Graphical abstract
Keywords: Mutations, Cancer, Hotspots, Recurrent mutations, Algorithms, Genomics, Sequencing, Exome, Whole genome sequencing
Abstract
Cancer mutations that are recurrently observed among patients are known as hotspots. Hotspots are highly relevant because they are, presumably, likely functional. Known hotspots in BRAF, PIK3CA, TP53, KRAS, IDH1 support this idea. However, hundreds of hotspots have never been validated experimentally. The detection of hotspots nevertheless is challenging because background mutations obscure their statistical and computational identification. Although several algorithms have been applied to identify hotspots, they have not been reviewed before. Thus, in this mini-review, we summarize more than 40 computational methods applied to detect cancer hotspots in coding and non-coding DNA. We first organize the methods in cluster-based, 3D, position-specific, and miscellaneous to provide a general overview. Then, we describe their embed procedures, implementations, variations, and differences. Finally, we discuss some advantages, provide some ideas for future developments, and mention opportunities such as application to viral integrations, translocations, and epigenetics.
1. Introduction
Mutations accumulate during tumor progression [1]. The number of mutations is highly heterogeneous, from a handful to thousands, even within the same tumor type [2]. This diversity leads to thinking that most observed mutations seem to appear due to random chance, known as passenger mutations [3], [4], and few due to positive selection, also referred to as driver mutations [4], [5], [6].
Many computational methods have been proposed to estimate which genes in cancer are under positive selection [7], [8], [9]. This determination is essential to characterize cancer behavior, the mechanisms of action, and, ultimately, to propose treatments [10], [11]. These gene-based methods focus on determining whether the number of observed mutations in a gene is higher than the number of mutations expected by random chance [2]. Nevertheless, the presence of random mutations obscures the estimation of those that yield a genuine functional impact. In this context, the hotspot mutations, which are specific mutations recurrently observed in different patients, are thought to be functionally important [12]. This is based on the unlikely expectation of observing recurrent mutations (when the number of patients is not so high or above expectations). Well-known hotspots support this idea, e.g., BRAF V600E in many cancer types [13], [14], [15], [16], [17], [18], IDH1 R132H in gliomas [10], KRAS G12/13 in lung cancer [19], or NRAS Q61 in melanomas [20].
International cancer sequencing projects have provided an increasing number of samples and tumor mutations [21], [22], whose aggregated data help to identify novel hotspots. Nevertheless, apparent non-functional hotspots also emerged, such as those generated by the APOBEC enzymes [23] or those observed in highly suspicious genes [2], generating the necessity for computational methods that identify functional hotspots.
Many methods have been proposed to identify likely functional hotspots. These tend to conceptually vary in aspects such as the region, coding or non-coding, the definition of a hotspot, point mutations or clustered in a small region, the databases used, the statistical distributions, and more. Despite a plethora of methods, to our knowledge, no efforts have been devoted to compare, summarize, and discuss the algorithms of this critical topic.
In this mini-review, we summarize more than 40 published approaches to detect cancer hotspots. First, we provide a conceptual summary classifying the methods in 4 groups. Then, we describe and compare each group of approaches. Finally, we discuss issues and future directions.
2. Cancer hotspots methods
To provide a comprehensive and updated view of hotspot methods, we searched literature in PubMed and the Internet using keywords related to “cancer”, “hotspots”, and “algorithms”. In particular, we used 3 main PubMed queries. (1) mutation*[TI] AND cancer[TIAB] AND (hotspot*[TI] OR recurren*[TI] OR positio*[TI] OR patter*[TI] OR clust*[TI] OR struct*[TI] OR 3D[TI]) AND (method[TIAB] OR algorithm*[TIAB] OR model*[TIAB]). (2) (hotspot*[TI] OR (recurren*[TI] AND mutat*[TI])) AND (cancer[TIAB]) AND (computa*[TIAB] OR algori*[TIAB]). (3) (hotspot*[TI] OR driver*[TI] OR ((recurren*[TI] OR clust*[TI]) AND (mutat*[TI] OR varian*[TI]))) AND (cancer[TIAB]) AND (computa*[TIAB] OR algori*[TIAB]). We curated the candidate list of 350 papers plus other known papers not listed. Finally, we revised 44 publications. The list is provided as a supplementary material.
3. Overview
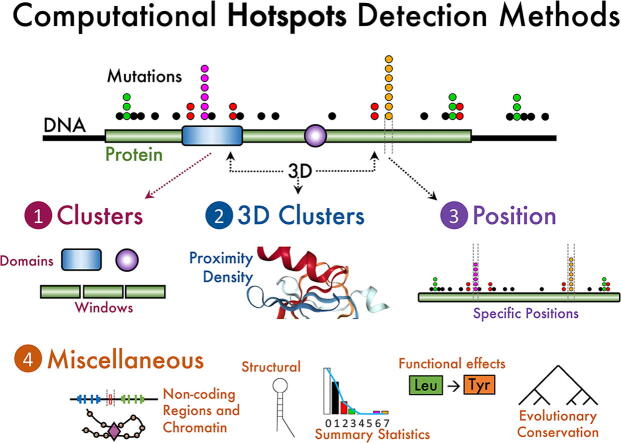
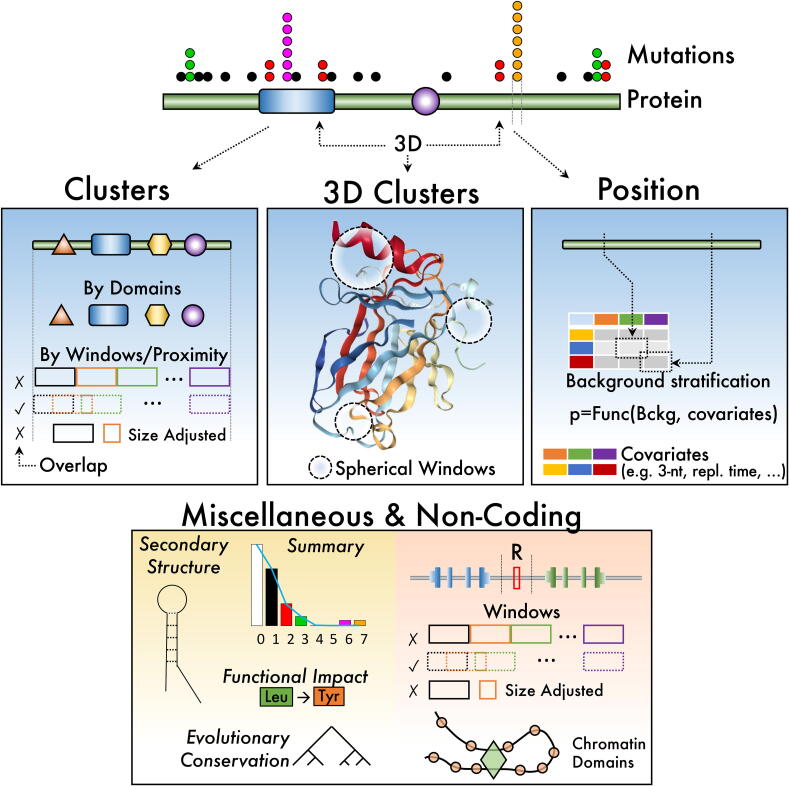
To provide a useful comparison, we first classified the algorithms in four types (Fig. 1). First, approaches considering a hotspot as a cluster of mutations within a small peptide or DNA region. Second, methods clustering mutations applied to 3D coordinates where amino acids can be distant within the protein sequence but close in 3D structure. Third, algorithms designed to compare mutations in specific amino acids or DNA positions. Fourth, procedures of other diverse origins or concepts applied to coding sequences. Finally, and except by the 3D protein coordinates, the approaches can also be applied to non-coding sequences plus some adaptations and considerations, which are referred to within the above four types. In the following sections, we describe the four types of algorithms and mention the differences applied to non-coding when needed.
Fig. 1.
Conceptual classification of hotspot methods reviewed. Clusters methods focus on aggregating mutations by local regions in residue coordinates while 3D Clusters approaches agglomerate mutations in 3D coordinates. Position methods consider the specific mutation position and, generally, its sequence context. Other methods are described as miscellaneous that include those approaches focusing on evolutionary conservation, functional impact of mutations, structural features, overall statistics of mutations, and other concepts impacting non-coding mutations such as chromatin domains.
4. Cluster-based methods
Cluster-based methods are perhaps the most frequent algorithms to detect mutation hotspots. These methods find hotspots mainly by grouping mutations within certain regions considering the frequency of mutations and other relevant biological features. The Fig. 2 provides an overview of the most common steps characterizing cluster-based methods, which is not exhaustive but sufficient to settle a general process.
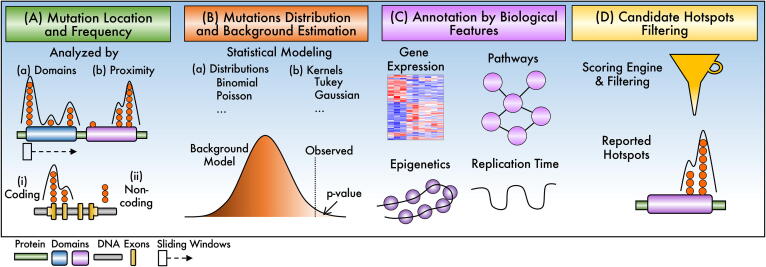
Fig. 2.
Overview of the steps characterizing clustering-based methods. (A) Location is used to count mutations by protein domains or proximity in both coding and non-coding regions. (B) A statistical model is used to estimate the probability of observing the number of mutations by random chance. (C) Highly unlikely mutations counts are further annotated or analyzed. (D) Filtering by annotation scores finally determine report hotspots.
Clustering Mutations by Location. Candidate hotspots are identified as mutations clustered by domain or by proximity (Fig. 2A). The domain criterion is stated as if mutations belong to the same protein domain, whose definition may vary depending on the database used [24], or into the same non-coding DNA motif such as transcription factor motifs [25]. Algorithms developed by Baeissa et al. [26], Jia et al. [27], and Porta-Pardo and Godzik [28] exemplify protein domains-based methods and those by Melton et al. [25], Kim et al. [29], and Juul et al. [30] for non-coding motifs. Proximity is defined as the distance measured in codons or bases and whether it is applied to coding or non-coding regions. Then, mutation clusters are regularly discovered using sliding, dynamic [31], [32], or fixed [33], [34] window algorithms, counting the mutations covered, and moving the window to a contiguous position [35]. The counting can be even [36] or weighted, for example, giving a higher score to positions having more mutations [37]. Sometimes, only “seed” positions might be considered, for example, those above a threshold [37]. The length of the window can be fixed [34], [37] or varied [35] and could also be applied to non-coding regions [34].
Identifying candidate hotspots above background expectations. Once a mutation cluster is proposed as a candidate hotspot, several algorithms model the number of mutations within clusters according to a statistical distribution or kernel-based estimations to calculate a probability above chance (Fig. 2B). As can be noticed, clustering-based algorithms use different probability distributions to model mutation clusters. The choice of the distribution appears to be related to the biological assumptions considered, such as mutation proximity [37] or domain affinity [26]. Logically, discrete event distributions as Poisson [38] or members of the binomial family (e.g., beta-binomial) are utilized [36], [37], [39]. Afterward, a p-value is calculated from a background distribution. These distributions can be generated by randomizing sequences [36], [40], by calculating the probability of occurrence for all possible candidate hotspots in order to provide enough measurements to create a distribution, or by using controls, such as silent mutations [37]. Other algorithms employ kernel-based estimations to estimate the probability of mutation clusters. The kernel density estimate function is used to calculate the probability density of mutation clusters [41]. Algorithms used different types of kernels, such as a Tukey [40] or Gaussian [42].
Candidate Hotspot Annotations. Besides the p-value, some algorithms incorporate biological features to enhance candidate hotspots evaluation (Fig. 2C). Among biological features, expression [38], [42], pathways [42], replication time [43], epigenetic context [38], sequence context [44], and others [2] are included. The importance of annotation is central in some algorithms. For instance, the works from Van den Eynden et al. [45] and Baeissa et al. [26], assume that mutation distribution depends on whether the gene is an oncogene or a tumor-suppressor gene (TSG). On the other hand, Jia et al. designed an algorithm inspired in GSEA, a well-known gene expression rank-based enrichment method, by calculating a mutation accumulation score (MAS), which depends on the proximity of mutations [27]. In addition, Araya and colleagues reported an algorithm that considers context-specific variables such as number and locations of mutations, gene expression, replication time, GC content, among others [43]. As it is observed, algorithms to detect mutation hotspots evolved their mutation cluster models from counting mutations to consider biological features as context variables.
Scoring and Filtering. Finally, an engine estimates a score for candidate hotspots (Fig. 2D). The score is used to rank candidate hotspots and select the most prominent.
For example, the OncodriveClust algorithm calculates a score based on the fraction of mutations composing the cluster or hotspot and the maximal distance within these mutations [37]. The score is used to select candidate hotspot whose score is higher than those from a background distribution of random sequences. On the other hand, Poole et al. score candidate hotspots with the logarithm of the ratio of the probability of occurring the hotspot and the probability of having a similar hotspot under the assumption of uniform distribution of mutations [42]. A filter can then be applied to select the most promising hotspots using a score scheme.
Coding and Non-Coding detection. Another distinction among clustering-based algorithms is the genome location, denoting finding hotspots in coding or non-coding regions. Above, we reviewed algorithms focusing on coding regions, nevertheless some algorithms center in non-coding regions [25], [29], [30], [31], [33], [34], [38], [39], [46]. To detect non-coding regions, algorithms need genomic annotation data such as mapping of enhancers-promoters, TF binding motifs, differential expression, protein–protein interactions, genetic and epigenetic features, and perhaps others [38]. These annotation procedures underscore the disadvantage of finding non-coding mutation hotspots. Moreover, non-coding detection is only feasible when mutations from whole-genome sequencing (WGS) data are available [43], [47], which are by far more scarce, but efforts and databases are available such as PCAWG [6] (https://dcc.icgc.org/pcawg).
To our knowledge, most of the methods for detecting hotspots have been developed using mutation data from cancer samples. Consequently, the majority of the studies reviewed exploited data from The Cancer Genome Atlas (TCGA) [21], COSMIC [9], and ICGC [22].
5. Hotspots as mutated 3D clusters
Finding hotspots directly in sequence, which is viewed in 1D, is limited by the fact that proteins tend to fold into three-dimensional structures. Therefore, positional clustering done in 1D will omit several 3D clusters after folding [48]. The increase in protein 3D structural data has led to the development of methods for the identification of cancer hotspots. Most of these methods have three fundamental steps (Fig. 3), which will be described next.
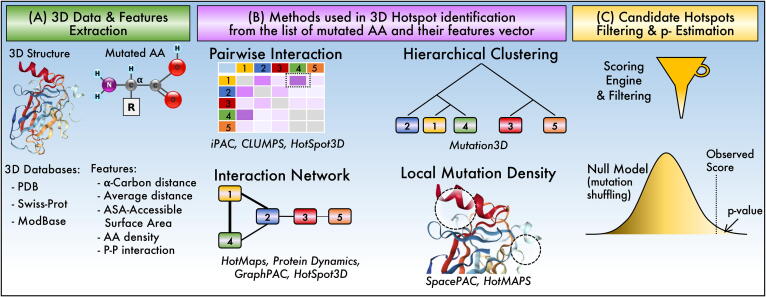
Fig. 3.
Summary of the procedure and methods to identify hotspots in 3D structures. (A) 3D residue or atom coordinates from databases are used to estimate proximity or interaction between mutated residues as shown in (B). (C) A scoring scheme is utilized to estimate statistical significance, generally calculated by shuffling sequence to estimate the null distribution of scores.
Obtaining mutational and 3D structural data. Only missense mutations registered as confirmed somatic variants are used in the reviewed methods; the most common primary sources are TCGA [21] and ICGC [22]. Some of the review methods also use other repositories to analyze mutation data, such as COSMIC [9], Human Gene Mutation [49], cBioPortal [50], or ENSEMBL [51]. Often, this step requires manual data curation for specific set genes or mutations [52], [53], [54].
The 3D structure information is needed to estimate neighborhood atoms or amino acids. The 3D structure is obtained primarily from the protein data bank (RCSB PDB) or from repositories, including variants, such as ModBase [55], Swiss-Model [56], mutation3D [57] and DrugPort [52] (Fig. 3A). Mainly, native, non-mutated proteins are used, then mutations are identified in the backbone of these structures. In exceptional cases, a 3D structure with a specific mutation is available (such as in TP53, and PIK3CA). This is based on the fact that observed mutations from genomics data are always ahead of protein structure where a mutant should be generated, crystalized, and analyzed. Commonly, the 3D structure of the protein is represented by the coordinates of all α-carbon atoms [58], [59], [60]. Some criteria must be applied when there are several structures of the same protein. Cristal conditions, solvents, included chains, and interacting partners are reviewed to choose the most appropriate.
Feature extraction. After choosing a 3D structure and its point mutations, some metrics are extracted (Fig. 3B). These features can be represented in the form of n-dimensional vectors. The most common feature is the distance between the 3D positions of the α-carbon belonging to the mutated residues [57], [58], [59], [60]. Besides, the average distance of all atoms is calculated as a centroid of the amino acid [48], [54], [56].
Some methods use 3D structural information to include features such as the Accessible Surface Area (ASA) [53], or the spatial proximity between any of the atoms of the molecular structure [52], [61] (Fig. 3A). Non-positional features, such as the density of mutated residues in local regions, are often used to represent possible underlying functionalities within a protein or in protein–protein interactions [48], [53], [60], [62].
Identification of significant 3D hotspots. The algorithms that make up the methods of this review can be classified into four sub-categories (Fig. 3B). Several methods combine these algorithms for more accurately hotspots identification.
Pairwise interaction. In algorithms of this type, the interaction between each pair of mutated amino acids is scored with respect to a set of rules applied to their feature vectors. For example, HotSpot3D [52], iPAC [58], iGraph [59], and CLUMPS [60] calculate scores from the Euclidian distances between each pair of mutated amino acids d(AAi, AAj). In iGraph and iPAC, the 3D structure is mapped to a 1D space to ease the identification of the clusters. These algorithms identified hotspots in EGFR, EIF2AK2, and HAO1 proteins not detected by non-3D methods. However, there is an inherent loss of information by reducing dimensionality, which is overcome by other algorithms. For example, SpacePAC [60] identified significant clusters in FGFR3 and CHRM2 missed by iPAC due to the remapping of the structure. Significant groups with rare mutations in the RAC1 and MAP2K1 proteins were also detected.
Local mutation density. Mutated residues in local 3D regions might form high-density clusters, commonly delimited by spheres, such as in SpacePAC, or in a proximity range such as the first step of HotMaps [48] and 3D Hotspot [61]. SpacePAC scores each sphere according to the number of mutations covered. HotMaps uses mutational diversity for the main score, which is based on Shannon entropy of the joint probability of a missense mutation occurring at a specific residue and having a specific mutant amino acid. Discoveries include relevant regions in CTNNB1, FGFR3, and FSHR proteins that have been implicated with cancer [48,60].
Interaction Networks. The interactions between the mutated residues can be informative to later identify the hotspots. Associative networks can estimate these interactions. Generally, the vertices of these graphs are vectors of features from the mutated amino acids, while the edges indicate the 3D distances. HotMaps and Hotspot3D [62] are examples of this concept. One of the advantages of interaction networks is the ability to form subnets whose interactions have higher biological significance. This allows, for example, the identification of novel and low-frequency putative driver genes with hotspot communities [63]. Another example is seen in HotMaps, where hotspots associated with oncogenes or tumor suppressor genes were detected in 30 cases due to the physicochemical properties of amino acids. There, regions containing oncogenic clusters are smaller, have less mutational diversity, and have greater solvent accessibility than those in tumor suppressors [48]. Hotspot3D uses graphs to prioritize clusters with a high degree of closeness and that are significantly enriched in mutations from multiple patient samples [52]. For instance, a cluster including A289, R108, and R222 in EGFR is enriched in lower-grade glioma and glioblastoma while a cluster including L858 and L861 is enriched in lung adenocarcinoma. Thus the feature vector helps to reveal enriched characteristics.
Hierarchical clustering (HC). The classical HC algorithm agglomerate elements by calculating a distance matrix among all elements from the feature vector to progressively cluster those elements that are close. For mutated residues, Mutation3D uses an HC algorithm to encapsulate all α-carbons that are within a given specified linkage distance [57]. This algorithm identifies known hotspots with high precision, such as TP53, KRAS, and PIK3CA, among others. However, the opposite occurs in types of cancer that have very high mutation rates, such as melanomas where no mutation hotspots were found, presumably due to high randomness that obscure clusters.
5.1. Candidate hotspot filtering and statistical estimation
In general, the significance of clustered distances is tested against the null hypothesis that amino acid mutations are distributed evenly throughout the polypeptide (Fig. 3C). The conventional procedure for this is to shuffle the mutations along the protein to estimate the distribution under the no clustering assumption [48], [52], [54], [57], [60], [61]. Some of these methods use scoring schemes (that depends on the cluster distance) or normalized scores rather than the distance alone. For filtering or prioritization, for instance, HotSpot3D sum closeness centrality (CC) of variants within the cluster and show that CC is higher among cancer-related genes than to other genes [52].
6. Hotspots as specific residue positions
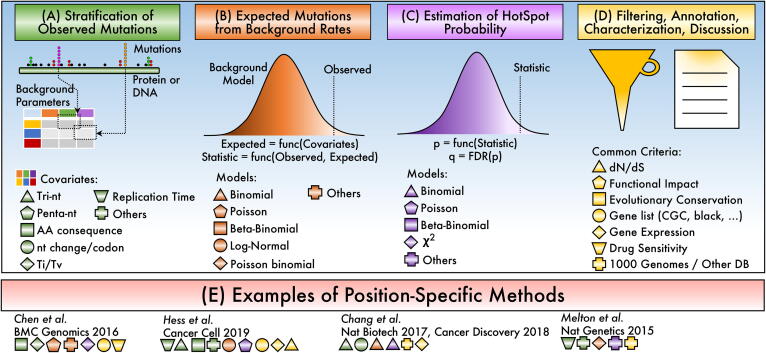
Specific residue positions focus on nucleotides or amino acids rather than defining regions. This avoids diluting the count of mutations along the region (cluster or window), facilitating the detection of recurrent positions. The assumption is that the residue rather than the region is subject to selection. This may be true for key residues in enzymatic reactions [64], alterations in the 3D structure, post-translational modifications [65], or degradation [66]. Details of the generic process shown in Fig. 4 are described below.
Fig. 4.
Schema of the steps to identify hotspot modeled as position specific. (A) Mutations are first contextualized relative to covariates. (B) Context is used to estimate specific background probabilities and a statistic representing the observed number of mutations. (C) The statistic is used to estimate a probability value. (D) The p-value and their annotation is considered to report significant hotspots. (E) Selected examples of methods with specific conceptual choices where symbols and colors represent implemented features.
Stratification of observed mutations. This procedure assumes that the probability of mutations is not uniform (Fig. 4A). The idea is based on the observation that the C residue is more prone to mutation [2] and highly influenced by the discovery of mutational signatures that were stratified by trinucleotide contexts [67]. Several criteria have been employed to stratify mutations. The most widely used are the tri-nucleotide (tri-nt) stratification and replication timing. Tri-nt assumes that the mutated nucleotide is different, and the flanking nucleotides provide a distinctive context. This strategy has been highly successful, generating more precise estimations [44], [68], [69]. The replication timing is based on the fact that there is a difference in mutations rates between early and late replicating regions of the genome [2], [44]. Other stratifications include pentanucleotide that include 2-nt in both sides from the mutated site [7], [44], gene expression levels [44], nucleosome positions or transcription factors binding sites [70], Ti/Tv (transitions/transversions) [68], nucleotide position within codon and amino acid [44], and consequence of mutation like missense, nonsense, stop codon, among others [44]. Commonly, one or more covariates are used for stratification. The result of the stratification is the estimation of more precise parameters for background estimations for the specific position being computed.
Estimation of expected mutations. The above stratification provides the parameters for background rate estimation, which can be different across the gene depending on covariates used. For example, if a binomial model is used for the estimation of expected mutations, the stratification provides the binomial parameter pk specific for the genomic position k (Fig. 4B). Different distributions can be employed depending on the assumptions, including Binomial [69], [71], Poisson [68], and Log-Normal [44] distributions. Other distributions could also be used such as the Beta-Binomial [46] and the Gamma [44], [72].
Estimating a hotspot probability. Most methods use a second procedure to estimate the probability of the hotspot given the expected and the observed mutations at the specific site (Fig. 4C). For example, Hess et al. use an advanced algorithmic estimation based on the Poisson distribution [44]. Chen et al. sum over all mutation-types probabilities using a Fisher method, then calculate the probability from a χ2 distribution [68]. Instead, Chang et al. truncate the binomial parameter of all position-specific p’s [69], [71]; thus, the expected mutations and the hotspot probability is done in the same step. The p-value is then corrected for multiple tests using, generally, by a false discovery rate (FDR) approach. Finally, significant hotspots are selected using a cut-off value.
Filtering, annotation, and characterization. The list of significant hotspots is usually filtered and accompanied by annotations or characterizations at the gene or hotspot level (Fig. 4D). For example, removing genes from a highly suspicious “blacklist” is common in which the list contains genes such as TTN because of its extremely large length, olfactory receptors due to association with late replication times, among other genes and gene families [2]. Other mutations included in common variation databases, such as 1000 genomes, are also removed mainly before the analysis to avoid germline contaminations [69]. It makes sense to annotate hotspots for common human variation, such as gnomAD [73]. Characterizations for drug responses are also interesting because it may highlight drug susceptibilities [68]. For example, some NRAS and KRAS hotspots are more susceptible to MEK inhibitors, while MAP3K4 hotspots are less sensitive to EGFR inhibitors [68]. Annotations for differential expressed genes [68], gene expression levels [69], functional impact [74], evolutionary conservation [34], or substitution rates dN/dS [75] are also common.
7. Other hotspots methods
Some methods may combine algorithms of the above categories, such as Melton et al., that combines cluster and position-specific methods [25] or apply similar concepts to non-coding or chromatin-domains [25], [29], [30], [31], [33], [34], [38], [39], [46]. Other methods that do not fit into the above categories are described next (Fig. 5). These methods are related to diverse functional effects [74], [76], to mesoscale DNA structures [23], and to deviations of summary statistics and are detailed below [75].
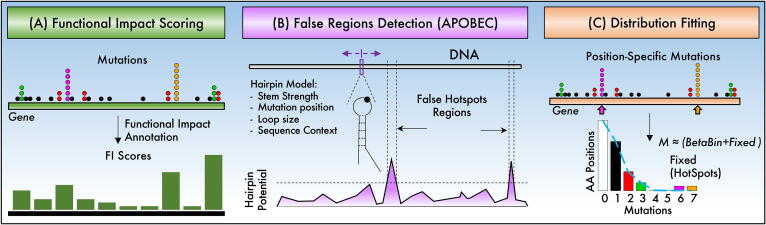
Fig. 5.
Other methods to detect hotspots or putatively false regions. (A) Functional impact methods quantify the effect of mutations in protein function, RNA structure, binding sites, or miRNA/lncRNA targets. (B) Estimation of structural features that increase the susceptibility of DNA mutations can be used to mark possible false regions. (C) Summary statistics, such as the distribution of mutated positions, can be used to detect positions that are over mutated.
The first method focuses on the possible functional impacts that mutations may incur on the protein function, RNA structure, binding sites, or miRNA/lncRNA targets [74], [76]. The estimation of the impact is achievable because, for protein-coding regions and promoters, estimations of functional effects are possible [77], and for 3′UTR and lncRNA, structural changes can be predicted [78]. Thus, the proposed methods highlight biases in the functional impact of observed mutations compared to the functional impact of random mutation in similar regions [74], [76]. The random mutations are used to estimate a p-value identifying hotspot in coding and non-coding regions. This can be similar to cluster-based methods with the additional concept of changing mutations counts by functional impact scores (Fig. 5A).
All the above methods focus on detecting positive hotspots. Nevertheless, another point of view is focusing on detecting regions that are potentially false functional, which may appear as hotspots [23]. Based on the fact that APOBEC enzymes are biases toward specific mutations, the proposed method by Buisson et al. sweep DNA detecting regions that form strong hairpins and are suitable for APOBEC activity suggesting that hotspots may not be due to positive selection but as a result of a mechanistic bias [23]. The estimation of the strength of the hairpin sweeps the DNA by a sliding window and considers the loop length, the position of the mutation within the loop, the size of the stem, and the sequence context (Fig. 5B). Loops of size 3 to 6 combined with mutation positions biased toward an end of the loop and located at sequences showing a strong stem (estimated by 3*GC + 1*AT) are more likely to be APOBEC spots [23]. Similar estimations have been included in HotSpotsAnnotations, a database of HotSpots, helping in recording hotspots close to APOBEC sensitive sites [79].
Instead of focusing on the domain, region, position, or local structure, a recent proposal focuses on the distribution of mutations per position of the whole gene, assuming they should fit relatively well a beta-binomial distribution [75]. Because most cancer genes do not follow a beta-binomial distribution well unless hotspots are removed, the method tries to remove candidate hotspots while beta-binomial fitting improves. High changes in fitting and a high number of either mutations or positions suggest genuine hotspots [75]. The beta-binomial fitting is then used to estimate a p-value of removed hotspots, then corrected by FDR.
8. Discussion
In this mini-review, we have revised most of the methods to detect mutational hotspots in cancer. We classified them into four main groups depending on the definition of a hotspot (Fig. 1) as Clusters, 3D Clusters, Position-Specific, and Miscellaneous.
By far, the most popular approach is the detection by clusters in the primary AA sequence followed by clusters in 3D. Clustering assumes mutation accumulation in local domains. This idea is a powerful concept proven to be highly successful. Nevertheless, clustering might not detect low- and mid- recurrent mutations in specific positions, which is the focus in position-specific methods. More robust methods can be designed if clusters may be size-adaptive up to the one-residue level. The miscellaneous can also be powerful and are less explored, presenting opportunities for development.
The large number of papers revised here demonstrates the enormous efforts made to detect hotspots. Little attention has been paid to the detection of false hotspots, such as those detected in the APOBEC mechanisms [23]. One way to avoid detecting false hotspots is by increasing the specificity of statistical models and algorithms, but these come at the risk of detecting less genuine hotspots [44]. Moreover, one of the problems to declare false hotspots is that they should have a precise biochemical mechanism behind it. Thus, novel algorithms focusing on mechanistic models, such as the proven APOBEC, are encouraged.
Most of the articles revised here commonly use the whole dataset (cancer or some cancer types). This practice intrinsically assumes that all samples have comparable underlying mutational processes. Moreover, it is well known that cancer subtypes within a specific cancer type may behave differently [2]. Thus, a question remains whether more hotspots are detected when applying the revised methods to biological-plausible subsets of cancer samples. Furthermore, it may also be possible that hotspots are specific for particular subsets of samples, which needs to be assessed. These analyses could be a direction for future studies.
Another question regarding mutation hotspots are their possible effects, for example, their relation to clinical variables or other genomic information. It is unknown whether there are hotspots that confer larger or shorter patient survival; that is, whether hotspots confer higher or lower risk where patient consequences differ compared to other mutations along the gene. It is also unknown whether a specific hotspot may increase downstream signaling, which can be assessed in gene expression, protein changes, methylation patterns, or even in small or large genomic rearrangements. Recent examples of these concepts are hotspots in PIK3CA and NOTCH1 [80]. Nevertheless, these issues need to be systematically studied.
There are other concepts that could be also useful for hotspots detections in cancer. For example, a co-evolution method considering epistasis has been shown to detect disease variants [81]. These concepts can be used, first, for annotation, but more importantly to quantify epistasis in cluster, 3D-clusters, or functional impact methods. We also noted methods like ReKINect focused in detection of mutations perturbing signaling networks [82], whose concept have not been applied to detect or to annotate and filter cancer hotspots.
It is known that cancer mutations are produced by the exposure to mutagenic processes generating mutational signatures [83]. Nevertheless, it is currently unknown whether there is a relationship between mutational signatures and hotspots. If exists, which comes first, the hotspot mutation or the mutational process, and what could be the intrinsic mechanism between them. A recent analysis noted an enrichment between hotspots in the TCG sequence context [75], pointing to APOBEC mutational signatures. Thus, the possible relationship between hotspots and mutational signatures needs to be methodically studied.
Although some methods used machine learning [29], methodologically, we did not observe intensive use of novel artificial intelligence algorithms, such as those having deep neural networks as core engines. Most methods were model-based, ad hoc or heuristic algorithms. The use of deep neural networks to detect cancer hotspots requires adapting current views of inputs and force to re-think on the problem. This can be attractive, for example, to explore and discover hidden features, genes, or regions not seen by model-based or heuristic approaches.
There are some pitfalls noted after reviewing all methods. First, there is a variety of datasets providers with diverse degrees of redundancy. The main sources of databases are TCGA, ICGC, and COSMIC, but there are others like TARGET (https://ocg.cancer.gov/programs/target), FM (foundation medicine, FM-AD within GDC data portal), and others emerging like AACR/GENIE (https://www.aacr.org/professionals/research/aacr-project-genie/), HM (hematological malignancy), among others. Consequently, the results are also varied and, if available, reported as supplementary information, which is not recorded in databases. These issues complicate comparisons of hotspots estimations among methods.
A coming scenario is the accumulation of cancer data. Current approaches are focused on whole-genome sequencing (WGS) [6], sequencing of less studied cancer types, and sequencing in specific cancer subtypes or subpopulation strata (young women in breast cancer, pre-malignant tissues, local cancers types, not sampled countries, among others). It is not clear whether the methods reviewed can be used “as is” in less studied cancer types or subtypes or whether the aggregation will reveal additional biases that need to be corrected in novel proposals. The increasing use of WGS will also demand further development of non-coding hotspots methods. Taking together the above ideas, it is expected that novel hotspots will emerge and that some current hotspot predictions are artifacts. Thus, the continuous use and development of hotspot methods are needed.
In this review, we focused on mutation hotspots. Nevertheless, many of the concepts shown here also apply to detect “hotspots” in other genomic contexts, which are perhaps the next big challenges to be solved regarding recurrent alterations. For example, there are reports of highly recurrent translocations, duplications, and deletions [84], [85], [86], [87]. Methylation dysregulation is also known in cancer [88] but not explored systematically as hotspots. In addition, there is evidence suggesting that viral integrations may also show recurrent patterns [89]. Thus, we believe that the concepts reviewed here can also be adapted to detect recurrent alterations in cancer genomes.
9. Conclusion
In this mini-review, we showed most of the published methods to detect recurrent mutations in cancer and classified into cluster, 3D, position-specific, and miscellaneous. The “hotspots” are likely functional but need validation, and therefore the use of hotspot methods is crucial in cancer research and clinics. The increasing availability of cancer genomics data will demand more specific and powerful methods. The identification of false hotspots and their underlying mechanisms are as important as the identification of novel hotspots. Standard databases for hotspots are needed to provide comparisons and improve methods. The concepts of the methods reviewed could also be applied to detect recurrent alterations from other genomic data in cancer.
CRediT authorship contribution statement
Emmanuel Martinez-Ledesma: Conceptualization, Formal analysis, Investigation, Methodology, Funding acquisition, Writing - original draft, Writing - review & editing. David Flores: Conceptualization, Formal analysis, Investigation, Methodology, Writing - original draft, Writing - review & editing. Victor Trevino: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.csbj.2020.11.020.
Contributor Information
Emmanuel Martinez-Ledesma, Email: juanemmanuel@tec.mx.
David Flores, Email: dflores@ucaribe.edu.mx.
Victor Trevino, Email: vtrevino@tec.mx.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hanahan D., Weinberg R. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sjoblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science:1133427. [DOI] [PubMed] [Google Scholar]
- 4.Wood L.D., Parsons D.W., Jones S., Lin J., Sjoblom T., Leary R.J. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science:1145720. [DOI] [PubMed] [Google Scholar]
- 5.Gerstung M., Jolly C., Leshchiner I., Dentro S.C., Yu K., Tarabichi M. The evolutionary history of 2,658 cancers. Nature. 2020;578:122–128. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell P.J., Getz G., Korbel J.O., Stuart J.M., Jennings J.L., Stein L.D. Pan-cancer analysis of whole genomes. Nature. 2020 doi: 10.1038/s41586-020-1969-6. [DOI] [Google Scholar]
- 7.Martincorena I., Raine K.M., Gerstung M., Dawson K.J., Haase K., Van Loo P. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171:1029–1041.e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porta-Pardo E., Kamburov A., Tamborero D., Pons T., Grases D., Valencia A. Comparison of algorithms for the detection of cancer drivers at subgene resolution. Nat. Methods. 2017;14:782–788. doi: 10.1038/nmeth.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019;47:D941–7. DOI:10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed]
- 10.Bass A.J., Thorsson V.V., Shmulevich I., Reynolds S.M., Miller M., Bernard B. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;513:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson B.E., Mazor T., Hong C., Barnes M., Aihara K., McLean C.Y. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science:1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller M.L., Reznik E., Gauthier N.P., Ciriello G., Schultz N., Miller M.L. Pan-cancer analysis of mutation hotspots in protein domains. Cell Systems. 2015;1:197–209. doi: 10.1016/j.cels.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer T, Atlas G, Agrawal N, Akbani R, Aksoy BA, Ally A, et al. Integrated Genomic Characterization of Papillary Thyroid Carcinoma. Cell 2014;159:676–90. DOI:10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed]
- 14.Hodis E., Watson I., Kryukov G., Arold S., Imielinski M., Theurillat J.-P. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tiacci E., Trifonov V., Schiavoni G., Holmes A., Kern W., Martelli M.P. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muzny D.M., Bainbridge M.N., Chang K., Dinh H.H., Drummond J.A., Fowler G. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammerman P.S., Voet D., Lawrence M.S., Voet D., Jing R., Cibulskis K. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012 doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salimian K.J., Fazeli R., Zheng G., Ettinger D., Maleki Z. V600E BRAF versus Non-V600E BRAF mutated lung adenocarcinomas: cytomorphology, histology, coexistence of other driver mutations and patient characteristics. Acta Cytol. 2018;62:79–84. doi: 10.1159/000485497. [DOI] [PubMed] [Google Scholar]
- 19.Collisson E.A., Campbell J.D., Brooks A.N., Berger A.H., Lee W., Chmielecki J. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, et al. Genomic Classification of Cutaneous Melanoma. Cell 2015;161:1681–96. DOI:10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed]
- 21.Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J., Bajari R., Andric D., Gerthoffert F., Lepsa A., Nahal-Bose H. The international cancer genome consortium data portal. Nat Biotechnol. 2019;37:367–369. doi: 10.1038/s41587-019-0055-9. [DOI] [PubMed] [Google Scholar]
- 23.Buisson R., Langenbucher A., Bowen D., Kwan E.E., Benes C.H., Zou L. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science. 2019;364:eaaw2872. doi: 10.1126/science:aaw2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 2019. DOI:10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed]
- 25.Melton C., Reuter J.A., Spacek D.V., Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet. 2015;47:710–716. doi: 10.1038/ng.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baeissa H., Benstead-Hume G., Richardson C.J., Pearl F.M.G. Identification and analysis of mutational hotspots in oncogenes and tumour suppressors. Oncotarget. 2017;8:21290–21304. doi: 10.18632/oncotarget.15514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia P., Wang Q., Chen Q., Hutchinson K.E., Pao W., Zhao Z. MSEA: detection and quantification of mutation hotspots through mutation set enrichment analysis. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porta-Pardo E, Godzik A. E-Driver: A novel method to identify protein regions driving cancer. Bioinformatics 2014;30:3109–14. DOI:10.1093/bioinformatics/btu499. [DOI] [PMC free article] [PubMed]
- 29.Kim K., Jang K., Yang W., Choi E.-Y., Park S.-M., Bae M. Chromatin structure–based prediction of recurrent noncoding mutations in cancer. Nat Genet. 2016;48:1321–1326. doi: 10.1038/ng.3682. [DOI] [PubMed] [Google Scholar]
- 30.Juul M, Bertl J, Guo Q, Nielsen MM, Świtnicki M, Hornshøj H, et al. Non-coding cancer driver candidates identified with a sample- and position-specific model of the somatic mutation rate. Elife 2017;6. DOI:10.7554/eLife.21778. [DOI] [PMC free article] [PubMed]
- 31.Feigin M.E., Garvin T., Bailey P., Waddell N., Chang D.K., Kelley D.R. Recurrent noncoding regulatory mutations in pancreatic ductal adenocarcinoma. Nat Genet. 2017;49:825–833. doi: 10.1038/ng.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhee J.-K., Yoo J., Kim K.R., Kim J., Lee Y.-J., Chul Cho B. Identification of local clusters of mutation hotspots in cancer-related genes and their biological relevance. IEEE/ACM Trans Comput Biol Bioinf. 2019;16:1656–1662. doi: 10.1109/TCBB.2018.2813375. [DOI] [PubMed] [Google Scholar]
- 33.Guo Y.A., Chang M.M., Huang W., Ooi W.F., Xing M., Tan P. Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piraino S.W., Furney S.J. Identification of coding and non-coding mutational hotspots in cancer genomes. BMC Genomics. 2017;18:1–17. doi: 10.1186/s12864-016-3420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fijal B.A., Idury R.M., Witte J.S. Analysis of mutational spectra: locating hotspots and clusters of mutations using recursive segmentation. Statist Med. 2002;21:1867–1885. doi: 10.1002/sim.1145. [DOI] [PubMed] [Google Scholar]
- 36.Ye J., Pavlicek A., Lunney E.A., Rejto P.A., Teng C.-H. Statistical method on nonrandom clustering with application to somatic mutations in cancer. BMC Bioinf. 2010;11 doi: 10.1186/1471-2105-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: Exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 2013;29:2238–44. DOI:10.1093/bioinformatics/btt395. [DOI] [PubMed]
- 38.Yang W., Bang H., Jang K., Sung M.K., Choi J.K. Predicting the recurrence of noncoding regulatory mutations in cancer. BMC Bioinf. 2016;17 doi: 10.1186/s12859-016-1385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lochovsky L., Zhang J., Fu Y., Khurana E., Gerstein M. LARVA: an integrative framework for large-scale analysis of recurrent variants in noncoding annotations. Nucleic Acids Res. 2015;43:8123–8134. doi: 10.1093/nar/gkv803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnedo-pac C, Mularoni L, Muiños F, Gonzalez-perez A, Lopez- N. OncodriveCLUSTL: a sequence-based clustering method to identify cancer drivers 1 Introduction 2018:1–6.
- 41.Lu X, Qian X, Li X, Miao Q, Peng S. DMCM: A Data-adaptive Mutation Clustering Method to identify cancer-related mutation clusters. Bioinformatics 2019. DOI:10.1093/bioinformatics/bty624. [DOI] [PubMed]
- 42.Poole W, Leinonen K, Shmulevich I, Knijnenburg TA, Bernard B. Multiscale mutation clustering algorithm identifies pan-cancer mutational clusters associated with pathway-level changes in gene expression. PLoS Comput Biol 2017;13:1–26. DOI:10.1371/journal.pcbi.1005347. [DOI] [PMC free article] [PubMed]
- 43.Araya C.L., Cenik C., Reuter J.A., Kiss G., Pande V.S., Snyder M.P. Identification of significantly mutated regions across cancer types highlights a rich landscape of functional molecular alterations. Nat Genet. 2016;48:117–125. doi: 10.1038/ng.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hess J.M., Bernards A., Kim J., Miller M., Taylor-Weiner A., Haradhvala N.J. Passenger hotspot mutations in cancer. Cancer Cell. 2019 doi: 10.1016/j.ccell.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van den Eynden J., Fierro A.C., Verbeke L.P.C., Marchal K. SomInaClust: detection of cancer genes based on somatic mutation patterns of inactivation and clustering. BMC Bioinf. 2015;16:1–12. doi: 10.1186/s12859-015-0555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rheinbay E., Parasuraman P., Grimsby J., Tiao G., Engreitz J.M., Kim J. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547:55–60. doi: 10.1038/nature22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rheinbay E, Nielsen MM, Abascal F, Wala JA. Analyses of non-coding somatic drivers in 2 , 658 cancer whole genomes 2020;578. DOI:10.1038/s41586-020-1965-x. [DOI] [PMC free article] [PubMed]
- 48.Tokheim C., Bhattacharya R., Niknafs N., Gygax D.M., Kim R., Ryan M. Exome-scale discovery of hotspot mutation regions in human cancer using 3D protein structure. Cancer Res. 2016;76:3719–3731. doi: 10.1158/0008-5472.CAN-15-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A.D., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012 doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, et al. Ensembl 2019. Nucleic Acids Res 2019. DOI:10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed]
- 52.Niu B., Scott A.D., Sengupta S., Bailey M.H., Batra P., Ning J. Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat Genet. 2016;48:827–837. doi: 10.1038/ng.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engin H.B., Hofree M., Carter H. Identifying mutation specific cancer pathways using a structurally resolved protein interaction network. Pacific Symp Biocomput. 2015 doi: 10.1142/9789814644730_0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamburov A., Lawrence M.S., Polak P., Leshchiner I., Lage K., Golub T.R. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc Natl Acad Sci USA. 2015;112:E5486–E5495. doi: 10.1073/pnas.1516373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pieper U., Webb B.M., Dong G.Q., Schneidman-Duhovny D., Fan H., Kim S.J. ModBase, a database of annotated comparative protein structure models and associated resources. Nucl Acids Res. 2014;42:D336–D346. doi: 10.1093/nar/gkt1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res 2018. DOI:10.1093/nar/gky427. [DOI] [PMC free article] [PubMed]
- 57.Meyer M.J., Lapcevic R., Romero A.E., Yoon M., Das J., Beltrán J.F. mutation3D: cancer gene prediction through atomic clustering of coding variants in the structural proteome. Hum Mutat. 2016;37:447–456. doi: 10.1002/humu.22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryslik G.A., Cheng Y., Cheung K.-H., Modis Y., Zhao H. Utilizing protein structure to identify non-random somatic mutations. BMC Bioinf. 2013;14 doi: 10.1186/1471-2105-14-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryslik G.A., Cheng Y., Cheung K.-H., Modis Y., Zhao H. A graph theoretic approach to utilizing protein structure to identify non-random somatic mutations. BMC Bioinf. 2014;15 doi: 10.1186/1471-2105-15-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryslik G.A., Cheng Y., Cheung K.-H., Bjornson R.D., Zelterman D., Modis Y. A spatial simulation approach to account for protein structure when identifying non-random somatic mutations. BMC Bioinf. 2014;15 doi: 10.1186/1471-2105-15-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao J., Chang M.T., Johnsen H.C., Gao S.P., Sylvester B.E., Sumer S.O. 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med. 2017;9 doi: 10.1186/s13073-016-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryslik G.A., Cheng Y., Modis Y., Zhao H. Leveraging protein quaternary structure to identify oncogenic driver mutations. BMC Bioinf. 2016;17 doi: 10.1186/s12859-016-0963-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar S., Clarke D., Gerstein M.B. Leveraging protein dynamics to identify cancer mutational hotspots using 3D structures. Proc Natl Acad Sci USA. 2019;116:18962–18970. doi: 10.1073/pnas.1901156116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arafeh R., Samuels Y. PIK3CA in cancer: The past 30 years. Semin Cancer Biol. 2019;59:36–49. doi: 10.1016/j.semcancer.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 65.Li S., Balmain A., Counter C.M. A model for RAS mutation patterns in cancers: finding the sweet spot. Nat Rev Cancer. 2018;18:767–777. doi: 10.1038/s41568-018-0076-6. [DOI] [PubMed] [Google Scholar]
- 66.Martínez-Jiménez F., Muiños F., López-Arribillaga E., Lopez-Bigas N., Gonzalez-Perez A. Systematic analysis of alterations in the ubiquitin proteolysis system reveals its contribution to driver mutations in cancer. Nat Cancer. 2020;1:122–135. doi: 10.1038/s43018-019-0001-2. [DOI] [PubMed] [Google Scholar]
- 67.Alexandrov L., Nik-Zainal S., Wedge D., Campbell P., Stratton M. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen T., Wang Z., Zhou W., Chong Z., Meric-Bernstam F., Mills G.B. Hotspot mutations delineating diverse mutational signatures and biological utilities across cancer types. BMC Genomics. 2016;17 doi: 10.1186/s12864-016-2727-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang M.T., Asthana S., Gao S.P., Lee B.H., Chapman J.S., Kandoth C. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sabarinathan R., Mularoni L., Deu-Pons J., Gonzalez-Perez A., López-Bigas N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature. 2016;532:264–267. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]
- 71.Chang M.T., Bhattarai T.S., Schram A.M., Bielski C.M., Donoghue M.T.A., Jonsson P. Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 2018;8:174–183. doi: 10.1158/2159-8290.CD-17-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith TCA, Carr AM, Eyre-Walker AC. Are sites with multiple single nucleotide variants in cancer genomes a consequence of drivers, hypermutable sites or sequencing errors? PeerJ 2016;2016. DOI:10.7717/peerj.2391. [DOI] [PMC free article] [PubMed]
- 73.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. BioRxiv. 2019 doi: 10.1101/531210. [DOI] [Google Scholar]
- 74.Mularoni L., Sabarinathan R., Deu-Pons J., Gonzalez-Perez A., López-Bigas N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 2016;17 doi: 10.1186/s13059-016-0994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trevino V. Modeling and analysis of site-specific mutations in cancer identifies known plus putative novel hotspots and bias due to contextual sequences. Comput Struct Biotechnol J. 2020;18:1664–1675. doi: 10.1016/j.csbj.2020.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic Acids Res 2012;40:1–10. DOI:10.1093/nar/gks743. [DOI] [PMC free article] [PubMed]
- 77.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sabarinathan R., Tafer H., Seemann S.E., Hofacker I.L., Stadler P.F., Gorodkin J. RNAsnp: efficient detection of local RNA secondary structure changes induced by SNPs. Hum Mutat. 2013;34:546–556. doi: 10.1002/humu.22273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trevino V. HotSpotAnnotations - A database for hotspot mutations and annotations in cancer. Database 2019:(In revision). [DOI] [PMC free article] [PubMed]
- 80.Saito Y., Koya J., Araki M., Kogure Y., Shingaki S., Tabata M. Landscape and function of multiple mutations within individual oncogenes. Nature. 2020;582:95–99. doi: 10.1038/s41586-020-2175-2. [DOI] [PubMed] [Google Scholar]
- 81.Hopf T.A., Ingraham J.B., Poelwijk F.J., Schärfe C.P.I., Springer M., Sander C. Mutation effects predicted from sequence co-variation. Nat Biotechnol. 2017;35:128–135. doi: 10.1038/nbt.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Creixell P., Schoof E., Simpson C., Longden J., Miller C., Lou H. Kinome-wide decoding of network-attacking mutations rewiring cancer signaling. Cell. 2015;163:202–217. doi: 10.1016/j.cell.2015.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alexandrov L.B., Kim J., Haradhvala N.J., Huang M.N. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar-Sinha C., Tomlins S.A., Chinnaiyan A.M. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang J.J., Park T.S., Wan T.S.K. Recurrent cytogenetic abnormalities in acute myeloid leukemia. Methods Mol Biol. 2017;1541:223–245. doi: 10.1007/978-1-4939-6703-2_19. [DOI] [PubMed] [Google Scholar]
- 86.Veeraraghavan J., Ma J., Hu Y., Wang X.-S. Recurrent and pathological gene fusions in breast cancer: current advances in genomic discovery and clinical implications. Breast Cancer Res Treat. 2016;158:219–232. doi: 10.1007/s10549-016-3876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tanaka H., Watanabe T. Mechanisms underlying recurrent genomic amplification in human cancers. Trends in Cancer. 2020;6:462–477. doi: 10.1016/j.trecan.2020.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Z, Yin J, Zhou W, Bai J, Xie Y, Xu K, et al. Complex impact of DNA methylation on transcriptional dysregulation across 22 human cancer types. Nucleic Acids Res 2020. DOI:10.1093/nar/gkaa041. [DOI] [PMC free article] [PubMed]
- 89.Ojesina A.I., Lichtenstein L., Freeman S.S., Pedamallu C.S., Imaz-Rosshandler I., Pugh T.J. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.