. 8D) suggesting presence of more number of G-quadruplexes as compared to control (Fig. 8D). This also indicates that in a cell, helicases such as WRN may play a significant role in maintaining balance of the number of G4 structures, thus ensuring proper cellular functions and maintaining genomic integrity.
Figure 8.
Modulation of G-quadruplex formation within cells following depletion of G4 resolvase, WRN in HeLa cells. (A) shRNA-mediated knockdown of Werner’s helicase, WRN was performed in HeLa. Ponceau-stained blot reveals equal loading of the proteins. (B) Immunoblotting to determine extent of WRN knockdown in cells incubated with shRNA. ACTIN served as loading control and was used for normalization. Bar diagram showing extent of knockdown is also shown. (C) Representative images after immunofluorescence studies showing BG4 foci following WRN knockdown in HeLa cells compared to cells transfected with scrambled shRNA controls. Cells were counterstained with DAPI and scored for G4 structure formation by quantifying BG4 foci, as described above. For other details refer Fig. 7 legend. (D) Scatter plot illustrating the comparison of G4 structure formation determined as a measure of BG4 foci in WRN knockdown cells.
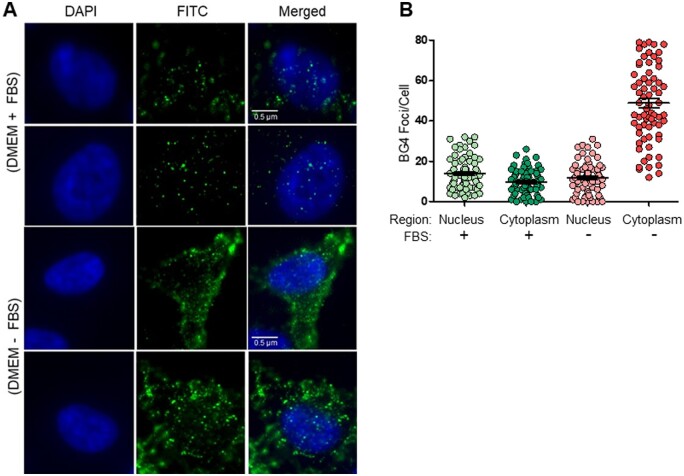
3.11 Nutrition deficiency can alter the extent of G-quadruplex formation within cellular regions
Absence of serum in the culture medium presents a form of nutrition deficiency to the cells. Under such conditions, we were interested in studying the formation of G4 structures within cells. HeLa cells were harvested after 48 h of serum starvation and then assessed by immunofluorescence following incubation with BG4 antibody (Fig. 9A and Supplementary Fig. S6). BG4 foci per cell in the nuclear region in presence of serum was significantly higher than that in cytoplasmic region (Fig. 9B and Supplementary Fig. 6); whereas in case of serum-starved cells, this count dropped in the nuclear region and significantly high for the cytoplasmic region (Fig. 9B and Supplementary Fig. S6). The presence of foci in the cytoplasm could be because of RNA G-quadruplexes.56,57 Higher number of foci in the nucleus of non-starved cells is understandable due to size of the genome being much greater than the RNA. The increase in cytoplasmic BG4 foci may also be attributed to increase in stress granules, which are formed upon stress induction owing to nutrition deficiency, suggesting presence of more G-quadruplexes in the cytoplasm (Fig. 9). This observation of BG4 localization in a serum-dependent manner warrants further investigation in future.
Figure 9.
Localization of BG4 binding within HeLa cells in nutrition proficient and deficient conditions. (A) Evaluation of BG4 binding to G4 structures in HeLa under nutrition proficient (presence of FBS) and deficient (absence of FBS, incubated for 48 h) conditions. Immunofluorescence was performed for BG4 as described above. BG4 foci are marked in both nuclear and cytoplasmic regions. Serum starvation of cells for 48 h, followed by BG4 staining, revealed more localization of the antibody towards the cytoplasm. (B) Scatter plot illustrates the comparison of G4 structure formation in nutrition proficient and deficient conditions in HeLa cells.
4. Discussion
Formation of non-canonical DNA structures that differ from Watson–Crick B-form has been investigated for past several years, albeit in more depth in in vitro conditions. Among different non-B DNA structures, G4 structures have gained significant attention due to their role in several biological processes. Although techniques like CD, NMR and X-ray crystallography have helped decipher several properties of the polymorphic G4 outside cell, its detection inside nucleus had been a challenge, given the limitations of available methods. Discovery of antibodies that can bind to G4 structures have provided a great momentum to intracellular studies associated with G-quadruplexes (Fig. 10). Particularly, the recent discovery of BG4 and its use in ChIP genome-wide sequencing, and evaluation of G-quadruplex formation in a cell-cycle dependent manner is important.3,58
Figure 10.
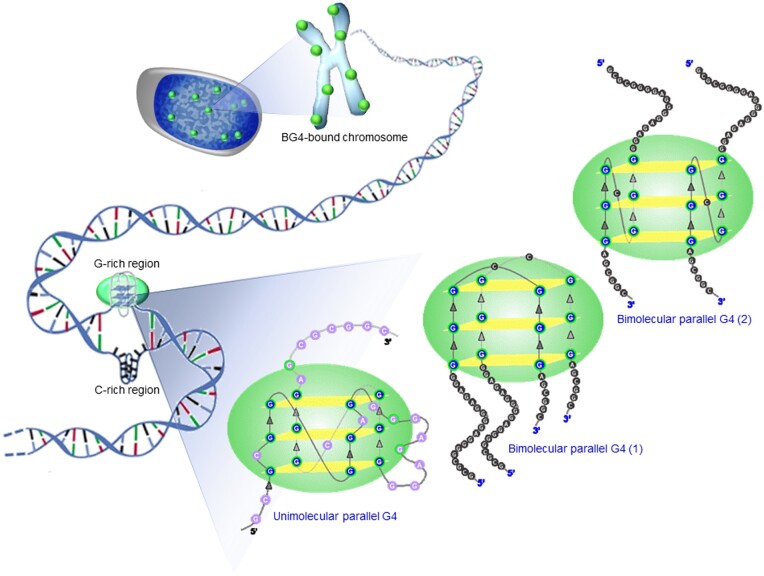
Model depicting BG4-binding preferences to G-quadruplex DNA structures both in vitro and in vivo. Several forms of G4 structures may exist in the chromatin context. The interaction pattern of BG4 seems to differ depending on the nature of the G4 structure. Bimolecular and unimolecular G4 molecules of parallel orientation appear to be bound by BG4 in a differential manner.
Other antibodies and probes reported so far have been incorporated in high-throughput studies as seen in case of hf243 and PDS.47,59 hf2 was used to enrich for genomic DNA fragments containing folded G-quadruplex structures from mechanically fragmented DNA derived from MCF7 breast cancer cells.43 These were experimentally mapped in sections of sub-telomeres, gene bodies and gene regulatory sites. DNA-damaged sites (γH2AX foci) induced by treatment with PDS were mapped by deep sequencing.59 These identified regions were enriched for computationally predicted G-quadruplex motifs. Staining of chromosomes and DNA fibres, higher foci count in FANCJ-depleted cells was demonstrated using 1H6 recently.42
In the present study, we overexpressed and purified BG4 antibody to homogeneity for further characterization. We have employed multiple biochemical and biophysical techniques to characterize BG4 binding preferences against, single-, double-stranded DNA sequences, G-quadruplexes (when present in different conformations) in the context of shorter oligomeric DNA, plasmid DNA and genome. Previously, BG4 specificity has been demonstrated primarily by ELISA, using oligomeric DNA substrates.3 We have resorted to gel mobility shift assays to demonstrate the nature of BG4 binding with oligomeric sequences. A titration of BG4 with G4 forming Hif1α substrate demonstrated a concentration-dependent increase of BG4-bound complexes. Importantly, we demonstrate that throughout the range of antibody concentrations tested, C-rich substrate remains unbound to BG4, confirming the antibody specificity towards G-rich sequences (Table 1 and Fig. 10). This holds true for random DNA substrates devoid of any non-B DNA motifs as well. DMS protection assay in presence of KCl confirmed that the G-rich DNA sequence (RT17) indeed folds into G-quadruplex structure. DMS footprinting further revealed protection towards DMS sensitivity upon incubation with BG4.
Table 1.
Sequence of the oligomers used for the study and their propensity to fold into G-quadruplex or i-motif structures
| Oligomer | Sequence (5′→ 3′) | Structure formation | G4 orientation | BG4 binding |
|---|---|---|---|---|
| RT17 | GCGCGGGGAGGGGAGAGGGGGCGGGAGCGGC | G4 | PARALLEL | ++++ |
| MN89 | GCCGCTCCCGCCCCCTCTCCCCTCCCCGCGC | i-motif | — | — |
| BTM6 | GAAGGACCACAGGATATCTCTATATAAATCTGT | NA | — | — |
| KD53 | GCTCCGCCCCGCGCCCCCGGAAACCTCC | i-motif | — | — |
| KD52 | GGAGGTTTCCGGGGGCGCGGGGCGGAGC | G4 | PARALLEL | +++ |
| SMJ36 | GTCCCAATTCCCATATTGAAATCCCCAATCCCAA | NA | — | — |
| SMJ37 | CAGGGTTAAGGGTATAACTTTAGGGGTTAGGGTT | G4 | HYBRID | — |
| KD16 | ACTGGGGCTGGGGTGGGGGGTAATCCAGAACTGGATCGGGGT | G4 | PARALLEL | ++ |
| KD17 | ACCCCGATCCAGTTCTGGATTACCCCCCACCCCAGCCCCAGT | NO | — | — |
| DG1 | GATCCCTCTAGACCGGTACTACTCGAGCTTGGTTGCAGATTAAGAGAGGCTCCTG | NA | — | — |
| KD50 | CTCCGCCCCGCCGGGACCCCGCCCCCGGCCCG | i-motif | — | |
| KD49 | CGGGCCGGGGGCGGGGTCCCGGCGGGGCGGAG | G4 | PARALLEL | ++ |
| KD80 | GCCCCGGCCCCTAGCGCGCGACTCCT | i-motif | — | |
| KD81 | AGGAGTCGCGCGCTAGGGGCCGGGGC | G4 | PARALLEL | +++ |
| KD83 | AGGCCGGAGGGTGGGCGGCGCGCGG | G4 | PARALLEL | + |
| SV23 | GGGTTAGGGTTAGGGTTAGGG | G4 | ANTIPARALLEL | — |
| SV24 | CCCTAACCCTAACCCTAACCC | i-motif | — | — |
| SV25 | GGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGG | G4 | ANTIPARALLEL | — |
| SV26 | CCCTAACCCTAACCCTAACCCTAACCCTAACCC | i-motif | — | — |
| SV27 | GGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGG | G4 | ANTIPARALLEL | — |
| SV28 | CCCTAACCCTAACCCTAACCCTAACCCTAACCCTAACCCTAACCC | i-motif | — | — |
| SMJ50 | TCGGGTTGCGGGCGCAGGGCACGGGCG | G4 | PARALLEL | — |
| SMJ51 | GTGGGGTTAGGGGTTAGGGGTTAGGGG | G4 | HYBRID | — |
| SMJ52 | TTGGGGCCGGGGCCGGGGCCGGGGTT | G4 | HYBRID | — |
| SMJ53 | GTAGGGTTAGGGTTAGGGTTAGGG | G4 | HYBRID | — |
| SMJ56 | TGGGTTTGGGTTTGGGTTTGGGT | G4 | HYBRID | — |
| SMJ58 | GGGGAGTAAAAGGGAGCGGGGTGCTGGG | G4 | HYBRID | — |
| SMJ59 | GGGCCTTGTGGGCCTTGTGGGCCTTGTGGG | G4 | ANTIPARALLEL | — |
| SMJ61 | GGGGTTTTGGGGTTTTGGGGTTTTGGGG | G4 | HYBRID | — |
| AKN11 | GACCTGAGGGCGAGCTTTTTTCGAGTAACTTAACAG | NA | — | — |
| AKN18 | CTGTTAAGTTACTCGAAAAAAGCTCGCCCTCAGGTC | NA | — | — |
Competition experiments confirmed BG4 specificity towards G4 structures. Further assessment of the interaction using BLI showed robust binding (Kd= 17.4 ± 0.588 nM). This was comparable to a previous study (Kd = 0.5–1.6 nM, and 2.0 nM for intramolecular and intermolecular DNA G-quadruplexes, respectively using ELISA).3
While we observed that oligomers that can fold into parallel G-quadruplexes bound efficiently to BG4, their respective double-stranded DNA counterparts did not bind with BG4 at all, which is in agreement with the observation that BG4 does not bind to dsDNA (Fig. 3C and D and Table 1). More importantly, we report that mere presence of G4 motifs within the double-stranded DNA sequence is not sufficient for recognition by BG4. This observation holds considerable significance in vivo, as it is possible that several G4 motifs, may not be in G4 conformation at a given time, when present in the genome.
Parallel, inter- and intramolecular structures show robust BG4 binding in our studies (Table 1). In a different study, it was observed that even a parallel G4 with a hairpin bulge was recognized by the BG4 antibody.29 Although previous studies suggest that BG4 may not have binding preference for any particular G4 DNA tested,3 in the panel of substrates investigated in the current study, we were unable to detect any BG4 binding to antiparallel and hybrid G-quadruplexes (Table 1). We also observed that BG4 can bind to GNG G4 structures. This finding is of interest as it suggests that the antibody could help identify structures that are not predicted by software, such as Non-B DNA Database and QGRS Mapper that use the classical empirical G4 formula.
Interestingly, we observed that the G4 structures of telomeric region in plasmid context can cause a shift in plasmid mobility, when electrophoresed on an agarose gel. However, such a shift was absent when the telomeric region was replaced with a random sequence in the same plasmid backbone, suggesting that formation of G4 complexes within plasmid DNA was responsible for BG4 binding and hence the mobility shift. Interestingly, in case of the supercoiled forms, the negative supercoiling and torsional stress promotes persistence of G4 in the telomeric repeats, and this form shifted significantly in presence of increasing amounts of BG4. This is understandable as G4 DNA formation requires energy for unwinding/melting of the DNA and hence such structures cannot be present on a nicked relaxed DNA.
Using cell lines derived from different lineages, we observed significant number of BG4 foci within the cells suggesting the formation of G4 DNA (Fig. 10). This was consistent with previous studies.3 Within the genome, G4 structures tend to form in conditions that cause single-strandedness in template or non-template strand. Thus, presumably, to a higher extent these would be intramolecular G4, though the occurrence of intermolecular G4 structures cannot be completely ruled out.60,61 These G4 structures act as epitopes for the antibody. In addition to the chromosomal G4 structures, RNA molecules have also been demonstrated to form G4 structures, which could additionally be potential epitopes for BG4 antibody.56,62 Helicases, a large family of ATP-dependent nucleic acid resolvases, play a critical role in G4 DNA formation and subsequent resolution within cells. Loss-of-function mutations in any distinct family of DNA resolvases are linked to various cancers and genetic disorders, thus linking G4 sequences and genome instability.63,64 It is emerging that G-quadruplex helicases play important roles in DNA replication and telomere maintenance.55,64 Knockdown of one of the helicase family members, WRN, revealed a significant increase in G4 structures, as determined from the foci counts. This observation is consistent with a recent study showing the role of WRN helicase in modulating gene expression in a discrete subpopulation of G4 DNA structures at many chromosomal sites. This population differs from that of Bloom syndrome RECQ helicase.65 Inflicting stress on cells can alter the gene expression pattern in many ways. Along these lines, serum starvation of the cells showed more localization of the antibody in the cytoplasm, which was over and above those in the nuclei, suggesting higher levels of G-quadruplexes, possibly of RNA origin, within the cytoplasm. Implications of this observation needs to be investigated further. It would be of interest to study how G-quadruplex structures regulate processes like transcription, replication, and how such antibodies could aid their presence within the cell. Additionally, it would be useful if such antibodies could be harnessed as tags for determining the existence of G4 within certain areas of interest in the genome, rendering them as probable diagnostic tools, and more ambitiously, as therapeutic agents.
Supplementary data
Supplementary data are available at DNARES online.
Supplementary Material
Acknowledgements
We thank S. Vartak and H. Siddiqua for help in cell culture, for technical guidance in immunofluorescence experiments, constructing the plasmid, pSV4 and critical reading of the manuscript. We thank M. Nambiar, V. Gopalakrishnan, S. Dahal and members of SCR laboratory for discussions and comments on the manuscript. We thank Biolayer Interferometry facility, IISc and S. Vaidya, Pall Life Sciences, for help in BLI data analyses.
Funding
This work was supported by grants from Department of Biotechnology, New Delhi [Grant No. BT/PR13616/GET/119/9/2015], Department of Atomic Energy [Grant No. 21/01/2016-BRNS/35068] to S.C.R. and financial assistance from IISc-DBT partnership programme [DBT/BF/PR/INS/2011-12/IISc]. S.M.J., N.M.N. and A.P. were supported by Senior Research fellowship from IISc, Bangalore, India.
Conflict of interest
None declared.
Author contributions
S.C.R. conceived and coordinated the study, S.C.R., S.M.J. and N.M.N. designed experiments; S.M.J., N.M.N., A.P. and K.D. performed experiments, S.C.R. and S.M.J. interpreted the data and wrote the manuscript.
References
- 1. Watson J.D., Crick F.H. 1953, Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid, Nature, 171, 737–8. [DOI] [PubMed] [Google Scholar]
- 2. Tsai Z.T., Chu W.Y., Cheng J.H., Tsai H.K. 2014, Associations between intronic non-B DNA structures and exon skipping, Nucleic Acids Res., 42, 739–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biffi G., Tannahill D., McCafferty J., Balasubramanian S. 2013, Quantitative visualization of DNA G-quadruplex structures in human cells, Nature Chem., 5, 182–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eddy J., Maizels N. 2008, Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes, Nucleic Acids Res., 36, 1321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huppert J.L., Balasubramanian S. 2007, G-quadruplexes in promoters throughout the human genome, Nucleic Acids Res., 35, 406–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Du Z., Zhao Y., Li N. 2009, Genome-wide colonization of gene regulatory elements by G4 DNA motifs, Nucleic Acids Res., 37, 6784–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lipps H.J., Gruissem W., Prescott D.M. 1982, Higher order DNA structure in macronuclear chromatin of the hypotrichous ciliate Oxytricha nova, Proc. Natl. Acad. Sci. U.S.A., 79, 2495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sundquist W.I., Klug A. 1989, Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops, Nature, 342, 825–9. [DOI] [PubMed] [Google Scholar]
- 9. Henderson E.R., Blackburn E.H. 1989, An overhanging 3′ terminus is a conserved feature of telomeres, Mol. Cell. Biol., 9, 345–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schroth G.P., Chou P.J., Ho P.S. 1992, Mapping Z-DNA in the human genome. Computer-aided mapping reveals a nonrandom distribution of potential Z-DNA-forming sequences in human genes, J. Biol. Chem., 267, 11846–55. [PubMed] [Google Scholar]
- 11. Strawbridge E.M., Benson G., Gelfand Y., Benham C.J. 2010, The distribution of inverted repeat sequences in the Saccharomyces cerevisiae genome, Curr. Genet., 56, 321–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Katapadi V.K., Nambiar M., Raghavan S.C. 2012, Potential G-quadruplex formation at breakpoint regions of chromosomal translocations in cancer may explain their fragility, Genomics, 100, 72–80. [DOI] [PubMed] [Google Scholar]
- 13. Sinclair P.B., Parker H., An Q., et al. 2011, Analysis of a breakpoint cluster reveals insight into the mechanism of intrachromosomal amplification in a lymphoid malignancy, Hum. Mol. Genet., 20, 2591–602. [DOI] [PubMed] [Google Scholar]
- 14. Inagaki H., Ohye T., Kogo H., et al. 2013, Two sequential cleavage reactions on cruciform DNA structures cause palindrome-mediated chromosomal translocations, Nat. Commun., 4, 1592. [DOI] [PubMed] [Google Scholar]
- 15. Nambiar M., Srivastava M., Gopalakrishnan V., Sankaran S.K., Raghavan S.C. 2013, G-quadruplex structures formed at the HOX11 breakpoint region contribute to its fragility during t(10; 14) translocation in T-cell leukemia, Mol. Cell Biol., 33, 4266–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bacolla A., Jaworski A., Connors T.D., Wells R.D. 2001, Pkd1 unusual DNA conformations are recognized by nucleotide excision repair, J. Biol. Chem., 276, 18597–604. [DOI] [PubMed] [Google Scholar]
- 17. Raghavan S.C., Swanson P.C., Wu X., Hsieh C.L., Lieber M.R. 2004, A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex, Nature, 428, 88–93. [DOI] [PubMed] [Google Scholar]
- 18. Nambiar M., Goldsmith G., Moorthy B.T., et al. 2011, Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14; 18) translocation in follicular lymphoma, Nucleic Acids Res., 39, 936–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kumari N., Vartak S.V., Dahal S., et al. 2019, G-quadruplex structures contribute to differential radiosensitivity of the human genome, iScience, 21, 288–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gopalakrishnan V., Raghavan S.C. 2012, Sequence and structural basis for chromosomal fragility during translocations in cancer, Future Oncol., 8, 1121–34. [DOI] [PubMed] [Google Scholar]
- 21. Sinden R.R. 1994, DNA Structure and Function. Academic Press: San Diego. [Google Scholar]
- 22. Huppert J.L. 2008, Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes, Chem. Soc. Rev., 37, 1375–84. [DOI] [PubMed] [Google Scholar]
- 23. Davis J.T. 2004, G-quartets 40 years later: from 5'-GMP to molecular biology and supramolecular chemistry, Angew. Chem. Int. Ed., 43, 668–98. [DOI] [PubMed] [Google Scholar]
- 24. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. 2006, Quadruplex DNA: sequence, topology and structure, Nucleic Acids Res., 34, 5402–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mukundan V.T., Phan A.T. 2013, Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences, J. Am. Chem. Soc., 135, 5017–28. [DOI] [PubMed] [Google Scholar]
- 26. Chung W.J., Heddi B., Schmitt E., Lim K.W., Mechulam Y., Phan A.T. 2015, Structure of a left-handed DNA G-quadruplex, Proc. Natl. Acad. Sci. U.S.A., 112, 2729–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huppert J.L., Balasubramanian S. 2005, Prevalence of quadruplexes in the human genome, Nucleic Acids Res., 33, 2908–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Konig S.L., Evans A.C., Huppert J.L. 2010, Seven essential questions on G-quadruplexes, Biomo. Concepts, 1, 197–213. [DOI] [PubMed] [Google Scholar]
- 29. Das K., Srivastava M., Raghavan S.C. 2016, GNG motifs can replace a GGG stretch during G-quadruplex formation in a context dependent manner, PLoS One, 11, e0158794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S. 2015, High-throughput sequencing of DNA G-quadruplex structures in the human genome, Nat. Biotechnol., 33, 877–81. [DOI] [PubMed] [Google Scholar]
- 31. Schiavone D., Guilbaud G., Murat P., et al. 2014, Determinants of G quadruplex-induced epigenetic instability in REV1-deficient cells, Embo J., 33, 2507–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miyoshi D., Karimata H., Sugimoto N. 2006, Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions, J. Am. Chem. Soc., 128, 7957–63. [DOI] [PubMed] [Google Scholar]
- 33. Lipps H.J., Rhodes D. 2009, G-quadruplex structures: in vivo evidence and function, Trends Cell Biol., 19, 414–22. [DOI] [PubMed] [Google Scholar]
- 34. Neidle S., Parkinson G. 2002, Telomere maintenance as a target for anticancer drug discovery, Nat. Rev. Drug Discov., 1, 383–93. [DOI] [PubMed] [Google Scholar]
- 35. Zahler A.M., Williamson J.R., Cech T.R., Prescott D.M. 1991, Inhibition of telomerase by G-quartet DNA structures, Nature, 350, 718–20. [DOI] [PubMed] [Google Scholar]
- 36. Besnard E., Babled A., Lapasset L., et al. 2012, Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs, Nat. Struct. Mol. Biol., 19, 837–44. [DOI] [PubMed] [Google Scholar]
- 37. Clark G.R., Pytel P.D., Squire C.J. 2012, The high-resolution crystal structure of a parallel intermolecular DNA G-4 quadruplex/drug complex employing syn glycosyl linkages, Nucleic Acids Res., 40, 5731–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Granotier C., Pennarun G., Riou L., et al. 2005, Preferential binding of a G-quadruplex ligand to human chromosome ends, Nucleic Acids Res., 33, 4182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Derenzini E., Agostinelli C., Imbrogno E., et al. 2015, Constitutive activation of the DNA damage response pathway as a novel therapeutic target in diffuse large B-cell lymphoma, Oncotarget, 6, 6553–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H.J., Pluckthun A. 2001, In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei, Proc. Natl. Acad. Sci. U.S.A., 98, 8572–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fernando H., Sewitz S., Darot J., Tavare S., Huppert J.L., Balasubramanian S. 2009, Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression, Nucleic Acids Res., 37, 6716–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Henderson A., Wu Y., Huang Y.C., et al. 2014, Detection of G-quadruplex DNA in mammalian cells, Nucleic Acids Res., 42, 860–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lam E.Y., Beraldi D., Tannahill D., Balasubramanian S. 2013, G-quadruplex structures are stable and detectable in human genomic DNA, Nat. Commun., 4, 1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang M., Cai E., Fujiwara N., et al. 2017, Odorant sensory input modulates DNA secondary structure formation and heterogeneous ribonucleoprotein recruitment on the tyrosine hydroxylase and glutamic acid decarboxylase 1 promoters in the olfactory bulb, J. Neurosci., 37, 4778–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tateishi-Karimata H., Kawauchi K., Sugimoto N. 2018, Destabilization of DNA G-quadruplexes by chemical environment changes during tumor progression facilitates transcription, J. Am. Chem. Soc., 140, 642–51. [DOI] [PubMed] [Google Scholar]
- 46. Hansel-Hertsch R., Beraldi D., Lensing S.V., et al. 2016, G-quadruplex structures mark human regulatory chromatin, Nat. Genet., 48, 1267–C. [DOI] [PubMed] [Google Scholar]
- 47. Di Antonio M., Ponjavic A., Radzevicius A., et al. 2020, Single-molecule visualization of DNA G-quadruplex formation in live cells, Nat Chem., 12, 832–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vartak S.V., Iyer D., Santhoshkumar T.R., et al. 2017, Novel BCL2 inhibitor, Disarib induces apoptosis by disruption of BCL2-BAK interaction, Biochem. Pharmacol., 131, 16–28. [DOI] [PubMed] [Google Scholar]
- 49. Nilavar N.M., Paranjape A.M., Raghavan S.C. 2020, Biochemical activity of RAGs is impeded by Dolutegravir, an HIV integrase inhibitor, Cell Death Discov., 6, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nishana M., Nilavar N.M., Kumari R., Pandey M., Raghavan S.C. 2017, HIV integrase inhibitor, Elvitegravir, impairs RAG functions and inhibits V(D)J recombination, Cell Death Dis., 8, e2852–e2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smargiasso N., Gabelica V., Damblon C., et al. 2009, Putative DNA G-quadruplex formation within the promoters of Plasmodium falciparum var genes, BMC Genomics, 10, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leroy J.L., Gueron M., Mergny J.L., Helene C. 1994, Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif, Nucl. Acids Res., 22, 1600–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Abou Assi H., Garavis M., Gonzalez C., Damha M.J. 2018, i-Motif DNA: structural features and significance to cell biology, Nucleic Acids Res., 46, 8038–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hanish J.P., Yanowitz J.L., de Lange T. 1994, Stringent sequence requirements for the formation of human telomeres, Proc. Natl. Acad. Sci. U.S.A., 91, 8861–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Crabbe L., Verdun R.E., Haggblom C.I., Karlseder J. 2004, Defective telomere lagging strand synthesis in cells lacking WRN helicase activity, Science, 306, 1951–3. [DOI] [PubMed] [Google Scholar]
- 56. Byrd A.K., Zybailov B.L., Maddukuri L., et al. 2016, Evidence that G-quadruplex DNA accumulates in the cytoplasm and participates in stress granule assembly in response to oxidative stress, J. Biol. Chem., 291, 18041–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu Y., Kaminaga K., Komiyama M. 2008, Human telomeric RNA in G-quadruplex structure, Nucleic Acids Symp. Ser., 52, 175–6. [DOI] [PubMed] [Google Scholar]
- 58. Hansel-Hertsch R., Spiegel J., Marsico G., Tannahill D., Balasubramanian S. 2018, Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing, Nat. Protoc., 13, 551–64. [DOI] [PubMed] [Google Scholar]
- 59. Rodriguez R., Miller K.M., Forment J.V., et al. 2012, Small-molecule-induced DNA damage identifies alternative DNA structures in human genes, Nat. Chem. Biol., 8, 301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mendoza O., Bourdoncle A., Boule J.B., Brosh R.M. Jr., Mergny J.L. 2016, G-quadruplexes and helicases, Nucleic Acids Res., 44, 1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Banuelos S., Lectez B., Taneva S.G., et al. 2013, Recognition of intermolecular G-quadruplexes by full length nucleophosmin. Effect of a leukaemia-associated mutation, FEBS Lett., 587, 2254–9. [DOI] [PubMed] [Google Scholar]
- 62. Cammas A., Millevoi S. 2017, RNA G-quadruplexes: emerging mechanisms in disease, Nucleic Acids Res., 45, 1584–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Johnson J.E., Cao K., Ryvkin P., Wang L.S., Johnson F.B. 2010, Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential, Nucleic Acids Res., 38, 1114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brosh R.M. Jr. 2013, DNA helicases involved in DNA repair and their roles in cancer, Nat. Rev. Cancer., 13, 542–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tang W., Robles A.I., Beyer R.P., et al. 2016, The Werner syndrome RECQ helicase targets G4 DNA in human cells to modulate transcription, Hum. Mol. Genet., 25, 2060–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kumari R., Nambiar M., Shanbagh S., Raghavan S.C. 2015, Detection of G-quadruplex DNA using primer extension as a tool, PLoS One, 10, e0119722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Javadekar S.M., Yadav R., Raghavan S.C. 2018, DNA structural basis for fragility at peak III of BCL2 major breakpoint region associated with t(14; 18) translocation, Biochim. Biophys. Acta, 1862, 649–59. [DOI] [PubMed] [Google Scholar]
- 68. Sharma S., Choudhary B., Raghavan S.C. 2011, Efficiency of nonhomologous DNA end joining varies among somatic tissues, despite similarity in mechanism, Cell. Mol. Life Sci., 68, 661–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kumar T.S., Kari V., Choudhary B., Nambiar M., Akila T.S., Raghavan S.C. 2010, Anti-apoptotic protein BCL2 down-regulates DNA end joining in cancer cells, J. Biol. Chem., 285, 32657–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dahal S., Dubey S., Raghavan S.C. 2018, Homologous recombination-mediated repair of DNA double-strand breaks operates in mammalian mitochondria, Cell. Mol. Life Sci., 75, 1641–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kavitha C.V., Nambiar M., Ananda Kumar C.S., et al. 2009, Novel derivatives of spirohydantoin induce growth inhibition followed by apoptosis in leukemia cells, Biochem. Pharmacol., 77, 348–63. [DOI] [PubMed] [Google Scholar]
- 72. Sebastian R., Raghavan S.C. 2016, Induction of DNA damage and erroneous repair can explain genomic instability caused by endosulfan, Carcinogenesis, 37, 929–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.