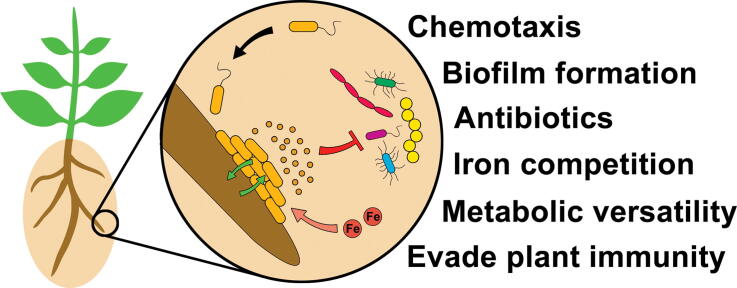
Graphical abstract
Keywords: Pseudomonas, Rhizosphere, Colonization, Determinants, PGPR, Biocontrol
Abstract
Plant growth-promoting rhizobacteria (PGPR) actively colonize the soil portion under the influence of plant roots, called the rhizosphere. Many plant-beneficial Pseudomonas spp. have been characterized as PGPR. They are ubiquitous rod-shaped motile Gram-negative bacteria displaying a high metabolic versatility. Their capacity to protect plants from pathogens and improve plant growth closely depends on their rhizosphere colonization abilities. Various molecular and cellular mechanisms are involved in this complex process, such as chemotaxis, biofilm formation, secondary metabolites biosynthesis, metabolic versatility, and evasion of plant immunity. The burst in Pseudomonas spp. genome sequencing in recent years has been crucial to better understand how they colonize the rhizosphere. In this review, we discuss the recent advances regarding these mechanisms and the underlying bacterial genetic factors required for successful rhizosphere colonization.
1. Introduction
The rhizosphere is defined as the soil portion under the influence of plant roots [1]. Roots release many compounds originating from the photosynthesis, such as exudates, but also from root debris. These carbon- and nitrogen-containing molecules provide valuable substrates for microbial growth [2]. This leads to the rhizosphere effect: a dramatic increase in microbial populations in the vicinity of roots where rhizodeposits are released, compared to the bulk soil. This abundance of energy sources also enhances acute microbial competition, making the rhizosphere both a challenging and heterogeneous environment for microbes [3].
The rhizosphere microbiome is highly diverse and notably contains plant-beneficial microbes. Among these microbes, plant growth-promoting rhizobacteria (PGPR) have been extensively studied [4]. Their potential to directly improve plant growth and/or inhibit plant pathogens is promising for the development of sustainable farming practices. PGPR include many bacterial genera such as Azospirillum, Azotobacter, Bacillus, Burkholderia, Enterobacter, Rhizobium, Serratia and Pseudomonas [5]. The latter has been broadly studied, especially the many biocontrol strains of interest discovered since the 1980′s [6].
Pseudomonas spp. are rod-shaped motile Gram-negative bacteria present in a wide array of environments and displaying a great metabolic diversity [7]. PGPR strains belonging to this genus are mainly found within the P. fluorescens complex [8]. They are able to inhibit the growth of several bacterial, fungal and oomycete plant pathogens, such as Streptomyces scabies, P. syringae, Fusarium oxysporum, Gaeumannomyces graminis, Rhizoctonia solani, Phytophthora infestans, and Pythium ultimum [9], [10], [11]. Pathogen inhibition primarily occurs through antibiosis, the stimulation of plant defense mechanisms, and competition for niches and nutrients, especially mediated by siderophores [12]. Direct plant growth promotion caused by Pseudomonas spp. instead relies on phosphate and iron solubilization, nitrogen fixation, phytohormone modulation and increased abiotic stress tolerance [4], [13], [14].
The efficiency of Pseudomonas spp. to control pathogens and to promote plant growth is closely related to their ability to competitively colonize the rhizosphere and persist in this compartment, which is defined as rhizocompetence [3], [10]. Some authors even suggest that a minimal colonization threshold of 105 bacteria per gram of root is required for antibiosis and the induction of the plant systemic resistance mechanisms [12], [15]. Rhizosphere colonization by phytobeneficial Pseudomonas spp. is therefore crucial to ensure their efficiency in agriculture. The breakthroughs in bacterial bioinformatics, especially in whole-genome sequencing and annotation, and in genome comparison tools, opened new paths of research enabling a better understanding of this complex process. This review aims to summarize these recent and exciting developments.
2. Settling in the rhizosphere
Pseudomonas spp. are ubiquitous in soils and can live under a planktonic or a sessile lifestyle [16], [17]. When they are in the vicinity of plant roots, they switch from the planktonic to the sessile lifestyle to better benefit from the rhizodeposits. This change is made possible by chemotaxis towards exudates, enabling the bacteria to get closer to the roots, and by biofilm formation, allowing them to attach to the roots and to develop fixed colonies [18].
2.1. Chemotaxis towards root exudates
Chemotaxis is a motility mechanism enabling the bacteria to move in response to a chemical gradient [19]. In the rhizosphere, it allows the bacteria to detect the presence of rhizodeposits, especially root exudates, and to get closer to their release sites such as root tips. The colonization sites can differ depending on the Pseudomonas strain. For example, two efficient avocado root tip colonizers, P. alcaligenes AVO73 and P. pseudoalcaligenes AVO110, display distinct colonization strategies: the latter colonizes root wounds and intercellular spaces between root epidermal cells, unlike the former [20]. There are three types of chemotaxis-driven motility: swimming, swarming, and twitching. Swimming and swarming depends on flagellar rotations, while twitching relies on the extension-retraction movements of type IV pili [19].
Flagella- and pili-driven chemotaxis are controlled by distinct but homologous signal transduction mechanisms. The detection of distinct rhizosphere compounds is mediated by chemoreceptors called methyl-accepting chemotaxis proteins (MCP). These are homodimers of transmembrane proteins displaying a periplasmic ligand-binding region, specific to a compound or a group of compounds, and a cytosolic methyl-accepting domain, conserved among MCP [21]. Most MCP described to date in rhizosphere-inhabiting Pseudomonas spp. enable the detection of amino acids, polyamines, or organic acids such as intermediates of the tricarboxylic acid cycle (TCA). In P. putida KT2440, these intermediates are the preferred energy source during the early growth phase, along with amino acids that can be converted into TCA intermediates [22].
The number of MCP closely depends on the strain lifestyle: bacteria able to colonize diverse environments, such as the rhizosphere, and to establish complex interactions with plant roots display more MCP [23]. Rhizosphere-inhabiting Pseudomonas strains have been shown to harbor between 27 and 37 MCP genes, but only few strains have been analyzed so far [24], [25]. The analysis of newly sequenced genomes could greatly increase the current MCP repertoire. Recently, López-Farfán et al. focused on MCP genes expression in P. putida KT2440 [26]. They demonstrated that their expression is inversely correlated with maize root exudates proximity: when the bacteria get closer to the roots, chemoreceptor genes expression decrease, probably because chemotaxis becomes less useful in the root vicinity.
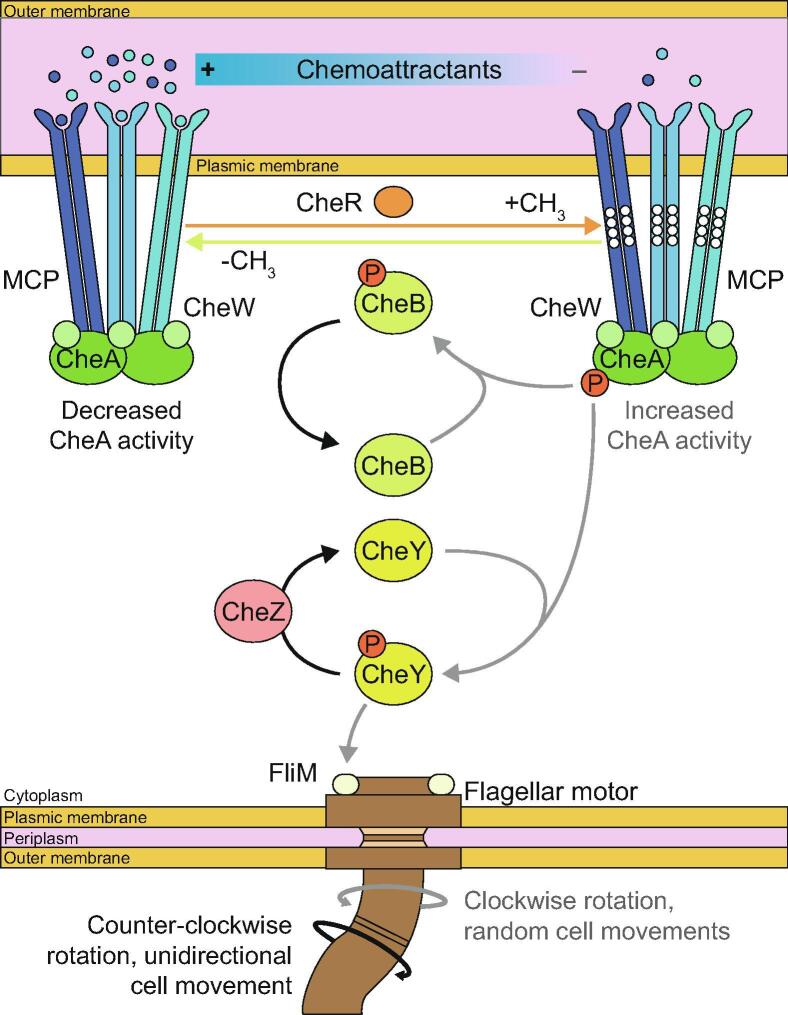
Regarding flagella-driven chemotaxis, MCP form complexes with an adaptor protein, CheW, and a histidine kinase, CheA [27]. Chemoeffectors like amino acids directly interact with the ligand-binding region of MCP, while others like sugars and dipeptides indirectly interact with MCP through periplasmic binding proteins [28]. When the amount of chemoattractants around the bacteria decreases, or when chemoreppellent concentrations increase, CheA is autophosphorylated (Fig. 1). Its phosphate group is then transferred to a response regulator protein, CheY, which interacts with the FliM protein belonging to the flagellum switch complex [29]. This triggers proton-driven clockwise rotation of the flagellum, causing the cell to randomly move [30]. When a chemoattractant such as a root exudate binds to MCP, CheA autophosphorylation is inhibited, which reduces CheY phosphorylation. In turn, counter-clockwise rotation of the flagellum is triggered, enabling the bacterium to advance in a given direction, towards exudates [28]. Rapid dephosphorylation of CheY is mediated by CheZ, allowing the bacterium to quickly change course depending on the detected chemical gradients. A methyltransferase called CheR is also involved in the switching mechanism. It constitutively transfers methyl groups to the MCP methyl-accepting domain, increasing CheA autophosphorylation rate. However, its activity is in competition with the methylesterase CheB, which is activated by phosphorylation by CheA. In the absence of chemoeffectors, the flagellum rotation switch frequency changes stochastically [31].
Fig. 1.
Mechanism involved in flagella-driven chemotaxis in Gram-negative bacteria. In the presence of chemoattractants, the flagellum preferably switches to counter-clockwise rotation, propelling the cell towards the chemoattractants. When chemoattractant concentrations is low, or when chemorepellents are detected by MCP, CheA is autophosphorylated and phosphorylates CheY and CheB. CheY induces in turn a flagellar rotation change through the switch complex component FliM, leading to clockwise rotation and random cell movements. Regulation of this mechanism is mediated by CheR, CheB, and CheZ, respectively through methylation, demethylation and dephosphorylation. MCP: Methyl-accepting chemotaxis proteins.
Twitching, which is pili-driven, relies on a chemosensory system similar to flagella-driven chemotaxis [32]. It has originally been described in the opportunistic human pathogen P. aeruginosa and is mainly enabled by the proteins encoded by the pilGHIJK and chpABC genes [33], [34]. These genes have also been identified in plant-beneficial Pseudomonas strains such as P. fluorescens F113 [35], P. stutzeri A1501 [36], and partly in P. putida KT2440 [37].
Several studies on Pseudomonas mutant strains lacking chemotaxis- or motility-related genes showed reduced rhizosphere colonization abilities [38], [39], [40], [41]. Oku et al. constructed several P. fluorescens Pf0-1 mutants for multiple MCP genes to assess their combined effect on tomato rhizosphere colonization [38], [41]. They showed that the ctaA ctaB ctaC triple mutant was less competitive than the wild type, and that the ctaA ctaB ctaC mcpS mcpT quintuple mutant was even less competitive. Those mutants were still better colonizers than the cheA mutant, lacking flagella-driven chemotaxis. This indicates that tomato rhizosphere colonization by P. fluorescens Pf0-1 relies on multiple MCP, more than the five proteins studied. In P. fluorescens F113, mutations in fliC, fliS, fleQ and fliT, involved in the flagellum synthesis, reduced or completely blocked motility, which resulted in a lower competitive alfalfa root colonization [40].
In order to improve rhizocompetence in wheat, Gao et al. recently tried to enhance the chemotaxis activity of Pseudomonas sp. UW4 towards 1-aminocyclopropane-1-carboxylic acid (ACC) [42]. This compound is a precursor of ethylene in plants and is exuded in the rhizosphere, where PGPR can use it as a carbon and nitrogen source using the ACC deaminase enzyme, encoded by the acdS gene [43]. ACC is known as a strong chemoattractant for the UW4 strain [44]. Gao et al. used an acdS-defective UW4 strain and complemented it with distinct bacterial promoters and acdS. They identified a linear and positive correlation between acdS expression levels, AcdS activity, chemotaxis towards ACC and wheat rhizosphere colonization. They also showed that the promoters inducing high acdS expression levels and high chemotactic responses towards ACC also strengthen chemotaxis responses to other chemoattractants such as amino and organic acids. These results demonstrate that increasing the bacterial metabolic rate of a single compound can lead, through chemotaxis and other mechanisms, to an enhanced rhizocompetence.
2.2. Biofilm lifecycle
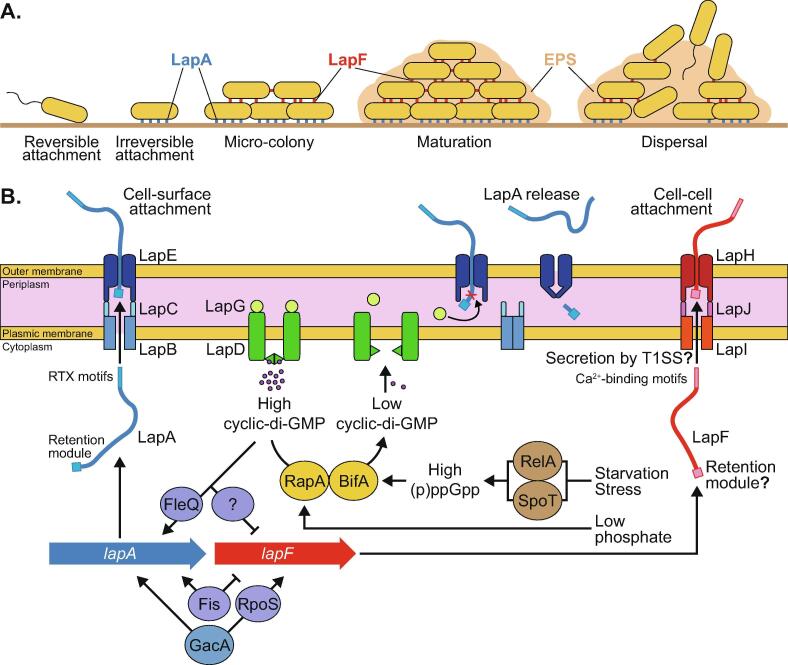
Once plant-beneficial Pseudomonas spp. are in the vicinity of the roots, they often form biofilms. These are bacterial aggregates embedded in a matrix made of highly hydrated extracellular polymeric substances (EPS) and adhering to biotic or abiotic surfaces [45]. Biofilms offer protection to the bacteria against various stresses, such as desiccation, antibiotics or protozoan predation, and can also improve nutrient assimilation [45]. They contribute to increasing bacterial densities required to produce secondary metabolites involved in plant–microbe and microbe-microbe interactions [18]. Biofilm development follows several steps: reversible attachment, irreversible attachment, micro-colony formation, maturation, and bacterial dispersal [46] (Fig. 2A).
Fig. 2.
A. Bacterial biofilm lifecycle. B. Mechanisms involved in the secretion of the large adhesion proteins LapA and LapF and in the regulation of the expression of their biosynthetic genes in P. putida. LapA is produced at the beginning of biofilm formation to promote cell-surface attachment. It is released from the outer membrane through the action of LapG in response to nutrient deficiency or other stresses. LapF is synthetized later in the process, allowing cell–cell attachment. Regulation is especially mediated by transcription factors such as FleQ, Fis and RpoS. Many aspects of LapF structure and secretion remain to be deciphered. The lapA and lapF genes are distant from each other in Pseudomonas putida genomes. Cyclic-di-GMP: cyclic dimeric guanosine monophosphate; EPS: extracellular polymeric substances; RTX: repeats-in-toxin; T1SS: type I secretion system.
Attachment to plant roots or soil particles is mediated by adhesins. They are adhesive structures, mostly proteins, expressed on the bacterial surface to promote attachment to a surface, a host, or to other bacteria [47]. Many adhesins have been identified in rhizosphere-associated Pseudomonas spp., such as the large adhesion proteins LapA and LapF [48], [49], [50], the medium adhesion protein MapA [51], and the flagellum itself [49].
LapA and the other Lap proteins have been extensively studied for their key role in biofilm formation in plant-beneficial Pseudomonas spp., recently reviewed by Collins et al. [52]. LapA belongs to the repeats-in-toxin (RTX) adhesion protein family and has been initially described by Hinsa et al. in P. fluorescens WCS365 [48], [53]. This is the largest protein identified in P. putida KT2440, in which it is encoded by a 26 kb gene resulting in a 8682-amino acid protein with an estimated molecular weight of about 888 kDa [48]. LapA is secreted by a type I secretion system (T1SS) ABC transporter encoded by lapEBC [48], [53] (Fig. 2B). LapE, an outer membrane TolC-like pore, is similar to the agglutinin AggA, needed for the attachment of P. putida strain Corvallis [48], [54]. Smith et al. showed that LapE plays a bigger role than just secreting LapA through the T1SS: it anchors this adhesin to the cell surface through the LapA N-terminal retention module [55]. Apart from this retention module, LapA consists of three other domains. Two domains display multiple repeats found in surface proteins in other bacterial genera. The C-terminal domain features a T1SS secretion signal as well as RTX repeats, which are Ca2+-binding motifs that can be found in secreted proteins involved in bacterial-eukaryotic interactions [56]. This domain could be responsible for the attachment to plant roots. Observations performed by Gjermansen et al. suggest that LapA is also able to bind to exopolysaccharides of the biofilm matrix [57]. LapA could then play multiple roles within biofilms [48]. Mutations in the lap genes show that they are required for irreversible attachment and biofilm formation [48], [49].
Once the bacterium is irreversibly attached to a biotic or an abiotic surface in the rhizosphere, it starts multiplying to form a micro-colony, evolving into a mature biofilm [46]. This lifestyle transition leads to a gene expression shift: the expression of genes involved in motility is inhibited, while the expression of genes related to EPS biosynthesis is promoted [58]. EPS are mainly polysaccharides, proteins, DNA and lipids providing structural stability to the biofilm and an external digestive system to the bacteria through the retention of extracellular enzymes [45]. In P. putida KT2440, four gene clusters are responsible for the production of exopolysaccharides forming the biofilm matrix: pea and peb (putida exopolysaccharide A and B), alg (alginate) and bcs (bacterial cellulose synthesis) [59], [60]. They are structural stabilizers of the biofilm, but a quadruple mutant lacking these four gene clusters is still able to form biofilms [60], indicating that other matrix components are involved in the biofilm structure. Recently, Marshall et al. showed that mutants of P. fluorescens Pf0-1 lacking alg genes displayed reduced soil colonization levels compared to the wild type, demonstrating the importance of this polysaccharide in the colonization process [61]. Other exopolysaccharide biosynthetic operons have been identified in rhizosphere-inhabiting Pseudomonas strains, especially in P. protegens: psl and pel [62], [63]. The associated polysaccharides, Psl and Pel, have been well characterized in P. aeruginosa [64]. Psl is a repeating pentamer containing mannose, glucose and rhamnose, while the structure of Pel remain unknown but could be a cellulose-like polymer of glucose [65]. These exopolysaccharides are redundant structural scaffolds in mature biofilms that can help the bacteria keep its ability to produce biofilms [64]. However, the contribution of Psl and Pel to biofilms varies between P. aeruginosa strains. Their role could then be different in rhizosphere-colonizing Pseudomonas strains. This remains to be elucidated. Extracellular DNA (eDNA) has been shown to play a critical role in Pseudomonas spp. biofilms [66]. eDNA is produced through cell lysis, releasing fragments ranging from 10 bp to 30 kbp [67]. These fragments binds to other biopolymers in the biofilm, such as exopolysaccharides or proteins, increasing biofilm integrity [68]. Another matrix component involved in the biofilm structure is LapF, a protein similar to LapA identified by Martínez-Gil et al. [50] (Fig. 2B). LapF is the second largest protein in P. putida KT2440 after LapA, with 6310 amino acid, and its encoding gene is distant from the lapA gene within KT2440 genome [69]. A lapF mutant is still able to irreversibly attach to a surface but is unable to form micro-colonies and mature biofilms, and to competitively colonize plant roots. It is presumably secreted to the cell surface by a putative T1SS encoded by the lapHIJ operon and seems to be involved in cell–cell attachment, contrary to LapA, which is involved in cell-surface attachment [50]. LapF is required for biofilm maturation and its three-dimensional development. Another LapA-like protein, MapA, has been recently identified for its structural role in biofilms [51]. Interestingly, mapA is only expressed at the bottom of large and thick biofilms, where oxygen and nutrients are less available than in the outer biofilm portions, suggesting a potential role in biofilm structural adaptation to improve access to oxygen and nutrients [47]. Its precise function however remains to be deciphered. The biofilm matrix also consists of mucigel produced by plant roots, mainly containing exopolysaccharides [70], [71].
When oxygen and nutrients become less available and when waste products and toxins accumulate in the biofilm, a dissolution process can be initiated to enable cell survival and dispersal [72], [73]. This allows the bacteria to find better conditions for their development and to form new biofilms. Within the rhizosphere, bacterial dispersal could occur to follow the root exudation sites, which are dynamically moving following the expansion of the root system [74]. During nutrient starvation, bacteria can adjust their physiology to rapidly stop their growth. This process is called the stringent response and is mediated by hyperphosphorylated guanine nucleotides called guanosine tetraphosphate and guanosine pentaphosphate (together abbreviated (p)ppGpp) [75]. Reacting to nutrient and oxygen deficiency, two proteins called RelA and SpoT, produce (p)ppGpp from guanosine diphosphate or triphosphate (GDP and GTP) and adenosine triphosphate (ATP) [76] (Fig. 2B). High (p)ppGpp concentrations lead to the repression of lapA through the expression of bifA, a gene encoding a phosphodiesterase decreasing the concentration of cyclic dimeric guanosine monophosphate (cyclic-di-GMP) [77]. Cyclic-di-GMP is a ubiquitous intracellular signalling molecule, especially mediating the bacterial lifestyle transition from motility to sessility [17]. When it binds to LapD, an inner membrane protein, LapD inhibits the periplasmic protease LapG [78]. This protein is responsible for the cleavage of the retention module of LapA [78]. When BifA decreases cyclic-di-GMP concentration, LapG is no longer inhibited by LapD and cleaves the periplasmic domain of LapA, which is then released by the T1SS that was anchoring it to the cell [57], [76]. MapA seems to be targeted by LapG as well [51]. Another phosphodiesterase, RapA (regulator of adherence by phosphate), is also responsible for lowering cyclic-di-GMP levels leading to the release of LapA [79]. Interestingly, RapA biosynthesis is controlled by the Pho regulon, a global regulatory mechanism sensing the extracellular level of inorganic phosphate [78]. Under low phosphate conditions, the Pho regulon activates rapA transcription, ultimately leading to a planktonic lifestyle and enabling the bacteria to find new favorable colonization sites. LapF, responsible for cell–cell attachment, does not seem to be cleavable by LapG [52]. Its fate during biofilm dissolution remains to be elucidated.
While forming mature biofilms has multiple benefits for the bacteria, it is not always the best strategy adopted by Pseudomonas spp. to efficiently colonize the rhizosphere. Barahona et al. have shown that a hypermotile variant of P. fluorescens F113 unable to develop a fully mature biofilm on abiotic surface and to form a biofilm matrix on the rhizoplane is not impaired in competitive alfalfa root tip colonization [80]. In the rhizosphere, cells of this variant were surrounded by plant mucigel, which may partly act as a biofilm matrix to protect the bacteria from multiple stresses. These results show that Pseudomonas spp. can deploy different biofilm strategies to colonize the rhizosphere.
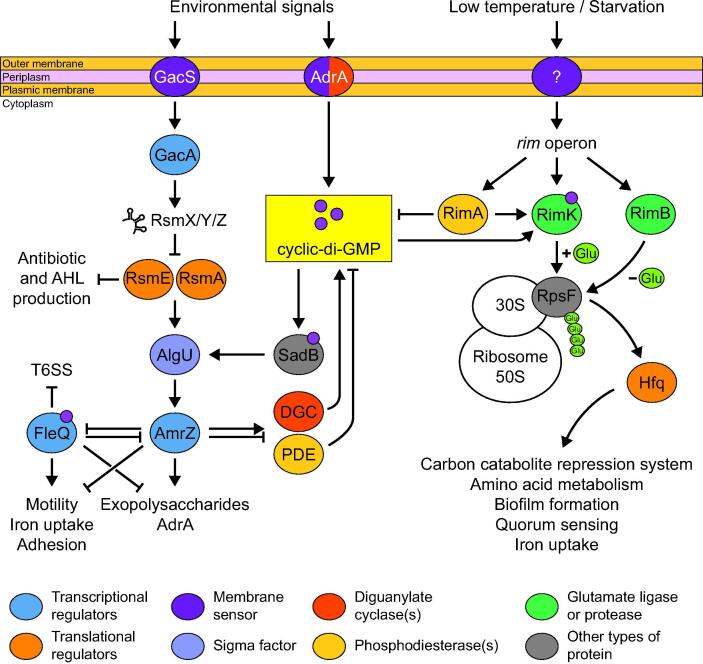
Flagellar motility and biofilm formation are together tightly controlled by two transcription factors: the flagellar expression protein FleQ and the alginate and motility regulator AmrZ (Fig. 3) [81]. Both proteins display more than a hundred putative binding sites [82], [83]. In P. fluorescens F113, 45 genes are targeted by both transcription factors, such as lapA, algD or MCP genes, as well as genes involved in iron homeostasis. AmrZ regulates most of the genes encoding proteins involved in cyclic-di-GMP biosynthesis or degradation, controlling the transition from motility to sessility [81], while FleQ regulation activity is determined by cyclic-di-GMP levels [84]. Interestingly, AmrZ and FleQ are also indirectly under the control of the Gac signal transduction pathway, which regulates antibiotics biosynthesis (see the dedicated section in this review), and of the surface attachment-defective protein SadB pathway [85]. These two pathways enable the bacteria to detect environmental signals. FleQ and AmrZ also repress the expression of each other, leading Blanco-Romero et al. to hypothesize that this reciprocal repression could work as an oscillator used as a general regulation system for environmental adaptation [82]. This complex regulation network illustrates the necessity for rhizosphere-colonizing Pseudomonas spp. to finely adjust their lifestyle according to environmental conditions.
Fig. 3.
Motility and metabolism regulation through the Gac/Rsm and Rim/Hfq pathways in plant-beneficial Pseudomonas spp. DGC: diguanylate cyclases; PDE: phosphodiesterases; RsmX/Y/Z: small non-coding RNA.
3. Coping with neighbors
Whether they are in a planktonic state or settled in biofilms, plant-beneficial Pseudomonas spp. have to compete against other rhizosphere-dwelling microbes to access rhizodeposits and niches, while dealing with plant defenses [86], [87]. To this effect, they display competitiveness-enhancing traits like antibiotic production and siderophores biosynthesis and uptake, and different strategies to evade the plant immune response, demonstrating the importance of the diversity of compounds they can secrete.
3.1. Antibiotics: More than defense compounds
Pseudomonas spp. are able to produce a wide array of antimicrobial compounds to better compete in the rhizosphere [88]. These compounds encompass phenazines, cyclic lipopeptides, polyketides, bacteriocins, type VI secretion system effectors, and other secondary metabolites such as tropolones.
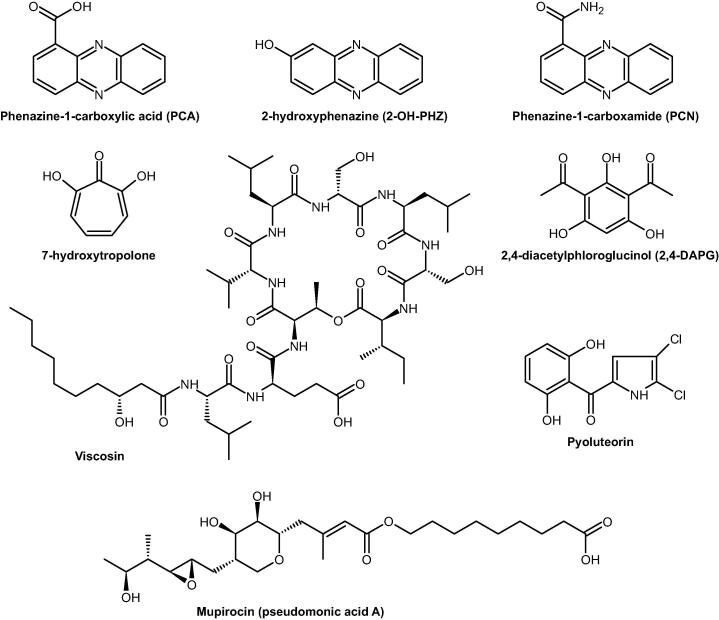
Phenazines are redox-active nitrogen-containing heterocyclic molecules able to inhibit many bacterial, fungal and oomycete plant pathogens [9]. The main phenazines found in phytobeneficial Pseudomonas spp. are phenazine-1-carboxylic acid (PCA), 2-hydroxyphenazine-1-carboxylic acid (2-OH-PCA), 2-hydroxyphenazine (2-OH-PHZ) and phenazine-1-carboxamide (PCN) (Fig. 4). Their biosynthesis, regulation and function have been recently reviewed by Biessy and Filion [9]. Their modes of action remain to be fully elucidated, but phenazines likely affect rival microbes through their redox activity, generating cytotoxic reactive oxygen species (ROS) [89]. Phenazine production has been shown to be directly involved in competitive rhizosphere colonization [90]. Mazzola et al. constructed P. chlororaphis 30–84 and P. synxantha 2–79 (formerly P. aureofaciens 30–84 and P. fluorescens 2–79) mutants defective in phenazine production and inoculated them in non-sterile and sterile soils planted with wheat. They showed that the mutant populations declined more rapidly than the wild type populations in non-sterile soil, but not in sterile soil. This suggests that phenazines are involved in the ability of the strains to compete with the soil microbiome. Yu et al. assessed the impact of the phenazine compounds produced on rhizocompetence in wheat over multiple growth cycles [91]. They did not identify any colonization differences between mutants producing distinct phenazine derivatives. However, they showed that the mutant unable to produce any phenazine colonized the rhizosphere to a lesser extent than those producing at least one phenazine compound, confirming the result obtained by Mazzola et al. They also investigated the role of phenazines in biofilm formation. Indeed, studies have shown that phenazines production directly affects biofilm formation and dispersion [92], [93]. Yu et al. demonstrated that the type of phenazine derivative impact on the amount of biofilm matrix [91]. They hypothesized that this was caused by differences in the amounts of eDNA released in the matrix. Phenazines are indeed responsible for cell lysis in biofilms, leading to the release of eDNA [94]. But in P. aeruginosa, another mechanism has been proposed to explain the effect of phenazines on biofilms: a phosphodiesterase called RmcA (redox modulator of c-di-GMP) could directly or indirectly detect phenazines and degrade cyclic-di-GMP, which plays an important role in the regulation of the biofilm structure [95]. Given that phenazines display different redox potentials, they may differentially affect eDNA release and RmcA activity, leading to the production of distinct biofilm structures, impacting rhizosphere colonization. Another interesting property of phenazines, especially pyocyanin, is their electron shuttle activity. They can accept electrons from NAD(P)H and transfer them outside the bacteria to reduce poorly crystalline iron (hydr)oxide, especially in environments where oxygen concentration is too low to use oxygen as a final electron acceptor, such as in biofilms [9], [96]. This activity may be used by the cells as a redox buffer and as an iron mobilization mechanism in iron-limiting conditions. In summary, phenazines play a role in rhizosphere colonization not only by inhibiting microbial rivals, but also by improving biofilm structure, iron availability, and anaerobic respiration.
Fig. 4.
Antibiotics produced by plant-beneficial Pseudomonas spp.
Cyclic lipopeptides (CLP) are highly diverse surfactants with antimicrobial activities. Their structures, biosynthesis and biological activities have recently been reviewed by Geudens and Martins [97] and by Götze and Stallforth [98]. CLP contain a cyclic oligopeptide (8–25 amino acids) and a linear fatty acid. They display a high structural diversity. Between 2009 and 2020, the number of identified CLP structural groups in Pseudomonas spp. almost doubled, increasing from 6 to 11 groups [88], [98]. This has been fueled by the identification of many new CLP, with around 100 CLP identified to date, enabled by whole-genome sequencing of many Pseudomonas strains coupled to the development of powerful bioinformatic analysis tools such as antiSMASH [99], [100]. CLP are produced by non-ribosomal peptide synthetases (NRPS), which are multi-modular megaenzymes displaying, in some cases, a molecular weight higher than 1.0 MDa [98]. CLP antimicrobial activity is thought to rely on membrane perturbation, especially pore formation, enabled by their amphiphilic nature [101]. However, their structural diversity suggests other potential antimicrobial modes of action that remain to be discovered [97]. Similarly to phenazines, the role of CLP extends beyond microbial inhibition: they can differently affect biofilm formation and dispersal depending on the produced CLP. For example, massetolide A improves biofilm formation in P. fluorescens SS101 [102] while viscosin (Fig. 4) facilitates cell dispersal in P. fluorescens SBW25 biofilms [103]. They can also contribute to swarming motility, i.e. rapid multicellular movements of bacteria across a surface, by reducing the critical surface tension of liquids [104]. This has been demonstrated in multiple rhizosphere-colonizing strains like in P. fluorescens SBW25 and in P. protegens Pf-5 with viscosin and orfamide A, respectively [97]. Therefore, CLP play several crucial roles in rhizosphere colonization.
Polyketides are a structurally and functionally diverse class of secondary metabolites produced by bacteria and fungi with antimicrobial, therapeutic and phytotoxic properties [88]. Polyketides biosynthesis is similar to fatty acid biosynthesis and is mediated by polyketide synthases (PKS), which are able to produce more diverse compounds than fatty acid synthases [105]. PKS have been divided into three categories: type I PKS are large and multifunctional proteins, type II PKS are complex-forming monofunctional proteins usually dedicated to the production of aromatic polyketides, and type III PKS are chalcone synthase-like proteins displaying a simple architecture [106], [88]. In plant-beneficial Pseudomonas, the main described polyketides are 2,4-diacetylphloroglucinol (2,4-DAPG), mupirocin and pyoluteorin [105] (Fig. 4). 2,4-DAPG plays an important role in the generation of suppressive soils against Gaeumannomyces graminis var. tritici, the wheat pathogen responsible for take-all [107]. It inhibits fungi by dissipating the mitochondrial proton gradient through its proton ionophore activity [108]. Mupirocin directly inhibits the bacterial isoleucyl-tRNA synthetase, leading to impaired protein biosynthesis [88]. To our knowledge, how pyoluteorin affects microbes, especially regarding the inhibition of the growth of the oomycete Pythium ultimum, remains to be deciphered [109]. Biosynthesis of these three polyketides is mediated by PKS and other proteins encoded by the phl, mmp/mup/macp and plt gene clusters, respectively [110], [111], [112]. Although the discovery of the mupirocin biosynthetic genes dates back to 2003, research is still underway to understand the role of each of the numerous genes involved, given the complexity of the biosynthetic pathway [113]. Interestingly, a co-regulation mechanism has been recently described in the biosynthesis of 2,4-DAPG and pyoluteorin in P. protegens Pf-5 [114]. Yan et al. have shown that a protein encoded by the pyoluteorin gene cluster converts an intermediate of the 2,4-DAPG biosynthetic pathway into intra- and intercellular signals able to induce the expression of pyoluteorin biosynthetic genes. This coregulation could allow the bacteria to produce either one or the other polyketide, depending on environmental conditions and especially on the presence of microbial rivals, instead of wasting energy by producing both [114]. Several bioinformatic tools have been developed to identify new polyketide biosynthetic gene clusters, such as antiSMASH or NaPDoS, which are also able to find NRPS [99], [115]. Using such tools, NRPS-PKS hybrid gene clusters have been discovered in plant-beneficial Pseudomonas spp. [9], [116]. For example, rhizoxin and its analogs are synthetized by PKS and mixed PKS-NRPS in P. protegens Pf-5 [116]. Rhizoxin analogs have been shown to play a role in the biocontrol activity of Pseudomonas sp. Os17 against the plant pathogens Fusarium oxysporum and Pythium ultimum [117]. Recently, Lozano et al. identified a new family of bacterial tetrahydropyridine alkaloids in P. koreensis: koreenceine A to D [118]. These are analogs of plant alkaloids produced by a type II PKS and selectively inhibit diverse rhizosphere Bacteroidetes. The associated biosynthetic gene cluster has been identified in numerous Pseudomonas spp. genomes.
Bacteriocins are secreted peptides and proteins deleterious for bacteria that are usually closely related to those producing them [119]. Most other antimicrobial compounds produced by plant-beneficial Pseudomonas spp. are effective against phylogenetically distant microbes and seldomly affect bacteria belonging to the same genus [120]. Hence, bacteriocins play an important role to control closely related bacteria in the rhizosphere, especially Pseudomonas spp. [121]. They include a wide array of structurally and functionally diverse compounds. Bacteriocin diversity, genomics, structure, transport and biological properties within the Pseudomonas genus have been extensively reviewed by Ghequire and De Mot [119]. The first identification of a bacteriocin in Pseudomonas spp. dates to 1954, in P. pyocyanea (now known as P. aeruginosa) by Jacob, who named the discovered substance pyocin, by analogy to the colicins, which are bacteriocins described in E. coli [122]. Since then, many bacteriocins have been described in the Pseudomonas genus, especially leading to a diversification of the pyocin family into different types: S-, R- and F-type pyocins [123]. R- and F-type pyocins are similar to bacteriophage tails and are called tailocins [124]. Pseudomonas spp. also harbor other bacteriocins, such as rearrangement hotspot proteins (Rhs), lectin-like bacteriocins, microcins and the contact-dependent inhibition system [119]. Bacteriocins affect cell survival through distinct modes of action such as nuclease activity against DNA, tRNA and rRNA, cell wall disorganization or membrane depolarization by pore formation [119]. They are usually coupled to an immunity protein to prevent self-inhibition [119]. Many bacteriocins have been identified in rhizosphere-associated Pseudomonas spp. genomes [125], [126], [127]. Recently, Sharp et al. developed a bioinformatics pipeline based on Hidden Markov Models to identify new nuclease bacteriocins in the genomes of multiple bacterial species isolated from diverse ecological niches [128]. They found more than 3000 bacteriocin genes, exclusively in γ-Proteobacteria, especially in Pseudomonas strains originating from soil or plant environments. These genes will require further characterization to elucidate their precise roles, but their identification already underlines their ubiquity in the genomes of underground Pseudomonas spp. Recently, Dorosky et al. investigated the role of two R-tailocins in the phytobeneficial strain P. chlororaphis 30–84 in the rhizosphere [129]. They showed that their killing spectra were distinct and limited to Pseudomonas spp. They also provided evidence that the loss of tailocin production impeded the strain competitiveness against other strains in biofilms and in the rhizosphere. In a subsequent study, they highlighted the importance of these two tailocins in competition with the native microflora in the wheat rhizosphere [130]. Mutants unable to produce them were not able to sustain populations as high as those of the wild type in the wheat rhizosphere over multiple harvest cycles.
Type VI secretion systems (T6SS) are contractile bacteriophage-like apparatuses able to inject toxic effectors into neighboring cells [131]. T6SS and their effectors often target other bacteria, from the same genus as the producer or not, but can also affect eukaryotic cells, contrary to bacteriocins [119]. Their main role in plant-associated bacteria seems to be in interbacterial competition [132]. T6SS genes are enriched in root and rhizosphere bacterial microbiomes in barley, indicating their importance in rhizosphere colonization and survival [133]. Multiple plant-beneficial Pseudomonas strains have been shown to carry at least one T6SS gene cluster, and up to four distinct clusters [132]. They also display multiple effector genes [126]. Those are usually followed by genes encoding immunity proteins, which protect the producing strains from the toxicity of their cognate effectors [119]. For example, in P. putida KT2440, Bernal et al. identified ten T6SS effector-immunity pairs, including putative nucleases, pore-forming colicins and a NAD(P)(+) glycohydrolase [134]. They also showed that a specific T6SS, called K1, was functional and responsible for the suppression of multiple bacterial plant pathogens. To our knowledge, the role of T6SS has not been explored in rhizosphere colonization by Pseudomonas spp. yet. However, this has recently been performed with rice endophytes belonging to the Kosakonia genus, which are also Gram-negative bacteria [135]. Mosquito et al. showed that a T6SS-deficient mutant displayed a reduced rice rhizoplane non-competitive colonization compared to the wild type, indicating a role for the T6SS in colonization and eventually, in plant-bacteria interactions. Several studies on the air isolate P. fluorescens strain MFE01 have highlighted the involvement of T6SS in motility and in biofilm formation [136], [137], [138]. A mutation in hcp1, encoding a component of the phage-like structure in T6SS, results in pleiotropic effects, especially in the loss of flagella, leading to the loss of motility. This could be the result of the accumulation of motility-related effectors in the cytoplasm of the producing strain [136]. This mutation also decreases exopolysaccharide accumulation, without impairing biofilm formation. On the contrary, a mutation in tssC, encoding a conserved T6SS cytoplasmic protein, severely affects biofilm formation [137]. Because this strain lacks the AHL quorum-sensing pathway, Gallique et al. hypothesized that the T6SS in this strain plays a cell–cell signalling role required for biofilm formation. This role remains to be confirmed, but it might occur in other Pseudomonas strains as well and affect rhizosphere colonization. T6SS are also involved in iron acquisition. Chen et al. have indeed shown that the T6SS of P. taiwanensis CMST played a role in the secretion of pyoverdine, an iron chelator [139]. The underlying mechanism is not elucidated yet. Many other questions about T6SS in plant-beneficial Pseudomonas spp. remain unanswered, such as regarding the underlying mechanisms of host specificity and of the recruitment of effectors by the T6SS [140]. Further characterization of the T6SS effectors in plant-associated Pseudomonas spp. is also required to better understand their roles in rhizosphere colonization [132].
Tropolones are non-benzenoid seven-membered aromatic compounds harboring a carbonyl group [141]. They have primarily been described in fungi and plants, but also in some Pseudomonas strains, especially for their interesting antibiotic and iron-chelating activities. While most of these strains have been isolated from habitats other than soils, like P. donghuensis DSM 101685 from lake water [142], a soil-inhabiting strain has been recently discovered: P. donghuensis SVBP6 [143]. This strain inhibits several fungal plant pathogens by producing 7-hydroxytropolone (HT) (Fig. 4). A 16-kb region has been shown to be involved in the biosynthesis of HT in this strain, but also in antibacterial activity in another Pseudomonas strain [143], [144]. Interestingly, HT was initially identified for its iron-chelating activity in P. donghuensis DSM 101685 [142]. Thus, the antifungal mechanism described by Muzio et al. in P. donghuensis SVBP6 could have been explained by this property contributing to iron depletion to the detriment of other microbes [143]. They however argued that another explanation was more plausible, especially given that iron supplementation did not suppress fungal growth inhibition and did not stop HT biosynthesis. They posited that the mechanism underlying HT antifungal activity could also be the direct inhibition of eukaryotic dinuclear metalloenzymes containing divalent ions like Cu2+, Mg2+ and Zn2+, as described in other tropolones [145]. The exact antibacterial modes of action of HT remain to be confirmed. The discovery of a soil-inhabiting Pseudomonas strain producing it shows that the biosynthesis of this compound could be an important trait for survival in soils and in the rhizosphere.
Finely tuned antibiotic production is essential for rhizosphere colonization, not only because of the metabolic cost of antibiotic biosynthesis, but also because producing too much antibiotics could lead to resistance emergence in microbial rivals, or even provide valuable substrates for antibiotic degraders [146]. Their production is often under the control of quorum sensing, a system regulating gene expression in response to cell density [147]. It relies on the constitutive production and secretion of the auto-induction signal molecules N-acyl-homoserine lactones (AHL) [148]. In biofilms, AHL concentration increases because of high cell densities and reaches a given threshold, triggering the transcription of target genes, especially responsible for secondary metabolites biosynthesis [149]. For example, phenazine production in Pseudomonas spp. is partly controlled by quorum sensing through the LuxI/LuxR homologues PhzI/PhzR [9]. PhzI is an AHL synthase leading to AHL accumulation. When the produced AHL reach a concentration threshold, they bind to and activate PhzR, a transcriptional regulator which in turn binds to the phenazine biosynthetic operon promoter to enhance the operon transcription. Phenazine production is also regulated by the highly conserved global activator of antibiotic and cyanide (Gac) signal transduction system, which also controls the production of other secondary metabolites such as 2,4-DAPG or cyclic lipopeptides [9], [150], [151]. The Gac system relies on two components: GacS, a transmembrane sensor kinase, and GacA, a cytoplasmic transcription regulator (Fig. 3) [152]. GacS reacts to an unknown environmental signal by phosphorylating GacA, which then promotes the expression of the small non-coding RNA (sRNA) rsmZ [153]. This sRNA likely binds and represses the regulator of secondary metabolism RsmE, which posttranscriptionally inhibits the expression of the phenazine biosynthetic operon and phzI by binding to specific mRNA motifs and preventing ribosome binding [154]. Another two-component signal transduction system is also involved in the regulation of phenazine production: RpeA/RpeB [155]. The transmembrane sensor kinase RpeA detects an environmental signal and activates RpeB, which enhances the expression of pip, encoding the phenazine inducing protein, a transcriptional regulator promoting PhzI and PhzR expression [153], [156]. The expression of pip is also positively regulated by the RNA polymerase sigma factor RpoS, the expression of which is promoted by GacA [153]. Interestingly, the Gac system also positively regulates the expression of the previously-mentioned biofilm-associated genes lapA and lapF, along with genes involved in the biosynthesis of many antibiotic compounds and extracellular enzymes [157], [158]. Spontaneous mutations in the GacS/GacA system have often been observed and are responsible for phenotypic variation in Pseudomonas spp. and the loss of antibiotics production [159], [160]. These mutations have been shown to be triggered by the site-specific recombinases encoded by sss and xerD [161]. Overexpression of these genes can lead to improved competitive rhizosphere colonization. Indeed, they contribute to the emergence of phenotypic variants that are able to colonize distinct root parts [161]. Interestingly, gac mutations are reversible, allowing bacterial subpopulations to switch from a phenotype to another [162]. Further research is still required to understand the underlying molecular mechanisms of these phenotypic variations.
3.2. Laying hands on iron
In addition to the direct inhibition of microbial rivals through antibiotic production, plant-beneficial Pseudomonas spp. are also able to efficiently compete for iron in the rhizosphere. Iron is a common element in soil, but is mostly unavailable for microbes due to the mediocre solubility of iron oxides [163]. Because it is essential for the primary metabolism of most organisms, including plants and microbes, competition for iron is exacerbated in the rhizosphere [13]. This competition can lead to high inhibition levels against plant pathogens like fungi, making iron starvation one of the main biocontrol mechanisms against fungi [12]. To gain access to iron, many microbes, including Pseudomonas spp., secrete low-molecular iron-chelating compounds called siderophores [164]. Rhizosphere Pseudomonas spp. generally produce the siderophore pyoverdine, which displays high affinity for Fe(III) [165]. Most Pseudomonas spp. are able to produce additional siderophores, such as achromobactin, enantio-pyochelin, pseudomonine, HT or the hemophore protein [8], [125], [126], [142]. Their biosynthesis can be mediated by NRPS, like pyoverdine, enantio-pyochelin and pseudomonine, or by other pathways, like achromobactin or the hemophore through the acs and has genes, respectively [164], [166], [167]. Once secreted in the rhizosphere, siderophores sequester ferric iron and the ferrisiderophores are actively taken back by Pseudomonas spp. through a TonB-dependent outer membrane receptor [168]. Iron is separated from the siderophore in the periplasm through reduction and transported alone into the cytoplasm, while the siderophore can be secreted again [169]. Pseudomonas spp. are also able to take up siderophores produced by other microbes in order to increase their iron supply and eventually control their competitors [170]. This is enabled by the presence of multiple TonB-dependent receptors genes in their genomes [168]. In the genome of P. protegens Pf-5, Hartney et al. identified 45 of these proteins [171]. They could be involved in broader resources acquisition systems than iron uptake only. Interestingly, some pyocins have been shown to enter bacteria through TonB-dependent receptors dedicated to the translocation of iron-bound siderophores inside P. aeruginosa [172]. While such mechanisms have not yet been identified in rhizosphere-inhabiting Pseudomonas spp., it highlights the importance of iron for Pseudomonas spp. survival. In the rhizosphere, the role of siderophore production in colonization, especially pyoverdine, has been demonstrated using mutants in multiple Pseudomonas species [173], [174].
3.3. Evading the plant immune response
The rhizosphere is shaped by plants through rhizodeposition, but also through their immune system: they are able to detect and react to potentially harmful microbes, especially through the recognition of microbe-associated molecular patterns (MAMP), in order to prevent infection [175]. Beneficial microbes are also identified by the plant immune system as potential threats, but they are able to repress or evade the immune response in order to efficiently colonize the rhizosphere [87], [147].
Evasion mostly consists in preventing pattern-triggered immunity (PTI), which occurs through the detection of MAMP such as the conserved bacterial flagellin epitope flg22 [176]. A major mechanism in plant immunity evasion is to decrease flagella synthesis [177]. In Pseudomonas spp., flagella synthesis is regulated by cyclic-di-GMP, which mediates the transition between the planktonic and sessile lifestyle [17]. High levels of cyclic-di-GMP inhibit flagellin biosynthesis, preventing ROS production by the plant [178]. The putative phosphodiesterase MorA could be involved in the process, as shown by Liu et al. [179]. Indeed, a Pseudomonas sp. WCS365 mutant defective in MorA induced PTI in Arabidopsis thaliana, contrary to the wild type. Because phosphodiesterases are known to be involved in cyclic-di-GMP regulation, MorA may play an important role in plant immunity evasion. Liu et al. also identified a potential role of putrescine in plant defense evasion through the putrescine aminotransferase SpuC [179]. Mutants lacking a functional SpuC triggered PTI in A. thaliana. The authors posited that the loss of SpuC caused physiological changes in the bacteria responsible for plant recognition, and that putrescine or its precursor, arginine, might be signalling molecules in the rhizosphere used by the bacteria to evade the plant immune response. This is consistent with recent results obtained by Barrientos‑Moreno et al. indicating that arginine promotes biofilm formation through an increase in cyclic-di-GMP concentration [180]. Phase variation may also be used by rhizosphere Pseudomonas spp. to evade plant immunity by generating bacterial subpopulations displaying different levels of flagellin production [87]. Another interesting flg22-recognition evasion mechanism has recently been demonstrated by Yu et al. in P. capeferrum WCS358 [181]. Mutants impaired in the genes responsible for gluconic acid and 2-keto gluconic acid production, pqqF and cyoB, respectively, were unable to suppress the flg22-induced root immune response and displayed reduced A. thaliana rhizosphere colonization levels. The authors demonstrated that this mechanism relied on a decrease in environmental pH through acid secretion. To prevent PTI, Pseudomonas spp. can also produce the alkaline protease AprA, which directly degrades monomeric flagellin required for recognition by plants [182]. While this mechanism has originally been deciphered in pathogenic species such as P. aeruginosa and P. syringae, homologs of AprA have also been identified in rhizosphere-inhabiting Pseudomonas spp. such as P. fluorescens strains WCS374 and WCS417 [183], [184]. It has also been found in P. brassicacearum, where it is repressed in the flagellin over-producing phenotypic variant, strengthening the hypothesis of a role of phase variation in immunity evasion [87]. Cole et al. recently identified mutants of P. simiae WCS417r lacking functional genes homologous to arnACDEFT displaying lower in vitro root colonization abilities than the wild type [185]. In Escherichia coli, these genes encode proteins responsible for the modification of lipid A, the lipophilic moiety of lipopolysaccharides anchoring them to the external layer of the outer membrane [186]. It protects the Gram-negative bacteria from antimicrobial compounds, but can also be recognized by plants and induce their defenses [187]. These genes may then be involved in plant immunity evasion.
Plant-beneficial Pseudomonas spp. could also directly repress the plant immune response using the type III secretion systems (T3SS). T3SS are transmembrane transport apparatuses found in Gram-negative bacteria and mediating direct interactions with eukaryotic cells by delivering effector proteins to affect host cellular functions [188]. The plant pathogen P. syringae has been shown to suppress the plant immune response during infection using its T3SS [189]. In the rhizosphere, many Pseudomonas strains display T3SS gene clusters, with sometimes distinct T3SS harbored by a single strain [125], [126], [190]. Their genomes also contain up to 15 T3SS effector genes [125], [184]. Mavrodi et al. investigated the role of T3SS effectors in P. fluorescens Q8r1-96 [190]. They showed that three effectors were able to suppress both the PAMP- and the effector-triggered immunity in Nicotiana benthamiana. However, mutants defective in a T3SS displayed similar rhizosphere colonization levels as the wild type. The role of T3SS in Pseudomonas spp. inhabiting the rhizosphere remains to be elucidated [191]. While they could be involved in the suppression of the root immune response, they could also play a role in interactions with other eukaryotes. It has been shown that the T3SS of P. fluorescens C7R12 was required to promote mycorrhization in Medicago truncatula, indicating a more complex role in the rhizosphere than plant immune evasion only [192]. To better understand how the plant immune system can be affected by beneficial Pseudomonas spp., Stringlis et al. recently performed RNA sequencing of A. thaliana in response to the inoculation with P. simiae WCS417 [193]. They demonstrated that half of the MAMP-triggered transcriptional responses was actively suppressed by the bacterium. Aside from T3SS and their effectors, a great diversity of bacterial molecules are known to impact the plant physiology and may take part in the immune suppression, as reviewed by Stringlis et al. [194]. Further research is needed to understand the underlying mechanisms allowing the bacteria to affect the plant immune responses.
4. Metabolic versatility
Members of the Pseudomonas genus are known for their great metabolic diversity, which allows them to colonize a vast range of environments, including the rhizosphere [7], [195]. Rhizodeposits released by plants, especially root exudates, offer to the rhizosphere microbiome a diversity of substrates to thrive on, such as organic acids, carbohydrates, fatty acids, amino acids and proteins [196]. The importance of root exudates use in competitive rhizosphere colonization has long been investigated [197], [198]. Latour et al. demonstrated that Pseudomonas spp. isolated from the rhizosphere have generally broader catabolic activities than those isolated from bulk soil, especially regarding specific sugars, polyols and amino acids that can be found in root exudates [199]. We recently performed a comparative study on 60 Pseudomonas strains showing a positive link between the ability to use some of these specific organic compounds (trehalose, sucrose and citrulline) and rhizosphere colonization in A. thaliana and in potato [200]. We also highlighted an association between the ability to use nitrous compounds, such as amines and amino acids, and high rhizosphere colonization levels. Several other studies have shown the importance of nitrous substrates for rhizosphere Pseudomonas spp., especially through the role of transport and catabolism genes [37], [185]. Cole et al. ingeniously used randomly barcoded transposon mutagenesis sequencing (RB-TnSeq) in P. simiae WCS417r to identify the genes required for A. thaliana competitive root colonization [185]. They found 115 genes with functions involved in high root colonization, especially in carbon and nitrogen transport and metabolism. Twenty-four of these genes were identified for their role in the metabolism of carbon-containing compounds: specific sugars (galactose, galacturonate, and glucose) and nucleosides (inosine and 2-deoxyribose). These compounds can be released by plants through exudation and are certainly used by rhizosphere Pseudomonas spp. to sustain their growth in the rhizosphere. Further investigation is required to explore the possibilities of substrate use preferences by Pseudomonas spp. to eventually manipulate soil composition and improve rhizosphere colonization by specific PGPR. Also, a better characterization of root exudates in diverse plant species could improve the identification of compatible bacterial strains, offering promising applications.
Nitrogen plays a major role in the metabolism of rhizosphere-dwelling Pseudomonas spp., especially through nitrate reduction [201]. This process primarily occurs through three mechanisms (and their associated nitrate reductase genes): dissimilation (nap) respiration (nar), and assimilation (nas). Nitrate dissimilation is a redox balancing mechanism occurring in the periplasm and thought to maintain the bacterial redox balance during the shift between aerobic and anaerobic conditions [202]. Nitrate respiration consists in using nitrate as a final electron acceptor, an alternative to dioxygen in the respiratory chain. Iron oxides can also be used as another electron acceptor in this process [199]. This is especially useful in anaerobic conditions such as in compact or moist soils. Nitrate assimilation enables the bacteria to use nitrate when other nitrogen sources are lacking. In this process, nitrate is reduced into nitrite, which is in turn reduced into ammonium to be directly used in anabolism. The genes encoding nitrate (and nitrite) reductases have been identified in many rhizosphere-associated Pseudomonas spp. [8], [126] and their roles in rhizocompetence have often been confirmed [203], [204]. However, the interplay between the three nitrate reduction mechanisms in root colonization remains unclear [201].
Sulfur is another essential element for microbes and plants, and occurs in soils mostly in the form of sulfate esters and carbon-bonded sulfur [205]. Sulfate esters are an important soil sulfur source for Pseudomonas spp., especially in aerobic soils [206]. In P. putida S-313, the use of sulfate esters is mediated by the enzymes encoded by the ats-sft gene cluster. It notably contains sftP, an outer membrane receptor belonging to the TonB-dependent receptor family, the protein family involved in siderophore uptake. Mutants unable to use sulfate esters show reduced survival in soils and in the tomato rhizosphere, as well as reduced plant-growth promotion abilities [206], [207], [208]. While sulfate use seems to be an important rhizosphere and soil survival trait, it is often overlooked in rhizosphere colonization studies and in genome analyses.
Metabolism regulation is crucial for Pseudomonas spp. to colonize the rhizosphere, which is a nutritionally dynamic niche. Their metabolism is under the control of a sophisticated global regulation system called the carbon catabolite repression (CCR) system, which has been recently reviewed by Bharwad et al. [209]. It is especially mediated by the catabolite repression control protein, which notably regulates the transport and assimilation of amino acids and sugars [210]. Carbon primary metabolism is partly controlled by two transcriptional regulator: HexR and RccR [211]. Mutants of P. fluorescens SBW25 lacking the hexR and/or rccR genes displayed reduced competitive rhizosphere colonization levels in wheat. These structurally close proteins finely regulate the synthesis of enzymes involved in carbon primary metabolism pathways, leading to optimal use of available nutrients by Pseudomonas spp. Little et al. have recently identified rccA, immediately located upstream of rccR in P. fluorescens SBW25 and encoding a putative phosphodiesterase [212]. They showed that RccA interacted with RccR and they postulated that it could degrade cyclic-di-GMP. Therefore, they hypothesized that RccA could be involved in the coordination between the bacterial metabolic state and the bacterial lifestyle transition associated with cyclic-di-GMP during rhizosphere colonization. Its precise functions remain to be deciphered. Another major metabolism regulation pathway involves the RimK (ribosomal modification) and Hfq (host factor required for phage Qβ RNA replication) proteins at the post-transcriptional level (Fig. 3) [213]. RimK is an ATP-dependent ligase adding glutamate residues to RpsF, the ribosomal protein S6, which is part of the 30S ribosomal subunit. This modification affects ribosome function and leads to an altered translational activity and proteome composition, mostly through Hfq [214]. Hfq is a hexameric RNA-binding protein that can bind to multiple sRNA and modulate the expression of their target mRNA [213]. How Hfq is affected by RimK and RpsF remains to be elucidated. Grenga et al. suggest that RpsF glutamation by RimK disturbs the binding of Hfq with another translation-required protein, S1, causing a redistribution of Hfq in the cytoplasm [215]. Hfq controls many essential bacterial processes involved in rhizosphere colonization, such as the catabolite repression system, biofilm formation, quorum sensing, iron homeostasis and amino acid utilization and transport [213]. Mutations in rimK or hfq affect the translation of several hundred genes and lead to impaired rhizosphere colonization abilities, demonstrating the importance of these proteins in this process [214]. RimK glutamation activity is controlled by other Rim proteins, by cyclic-di-GMP level and by environmental cues, which enable the bacteria to quickly tune their translation processes according to environmental changes.
5. Summary and outlook
The burst in both bacterial genome sequencing and bioinformatic tools development over the last 15 years enabled researchers to identify formerly inaccessible genes, proteins and mechanisms involved in rhizosphere colonization by phytobeneficial Pseudomonas spp. This is a complex and dynamic process essential for plant growth promotion and biocontrol of plant pathogens. It is driven by multiple interacting bacterial factors under the control of global regulators such as the Gac, FleQ/AmrZ, RimK/Hfq and CCR pathways, often involving the second messenger cyclic-di-GMP. This process requires lifestyle transitions, antibiotics biosynthesis, iron sequestration, plant immune system evasion, and a finely tuned versatile metabolism. These are the main bacterial determinants of rhizosphere colonization by Pseudomonas spp. Successful colonization also relies on external factors such as the soil type and condition, the microbiome composition, and the host plant. They should also be considered to better understand rhizosphere ecology and to develop commercial applications of single high-efficiency biocontrol or plant growth-promoting strains. Future research on this topic should especially focus on the modes of action of antibiotics such as phenazines and T6SS effectors, the biofilm dispersal mechanisms in the rhizosphere, the effects of beneficial Pseudomonas spp. on the physiology and immune system of plants, and the role of cyclic-di-GMP in the fine regulation of rhizosphere colonization. The availability of complete genomes and bioinformatic tools will be crucial to investigate these issues.
CRediT authorship contribution statement
Antoine Zboralski: Conceptualization, Investigation, Writing - original draft, Writing - review & editing. Martin Filion: Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Natural Sciences and Engineering Research Council of Canada and New Brunswick Innovation Foundation grants to M.F. We thank Adrien Biessy for proofreading the manuscript and the reviewers for their insightful comments and suggestions.
References
- 1.Hiltner L. Über neuere Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie unter besonderer Berücksichtigung der Gründüngung und Brache. Arbeiten Der Deutschen Landwirtschaftlichen Gesellschaft. 1904;98:59–78. [Google Scholar]
- 2.Dennis PG, Miller AJ, Hirsch PR. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 2010;72:313–27. https://doi.org/10.1111/j.1574-6941.2010.00860.x. [DOI] [PubMed]
- 3.Zboralski A, Biessy A, Filion M. Rhizosphere colonization by plant-beneficial Pseudomonas spp.: thriving in a heterogeneous and challenging environment. In: Singh HB, editor. Advances in PGPR research, Oxfordshire, UK: CAB International; 2017, p. 197–217.
- 4.Lugtenberg B., Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol. 2009;63(1):541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 5.Backer R, Rokem JS, Ilangumaran G, Lamont J, Praslickova D, Ricci E, et al. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front Plant Sci 2018;9:1473. https://doi.org/10.3389/fpls.2018.01473. [DOI] [PMC free article] [PubMed]
- 6.Weller D.M. Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology®. 2007;97(2):250–256. doi: 10.1094/PHYTO-97-2-0250. [DOI] [PubMed] [Google Scholar]
- 7.Stanier R.Y., Palleroni N.J., Doudoroff M. The Aerobic pseudomonads a taxonomic study. J Gen Microbiol. 1966;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 8.Garrido-Sanz D, Meier-Kolthoff JP, Göker M, Martín M, Rivilla R, Redondo-Nieto M. Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS One 2016;11:e0150183. https://doi.org/10.1371/journal.pone.0150183. [DOI] [PMC free article] [PubMed]
- 9.Biessy A., Filion M. Phenazines in plant-beneficial Pseudomonas spp.: biosynthesis, regulation, function and genomics : phenazines in plant-beneficial Pseudomonas spp. Environ Microbiol. 2018;20(11):3905–3917. doi: 10.1111/1462-2920.14395. [DOI] [PubMed] [Google Scholar]
- 10.Haas D., Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3(4):307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 11.Sosa M.F., Sobrero P., Valverde C., Agaras B. A black-pigmented pseudomonad isolate with antibacterial activity against phyllospheric pathogens. Rhizosphere. 2020;15:100207. doi: 10.1016/j.rhisph.2020.100207. [DOI] [Google Scholar]
- 12.Thomashow L., Bakker P.A.H.M. Microbial Control of Root-Pathogenic Fungi and Oomycetes. In: Lugtenberg B., editor. Principles of Plant-Microbe Interactions. Springer International Publishing; Cham, Switzerland: 2015. pp. 165–173. [DOI] [Google Scholar]
- 13.Robin A, Vansuyt G, Hinsinger P, Meyer JM, Briat JF, Lemanceau P. Iron dynamics in the rhizosphere: consequences for plant health and nutrition. In: Sparks DL, editor. Advances in agronomy, vol. 99, London, UK: Academic Press; 2008, p. 183–225.
- 14.Richardson A.E., Barea J.-M., McNeill A.M., Prigent-Combaret C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil. 2009;321(1-2):305–339. doi: 10.1007/s11104-009-9895-2. [DOI] [Google Scholar]
- 15.Pieterse C.M.J., Zamioudis C., Berendsen R.L., Weller D.M., Van Wees S.C.M., Bakker P.A.H.M. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52(1):347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 16.Sørensen J., Nybroe O. Pseudomonas. Springer US; Boston, MA: 2004. pp. 369–401. [DOI] [Google Scholar]
- 17.Valentini M., Filloux A. Biofilms and Cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 2016;291(24):12547–12555. doi: 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danhorn T., Fuqua C. Biofilm formation by plant-associated bacteria. Annu Rev Microbiol. 2007;61(1):401–422. doi: 10.1146/annurev.micro.61.080706.093316. [DOI] [PubMed] [Google Scholar]
- 19.Sampedro I., Parales R.E., Krell T., Hill J.E. Pseudomonas chemotaxis. FEMS Microbiol Rev. 2014:n/a–n/a. doi: 10.1111/1574-6976.12081. [DOI] [PubMed] [Google Scholar]
- 20.Pliego C, de Weert S, Lamers G, de Vicente A, Bloemberg G, Cazorla FM, et al. Two similar enhanced root-colonizing Pseudomonas strains differ largely in their colonization strategies of avocado roots and Rosellinia necatrix hyphae. Environ Microbiol 2008;10:3295–304. https://doi.org/10.1111/j.1462-2920.2008.01721.x. [DOI] [PubMed]
- 21.Wadhams G.H., Armitage J.P. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5(12):1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 22.Molina L., Rosa R.L., Nogales J., Rojo F. Pseudomonas putida KT2440 metabolism undergoes sequential modifications during exponential growth in a complete medium as compounds are gradually consumed. Environ Microbiol. 2019;21(7):2375–2390. doi: 10.1111/1462-2920.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacal J, García-Fontana C, Muñoz-Martínez F, Ramos J-L, Krell T. Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ Microbiol 2010;12:2873–84. https://doi.org/10.1111/j.1462-2920.2010.02325.x [DOI] [PubMed]
- 24.Parales RE, Luu RA, Chen GY, Liu X, Wu V, Lin P, et al. Pseudomonas putida F1 has multiple chemoreceptors with overlapping specificity for organic acids. Microbiology 2013;159:1086–96. https://doi.org/10.1099/mic.0.065698-0. [DOI] [PMC free article] [PubMed]
- 25.Muriel C, Jalvo B, Redondo-Nieto M, Rivilla R, Martín M. Chemotactic motility of Pseudomonas fluorescens F113 under aerobic and denitrification conditions. PLoS One 2015;10:e0132242. https://doi.org/10.1371/journal.pone.0132242. [DOI] [PMC free article] [PubMed]
- 26.López-Farfán D, Reyes-Darias JA, Matilla MA, Krell T. Concentration dependent effect of plant root exudates on the chemosensory systems of Pseudomonas putida KT2440. Front Microbiol 2019;10. https://doi.org/10.3389/fmicb.2019.00078. [DOI] [PMC free article] [PubMed]
- 27.Li M., Hazelbauer G.L. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci. 2011;108(23):9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sourjik V., Wingreen N.S. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol. 2012;24(2):262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarkar M.K., Paul K., Blair D. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc Natl Acad Sci. 2010;107(20):9370–9375. doi: 10.1073/pnas.1000935107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bi S., Lai L. Bacterial chemoreceptors and chemoeffectors. Cell Mol Life Sci. 2015;72(4):691–708. doi: 10.1007/s00018-014-1770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamer R., Chen P.-Y., Armitage J.P., Reinert G., Deane C.M. Deciphering chemotaxis pathways using cross species comparisons. BMC Syst Biol. 2010;4(1):3. doi: 10.1186/1752-0509-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertrand J.J., West J.T., Engel J.N. Genetic analysis of the regulation of Type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa. JB. 2010;192(4):994–1010. doi: 10.1128/JB.01390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11(1):137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 34.Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, Bertrand JJ, et al. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol 2004;52:873–93. https://doi.org/10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed]
- 35.Redondo-Nieto M., Barret M., Morrissey J., Germaine K., Martínez-Granero F., Barahona E., Navazo A., Sánchez-Contreras M., Moynihan J.A., Muriel C., Dowling D., O’Gara F., Martín M., Rivilla R. Genome sequence reveals that Pseudomonas fluorescens F113 possesses a large and diverse array of systems for rhizosphere function and host interaction. BMC Genomics. 2013;14(1):54. doi: 10.1186/1471-2164-14-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan Y., Yang J., Dou Y., Chen M., Ping S., Peng J., Lu W., Zhang W., Yao Z., Li H., Liu W., He S., Geng L., Zhang X., Yang F., Yu H., Zhan Y., Li D., Lin Z., Wang Y., Elmerich C., Lin M., Jin Q. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc Natl Acad Sci. 2008;105(21):7564–7569. doi: 10.1073/pnas.0801093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matilla MA, Espinosa-Urgel M, Rodríguez-Herva JJ, Ramos JL, Ramos-González M. Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol 2007;8:R179. https://doi.org/10.1186/gb-2007-8-9-r179. [DOI] [PMC free article] [PubMed]
- 38.Oku S., Komatsu A., Nakashimada Y., Tajima T., Kato J. Identification of Pseudomonas fluorescens chemotaxis sensory proteins for malate, succinate, and fumarate, and their involvement in root colonization. Microb Environ. 2014;29(4):413–419. doi: 10.1264/jsme2.ME14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Weert S., Vermeiren H., Mulders I.H.M., Kuiper I., Hendrickx N., Bloemberg G.V., Vanderleyden J., De Mot R., Lugtenberg B.J.J. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. MPMI. 2002;15(11):1173–1180. doi: 10.1094/MPMI.2002.15.11.1173. [DOI] [PubMed] [Google Scholar]
- 40.Capdevila S, Martínez-Granero FM, Sánchez-Contreras M, Rivilla R, Martín M. Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 2004;150:3889–97. https://doi.org/10.1099/mic.0.27362-0. [DOI] [PubMed]
- 41.Oku S., Komatsu A., Tajima T., Nakashimada Y., Kato J. Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microb Environ. 2012;27(4):462–469. doi: 10.1264/jsme2.ME12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao X, Li T, Liu W, Zhang Y, Shang D, Gao Y, et al. Enhancing the 1-aminocyclopropane-1-carboxylate metabolic rate of Pseudomonas sp. UW4 intensifies chemotactic rhizocompetence. Microorganisms 2020;8:71. https://doi.org/10.3390/microorganisms8010071. [DOI] [PMC free article] [PubMed]
- 43.Blaha D, Prigent-Combaret C, Mirza MS, Moënne-Loccoz Y. Phylogeny of the 1-aminocyclopropane-1-carboxylic acid deaminase-encoding gene acdS in phytobeneficial and pathogenic Proteobacteria and relation with strain biogeography. FEMS Microbiol Ecol 2006;56:455–70. https://doi.org/10.1111/j.1574-6941.2006.00082.x. [DOI] [PubMed]
- 44.Li T., Zhang J., Shen C., Li H., Qiu L. 1-Aminocyclopropane-1-carboxylate: a novel and strong chemoattractant for the plant beneficial rhizobacterium Pseudomonas putida UW4. MPMI. 2019;32(6):750–759. doi: 10.1094/MPMI-11-18-0317-R. [DOI] [PubMed] [Google Scholar]
- 45.Flemming H.-C., Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 46.Castiblanco L.F., Sundin G.W. New insights on molecular regulation of biofilm formation in plant-associated bacteria: molecular regulation of biofilm formation. J. Integr. Plant Biol. 2016;58(4):362–372. doi: 10.1111/jipb.12428. [DOI] [PubMed] [Google Scholar]
- 47.Monds R.D., O’Toole G.A. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17(2):73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Hinsa SM, Espinosa-Urgel M, Ramos JL, O’Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol 2003;49:905–18. https://doi.org/10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed]
- 49.Yousef-Coronado F, Travieso ML, Espinosa-Urgel M. Different, overlapping mechanisms for colonization of abiotic and plant surfaces by Pseudomonas putida. FEMS Microbiol Lett 2008;288:118–24. https://doi.org/10.1111/j.1574-6968.2008.01339.x. [DOI] [PubMed]
- 50.Martínez-Gil M, Yousef-Coronado F, Espinosa-Urgel M. LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol Microbiol 2010;77:549–61. https://doi.org/10.1111/j.1365-2958.2010.07249.x. [DOI] [PubMed]
- 51.Collins AJ, Pastora AB, Smith TJ, O’Toole GA. MapA, a second large RTX adhesin conserved across the Pseudomonads, contributes to biofilm formation by Pseudomonas fluorescens. J Bacteriol 2020:JB.00277-20, jb;JB.00277-20v1. https://doi.org/10.1128/JB.00277-20. [DOI] [PMC free article] [PubMed]
- 52.Collins A.J., Smith T.J., Sondermann H., O'Toole G.A. From input to output: the Lap/c-di-GMP biofilm regulatory circuit. Annu Rev Microbiol. 2020;74(1):607–631. doi: 10.1146/annurev-micro-011520-094214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo S., Vance T.D.R., Stevens C.A., Voets I., Davies P.L. RTX adhesins are key bacterial surface megaproteins in the formation of biofilms. Trends Microbiol. 2019;27(5):453–467. doi: 10.1016/j.tim.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 54.Buell C.R., Anderson A.J. Genetic analysis of the aggA locus involved in agglutination and adherence of Pseudomonas putida, a beneficial fluorescent pseudomonad. Mol Plant Microbe Interact. 1992;5:154–162. doi: 10.1094/mpmi-5-154. [DOI] [PubMed] [Google Scholar]
- 55.Smith TJ, Font ME, Kelly CM, Sondermann H, O’Toole GA. An N-terminal retention module anchors the giant adhesin LapA of Pseudomonas fluorescens at the cell surface: a novel subfamily of type I secretion systems. J Bacteriol 2018;200:e00734-17. https://doi.org/10.1128/JB.00734-17. [DOI] [PMC free article] [PubMed]
- 56.Satchell K.J.F. Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu Rev Microbiol. 2011;65(1):71–90. doi: 10.1146/annurev-micro-090110-102943. [DOI] [PubMed] [Google Scholar]
- 57.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 2010;75:815–26. https://doi.org/10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed]
- 58.Ansari FA, Jafri H, Ahmad I, Abulreesh HH. Factors affecting biofilm formation in in vitro and in the rhizosphere. In: Ahmad I, Husain FM, editors. Biofilms in plant and soil health, Chichester, UK: John Wiley & Sons, Ltd; 2017, p. 275–90. https://doi.org/10.1002/9781119246329.ch15.
- 59.Nielsen L, Li X, Halverson LJ. Cell-cell and cell-surface interactions mediated by cellulose and a novel exopolysaccharide contribute to Pseudomonas putida biofilm formation and fitness under water-limiting conditions. Environ Microbiol 2011;13:1342–56. https://doi.org/10.1111/j.1462-2920.2011.02432.x. [DOI] [PubMed]
- 60.Nilsson M, Chiang W-C, Fazli M, Gjermansen M, Givskov M, Tolker-Nielsen T. Influence of putative exopolysaccharide genes on Pseudomonas putida KT2440 biofilm stability. Environ Microbiol 2011;13:1357–69. https://doi.org/10.1111/j.1462-2920.2011.02447.x. [DOI] [PubMed]
- 61.Marshall D.C., Arruda B.E., Silby M.W. Alginate genes are required for optimal soil colonization and persistence by Pseudomonas fluorescens Pf0-1. Access Microbiol. 2019;1:e000021. doi: 10.1099/acmi.0.000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ueda A., Saneoka H. Characterization of the ability to form biofilms by plant-associated Pseudomonas species. Curr Microbiol. 2015;70(4):506–513. doi: 10.1007/s00284-014-0749-7. [DOI] [PubMed] [Google Scholar]
- 63.Wu L., Wang Z., Guan Y., Huang X., Shi H., Liu Y., Zhang X. The (p)ppGpp-mediated stringent response regulatory system globally inhibits primary metabolism and activates secondary metabolism in Pseudomonas protegens H78. Appl Microbiol Biotechnol. 2020;104(7):3061–3079. doi: 10.1007/s00253-020-10421-5. [DOI] [PubMed] [Google Scholar]
- 64.Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol 2012;14:1913–28. https://doi.org/10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed]
- 65.Franklin M.J., Nivens D.E., Weadge J.T., Howell P.L. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, pel, and Psl. Front Microbiol. 2011;2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitchurch C.B., Tolker-Nielsen T., Ragas P.C., Mattick J.S. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 67.Sarkar S. Release mechanisms and molecular interactions of Pseudomonas aeruginosa extracellular DNA. Appl Microbiol Biotechnol. 2020;104(15):6549–6564. doi: 10.1007/s00253-020-10687-9. [DOI] [PubMed] [Google Scholar]
- 68.Das T., Sehar S., Manefield M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development: the roles of eDNA in the bacterial biofilm development. Environ Microbiol Rep. 2013;5(6):778–786. doi: 10.1111/1758-2229.12085. [DOI] [PubMed] [Google Scholar]
- 69.Fuqua C. Passing the baton between laps: adhesion and cohesion in Pseudomonas putida biofilms. Mol Microbiol 2010;77:533–6. https://doi.org/10.1111/j.1365-2958.2010.07250.x. [DOI] [PMC free article] [PubMed]
- 70.Chin-A-Woeng T.F.C., de Priester W., van der Bij A.J., Lugtenberg B.J.J. Description of the colonization of a gnotobiotic tomato rhizosphere by Pseudomonas fluorescens biocontrol strain WCS365, using scanning electron microscopy. MPMI. 1997;10(1):79–86. doi: 10.1094/MPMI.1997.10.1.79. [DOI] [Google Scholar]
- 71.Walker T.S., Bais H.P., Grotewold E., Vivanco J.M. Root exudation and rhizosphere biology. Plant Physiol. 2003;132:44–51. doi: 10.1104/pp.102.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gjermansen M, Ragas P, Sternberg C, Molin S, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ Microbiol 2005;7:894–904. https://doi.org/10.1111/j.1462-2920.2005.00775.x. [DOI] [PubMed]
- 73.Petrova O.E., Sauer K. Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr Opin Microbiol. 2016;30:67–78. doi: 10.1016/j.mib.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Badri DV, Vivanco JM. Regulation and function of root exudates. Plant Cell Environ 2009;32:666–81. https://doi.org/10.1111/j.1365-3040.2009.01926.x. [DOI] [PubMed]
- 75.Dalebroux Z.D., Swanson M.S. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10(3):203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 76.Díaz-Salazar C., Calero P., Espinosa-Portero R., Jiménez-Fernández A., Wirebrand L., Velasco-Domínguez M.G., López-Sánchez A., Shingler V., Govantes F. The stringent response promotes biofilm dispersal in Pseudomonas putida. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-18518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiménez-Fernández A., López-Sánchez A., Calero P., Govantes F. The c-di-GMP phosphodiesterase BifA regulates biofilm development in Pseudomonas putida: c-di-GMP and BifA regulation of biofilm development. Environ Microbiol Rep. 2015;7(1):78–84. doi: 10.1111/1758-2229.12153. [DOI] [PubMed] [Google Scholar]
- 78.Newell PD, Boyd CD, Sondermann H, O’Toole GA. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 2011;9:e1000587. https://doi.org/10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed]
- 79.Newell P.D., Monds R.D., O'Toole G.A. LapD is a bis-(3',5')-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci. 2009;106(9):3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barahona E, Navazo A, Yousef-Coronado F, Aguirre de Cárcer D, Martínez-Granero F, Espinosa-Urgel M, et al. Efficient rhizosphere colonization by Pseudomonas fluorescens f113 mutants unable to form biofilms on abiotic surfaces. Environ Microbiol 2010;12:3185–95. https://doi.org/10.1111/j.1462-2920.2010.02291.x. [DOI] [PubMed]
- 81.Muriel C., Arrebola E., Redondo-Nieto M., Martínez-Granero F., Jalvo B., Pfeilmeier S., Blanco-Romero E., Baena I., Malone J.G., Rivilla R., Martín M. AmrZ is a major determinant of c-di-GMP levels in Pseudomonas fluorescens F113. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-20419-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blanco-Romero E., Redondo-Nieto M., Martínez-Granero F., Garrido-Sanz D., Ramos-González M.I., Martín M., Rivilla R. Genome-wide analysis of the FleQ direct regulon in Pseudomonas fluorescens F113 and Pseudomonas putida KT2440. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-31371-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martínez-Granero F., Redondo-Nieto M., Vesga P., Martín M., Rivilla R. AmrZ is a global transcriptional regulator implicated in iron uptake and environmental adaption in P. fluorescens F113. BMC Genomics. 2014;15(1):237. doi: 10.1186/1471-2164-15-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Molina-Henares M.A., Ramos-González M.I., Daddaoua A., Fernández-Escamilla A.M., Espinosa-Urgel M. FleQ of Pseudomonas putida KT2440 is a multimeric cyclic diguanylate binding protein that differentially regulates expression of biofilm matrix components. Res Microbiol. 2017;168(1):36–45. doi: 10.1016/j.resmic.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 85.Muriel C., Blanco-Romero E., Trampari E., Arrebola E., Durán D., Redondo-Nieto M., Malone J.G., Martín M., Rivilla R. The diguanylate cyclase AdrA regulates flagellar biosynthesis in Pseudomonas fluorescens F113 through SadB. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-44554-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bloemberg G.V., Lugtenberg B.J.J. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr Opin Plant Biol. 2001;4(4):343–350. doi: 10.1016/S1369-5266(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 87.Zamioudis C., Pieterse C.M.J. Modulation of host immunity by beneficial microbes. MPMI. 2012;25(2):139–150. doi: 10.1094/MPMI-06-11-0179. [DOI] [PubMed] [Google Scholar]
- 88.Gross H., Loper J.E. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep. 2009;26(11):1408. doi: 10.1039/b817075b. [DOI] [PubMed] [Google Scholar]
- 89.Pierson L.S., III, Pierson E.A. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol. 2010;86(6):1659–1670. doi: 10.1007/s00253-010-2509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mazzola M, Cook RJ, Thomashow LS, Weller DM, Pierson LS. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl Environ Microbiol 1992;58:2616–2624. [DOI] [PMC free article] [PubMed]
- 91.Yu J.M., Wang D., Pierson L.S., III, Pierson E.A. Effect of producing different phenazines on bacterial fitness and biological control in Pseudomonas chlororaphis 30–84. Plant Pathol J. 2018;34:44–58. doi: 10.5423/PPJ.FT.12.2017.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maddula V.S.R.K., Pierson E.A., Pierson L.S., III Altering the ratio of phenazines in Pseudomonas chlororaphis (aureofaciens) strain 30-84: effects on biofilm formation and pathogen inhibition. JB. 2008;190(8):2759–2766. doi: 10.1128/JB.01587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.LeTourneau M.K., Marshall M.J., Cliff J.B., Bonsall R.F., Dohnalkova A.C., Mavrodi D.V., Devi S.I., Mavrodi O.V., Harsh J.B., Weller D.M., Thomashow L.S. Phenazine‐1‐carboxylic acid and soil moisture influence biofilm development and turnover of rhizobacterial biomass on wheat root surfaces. Environ Microbiol. 2018;20(6):2178–2194. doi: 10.1111/1462-2920.14244. [DOI] [PubMed] [Google Scholar]
- 94.Wang D, Yu JM, Dorosky RJ, Pierson III LS, Pierson EA. The phenazine 2-hydroxy-phenazine-1-carboxylic acid promotes extracellular DNA release and has broad transcriptomic consequences in Pseudomonas chlororaphis 30–84. PLoS One 2016;11:e0148003. https://doi.org/10.1371/journal.pone.0148003. [DOI] [PMC free article] [PubMed]
- 95.Okegbe C, Fields BL, Cole SJ, Beierschmitt C, Morgan CJ, Price-Whelan A, et al. Electron-shuttling antibiotics structure bacterial communities by modulating cellular levels of c-di-GMP. Proc Natl Acad Sci U S A 2017;114:E5236–45. https://doi.org/10.1073/pnas.1700264114. [DOI] [PMC free article] [PubMed]
- 96.Wang Y., Newman D.K. Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ Sci Technol. 2008;42(7):2380–2386. doi: 10.1021/es702290a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Geudens N, Martins JC. Cyclic lipodepsipeptides from Pseudomonas spp. – biological swiss-army knives. Front Microbiol 2018;9:1867. https://doi.org/10.3389/fmicb.2018.01867. [DOI] [PMC free article] [PubMed]
- 98.Götze S., Stallforth P. Structure, properties, and biological functions of nonribosomal lipopeptides from pseudomonads. Nat Prod Rep. 2020;37(1):29–54. doi: 10.1039/C9NP00022D. [DOI] [PubMed] [Google Scholar]
- 99.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 2019;47:W81–7. https://doi.org/10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed]
- 100.Pupin M., Flissi A., Jacques P., Leclère V. Bioinformatics tools for the discovery of new lipopeptides with biocontrol applications. Eur J Plant Pathol. 2018;152(4):993–1001. doi: 10.1007/s10658-018-1544-2. [DOI] [Google Scholar]
- 101.Raaijmakers J.M., de Bruijn I., de Kock M.J.D. Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. MPMI. 2006;19(7):699–710. doi: 10.1094/MPMI-19-0699. [DOI] [PubMed] [Google Scholar]
- 102.de Bruijn I., de Kock M.J.D., de Waard P., van Beek T.A., Raaijmakers J.M. Massetolide A biosynthesis in Pseudomonas fluorescens. JB. 2008;190(8):2777–2789. doi: 10.1128/JB.01563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bonnichsen L, Svenningsen NB, Rybtke M, de Bruijn I, Raaijmakers JM, Tolker-Nielsen T, et al. Lipopeptide biosurfactant viscosin enhances dispersal of Pseudomonas fluorescens SBW25 biofilms. Microbiology 2015;161:2289. https://doi.org/10.1099/mic.0.000191. [DOI] [PMC free article] [PubMed]
- 104.Kearns D.B. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8(9):634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bender C.L., Rangaswamy V., Loper J. Polyketide production by plant-associated pseudomonads. Annu Rev Phytopathol. 1999;37:175–196. doi: 10.1146/annurev.phyto.37.1.175. [DOI] [PubMed] [Google Scholar]
- 106.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep 2003;20:79–110. https://doi.org/10.1039/b100917f. [DOI] [PubMed]
- 107.Weller DM, Landa BB, Mavrodi OV, Schroeder KL, De La Fuente L, Blouin Bankhead S, et al. Role of 2,4-Diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biol (Stuttg) 2007;9:4–20. https://doi.org/10.1055/s-2006-924473. [DOI] [PubMed]
- 108.Troppens D.M., Dmitriev R.I., Papkovsky D.B., O'Gara F., Morrissey J.P. Genome-wide investigation of cellular targets and mode of action of the antifungal bacterial metabolite 2,4-diacetylphloroglucinol in Saccharomyces cerevisiae. FEMS Yeast Res. 2013;13(3):322–334. doi: 10.1111/1567-1364.12037. [DOI] [PubMed] [Google Scholar]
- 109.Maurhofer M., Keel C., Schnider U., Voisard C., Haas D., Defao G. Influence of enhanced antibiotic production in Pseudomanas fluorescens strain CHA0 on its disease suppressive capacity. Phytopathology. 1992;82:190–195. doi: 10.1094/Phyto-82-190. [DOI] [Google Scholar]
- 110.Nowak-Thompson B., Chaney N., Wing J.S., Gould S.J., Loper J.E. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J Bacteriol. 1999;181(7):2166–2174. doi: 10.1128/JB.181.7.2166-2174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bangera MG, Thomashow LS. Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J Bacteriol 1999;181:3155–63. https://doi.org/10.1128/JB.181.10.3155-3163.1999. [DOI] [PMC free article] [PubMed]
- 112.El-Sayed A.K., Hothersall J., Cooper S.M., Stephens E., Simpson T.J., Thomas C.M. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem Biol. 2003;10(5):419–430. doi: 10.1016/S1074-5521(03)00091-7. [DOI] [PubMed] [Google Scholar]
- 113.Connolly J.A., Wilson A., Macioszek M., Song Z., Wang L., Mohammad H.H., Yadav M., di Martino M., Miller C.E., Hothersall J., Haines A.S., Stephens E.R., Crump M.P., Willis C.L., Simpson T.J., Winn P.J., Thomas C.M. Defining the genes for the final steps in biosynthesis of the complex polyketide antibiotic mupirocin by Pseudomonas fluorescens NCIMB10586. Sci Rep. 2019;9(1) doi: 10.1038/s41598-018-38038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yan Q, Philmus B, Chang JH, Loper JE. Novel mechanism of metabolic co-regulation coordinates the biosynthesis of secondary metabolites in Pseudomonas protegens. Elife 2017;6:e22835. https://doi.org/10.7554/eLife.22835. [DOI] [PMC free article] [PubMed]
- 115.Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One 2012;7:e34064. https://doi.org/10.1371/journal.pone.0034064. [DOI] [PMC free article] [PubMed]
- 116.Loper J.E., Henkels M.D., Shaffer B.T., Valeriote F.A., Gross H. Isolation and identification of rhizoxin analogs from Pseudomonas fluorescens Pf-5 by using a genomic mining strategy. AEM. 2008;74(10):3085–3093. doi: 10.1128/AEM.02848-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Takeuchi K., Noda N., Katayose Y., Mukai Y., Numa H., Yamada K., Someya N. Rhizoxin analogs contribute to the biocontrol activity of a newly isolated Pseudomonas strain. MPMI. 2015;28(3):333–342. doi: 10.1094/MPMI-09-14-0294-FI. [DOI] [PubMed] [Google Scholar]
- 118.Lozano GL, Park HB, Bravo JI, Armstrong EA, Denu JM, Stabb EV, et al. Bacterial analogs of plant tetrahydropyridine alkaloids mediate microbial interactions in a rhizosphere model system. Appl Environ Microbiol 2019;85:e03058-18. https://doi.org/10.1128/AEM.03058-18. [DOI] [PMC free article] [PubMed]
- 119.Ghequire M.G.K., De Mot R. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev. 2014;38(4):523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 120.Li W, Estrada-de los Santos P, Matthijs S, Xie G-L, Busson R, Cornelis P, et al. Promysalin, a salicylate-containing Pseudomonas putida antibiotic, promotes surface colonization and selectively targets other Pseudomonas. Chem Biol 2011;18:1320–30. https://doi.org/10.1016/j.chembiol.2011.08.006. [DOI] [PubMed]
- 121.Bruce J.B., West S.A., Griffin A.S. Bacteriocins and the assembly of natural Pseudomonas fluorescens populations. J Evol Biol. 2017;30(2):352–360. doi: 10.1111/jeb.13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jacob F. Biosynthèse induite et mode d’action d’une pyocine, antibiotique de Pseudomonas pyocyanea. Ann Inst Pasteur (Paris) 1954;86:149–160. [PubMed] [Google Scholar]
- 123.Chavan MA, Riley MA. Molecular evolution of bacteriocins in gram-negative bacteria. In: Riley MA, Chavan MA, editors. Bacteriocins: Ecology and Evolution, Berlin, Heidelberg, Germany: Springer; 2007, p. 19–43.
- 124.Ghequire M.G.K., De Mot R. The Tailocin Tale: peeling off phage tails. Trends Microbiol. 2015;23(10):587–590. doi: 10.1016/j.tim.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 125.Loper JE, Hassan KA, Mavrodi DV, Davis EW, Lim CK, Shaffer BT, et al. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 2012;8:e1002784. https://doi.org/10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed]
- 126.Biessy A., Novinscak A., Blom J., Léger G., Thomashow L.S., Cazorla F.M. Diversity of phytobeneficial traits revealed by whole-genome analysis of worldwide-isolated phenazine-producing Pseudomonas spp. Environ Microbiol. 2019;21:437–455. doi: 10.1111/1462-2920.14476. [DOI] [PubMed] [Google Scholar]
- 127.Ghequire MGK, Dillen Y, Lambrichts I, Proost P, Wattiez R, De Mot R. Different Ancestries of R Tailocins in Rhizospheric Pseudomonas Isolates. Genome Biol Evol 2015;7:2810–28. https://doi.org/10.1093/gbe/evv184. [DOI] [PMC free article] [PubMed]
- 128.Sharp C, Bray J, Housden NG, Maiden MCJ, Kleanthous C. Diversity and distribution of nuclease bacteriocins in bacterial genomes revealed using Hidden Markov Models. PLoS Comput Biol 2017;13:e1005652. https://doi.org/10.1371/journal.pcbi.1005652. [DOI] [PMC free article] [PubMed]
- 129.Dorosky R.J., Yu J.M., Pierson L.S., III, Pierson E.A., Kivisaar M. Pseudomonas chlororaphis produces two distinct R-tailocins that contribute to bacterial competition in biofilms and on roots. Appl Environ Microbiol. 2017;83(15) doi: 10.1128/AEM.00706-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dorosky RJ, Pierson LS, Pierson EA. Pseudomonas chlororaphis produces multiple R-tailocin particles that broaden the killing spectrum and contribute to persistence in rhizosphere communities. Appl Environ Microbiol 2018;84:e01230-18. https://doi.org/10.1128/AEM.01230-18. [DOI] [PMC free article] [PubMed]
- 131.Silverman J.M., Brunet Y.R., Cascales E., Mougous J.D. Structure and regulation of the Type VI secretion system. Annu Rev Microbiol. 2012;66(1):453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bernal P., Llamas M.A., Filloux A. Type VI secretion systems in plant-associated bacteria: T6SS in phytobacteria. Environ Microbiol. 2018;20(1):1–15. doi: 10.1111/1462-2920.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bulgarelli D., Garrido-Oter R., Münch P., Weiman A., Dröge J., Pan Y., McHardy A., Schulze-Lefert P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17(3):392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bernal P., Allsopp L.P., Filloux A., Llamas M.A. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 2017;11(4):972–987. doi: 10.1038/ismej.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mosquito S., Bertani I., Licastro D., Compant S., Myers M.P., Hinarejos E., Levy A., Venturi V. In planta colonization and role of T6SS in two rice Kosakonia endophytes. MPMI. 2020;33(2):349–363. doi: 10.1094/MPMI-09-19-0256-R. [DOI] [PubMed] [Google Scholar]
- 136.Decoin V., Gallique M., Barbey C., Le Mauff F., Poc C.D., Feuilloley M.GJ., Orange N., Merieau A. A Pseudomonas fluorescens type 6 secretion system is related to mucoidy, motility and bacterial competition. BMC Microbiol. 2015;15(1) doi: 10.1186/s12866-015-0405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gallique M, Decoin V, Barbey C, Rosay T, Feuilloley MGJ, Orange N, et al. Contribution of the Pseudomonas fluorescens MFE01 Type VI secretion system to biofilm formation. PLoS One 2017;12:e0170770. https://doi.org/10.1371/journal.pone.0170770. [DOI] [PMC free article] [PubMed]
- 138.Bouteiller M, Gallique M, Bourigault Y, Kosta A, Hardouin J, Massier S, et al. Crosstalk between the Type VI Secretion system and the expression of class IV flagellar genes in the Pseudomonas fluorescens MFE01 strain. Microorganisms 2020;8:622. https://doi.org/10.3390/microorganisms8050622. [DOI] [PMC free article] [PubMed]
- 139.Chen W.-J., Kuo T.-Y., Hsieh F.-C., Chen P.-Y., Wang C.-S., Shih Y.-L., Lai Y.-M., Liu J.-R., Yang Y.-L., Shih M.-C. Involvement of type VI secretion system in secretion of iron chelator pyoverdine in Pseudomonas taiwanensis. Sci Rep. 2016;6(1) doi: 10.1038/srep32950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cianfanelli F.R., Monlezun L., Coulthurst S.J. Aim, load, fire: the Type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016;24(1):51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 141.Liu N.a., Song W., Schienebeck C.M., Zhang M., Tang W. Synthesis of naturally occurring tropones and tropolones. Tetrahedron. 2014;70(49):9281–9305. doi: 10.1016/j.tet.2014.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jiang Z., Chen M., Yu X., Xie Z. 7-Hydroxytropolone produced and utilized as an iron-scavenger by Pseudomonas donghuensis. Biometals. 2016;29(5):817–826. doi: 10.1007/s10534-016-9954-0. [DOI] [PubMed] [Google Scholar]
- 143.Muzio F.M., Agaras B.C., Masi M., Tuzi A., Evidente A., Valverde C. 7‐hydroxytropolone is the main metabolite responsible for the fungal antagonism of Pseudomonas donghuensis strain SVBP6. Environ Microbiol. 2020;22(7):2550–2563. doi: 10.1111/1462-2920.14925. [DOI] [PubMed] [Google Scholar]
- 144.Krzyżanowska DM, Ossowicki A, Rajewska M, Maciąg T, Jabłońska M, Obuchowski M, et al. When genome-based approach meets the “old but good”: revealing genes involved in the antibacterial activity of Pseudomonas sp. P482 against soft rot pathogens. Front Microbiol 2016;7:782. https://doi.org/10.3389/fmicb.2016.00782. [DOI] [PMC free article] [PubMed]
- 145.Guo H., Roman D., Beemelmanns C. Tropolone natural products. Nat Prod Rep. 2019;36(8):1137–1155. doi: 10.1039/C8NP00078F. [DOI] [PubMed] [Google Scholar]
- 146.Garbeva P, Tyc O, Remus-Emsermann MNP, van der Wal A, Vos M, Silby M, et al. No apparent costs for facultative antibiotic production by the soil bacterium Pseudomonas fluorescens Pf0-1. PLoS One 2011;6:e27266. https://doi.org/10.1371/journal.pone.0027266. [DOI] [PMC free article] [PubMed]
- 147.Venturi V., Keel C. Signaling in the rhizosphere. Trends Plant Sci. 2016;21(3):187–198. doi: 10.1016/j.tplants.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 148.Fuqua C., Parsek M.R., Greenberg E.P. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35(1):439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 149.Rieusset L., Rey M., Muller D., Vacheron J., Gerin F., Dubost A., Comte G., Prigent‐Combaret C. Secondary metabolites from plant‐associated Pseudomonas are overproduced in biofilm. Microb Biotechnol. 2020;13(5):1562–1580. doi: 10.1111/1751-7915.13598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lapouge K, Schubert M, Allain FH-T, Haas D. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 2007;67:241–53. https://doi.org/10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed]
- 151.Jahanshah G., Yan Q., Gerhardt H., Pataj Z., Lämmerhofer M., Pianet I., Josten M., Sahl H.-G., Silby M.W., Loper J.E., Gross H. Discovery of the cyclic lipopeptide gacamide A by genome mining and repair of the defective GacA regulator in Pseudomonas fluorescens Pf0-1. J Nat Prod. 2019;82(2):301–308. doi: 10.1021/acs.jnatprod.8b00747.s001. [DOI] [PubMed] [Google Scholar]
- 152.Heeb S., Haas D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. MPMI. 2001;14(12):1351–1363. doi: 10.1094/MPMI.2001.14.12.1351. [DOI] [PubMed] [Google Scholar]
- 153.Wang D., Lee S.-H., Seeve C., Yu J.M., Pierson L.S., Pierson E.A. Roles of the Gac-Rsm pathway in the regulation of phenazine biosynthesis in Pseudomonas chlororaphis 30–84. Microbiologyopen. 2013;2:505–524. doi: 10.1002/mbo3.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yu JM, Wang D, Ries TR, Pierson LS, Pierson EA. An upstream sequence modulates phenazine production at the level of transcription and translation in the biological control strain Pseudomonas chlororaphis 30-84. PLoS One 2018;13:e0193063. https://doi.org/10.1371/journal.pone.0193063. [DOI] [PMC free article] [PubMed]
- 155.Wang D, Yu JM, Pierson LS, Pierson EA. Differential regulation of phenazine biosynthesis by RpeA and RpeB in Pseudomonas chlororaphis 30-84. Microbiology 2012;158:1745–57. https://doi.org/10.1099/mic.0.059352-0. [DOI] [PubMed]
- 156.Girard Geneviève, Barends S., Rigali Sébastien, van Rij E.T., Lugtenberg B.J.J., Bloemberg G.V. Pip, a novel activator of phenazine biosynthesis in Pseudomonas chlororaphisPCL1391. JB. 2006;188(23):8283–8293. doi: 10.1128/JB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Martinez-Gil M., Ramos-Gonzalez M.I., Espinosa-Urgel M. Roles of cyclic Di-GMP and the gac system in transcriptional control of the genes coding for the Pseudomonas putida adhesins LapA and LapF. J Bacteriol. 2014;196(8):1484–1495. doi: 10.1128/JB.01287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sobrero PM, Valverde C. Comparative genomics and evolutionary analysis of RNA-binding proteins of the CsrA family in the genus Pseudomonas. Front Mol Biosci 2020;7:127. https://doi.org/10.3389/fmolb.2020.00127. [DOI] [PMC free article] [PubMed]
- 159.Bull C.T., Duffy B., Voisard C., Défago G., Keel C., Haas D. Characterization of spontaneous gacS and gacA regulatory mutants of Pseudomonas fluorescens biocontrol strain CHA0. Antonie Van Leeuwenhoek. 2001;79:327–336. doi: 10.1023/A:1012061014717. [DOI] [PubMed] [Google Scholar]
- 160.Lalaouna D., Fochesato S., Sanchez L., Schmitt-Kopplin P., Haas D., Heulin T., Achouak W. Phenotypic switching in Pseudomonas brassicacearum involves GacS- and GacA-dependent Rsm small RNAs. Appl Environ Microbiol. 2012;78(6):1658–1665. doi: 10.1128/AEM.06769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Martínez-Granero F, Capdevila S, Sánchez-Contreras M, Martín M, Rivilla R. Two site-specific recombinases are implicated in phenotypic variation and competitive rhizosphere colonization in Pseudomonas fluorescens. Microbiology 2005;151:975–83. https://doi.org/10.1099/mic.0.27583-0. [DOI] [PubMed]
- 162.van den Broek D., Chin-A-Woeng T.F.C., Bloemberg G.V., Lugtenberg B.J.J. Molecular nature of spontaneous modifications in gacS which cause colony phase variation in Pseudomonas sp. strain PCL1171. JB. 2005;187(2):593–600. doi: 10.1128/JB.187.2.593-600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Colombo C., Palumbo G., He J.-Z., Pinton R., Cesco S. Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. J Soils Sediments. 2014;14(3):538–548. doi: 10.1007/s11368-013-0814-z. [DOI] [Google Scholar]
- 164.Wandersman C., Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58(1):611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 165.Meyer J.-M. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch Microbiol. 2000;174(3):135–142. doi: 10.1007/s002030000188. [DOI] [PubMed] [Google Scholar]
- 166.Berti A.D., Thomas M.G. Analysis of achromobactin biosynthesis by Pseudomonas syringae pv. syringae B728a. JB. 2009;191(14):4594–4604. doi: 10.1128/JB.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cornelis P. Iron uptake and metabolism in pseudomonads. Appl Microbiol Biotechnol. 2010;86(6):1637–1645. doi: 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 168.Cornelis P, Bodilis J. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ Microbiol Rep 2009;1:256–62. https://doi.org/10.1111/j.1758-2229.2009.00041.x. [DOI] [PubMed]
- 169.Imperi F., Tiburzi F., Visca P. Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc Natl Acad Sci. 2009;106(48):20440–20445. doi: 10.1073/pnas.0908760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Raaijmakers J.M., Sluis L.V.D., Bakker P.A.H.M., Schippers B., Koster M., Weisbeek P.J. Utilization of heterologous siderophores and rhizosphere competence of fluorescent Pseudomonas spp. Can J Microbiol. 1995;41(2):126–135. doi: 10.1139/m95-017. [DOI] [Google Scholar]
- 171.Hartney S.L., Mazurier S., Kidarsa T.A., Quecine M.C., Lemanceau P., Loper J.E. TonB-dependent outer-membrane proteins and siderophore utilization in Pseudomonas fluorescens Pf-5. Biometals. 2011;24(2):193–213. doi: 10.1007/s10534-010-9385-2. [DOI] [PubMed] [Google Scholar]
- 172.Ghequire M.G.K., Kemland L., Anoz-Carbonell E., Buchanan S.K., De Mot R., Vidaver A.K. A natural chimeric pseudomonas bacteriocin with novel pore-forming activity parasitizes the ferrichrome transporter. mBio. 2017;8(1) doi: 10.1128/mBio.01961-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lemanceau P, Robin A, Mazurier S, Vansuyt G. Implication of pyoverdines in the interactions of fluorescent pseudomonads with soil microflora and plant in the rhizosphere. In: Varma A, Chincholkar SB, editors. Microbial siderophores, vol. 12, Berlin, Heidelberg, Germany: Springer; 2007, p. 165–92.
- 174.Drehe I., Simonetti E., Ruiz J.A. Contribution of the siderophores pyoverdine and enantio-pyochelin to fitness in soil of Pseudomonas protegens Pf-5. Curr Microbiol. 2018;75(12):1560–1565. doi: 10.1007/s00284-018-1560-7. [DOI] [PubMed] [Google Scholar]
- 175.Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 176.Yu K.e., Pieterse C.M.J., Bakker P.A.H.M., Berendsen R.L. Beneficial microbes going underground of root immunity. Plant Cell Environ. 2019;42(10):2860–2870. doi: 10.1111/pce:13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rossez Y, Wolfson EB, Holmes A, Gally DL, Holden NJ. Bacterial Flagella: twist and stick, or dodge across the kingdoms. PLoS Pathog 2015;11:e1004483. https://doi.org/10.1371/journal.ppat.1004483. [DOI] [PMC free article] [PubMed]
- 178.Pfeilmeier S., Saur I.-L., Rathjen J.P., Zipfel C., Malone J.G. High levels of cyclic-di-GMP in plant-associated Pseudomonas correlate with evasion of plant immunity: Pseudomonas cyclic-di-GMP and plant immune evasion. Mol Plant Pathol. 2016;17(4):521–531. doi: 10.1111/mpp.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Liu Z, Beskrovnaya P, Melnyk RA, Hossain SS, Khorasani S, O’Sullivan LR, et al. A genome-wide screen identifies genes in rhizosphere-associated Pseudomonas required to evade plant defenses. MBio 2018;9:e00433-18. https://doi.org/10.1128/mBio.00433-18. [DOI] [PMC free article] [PubMed]
- 180.Barrientos-Moreno L., Molina-Henares M.A., Ramos-González M.I., Espinosa-Urgel M. Arginine as an environmental and metabolic cue for cyclic diguanylate signalling and biofilm formation in Pseudomonas putida. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-70675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yu K.e., Liu Y., Tichelaar R., Savant N., Lagendijk E., van Kuijk S.J.L., Stringlis I.A., van Dijken A.J.H., Pieterse C.M.J., Bakker P.A.H.M., Haney C.H., Berendsen R.L. Rhizosphere-associated Pseudomonas suppress local root immune responses by gluconic acid-mediated lowering of environmental pH. Curr Biol. 2019;29(22):3913–3920.e4. doi: 10.1016/j.cub.2019.09.015. [DOI] [PubMed] [Google Scholar]
- 182.Bardoel BW, van der Ent S, Pel MJC, Tommassen J, Pieterse CMJ, van Kessel KPM, et al. Pseudomonas evades immune recognition of flagellin in both mammals and plants. PLoS Pathog 2011;7:e1002206. https://doi.org/10.1371/journal.ppat.1002206. [DOI] [PMC free article] [PubMed]
- 183.Pel M.J.C., van Dijken A.J.H., Bardoel B.W., Seidl M.F., van der Ent S., van Strijp J.A.G., Pieterse C.M.J. Pseudomonas syringae evades host immunity by degrading flagellin monomers with alkaline protease AprA. MPMI. 2014;27(7):603–610. doi: 10.1094/MPMI-02-14-0032-R. [DOI] [PubMed] [Google Scholar]
- 184.Berendsen R.L., van Verk M.C., Stringlis I.A., Zamioudis C., Tommassen J., Pieterse C.M.J., Bakker P.A.H.M. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genomics. 2015;16(1) doi: 10.1186/s12864-015-1632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cole BJ, Feltcher ME, Waters RJ, Wetmore KM, Mucyn TS, Ryan EM, et al. Genome-wide identification of bacterial plant colonization genes. PLoS Biol 2017;15:e2002860. https://doi.org/10.1371/journal.pbio.2002860. [DOI] [PMC free article] [PubMed]
- 186.Gatzeva-Topalova P.Z., May A.P., Sousa M.C. Crystal structure and mechanism of the Escherichia coli ArnA (PmrI) transformylase domain. An enzyme for lipid A modification with 4-amino-4-deoxy-l-arabinose and polymyxin resistance. Biochemistry. 2005;44:5328–5338. doi: 10.1021/bi047384g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Newman M.-A., Dow J.M., Molinaro A., Parrilli M. Priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J Endotoxin Res. 2007;13:69–84. doi: 10.1177/0968051907079399. [DOI] [PubMed] [Google Scholar]
- 188.Deng W., Marshall N.C., Rowland J.L., McCoy J.M., Worrall L.J., Santos A.S., Strynadka N.C.J., Finlay B.B. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. 2017;15(6):323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 189.Block A., Alfano J.R. Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr Opin Microbiol. 2011;14(1):39–46. doi: 10.1016/j.mib.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mavrodi D.V., Joe A., Mavrodi O.V., Hassan K.A., Weller D.M., Paulsen I.T., Loper J.E., Alfano J.R., Thomashow L.S. Structural and functional analysis of the Type III secretion system from Pseudomonas fluorescens Q8r1-96. J Bacteriol. 2011;193(1):177–189. doi: 10.1128/JB.00895-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Stringlis IA, Zamioudis C, Berendsen RL, Bakker PAHM, Pieterse CMJ. Type III secretion system of beneficial rhizobacteria Pseudomonas simiae WCS417 and Pseudomonas defensor WCS374. Front Microbiol 2019;10:1631. https://doi.org/10.3389/fmicb.2019.01631. [DOI] [PMC free article] [PubMed]
- 192.Viollet A., Pivato B., Mougel C., Cleyet-Marel J.-C., Gubry-Rangin C., Lemanceau P., Mazurier S. Pseudomonas fluorescens C7R12 type III secretion system impacts mycorrhization of Medicago truncatula and associated microbial communities. Mycorrhiza. 2017;27(1):23–33. doi: 10.1007/s00572-016-0730-3. [DOI] [PubMed] [Google Scholar]
- 193.Stringlis I.A., Proietti S., Hickman R., Van Verk M.C., Zamioudis C., Pieterse C.M.J. Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 2018;93(1):166–180. doi: 10.1111/tpj.13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Stringlis I.A., Zhang H., Pieterse C.M.J., Bolton M.D., de Jonge R. Microbial small molecules – weapons of plant subversion. Nat Prod Rep. 2018;35(5):410–433. doi: 10.1039/C7NP00062F. [DOI] [PubMed] [Google Scholar]
- 195.Latour X., Lemanceau P. Métabolisme carboné et énergétique des Pseudomonas spp fluorescents saprophytes à oxydase positive. Agronomie. 1997;17(9–10):427–443. doi: 10.1051/agro:19970901. [DOI] [Google Scholar]
- 196.Vives-Peris V., de Ollas C., Gómez-Cadenas A., Pérez-Clemente R.M. Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 2020;39(1):3–17. doi: 10.1007/s00299-019-02447-5. [DOI] [PubMed] [Google Scholar]
- 197.Lugtenberg B.J.J., Kravchenko L.V., Simons M. Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ Microbiol. 1999;1(5):439–446. doi: 10.1046/j.1462-2920.1999.00054.x. [DOI] [PubMed] [Google Scholar]
- 198.Kamilova F., Kravchenko L.V., Shaposhnikov A.I., Azarova T., Makarova N., Lugtenberg B. Organic acids, sugars, and l-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol Plant Microbe Interact. 2006;19:250–256. doi: 10.1094/MPMI-19-0250. [DOI] [PubMed] [Google Scholar]
- 199.Latour X., Delorme S., Mirleau P., Lemanceau P. Identification of traits implicated in the rhizosphere competence of fluorescent pseudomonads: description of a strategy based on population and model strain studies. Agronomie. 2003;23(5-6):397–405. doi: 10.1051/agro:2003015. [DOI] [Google Scholar]
- 200.Zboralski A, Biessy A, Savoie M-C, Novinscak A, Filion M. Metabolic and genomic traits of phytobeneficial phenazine-producing Pseudomonas spp. are linked to rhizosphere colonization in Arabidopsis thaliana and Solanum tuberosum. Appl Environ Microbiol 2020;86:e02443-19. https://doi.org/10.1128/AEM.02443-19. [DOI] [PMC free article] [PubMed]
- 201.Rediers H., Vanderleyden J., De Mot R. Nitrate respiration in Pseudomonas stutzeri A15 and its involvement in rice and wheat root colonization. Microbiol Res. 2009;164(4):461–468. doi: 10.1016/j.micres.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 202.Philippot L., Højberg O. Dissimilatory nitrate reductases in bacteria. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1999;1446(1-2):1–23. doi: 10.1016/S0167-4781(99)00072-X. [DOI] [PubMed] [Google Scholar]
- 203.Ghiglione J.-F., Gourbiere F., Potier P., Philippot L., Lensi R. Role of respiratory nitrate reductase in ability of Pseudomonas fluorescens YT101 to colonize the rhizosphere of maize. Appl Environ Microbiol. 2000;66(9):4012–4016. doi: 10.1128/AEM.66.9.4012-4016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mirleau P., Philippot L., Corberand Thérèse, Lemanceau P. Involvement of nitrate reductase and pyoverdine in competitiveness of Pseudomonas fluorescens strain C7R12 in soil. Appl Environ Microbiol. 2001;67(6):2627–2635. doi: 10.1128/AEM.67.6.2627-2635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bünemann E.K., Condron L.M. Soil biologynutrient cycling in terrestrial ecosystems. Springer Berlin Heidelberg; Berlin, Heidelberg: 2007. pp. 65–92. [DOI] [Google Scholar]
- 206.Kahnert A., Mirleau P., Wait R., Kertesz M.A. The LysR-type regulator SftR is involved in soil survival and sulphate ester metabolism in Pseudomonas putida. Environ Microbiol. 2002;4(4):225–237. doi: 10.1046/j.1462-2920.2002.00289.x. [DOI] [PubMed] [Google Scholar]
- 207.Kertesz MA. The role of soil microbes in plant sulphur nutrition. J Exp Bot 2004;55:1939–45. https://doi.org/10.1093/jxb/erh176. [DOI] [PubMed]
- 208.Mirleau P., Wogelius R., Smith A., Kertesz M.A. Importance of organosulfur utilization for survival of Pseudomonas putida in soil and rhizosphere. AEM. 2005;71(11):6571–6577. doi: 10.1128/AEM.71.11.6571-6577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bharwad K., Rajkumar S. Rewiring the functional complexity between Crc, Hfq and sRNAs to regulate carbon catabolite repression in Pseudomonas. World J Microbiol Biotechnol. 2019;35(9) doi: 10.1007/s11274-019-2717-7. [DOI] [PubMed] [Google Scholar]
- 210.Moreno R., Martínez-Gomariz M., Yuste L., Gil C., Rojo F. The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics. 2009;9(11):2910–2928. doi: 10.1002/pmic.200800918. [DOI] [PubMed] [Google Scholar]
- 211.Campilongo R, Fung RKY, Little RH, Grenga L, Trampari E, Pepe S, et al. One ligand, two regulators and three binding sites: how KDPG controls primary carbon metabolism in Pseudomonas. PLoS Genet 2017;13:e1006839. https://doi.org/10.1371/journal.pgen.1006839. [DOI] [PMC free article] [PubMed]
- 212.Little RH, Woodcock SD, Campilongo R, Fung RKY, Heal R, Humphries L, et al. Differential regulation of genes for cyclic-di-GMP metabolism orchestrates adaptive changes during rhizosphere colonization by Pseudomonas fluorescens. Front Microbiol 2019;10:1089. https://doi.org/10.3389/fmicb.2019.01089. [DOI] [PMC free article] [PubMed]
- 213.Grenga L, Little RH, Malone JG. Quick change: post-transcriptional regulation in Pseudomonas. FEMS Microbiol Lett 2017;364. https://doi.org/10.1093/femsle/fnx125. [DOI] [PMC free article] [PubMed]
- 214.Little RH, Grenga L, Saalbach G, Howat AM, Pfeilmeier S, Trampari E, et al. Adaptive remodeling of the bacterial proteome by specific ribosomal modification regulates Pseudomonas infection and niche colonisation. PLoS Genet 2016;12:e1005837. https://doi.org/10.1371/journal.pgen.1005837. [DOI] [PMC free article] [PubMed]
- 215.Grenga L, Little RH, Chandra G, Woodcock SD, Saalbach G, Morris RJ, et al. Control of mRNA translation by dynamic ribosome modification. PLoS Genet 2020;16:e1008837. https://doi.org/10.1371/journal.pgen.1008837. [DOI] [PMC free article] [PubMed]