Highlights
-
•
An implemented high cell density cultivation process for biotechnological polysialic acid production is shown.
-
•
Efficiency of polysialic acid production using E. coli K1 is significantly improved compared to state-of-the-art processes.
-
•
Non-pathogenic E. coli BL21 is successfully engineered for production of α2,8- and α2,9-linked polySia.
-
•
Differences of the α2,8- and α2,9-linked polysialic acid can be markedly visualized by the recorded NMR spectra.
Keywords: Escherichia coli, Polysialic acid, High cell density, NMR spectroscopy
Abstract
Polysialic acid (polySia) are α2,8- and/or α2,9-linked homopolymers with interesting properties for meningococcal vaccine development or the cure of human neurodegenerative disorders.
With the goal to avoid large scale production of pathogenic bacteria, we compare in the current study the efficacy of conventional polySia production to recombinant approaches using the engineered laboratory safety strain E. coli BL21. High cell density cultivation (HCDC) experiments were performed in two different bioreactor systems. Increased cell densities of up to 11.3 (±0.4) g/L and polySia concentrations of up to 774 (±18) mg/L were reached in E. coli K1. However, cultivation of engineered E. coli BL21 strains delivered comparable cell densities but a maximum of only 133 mg/L polySia. Using established downstream procedures, host cell DNA and proteins were removed. All recombinant polySia products showed an identical degree of polymerization >90. Polymers with different glycosidic linkages could be successfully differentiated by nuclear magnetic resonance spectroscopy.
1. Introduction
Many bacteria express a thick layer of surface associated polysaccharides known as the capsule or K-antigen (from the German word Kapsel) [1]. Forming the outermost surface of the bacterium, the capsules define interactions with the external environment such as mediating cell-cell interactions [2] and protect bacteria against harsh environmental conditions, including a critical role in the virulence of invasive pathogens [1]. In some neuroinvasive strains of E. coli and Neisseria meningitidis (Nm), the capsular polysaccharides (CPS) provide linear homopolymers of the negatively charged nona-sugar sialic acid (Sia), known as polysialic acid (polySia) [3]. With a degree of polymerisation (DP) of 100 sialyl residues [4] the CPS form protective layers that prevent bacteria from desiccation and impair host immune attack. Indeed, meningococcal survival in human serum has been demonstrated to essentially depend on the presence of the CPS [5].
However, due to the existence of different glycosidic linkages, the polySia chains exhibit very different chemical, structural and immunological properties [6,7]. In E. coli K1 and Nm serogroup B (NmB), the Sia-monomers are α2,8-glycosidically linked (Fig. 1A), while the inter connection is α2,9-glycosidic in NmC (Fig. 1B) [6]. The anchoring of CPS to the outer bacterial membrane is mediated by a phospholipid structure [8]. The negative charge of sialic acids is due to a carboxyl residue at C1 of the sugar molecule. Although a broad panel of modifications, including phosphorylation, sulfatation, methylation and acetylation has been described to give rise to more than 50 naturally occurring variants [9,10], no other modification than O-acetylation has so far been described in bacterial polySia [11].
Fig. 1.
Schematic representation of the molecular structure of polysialic acid and the capsular biosynthesis (kps) gene cluster (C). E. coli K1 and serogroup B N. meningitidis synthesize human-identical α2,8-linked polySia (A) via the kps gene cluster. Serogroup C N. meningitidis expresses the polysialyltransferase synE instead of the polysialyltransferase neuS. This leads to a change in the linking pattern, whereby the Neu5Ac residues are linked α2,9-glycosidically (B).
The polysialyltransferases (polySTs) that catalyze the biosynthesis of these CPS constitute their own CAZy family (GT-38) [12] and are part of a so called group 2 capsule biosynthesis cluster [13] (in Fig. 1C the region is shown for E. coli K1). Region 1 and 3 of this cluster encode (I) enzymes assembling a glycolipid precursor on which the polymer is built, and (II) proteins of an ABC-transporter dependent export complex that spans the periplasm and allows the translocation of the CPS from the cytoplasm to the outside of the cell. Both regions are highly conserved among bacteria using group 2 biosynthesis systems. In contrast, region 2 harbors the enzymatic machinery that assembles the polysaccharide (capsule polymerase, polysaccharide modifying enzymes, etc.) and is thus highly variable and serotype specific.
In humans, α2,8-polySia provides a post-translational modification of a limited number of glycoproteins [3], the most prominent of which is the neural cell adhesion molecule (NCAM). While the functions of polySia have long been limited to the regulation of cell-cell contacts during the development of the nervous system, more recent studies demonstrate that both, protein bound and soluble α2,8-polySia have essential functions in the control of immune reactions, e.g. by exerting anti-inflammatory effects in macrophages and microglia and by interfering with complement activation [[14], [15], [16], [17]]. Remarkably these later studies clearly demonstrated that polySia-functions can strictly depend on the DP. For instance, in a model of Macula degeneration exogenously added polySia demonstrated a therapeutic potential if added as a fraction of average DP20 (avDP20) [18], while binding of neurotrophic factors by polySia was found to require a minimal chain length of DP12 or DP17, respectively [19,20]. Finally, polySia attracted considerable attention for the stabilization of protein drugs [10,21,22].
Given the wide range of medical applications, it is obvious, that the interest in producing polySia biotechnologically has increased massively over the past decades. Many research groups have favored production by cultivating E. coli strains [[23], [24], [25]] over chemical synthesis. The yields depend mainly on the cell densities achieved. Even a production process based on fermentation in a wave-induced disposable bag reactor system which facilitates GMP-compliant production was developed [26]. Cultures on the used synthetic medium resulted in moderate polySia yields and cell densities. Long chain polySia can subsequently be isolated from the culture broth. Contaminants such as host DNA and proteins (HCP) as well as endotoxins can be completely depleted by ethanol precipitation and clay minerals [27]. With a focus on a later medical application, these systems offer simple possibilities to produce polySia according to the quality requirements of "good manufacturing practice" (GMP) [28]. Nevertheless, the pathogenicity of E. coli K1 use makes it difficult to comply with and document the safety standards for patient protection according to GMP. The use of laboratory safety E. coli derivatives, such as the well-known E. coli BL21 strain, could overcome these problems [29].
In previous work, E. coli BL21, which does not naturally produce K-antigens [30], was engineered for the recombinant production of polySia. The strain was transformed with the pKT274 plasmid, which contains the complete capsular biosynthesis cluster (kps) of E. coli K1 (Fig. 1C). By exchanging the K1 polysialyltransferase (neuS) for the NmC polysialyltransferase (synE) [31,32] a α2,9-polySia producing variant was produced.
In this study, a high cell density cultivation process of E. coli K1 was validated to improve the production of long chain polySia by achieving increased cell densities. The strain was cultivated in a classical 2 Lstirred tank and a 50 L disposable bag reactor with wave-induced mixing to investigate its potential compared to state-of-the-art processes. Subsequently, the efficiency of conventional polySia production was compared with a recombinant approach using the engineered laboratory safety strain E. coli BL21 to avoid large-scale production of α2,8- and α2,9-polySia in pathogenic bacteria. The produced polySia was then purified using a validated process and characterized in detail by HPLC analysis and nuclear magnetic resonance (NMR) spectroscopy.
2. Material and methods
2.1. Material
2.1.1. Chemicals
Bulk chemicals were purchased from Sigma-Aldrich (Taufkirchen, Germany) or Carl Roth GmbH & Co.KG (Karlsruhe, Germany). Deionized water was prepared with ARIUM® (Sartorius Stedim Biotech, Göttingen, Germany).
2.1.2. Culture medium
In this study, for the bioreactor cultivations a basic defined medium was used and prepared essentially as described in previous studies [33]. The composition of the batch medium is given in Table 1.
Table 1.
Medium composition for high-cell density cultivations (HCDC).
| Component | Batch medium |
|---|---|
| Glucose | 25 g/L |
| KH2PO4 | 13.3 g/L |
| (NH4)2HPO4 | 4 g/L |
| MgSO4 x 7H2O | 1.2 g/L |
| Citric acid | 1.7 g/L |
| EDTA | 8.4 mg/L |
| CoCl2 | 1.37 mg/L |
| CuCl2 | 1.83 mg/L |
| H3BO3 | 3 mg/L |
| MnCl2 | 9.54 mg/L |
| Na2MoO4 x 2H2O | 2.5 mg/L |
| Zn(CH3-COO)2 x 2H2O | 13 mg/L |
| Fe(III)-citrat Hydrat | 100 mg/L |
| Thiamine-HCl | 4.5 mg/L |
2.1.3. Bacterial strains
All bacterial strains used in this study as well as their relevant characteristics are listed in Table 2. For preparation of stock culture, the bacteria were grown in a complex medium at 37 °C until the optical density at 600 nm (OD600) was between a value of 0.7 and 1 relative absorption units (rel. AU). Half of the culture broth was mixed with 50 % (v/v) glycerol and stored at −80 °C.
Table 2.
Bacterial strains and their relevant characteristics.
| Strains | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli B2032/82 K1 | wild type DSM-Nr. 107164 | [34] |
| E. coli BL21 pKT274 | AmpR, pHC79 derivate containing the kps gene cluster | Institute of Clinical Chemistry (Hannover, Germany) [35] |
| E. coli BL21 pKT774 | KanR, pKT274 derivate | Institute of Clinical Chemistry (Hannover, Germany) |
| E. coli BL21 pKT274 NmC-CAT | ClpR, pKT274 containing the polysialyltransferase gene from N. meningitidis Serogroup C; ΔneuS::synE | Institute of Clinical Chemistry (Hannover, Germany) |
2.2. Methods
2.2.1. Precultivation in shake flasks
For precultivation, E. coli strains were grown in 500 mL baffled shake flasks with 50 mL complex medium consisting of yeast extract (10 g/L), tryptone (10 g/L), and NaCl (5 g/L) at pH 7.3. Where appropriate, kanamycin (50 μg/mL), ampicillin (100 μg/mL) or chloramphenicol (34 μg/mL) was added to the medium. Incubation was carried out on a rotary shaker at 37 °C and 200 rpm. When the OD600 was around 6–8 rel. AU, the complex medium cell suspension was transferred into 100 mL defined salt medium (Table 1) with respective antibiotics and incubated for 12–15 h at 37 °C and 150 rpm. These cells were used as inoculum for the bioreactor cultivation.
2.2.2. Stirred tank reactor (STR) cultivation
First cultivation experiments were carried out in a 2 Liter glass reactor of the Biostat A fermenter system (Sartorius Stedim Biotech, Göttingen, Germany). The reactor was filled with defined HCDC medium (composition given in Table 1). Where appropriate, kanamycin (50 μg/mL), ampicillin (100 μg/mL) or chloramphenicol (34 μg/mL) was added to the medium. To prevent foam formation Desmophen 3900 (Bayer AG, Leverkusen, Germany) was added. The main culture in the bioreactor was inoculated with defined medium preculture cells in the mid exponential phase (OD600 = 8–11 rel. AU) to an OD600 of 0.4 rel. AU.
The cultivation temperature was kept at 37 °C. The pH was kept constant at pH 7.5 by automatic addition of 25 % (v/v) NH4OH and 1 M HCl. The stirrer speed was set to 600 rpm. The culture was aerated with a constant rate of 2 L/min (1 vvm, volume per volume per minute) with compressed air. The set point for dissolved oxygen (DO) was 30 %. When necessary, compressed air used for aeration was mixed with pure oxygen to provide sufficient oxygen supply.
2.2.3. Disposable bioreactor cultivation
Further upscaling cultivation experiments were carried out on a rocking platform (Biostat® CultiBag RM, Sartorius Stedim Biotech, Göttingen, Germany) in a 50 L disposable bag reactor (Flexsafe® RM 50 optical, Sartorius Stedim Biotech, Göttingen, Germany). 24 Liter defined HCDC medium was filled in the reactor via a sterile filter (Sartobran® P 300, Sartorius Stedim Biotech, Göttingen, Germany). The remaining 25 L reactor volume was used as head space. The reactor was inoculated with 4 % (v/v) of the total cultivation volume. Cultivation conditions were used as previously reported [26].
2.2.4. Offline sample measurement
For offline analysis 10 mL sample was taken every 1 h during the cultivation. Measurement of the OD600, cell dry weight (CDW) and glucose concentration were performed as previously reported [26]. Acetate concentrations of the culture supernatant were determined as described elsewhere [25].
2.2.5. Downstream processing
A maturation process and the separation of the cells was performed as previously reported [36]. The cell-free supernatant was concentrated to a final working volume of 800 mL by crossflow ultrafiltration (Sartoflow® Smart, Sartorius Stedim Biotech, Göttingen, Germany). The filter cassette had a molecular weight cut off (MWCO) of 30 kDa (Sartocon Slice Hydrosart, Sartorius Stedim Biotech, Göttingen, Germany). The inlet pressure towards the filter cassette was regulated to maximal 2 bar by controlling the pump flow rate.
The crossflow retentate was used for ethanol precipitation. The retentate was mixed with 96 % (v/v) ethanol to a final concentration of 80 % (v/v). The precipitate was centrifuged at 4816 × g and 4 °C for 30 min and dissolved in deionized water. The procedure was repeated twice. Afterwards the sample was further polished by treatment with clay minerals EX M 1753 as previously reported [26]. The treatment with clay minerals was repeated once followed by a sterile filtration the crossflow ultrafiltration device using a filter cassette with a MWCO of 0.45 μm. The product sample was subsequently dialyzed against deionized water. The filter cassette had a MWCO of 5 kDa. Finally, the product was recovered after 72 h of lyophilization at 0.1 mbar (Alpha 2–4 LSCplus, Martin Christ Gefriertrocknungsanlagen GmbH, Osterode, Germany).
2.2.6. Product analysis
2.2.6.1. Thiobarbituric acid (TBA)-assay for polySia quantification
The polySia concentration during the cultivation and downstream processing was measured colorimetrically using a modified TBA assay as previously reported [23]. For calibration, commercially available colominic acid (Carbosynth, Compton, United Kingdom) was used in concentrations ranging from 0.05 to 1 mg/mL and deionized water as blank.
2.2.6.2. Protein determination by the Bradford method
The protein concentration during the cultivation and downstream processing was determined using the Bradford quantification method. For quantification, 20 μL sample was mixed with 300 μL Bradford reagent (Quick Start™ Bradford dye, Bio-Rad Laboratories GmbH, Feldkirchen, Germany). The samples were incubated for 10 min at room temperature and the protein absorption from which the protein concentration was calculated was measured at 595 nm (Multiskan Spectro, Thermo Scientific). For calibration, commercially available Bovine Serum Albumin (BSA) was used in concentrations ranging from 0.025 to 2 mg/mL and deionized water as blank.
2.2.6.3. DNA analysis
The DNA concentration was determined by a UV–vis Spectrometer based on the extinction at 260 nm (NanoDrop 200, Thermo Scientific), whereby deionized water was used as blank.
2.2.6.4. Endotoxin determination
The endotoxin concentration was determined using the Endosafe nexgen-PTS™ system (Endosafe nexgen-PTS™, Charles River Laboratories, Boston, MA, USA).
2.2.6.5. Chain length characterization of polySia by DMB-HPLC analysis
The DP of the produced polySia was analyzed using common 1,2-diamino-4,5-methylenedioxybenzene (DMB) anion exchange (AEX) HPLC analysis [37]. Sample preparation through DMB-derivatization and subsequent HPLC analysis was conducted as described previously [38].
2.2.6.6. Structure analysis by 1H- and 13C-NMR Spectroscopy
The glycosidic linkage patterns of the polySia produced were investigated by NMR spectroscopy. Sample preparation and measurement were performed as previously reported [28].
3. Results
In the following chapters, the cultivation work for polySia production is described first. The strains used in this study were cultivated in two different bioreactor systems on a synthetic medium that allows fermentations with high cell density. Subsequently, the synthesized polySia was purified according to a validated process scheme [26].
3.1. Stirred bioreactor cultivation of E. coli K1
Stirred tank reactors (STR) are established cultivation systems that have been used in both industrial production and research for several decades. The commonly used wild type strain E. coli K1 was cultivated using the alternative synthetic HCDC medium in a 2 Liter glass STR. A total of three cultivations were performed in batch mode. The cultivation data of an exemplary cultivation in the STR are shown in Fig. 2.
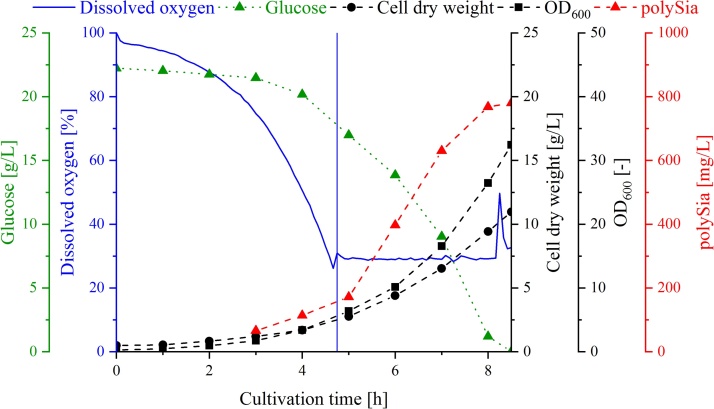
Fig. 2.
Time course of a typical batch cultivation of E. coli K1 in synthetic HCDC medium in a 2 L STR. Displayed are the dissolved oxygen content (blue line), the decrease in glucose concentration (green triangle) and the increase in cell density (square), CDW (circle) and polySia concentration (red triangle). The vertical blue line symbolizes the time after 4.6 h, from which the dissolved oxygen concentration in the medium was regulated to 30 % by adding pure oxygen to the air supply (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
Cultivation was stopped after 7.8 (±0.8) h after the glucose was completely consumed. The OD600 was determined to be 30.8 (±2.8) rel. AU, which correlates with a CDW of 10.6 (±0.7) mg/L. The measured polySia concentration in the supernatant was 774 (±17) mg/L. During the cultivation, an accumulation of acetate was also observed, and the final concentration was determined to be 0.9 (±0.2) g/L. Compared to previous results [23], this value is much lower and is due to the continuous oxygen supply to the cells, which indicated the absence of oxygen limitation during the cultivation. The use of the synthetic HCDC medium resulted in a total biomass yield (YX/S) of 0.452 (±0.071) g CDW/g Glucose. The product yield of polySia (YP/S) in the culture supernatant was 0.032 (±0.003) g polySia/g Glucose.
To better assess the efficacy of the new medium in the STR, all relevant cultivation results are summarized in Table 3 and compared to literature data [39]. In previous published studies, a synthetic minimal medium with cost-efficient components was used [23,25,27,39], which was optimized for polySia production [40]. Nevertheless, with the HCDC medium a higher biomass (22 %) was achieved, which additionally led to a significantly higher product yield (YP/S) in the culture supernatant (128 %). The maximum growth rate (μmax) was determined at 0.69 (±0.01) /h, which corresponds to a doubling time of 61 min. At the same time the amount of proteinogenic impurities (28 %) and the process time (-22 %) were reduced. The economically most relevant key figure, the space-time yield (STY), was calculated to be 0.099 (±0.011) g polySia/L/h, which corresponds to an increase of 196 % in comparison to the commonly applied medium.
Table 3.
Comparison of the cultivation results of E. coli K1 from media comparison in the STR.
| Parameter | This study | Literature [39] | Deviation |
|---|---|---|---|
| Time | 7.8 (±0.8) h | 10.1 (±1.8) h | −23 % |
| CDW | 10.6 (±0.7) g/L | 8.7 (±1.7) g/L | +22 % |
| polySia | 774 (±18) mg/L | 339 (±23) mg/L | +128 % |
| Protein | 129 (±25) mg/L | 181 (±51) mg/L | −28 % |
| μmax | 0.69 (±0.01) /h | 0.65 (±0.05) /h | +7 % |
| YP/S | 0.032 (±0.003) g polySia/g Glucose | 0.017 (±0.001) g polySia/g Glucose | +90 % |
| YX/S | 0.452 (±0.071) g CDW/g Glucose | 0.435 (±0.08) g CDW/g Glucose | +4 % |
| YP/X | 0.074 (±0.011) g polySia/g CDW | 0.039 (±0.005) g polySia/g CDW | +89 % |
| STY | 0.099 (±0.011) g polySia/L/h | 0.034 (±0.007) g polySiaL/h | +196 % |
3.2. Stirred bioreactor cultivation of recombinant E. coli BL21 strains
The three recombinant E. coli BL21 strains expressing α2,8- or α2,9polySia were cultivated under identical conditions in a 2 Liter STR. The experimental results were compared to the results of E. coli K1 cultures and are summarized in Table 4. Under the selected conditions, bacterial growth of the two α2,8-polySia expressing strains, E. coli BL21 pKT274 and E. coli BL21 pKT774, was stopped after 10.7 h and 10.9 h respectively. In the cultivation of E. coli BL21 pKT274 NmC-CAT expressing α2,9-polySia, the glucose was only completely metabolized after 18.2 h. Here, a 9 -h lag-phase was observed before the strain entered the exponential phase. However, longer lag-phases were already observed in earlier cultivation experiments of Nm strains that synthesize α2,9-polySia as a natural product [22]. All E. coli BL21-based strains achieved a CDW between 11.3 g/L and 11.9 g/L. Interestingly, despite the slightly higher biomass yields (YX/S), significantly lower polySia concentrations were measured in each culture supernatant. The highest measured polySia concentration reached only 133 mg/L, which also led to correspondingly lower product yields (YP/S) in the range from 0.003−0.005 g polySia/g Glucose. In comparison, the product yield of E. coli K1 was ten times higher. The different μmax were in a range from 0.4 /h - 0.42 /h, corresponding to doubling times of 90–98 min. The slower cell growth and lower growth rates are likely due to the additional exposure to the antibiotic selection markers in the medium. The space-time yields (STY) were calculated with 0.008 g polySia/L/h for E. coli pKT274, 0.012 g polySia/L/h for E. coli pKT774, and 0.007 g polySia/L/h E. coli pKT274 NmC-CAT.
Table 4.
Comparison of the cultivation results of different recombinant E. coli strains in the stirred tank reactor. The results for E. coli K1 are derived from the mean values of a total of three batch cultivations while the cultivation of each recombinant E. coli BL21 strain was performed exemplary.
| Parameter | E. coli K1 | E. coli pKT274 | E. coli pKT774 |
E. coli pKT274 NmC-CAT |
|---|---|---|---|---|
| Time [h] | 7.8 (±0.8) | 10.7 | 10.9 | 18.2 |
| CDW [g/L] | 10.6 (±0.7) | 11.3 | 11.8 | 11.9 |
| polySia [mg/L] | 774 (±18) | 82 | 133 | 127 |
| Protein [mg/L] | 129 (±25) | 304 | 195 | 209 |
| μmax [/h] | 0.69 (±0.01) | 0.42 | 0.42 | 0.40 |
| YP/S [g polySia/g Glucose] | 0.032 (±0.003) | 0.003 | 0.005 | 0.005 |
| YX/S [g CDW/g Glucose] | 0.452 (±0.071) | 0.451 | 0.471 | 0.475 |
| YP/X [g polySia/g CDW] | 0.074 (±0.011) | 0.007 | 0.011 | 0.011 |
| STY [g polySia/L/h] | 0.099 (±0.011) | 0.008 | 0.012 | 0.007 |
3.3. Disposable bag reactor cultivation of E. coli K1
In the production of medically relevant products, certified disposable bioreactor systems are increasingly used, which facilitate subsequent transfer to a GMP-compliant environment. The biotechnological production of polySia was also established in a wave-induced disposable bag reactor [26]. Aeration was carried out via the headspace and the gas input was controlled by regulating the rocking motion and dosing the oxygen content. In this work E. coli K1 was cultivated in a 50 Liter disposable bag reactor with a working volume of 25 liters. A total of three cultivations were performed in batch mode. The cultivation parameters were taken from previous work [26], but the defined HCDC medium was used. Fig. 3 shows the exemplary course of a typical cultivation of E. coli K1.
Fig. 3.
Time course of a typical batch cultivation of E. coli K1 in synthetic HCDC medium in a 25 Liter disposable bag reactor. Mixing was performed by wave induction. Displayed are the content of dissolved oxygen content (blue line), the rocking rate (black line), the decrease of glucose concentration (green triangle) and the increase of CDW (circle), acetate (square) and polySia concentration (red triangle). The vertical blue line symbolizes the time after 1.5 h, from which the dissolved oxygen concentration in the medium was regulated above 40 % by adding pure oxygen to the air supply (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
Immediately after inoculation, the DO content dropped below 50 %, so that the rocking frequency was automatically adjusted. Already after 1.6 h at a low cell density (CDW approximately 1.4 g/L) the airflow was supplied with pure oxygen. After five hours the system was aerated with an aeration flow of pure oxygen. Since the cell growth was not yet completed at this point, an oxygen limitation was observed from this time until the end of the cultivation. This is due to the poor oxygen transfer coefficient (kLa) and the resulting poor oxygen input by the headspace aeration [41,42].
The cultivation experiments were terminated after 7.5 (±1.3) h when the glucose was completely metabolized. A final CDW of 11.3 (±0.4) g/L and polySia concentration of 589 (±23) mg/L were achieved at the end of cultivation. Interestingly, the polySia concentration in the supernatant increased only slightly after the onset of oxygen limitation. The acetate concentration under non-limiting conditions was in the range of 2.1 g/L. Under anaerobic conditions, the concentration increased 5-fold to a final value of 10.2 g/L. In these amounts acetate also limits the growth of E. coli [43], which can be clearly seen in the CDW curse (Fig. 3). The YX/S achieved 0.451 (±0.019) g CDW/g Glucose and YP/S was determined at 0.024 (±0.001) g polySia/g Glucose.
Table 5 gives a complete overview of all cultivation results obtained in the wave-induced disposable bag reactor. In the following, the cultivation data using the HCDC medium were compared with previous literature values [39]. The change of the medium also reduced the cultivation time by 35 % in the bag reactor system used. The cell density (50 %) and polySia concentration (96 %) could be significantly increased at the same time. Unfortunately, the percentage of proteinogenic impurities (90 %) also increased. The growth rate was calculated with 0.72 (±0.02) /h, which corresponds to a doubling time of 58 min. The yield coefficients also show that more biomass and product is formed per substrate (see Table 5). The STY was also significantly increased to 0.079 (±0.011) g polySia/L/h (206 %).
Table 5.
Comparison of the cultivation results of E. coli K1 from media comparison in the disposable bag reactor.
| Parameter | This study | Literature [39] | Deviation |
|---|---|---|---|
| Time | 7.5 (±1.3) h | 11.5 (±1.8) h | −35 % |
| CDW | 11.3 (±0.4) g/L | 7.5 (±0.8) g/L | +50 % |
| polySia | 589 (±23) mg/L | 301 (±20) mg/L | +96 % |
| Protein | 207 (±38) mg/L | 109 (±42) mg/L | +90 % |
| μmax | 0.72 (±0.02) /h | 0.64 (±0.1) /h | +12 % |
| YP/S | 0.024 (±0.001) g polySia/g Glucose | 0.015 (±0.001) g polySia/g Glucose | +57 % |
| YX/S | 0.451 (±0.019) g CDW/g Glucose | 0.375 (±0.04) g CDW/g Glucose | +20 % |
| YP/X | 0.052 (±0.004) g polySia/g CDW | 0.04 (±0.003) g polySia/g CDW | +30 % |
| STY | 0.079 (±0.011) g polySia/L/h | 0.026 (±0.004) g polySia/L/h | +206 % |
3.4. Disposable bag reactor cultivation of recombinant E. coli BL21 strains
The recombinant polySia production was also performed in the disposable bag bioreactor. The exemplary time course of the cultivation of E. coli BL21 pKT274 NmC-CAT expressing α2,9-polySia is shown in Fig. 4.
Fig. 4.
Time course of the batch cultivation of E. coli BL21 pKT274 NmC-CAT on the synthetic HCDC medium in a 25 Liter disposable bag reactor. Mixing was performed by wave induction. Displayed are the content of dissolved oxygen content (blue line), the rocking rate (black line), the decrease of glucose concentration (green triangle) and the increase of CDW (circle), acetate (square) and polySia concentration (red triangle). The vertical blue line symbolizes the time after 9.5 h, from which the dissolved oxygen concentration in the medium was regulated above 40 % by adding pure oxygen to the air supply (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
After inoculating the bioreactor, the rocker rate increased to 32 rocks per minute (Rpm) in order to regulate the DO content at 50 %. The rate is only further adjusted automatically after 6.5 h. After 9.5 the maximum rate of 42 Rpm is reached and pure oxygen is partially supplied to the aeration flow. Sampling takes place after 10 h at the beginning of the exponential phase. As before in the cultivation of E. coli K1, an oxygen limitation occured after 15 h, since the aeration flow was supplied with pure oxygen, but the cells were still growing. After 17.8 h the glucose was completely consumed. Then, a cell density of 9.1 g/L and a polySia concentration of 93 mg/L were reached. Acetate accumulation increased from 2.2 g/L to 7.1 g/L during anaerobic growth, which slowed cell growth. The time courses of both α2,8-polySia producing recombinant strains were comparable except for the much shorter lag phase.
The experimental results were compared with the results of E. coli K1 cultures and are summarized in Table 6. All recombinant strains showed comparable maximum growth rates of 0.49−0.53 /h, which were, however, significantly lower than those of E. coli K1 (0.72 /h). This resulted in a much longer cultivation time for all recombinant strains. Due to the oxygen limitation occurring, low final cell densities in the range of 8.1 g/L and 9.3 g/L were achieved. The yields of product and biomass per substrate and product per biomass are comparable among the recombinant strains. However, the values are significantly lower than the yields of E. coli K1. The STY was calculated at 0.01 g polySia/L/h and 0.099 g polySia/L/h for the α2,8-polySia producing strains and 0.005 g polySia/L/h for the α2,9-polySia producing strain.
Table 6.
Comparison of the cultivation results of different recombinant E. coli strains in the disposable bag reactor. The results for E. coli K1 are derived from the mean values of a total of three batch cultivations while the cultivation of each recombinant E. coli BL21 strain was performed exemplary.
| Parameter | E. coli K1 | E. coli pKT274 | E. coli pKT774 |
E. coli pKT274 NmC-CAT |
|---|---|---|---|---|
| Time [h] | 7.5 (±1.3) | 10.4 | 9.3 | 17.8 |
| CDW [g/L] | 11.3 (±0.4) | 9.3 | 8.1 | 9.1 |
| polySia [mg/L] | 589 (±23) | 98 | 86 | 93 |
| Protein [mg/L] | 207 (±38) | 71 | 56 | 72 |
| μmax [/h] | 0.72 (±0.02) | 0.52 | 0.53 | 0.49 |
| YP/S [g polySia/g Glucose] | 0.024 (±0.001) | 0.004 | 0.003 | 0.004 |
| YX/S [g CDW/g Glucose] | 0.452 (±0.019) | 0.359 | 0.323 | 0.363 |
| YP/X [g polySia/g CDW] | 0.052 (±0.004) | 0.011 | 0.011 | 0.010 |
| STY [g polySia/L/h] | 0.079 (±0.011) | 0.01 | 0.009 | 0.005 |
3.5. Downstream processing for purification of polySia
After the upstream process, the produced polySia was isolated from the culture broth to obtain a pure product. The performance of the purification process is described using the data of E. coli K1 (Fig. 5). The culture broth was stored for 17 h at 8 °C to release bound material from the cell surface. This maturation process increased the amount of polySia in the culture supernatant by 10 % [36]. The final polySia concentration after maturation was 634 (±13) mg/L while the protein concentration reached 275 (±31) mg/L. Cell separation was then performed by continuous centrifugation as described before [26]. The supernatant was subsequently concentrated 50-fold to 500 mL using the crossflow ultrafiltration system with a filter cassette with MWCO of 30 kDa for the further process steps. Approximately 48 % of the polySia got lost during this process step. The larger MWCO led to a comparable product loss [26], but at the same time was able to reduce the working time. During these process steps the ratio was approximately 0.44 g HCP/g polySia. To remove proteins, the retentate was first precipitated three times with 80 % (v/v) ethanol, which reduced the ratio to 0.3 g HCP/g polySia. For further HCP removal, the retentate was treated with clay minerals, which adsorb HCP non-specifically. Due to the higher sample concentrations, the clay mineral treatment was repeated, and the supernatant was sterile filtered using the crossflow ultrafiltration system with a filter cassette with MWCO of 0.45 μm. After this process step, no protein was clearly detectable. The salts introduced during the process were finally removed by diafiltration with a filter cassette with MWCO of 5 kDa before the product was freeze-dried, resulting in similar purities and yields as described before [26]. The yield of pure polySia after lyophilization was 29 (±5) % as shown in Fig. 5. This corresponds to an absolute quantity of 4.5 (± 0.5) g of pure polySia in the bag reactor per 25 L-batch. The bacterial DNA concentration was determined with 1.3 (±0.2) mg DNA/g polySia. Interestingly, the endotoxin burden due to the higher cell quantities after cultivation was also significantly higher than described in the literature. The final product had a concentration of over 100,000 EU/mg. However, in this DSP no specific methods were applied to specifically reduce the endotoxin burden.
Fig. 5.
Protein and polySia concentrations during the entire downstream process. The amount of proteinogenic impurities (white bars) was efficiently reduced using a common validated purification process. After completing the purification process no protein was detectable in the final lyophilized product. The recovery yield of pure polySia (black bars) was 29 (±5) %.
The purification of recombinantly produced polySia was also carried out from disposable bioreactor culture broth using the described process. Interestingly, it was shown that treatment with clay minerals led to a significant product loss. Although DNA and HCP were also reduced to below the detection limit, the yield of the final product was only between 8 and 15 %. As the concentrations in the culture supernatant were already much lower compared to the yield of E. coli K1, only 91–300 mg polySia were produced. As the small quantities are not yet of economic relevance, cost-expensive analysis for endotoxins was not performed.
3.6. Product analysis
To confirm their identity the final products were subjected to NMR spectroscopy. 1H spectra were recorded and compared with the spectrum of α2,8-polySia from E. coli K1. Comparison with the published chemical shifts of colominic acid (α2,8) and NmC capsular polysaccharide (α2,9) allowed the assignment of all observed signals [11,28,44]. All 1H-NMR (Fig. 6) show the signals for H3eq and H3ax of sialic acid. In the spectrum of recombinant polySia from E. coli BL21 pKT274 and pKT774, chemical shifts for H9b, H8 and H7, which are characteristic for colominic acid, corroborate the α2,8 linkage. The spectrum of E. coli BL21 pKT274 polySia contains some unknown signals at a chemical shift of 3.6 ppm and in the range of 3.4 ppm and 3.1 ppm, which are most likely due to process related impurities. In summary, the high identity to the E. coli K1 reference spectrum and published spectra confirm that both polymers consist of α2,8-linked polySia.
Fig. 6.
Comparison of the 1H-NMR spectra of the produced polySia from E. coli K1 (1) and the three recombinant E. coli BL21 strains (2-4). The spectra of the recombinant strain carrying the pKT274 (2) and pKT774 (3) plasmid, respectively, showed a high similarity towards the reference spectrum, indicating α2,8-glycosidic linked monomers. However, the spectrum of the NmC-CAT variant (4) shows signal differences at a chemical shift of 3.5 - 4 ppm, which indicates a α2,9-glycosidic linkage of the monomers.
The spectrum obtained for polySia from E. coli BL21 pKT274 NmC-CAT markedly differs from the α2,8-linked polymer (compare Fig. 6.1-3 and Fig. 6.4), but is in very good agreement with the chemical shifts reported for de-O-acetylated α2,9-linked PolySia harvested from Nm serogroup C [11], which was used as basis for peak assignment. Most importantly, the signals observed for H9b, H8 and H7 are shifted upfield, further corroborating the different placement of the glycosidic linkage.
Another important characteristic for the assignment of a specific application area of polySia is the maximum chain length. The maximum chain lengths of the different polysialic acids produced in this study were determined by HPLC analysis. Subsequently, the samples were first labelled with the fluorescent dye 1.2-diamino-4.5-methylenedioxybenzene (DMB). The chain length of polySia from E. coli K1 was clearly detectable up to a DP of 92 (Fig. 7A). The fluorescence signal in the chromatogram suggests that the actual maximum DP is still higher. These results are comparable to previous works [28,38]. The chromatograms of the recombinant strains show that the maximum chain length of the reference is comparable. The maximum DP of the recombinant α2,8-polySia was determined to be DP > 90 (Fig. 7B). A maximum DP of >80 was measured for the recombinant α2,9-polySia (Fig. 7C).
Fig. 7.
Characterization and comparison of the maximal polySia chain length. Endogenous polySia from E. coli K1 (A) was analyzed and compared to recombinant α2,8-polySia expressed by E. coli BL21 pKT274 (B) and α2,9-polySia expressed by E. coli BL21 pKT274 NmC-CAT (C). The degree of polymerization (DP) was measured using HPLC analysis with subsequent DMB-derivatization.
4. Discussion
Polysialic acid is an interesting candidate for various medical and biotechnological fields and can be produced very easily by fermentation of E. coli K1. However, state-of-art cultivations only achieve moderate final concentrations of polySia. This study describes the establishment of a high cell density process to improve the biotechnological production of long chain polySia by cultivation of E. coli K1. In addition, α2,8- and α2,9-polySia polySia is produced via three engineered laboratory safety E. coli BL21 strains to overcome risks of large-scale cultivations of pathogenic bacteria. The application of an alternative defined medium [33] for high cell density cultivation of E. coli K1 significantly improved the polySia yield due to higher cell densities in comparison to the commonly used medium [40]. The production was carried out in a conventional STR and a GMP-compliant wave-induced disposable bag reactor. In both systems the cultivation time was efficiently reduced while higher growth rates were achieved. Additionally, the space-time yield was tripled. A comparison of the medium composition with the medium previously used for polySia production [40] shows that the HCDC medium contains significantly more trace elements (Table 1). In addition, the glucose concentration is 25 g/L instead of 20 g/L, which supports cell growth and polySia production.
In direct comparison of the cultivation systems, the STR achieved the highest product yields, mainly because polysialic acid production was not negatively influenced by oxygen limitation. Compared to the STR, the oxygen transfer in the bag reactor is poor due to the headspace aeration [41]. The resulting anaerobic conditions increase the production of acetate while the polysialic acid metabolism is suppressed. In the bag reactor, the acetate concentration even reached critical concentrations that slowed down cell growth [23,43]. The establishment of a fed-batch process via DO-based control and reduction of the initial concentration of glucose to approximately 10 g/L could likely further increase the production yield while oxygen limitation could be prevented.
The selected conditions were successfully transferred to laboratory safety E. coli BL21 strains to produce α2,8- and α2,9-polySia recombinantly. Despite comparable cell densities, the maximal measured polySia concentration in the culture supernatant was considerably lower. One possible explanation for this observation might be the presence of two functional capsule export machineries in the engineered strains. E. coli BL21(DE3) was shown to encode functional regions 1 and 3 of its capsule gene cluster, while an IS1 element in region 2 was hypothesized to prevent capsule expression [30]. Although genes of region 1 and 3 are highly conserved and were shown to be interchangeable between serotypes and even species [30,45], a previous study demonstrated that KpsCS from BL21(DE3) was less suitable to promote E. coli K1 polysialic acid synthesis if compared to endogenous KpsCS [30]. Consequently, BL21 KpsCS as well as other BL21 capsule biosynthesis enzymes might interfere with capsule assembly or export.
The polySia produced was successfully isolated using a validated purification process. The yields for endogenous polySia from E. coli K1 were 29 (±5) %, which corresponds to a total of 4.5 (± 0.5) g in the disposable bag reactor. Purification of the recombinant polySia, however, resulted in yields of 15 % and less. Above all, the adsorption on clay minerals increased the product loss, which indicates that the downstream processing must be further optimized for the recombinant strains. Product analysis showed that bacterial DNA and proteinogenic contaminants were successfully removed below detectable limits. Since no targeted methods for endotoxin depletion were carried out in this process, the endotoxin load in the final product was also >100,000 EU/mg due to the higher cell densities after cultivation. The integration of alkaline incubation and subsequent anion exchange fractionation enables the production of highly pure polySia [28,36].
1H-NMR spectroscopy confirmed that two of the recombinant strains produce α2,8-linked polySia. However, the exchange of neuS with synE in the kps gene cluster also enables the synthesis of α2,9-linked polySia in E. coli [32,46]. The maximum chain lengths of endogenous and recombinant polySia were >90 monomer residues. These values are comparable to the results of previous work [28,39]. Despite the low yields after downstream processing, the recombinant production of α2,8- and α2,9-linked polySia in E. coli was successful.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
CRediT authorship contribution statement
Bastian Bartling: Conceptualization, Methodology, Validation, Investigation, Resources, Data curation, Writing - original draft, Writing - review & editing, Visualization. Nora C. Brüchle: Methodology, Validation, Investigation, Data curation, Visualization. Johanna S. Rehfeld: Methodology, Validation, Investigation, Data curation, Visualization. Daniel Boßmann: Conceptualization, Methodology, Investigation. Timm Fiebig: Resources, Writing - review & editing. Christa Litschko: Validation, Resources, Writing - review & editing. Jörg Fohrer: Methodology, Formal analysis, Investigation, Data curation. Rita Gerardy-Schahn: Resources, Writing - review & editing. Thomas Scheper: Resources, Writing - review & editing, Funding acquisition. Sascha Beutel: Conceptualization, Resources, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
This work was financially supported by the German Federal Ministry of Education and Research (BMBF-VIP+, grant numbers: BMBF-03VP00271 and BMBF-03VP00273). Furthermore, we would like to thank Monika Berger (Institute of Clinical Chemistry) for excellent technical assistance.
Contributor Information
Bastian Bartling, Email: Bartling@iftc.uni-hannover.de.
Nora C. Brüchle, Email: Bruechle@iftc.uni-hannover.de.
Johanna S. Rehfeld, Email: Rehfeld@iftc.uni-hannover.de.
Daniel Boßmann, Email: Bossmann@iftc.uni-hannover.de.
Timm Fiebig, Email: Fiebig.Timm@mh-hannover.de.
Christa Litschko, Email: Litschko.Christa@mh-hannover.de.
Jörg Fohrer, Email: Joerg.Fohrer@oci.uni-hannover.de.
Rita Gerardy-Schahn, Email: Gerardy-Schahn.Rita@mh-hannover.de.
Thomas Scheper, Email: Scheper@iftc.uni-hannover.de.
Sascha Beutel, Email: Beutel@iftc.uni-hannover.de.
References
- 1.Whitfield C., Roberts I.S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 2.Weisgerber F.A., Christoph Troy. Biosynthesis of the polysialic acid capsule in Escherichia coli K1. J. Biol. Chem. 1990;265:1578–1587. [PubMed] [Google Scholar]
- 3.Schnaar R.L., Gerardy-Schahn R., Hildebrandt H. Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 2014;94:461–518. doi: 10.1152/physrev.00033.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyata S., Sato C., Kumita H., Toriyama M., Vacquier V.D., Kitajima K. Flagellasialin: a novel sulfated α2,9-linked polysialic acid glycoprotein of sea urchin sperm flagella. Glycobiology. 2006;16:1229–1241. doi: 10.1093/glycob/cwl036. [DOI] [PubMed] [Google Scholar]
- 5.Tzeng Y.L., Stephens D.S. Epidemiology and pathogenesis of Neisseria meningitidis. Microbes Infect. 2000;2:687–700. doi: 10.1016/s1286-4579(00)00356-7. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharjee A.K., Jennings H.J., Kenny C.P., Martin A., Smith I.C. Structural determination of the sialic acid polysaccharide antigens of Neisseria meningitidis serogroups B and C with carbon 13 nuclear magnetic resonance. J. Biol. Chem. 1975;250:1926–1932. [PubMed] [Google Scholar]
- 7.Lindon J.C., Vinter J.G., Lifely M.R., Moreno C. Conformational and dynamic differences between N. meningitidis serogroup B and C polysaccharides, using n.m.r. spectroscopy and molecular mechanics calculations. Carbohydr. Res. 1984;133:59–74. doi: 10.1016/0008-6215(84)85183-6. [DOI] [PubMed] [Google Scholar]
- 8.Willis L.M., Stupak J., Richards M.R., Lowary T.L., Li J., Whitfield C. Conserved glycolipid termini in capsular polysaccharides synthesized by ATP-binding cassette transporterdependent pathways in Gram-negative pathogens. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7868–7873. doi: 10.1073/pnas.1222317110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colley K.J., Kitajima K., Sato C. Polysialic acid: Biosynthesis, novel functions and applications. Crit. Rev. Biochem. Mol. Biol. 2014;49:498–532. doi: 10.3109/10409238.2014.976606. [DOI] [PubMed] [Google Scholar]
- 10.Wu J., Zhan X., Liu L., Xia X. Bioproduction, purification, and application of polysialic acid. Appl. Microbiol. Biotechnol. 2018;102:9403–9409. doi: 10.1007/s00253-018-9336-3. [DOI] [PubMed] [Google Scholar]
- 11.Lemercinier X., Jones C. Full 1H NMR assignment and detailed O-acetylation patterns of capsular polysaccharides from Neisseria meningitidis used in vaccine production. Carbohydr. Res. 1996;296:83–96. doi: 10.1016/S0008-6215(96)00253-4. [DOI] [PubMed] [Google Scholar]
- 12.Cantarel B.I., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willis L.M., Whitfield C. Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by ABC transporter-dependent pathways. Carbohydr. Res. 2013;378:35–44. doi: 10.1016/j.carres.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Shahraz A., Kopatz J., Mathy R., Kappler J., Winter D., Kapoor S., Schütza V., Scheper T., Gieselmann V., Neumann H. Anti-inflammatory activity of low molecular weight polysialic acid on human macrophages. Sci. Rep. 2015;5:1–17. doi: 10.1038/srep16800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werneburg S., Buettner F.F.R., Mühlenhoff M., Hildebrandt H. Polysialic acid modification of the synaptic cell adhesion molecule SynCAM 1 in human embryonic stem cell-derived oligodendrocyte precursor cells. Stem Cell Res. 2015;14:339–346. doi: 10.1016/j.scr.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Werneburg S., Buettner F.F.R., Erben L., Mathews M., Neumann H., Mühlenhoff M., Hildebrandt H. Polysialylation and lipopolysaccharide-induced shedding of E-selectin ligand-1 and neuropilin-2 by microglia and THP-1 macrophages. Glia. 2016;64:1314–1330. doi: 10.1002/glia.23004. [DOI] [PubMed] [Google Scholar]
- 17.Kallolimath S., Castilho A., Strasser R., Grünwald-Gruber C., Altmann F., Strubl S., Galuska C.E., Zlatina K., Galuska S.P., Werner S., Thiesler H., Werneburg S., Hildebrandt H., Gerardy-Schahn R., Steinkellner H. Engineering of complex protein sialylation in plants. Proc. Natl. Acad. Sci. U. S. A. 2016;113:9498–9503. doi: 10.1073/pnas.1604371113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlstetter M., Kopatz J., Aslanidis A., Shahraz A., Caramoy A., Linnartz‐Gerlach B., Lin Y., Lückoff A., Fauser S., Düker K., Claude J., Wang Y., Ackermann J., Schmidt T., Hornung V., Skerka C., Langmann T., Neumann H. Polysialic acid blocks mononuclear phagocyte reactivity, inhibits complement activation, and protects from vascular damage in the retina. EMBO Mol. Med. 2017;9:154–166. doi: 10.15252/emmm.201606627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanato Y., Kitajima K., Sato C. Direct binding of polysialic acid to a brain-derived neurotrophic factor depends on the degree of polymerization. Glycobiology. 2008;18:1044–1053. doi: 10.1093/glycob/cwn084. [DOI] [PubMed] [Google Scholar]
- 20.Ono S., Hane M., Kitajima K., Sato C. Novel regulation of Fibroblast Growth Factor 2 (FGF2)-mediated cell growth by polysialic acid. J. Biol. Chem. 2012;287:3710–3722. doi: 10.1074/jbc.M111.276618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregoriadis G., Fernandes A., Mital M., McCormack B. Polysialic acids: potential in improving the stability and pharmacokinetics of proteins and other therapeutics. Cell. Mol. Life Sci. 2000;57:1964–1969. doi: 10.1007/PL00000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma S., Hanif S., Kumar N., Joshi N., Rana R., Dalal J., Singh D., Chhikara M.K. Rapid processes for purification of capsular polysaccharides from Neisseria meningitidis serogroups A and C. Biologicals. 2015;43:383–389. doi: 10.1016/j.biologicals.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Rode B., Endres C., Ran C., Stahl F., Beutel S., Kasper C., Galuska S., Geyer R., Mühlenhoff M., Gerardy-Schahn R., Scheper T. Large-scale production and homogenous purification of long chain polysialic acids from E. coli K1. J. Biotechnol. 2008;135:202–209. doi: 10.1016/j.jbiotec.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Z.Y., Wang S.Z., Li G.S., Zhan X.B., Lin C.C., Wu J.R., Zhu L. A new polysialic acid production process based on dual-stage pH control and fed-batch fermentation for higher yield and resulting high molecular weight product. Appl. Microbiol. Biotechnol. 2013;97:2405–2412. doi: 10.1007/s00253-012-4503-4. [DOI] [PubMed] [Google Scholar]
- 25.Chen R., John J., Rode B., Hitzmann B., Gerardy-Schahn R., Kasper C., Scheper T. Comparison of polysialic acid production in Escherichia coli K1 during batch cultivation and fed-batch cultivation applying two different control strategies. J. Biotechnol. 2011;154:222–229. doi: 10.1016/j.jbiotec.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 26.de Vries I., Busse C., Kopatz J., Neumann H., Beutel S., Scheper T. Polysialic acid production using Escherichia coli K1 in a disposable bag reactor. Eng. Life Sci. 2017;17:723–731. doi: 10.1002/elsc.201600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bice I., Celik H., Wolff C., Beutel S., Zahid M., Hitzmann B., Rinas U., Kasper C., Gerardy-Schahn R., Scheper T. Downstream processing of high chain length polysialic acid using membrane adsorbers and clay minerals for application in tissue engineering. Eng. Life Sci. 2013;13:140–148. doi: 10.1002/elsc.201200041. [DOI] [Google Scholar]
- 28.Bartling B., Rehfeld J.S., Boßmann D., De Vries I., Fohrer J., Lammers F., Scheper T., Beutel S. Determination of the structural integrity and stability of polysialic acid during alkaline and thermal treatment. Molecules. 2019;25:1–15. doi: 10.3390/molecules25010165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chart H., Smith H.R., La Ragione R.M., Woodward M.J. An investigation into the pathogenic properties of Escherichia coli strains BLR, BL21, DH5α and EQ1. J. Appl. Microbiol. 2000;89:1048–1058. doi: 10.1046/j.1365-2672.2000.01211.x. [DOI] [PubMed] [Google Scholar]
- 30.Andreishcheva E.N., Vann W.F. Escherichia coli BL21(DE3) chromosome contains a group II capsular gene cluster. Gene. 2006;384:113–119. doi: 10.1016/j.gene.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 31.Navasa N., Rodríguez‑Aparicio L., Ferrero M.Á., Monteagudo‑Mera A., Martínez‑Blanco H. Polysialic and colanic acids metabolism in Escherichia coli K92 is regulated by RcsA and RcsB. Biosci. Rep. 2013;33:405–415. doi: 10.1042/bsr20130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Chen X. Sialic acid metabolism and syaliltransferases: natural functions and aplications. Appl. Microbiol. Biotechnol. 2012;29:997–1003. doi: 10.1016/j.biotechadv.2011.08.021.Secreted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korz D.J., Rinas U., Hellmuth K., Sanders E.A., Deckwer W.D. Simple fed-batch technique for high cell density cultivation of Escherichia coli. J. Biotechnol. 1995;39:59–65. doi: 10.1016/0168-1656(94)00143-Z. [DOI] [PubMed] [Google Scholar]
- 34.Frosch M., Roberts I., Görgen I., Metzger S., Boulnois G.J., Bitter-Suermann D. Serotyping and genotyping of encapsulated Escherichia coli K1 sepsis isolates with a monoclonal IgG anti K1 antibody and K1 gene probes. Microb. Pathog. 1987;2:319–326. doi: 10.1016/0882-4010(87)90074-X. [DOI] [PubMed] [Google Scholar]
- 35.Boulnois G.J., Roberts I.S., Hodge R., Hardy K.R., Jann K.B., Timmis K.N. Definition of three functional regions for capsule production. Mol. Genet. Genomics. 1987;208:242–246. doi: 10.1007/BF00330449. [DOI] [PubMed] [Google Scholar]
- 36.de Vries I., Schreiber S., Boßmann D., Hellmann Z., Kopatz J., Neumann H., Beutel S. Single-use membrane adsorbers for endotoxin removal and purification of endogenous polysialic acid from Escherichia coli K1. Biotechnol. Rep. 2018;17:110–116. doi: 10.1016/j.btre.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue S., Lin S.G., Lee Y.C., Inoue Y. An ultrasensitive chemical method for polysialic acid analysis. Glycobiology. 2001;11:759–767. doi: 10.1093/glycob/11.9.759. [DOI] [PubMed] [Google Scholar]
- 38.Boßmann D., Bartling B., de Vries I., Winkler J., Neumann H., Lammers F., Beutel S., Scheper T. Charged aerosol detector HPLC as a characterization and quantification application of biopharmaceutically relevant polysialic acid from E. coli K1. J. Chromatogr. A. 2019;20:1–10. doi: 10.1016/j.chroma.2019.03.069. [DOI] [PubMed] [Google Scholar]
- 39.de Vries I. 2018. Herstellung zelleigener Produkte mittels Escherichia coli in unterschiedlichen Bioreaktorsystemen. [Google Scholar]
- 40.Rodríguez-Aparicio L.B., Reglero A., Ortiz A.I., Luengo J.M. Effect of physical and chemical conditions on the production of colominic acid by E. coli in defined medium. Appl. Microbiol. Biotechnol. 1988;27:474–483. doi: 10.1007/BF00451616. [DOI] [Google Scholar]
- 41.Dreher T., Husemann U., Zahnow C., de Wilde D., Adams T., Greller G. High cell density Escherichia coli cultivation in different single-use bioreactor systems. Chem. Ing. Tech. 2013;85:162–171. doi: 10.1002/cite.201200122. [DOI] [Google Scholar]
- 42.Jonczyk P., Takenberg M., Hartwig S., Beutel S., Berger R.G., Scheper T. Cultivation of shear stress sensitive microorganisms in disposable bag reactor systems. J. Biotechnol. 2013;167:370–376. doi: 10.1016/J.JBIOTEC.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Luli G.W., Strohl W.R. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 1990;56:1004–1011. doi: 10.1128/aem.56.4.1004-1011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michon F., Brisson J.R., Jennings H.J. Conformational differences between linear α(2→8)-linked homosialooligosaccharides and the epitope of the group B meningococcal polysaccharide. Biochemistry. 1987;26:8399–8405. doi: 10.1021/bi00399a055. [DOI] [PubMed] [Google Scholar]
- 45.Willis L.M., Whitfield C. KpsC and KpsS are retaining 3-deoxy-D-manno-oct-2- ulosonic acid (Kdo) transferases involved in synthesis of bacterial capsules. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20753–20758. doi: 10.1073/pnas.1312637110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrero M.Á., Aparicio L.R. Biosynthesis and production of polysialic acids in bacteria. Appl. Microbiol. Biotechnol. 2010;86:1621–1635. doi: 10.1007/s00253-010-2531-5. [DOI] [PubMed] [Google Scholar]