Abstract
In the functional proteome era, the proteomic profiling of clinicopathologic annotated tissues is an essential step for mining and evaluations of candidate biomarkers for disease. Previously, application of routine proteomic methodologies to clinical tissue specimens has provided unsatisfactory results. Multiplex tissue immunoblotting is a method of transferring proteins from a formalin-fixed, paraffin-embedded tissue section to a stack of membranes which can be applied to a conventional immunoblotting method. A single tissue section can be transferred to up to ten membranes, each of which is probed with antibodies and detected with fluorescent tags. By this approach, total protein and target signals can be simultaneously determined on each membrane; hence each antibody is internally normalized. Phosphorylation specific antibodies as well as antibodies that do not readily work well with paraffin-embedded tissue are applicable to the membranes, expanding the menu of antibodies that can be utilized with formalin-fixed tissue. This novel platform can provide quantitative detection retaining histomorphologic detail in clinical samples and has great potential to facilitate discovery and development of new diagnostic assays and therapeutic agents.
Keywords: Formalin-fixed, Paraffin-embedded, Tissue, Immunodetection, Histomorphology, Proteomics, Expressional profiling
1. Introduction
Proteomic profiling of tissue specimens, having pathologic and histologic relevance, promises to the development of biomarkers to guide diagnosis and therapy in biomedicine [1, 2]. Many of the traditional approaches such as western blots are based on a “grind and bind” means of isolating proteins from tissue. This “grind and bind” approach fails to provide a histomorphologic perspective of protein expression. The only means of gaining a histomorphologic understanding of protein expression have been immunohistochemistry and laser capture microdissection based collection of samples for traditional analysis. Laser capture microdissection does provide the capacity to perform a directed western blot on tissue [3–5]; however, it is time-consuming and does not provide a global expression view of a target protein. Immunohistochemistry, yet providing excellent localization, lacks quantification without sophisticated instrumentation [6] and lacks normalization. However, formalin-fixed and paraffin-embedded tissue, the gold standard of diagnostic histopathology, is not routinely applicable to the “grind and bind” approach due to the high level of covalently cross-linked proteins arising from formalin fixation. In translational research, there is the great desire to utilize the vast archive of formalin-fixed and paraffin-embedded tissue that has been collected [7]. Most research antibodies do not perform well in paraffin-embedded tissues. This failure is thought to be related to protein cross-linking, inadequate deparaffinization and issues of epitope presentation. This problem has become a bottleneck in translational research [2].
To address these challenges, a number of protein-based arrays have been developed and evaluated in clinical research fields. Although these techniques are generally superior in expression profiling and quantitation of protein changes associated with disease states, each still has significant limitations [8–10]. Investigators continue to seek a convenient and reliable proteomic tool, which can detect protein in tissue, while having retention of both quantitative and histomorphologic features. Multiplex tissue immunoblotting meets the criteria. This method provides a level of histomorphologic correlation, but at the same time, presents the proteins on a membrane platform that widens the number of antibodies that can be utilized. Additionally the total amount of protein present on each membrane can be determined and used for normalization [11–15]. As a research tool, this method expands the capacity of a tissue microarray to a protein array with quantitative data that can be normalized and directly composed for different antigens detected on a single stack of membranes [13–15]. When applied to a whole section of tissue [11, 12], it allows the ability to profile a tumor for multiple antigens with the use of single paraffin-embedded slide.
As an example of the utility of multiplex tissue immunoblotting, we have quantified the change in seven proteins in the transition from normal epithelium to invasive tumor [12]. This approach allows the quantification of changes in the expression of potential biomarkers in normal, in situ and invasive disease. This approach provided insight into the timing and magnitude of protein changes seen in this transition of from benign to invasive cancer as is useful in the development of novel biomarkers for prevention and screening of cancer. Additionally, the application of this method can be expanded to studying the proteomic profile of a signaling pathway, and is a promising means of assessment of molecular-targeting therapies as well. We previously demonstrated the feasibility of this method in studying PI3K/AKT pathway in extrahepatic cholangiocarcinoma and premalignant lesions. Finally, this approach can be applied to tissue microarrays, creating a form of protein array [15].
2. Materials
2.1. Deparaffinization and Enzyme Treatment
Xylene or dewaxing reagents.
100, 95, and 70 % ethanol (EtOH) (molecular biology grade).
50 mM ammonium bicarbonate (NH4HCO3) buffer (pH 8.2). Store room temperature (RT).
Stock solution of trypsin (200×): 0.2 % trypsin solution in 50 mM ammonium bicarbonate solution (pH 8.2). Immediately freeze in single use (200 μL) aliquots at −20 °C.
Stock solution of Proteinase-K (400×): ready-to-use Proteinase-K, freeze in single use (50 μL) aliquots at −20 °C (see Note 1).
Enzyme cocktail solution (prepare freshly before use): 0.001 % trypsin plus 1:400-fold diluted the ready-to-use Proteinase-K in 50 mM ammonium bicarbonate solution, pH 8.2 (see Note 2).
ProBuffer: One tablet of complete protease inhibitor, 0.5 mL phosphatase inhibitor I, 0.5 mL phosphatase inhibitor II, in 50 mL PBS (pH 7.2). Store at 4 °C.
Phosphate-buffered saline (PBS, pH 7.2).
Plastic or glass coplin jars for slide processing.
Incubation chamber for enzyme reaction (five-slides mailer box).
2.2. Transferring from the Tissue Slide to Membrane
Transfer buffer (5×): 250 mM Tris base (do not adjust pH), 1,900 mM glycine. Store at RT. Dilute 200 mL with 800 mL distilled water for use.
A stack of membranes (five or ten sheets) (P-Film, 20/20 GeneSystems, Rockville, MD, USA) (see Note 3).
Spacer membrane (GE polycarbonate PVPF membrane, GE Osmonics Labstore, Minnetonka, MN, USA) (see Note 4).
Nitrocellulose membrane.
Absorbent pads (Blot Absorbent Filter paper).
Slide glass (Opticlear® Microscope slide).
Kapak SealPAK pouches.
Impulse sealer.
Heat block (Dry bath).
2.3. Immunoblotting
Biotinylation solution (1 μg/mL, EZ-link Sulfo-NHS-Biotin, Pierce, Rockford, IL, USA): Prepare freshly solution before use in PBS.
Blot FastStain kit (Chemicon International, Temecula, CA, USA).
Tris-buffered saline (TBS, 10×): 1.5 M NaCl, 0.5 M Tris–HCl, pH 8. Store at RT.
Tris-buffered saline with Tween-20 (TBST): TBS plus 0.05 % (w/v) Tween-20. Store at RT.
Primary and secondary antibody dilution buffer: TBST supplemented with 0.5 % (w/v) fraction V bovine serum albumin (BSA).
Primary antibodies (see Note 5).
Streptavidin linked Cy5.
FITC conjugated anti-rabbit IgG or anti-mouse IgG.
2.4. Image Scanning and Quantitation
GE Typhoon 9410 Imager (GE Healthcare Life Sciences, Pittsburgh, PA, USA).
ImageQuant 5.2 software (GE Healthcare Life Sciences, Pittsburgh, PA, USA).
3. Methods
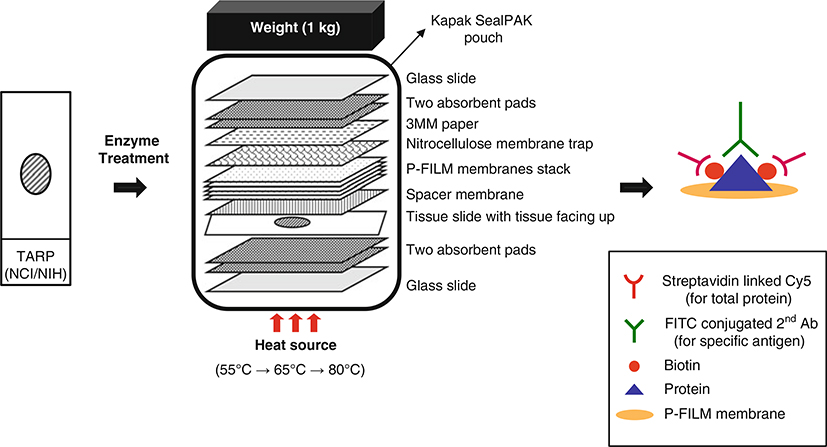
This method begins with routinely processed formalin-fixed, paraffin-embedded tissue section on a regular glass slide and utilizing routine laboratory procedures for microtomy. Initial retrieval of protein from the FFPE tissue section involves enzymatic digestion using trypsin and proteinase K. Subsequently, the tissue section is placed in an assembled heat-facilitated capillary transfer unit (Fig. 1). The unit is sealed in a pouch and applied multiple-serial heating system so that protein is transferred from the slide to a stack of membrane. Typically, we can obtain five replicate membranes from a 5 μM-thick formalin-fixed, paraffin-embedded tissue section. After transfer, the membranes stack is disassembled and each membrane can be probed using conventional Western blotting or immunoblotting method. This method allows quantification of the specific target signal based on total protein normalization, which is expressed as an intensity ratio between the protein of interest and total protein. In addition, post-transfer tissue on the original slide can be stained with routine Hematoxylin and Eosin (H and E) for direct correlation of histopathology with the immunoblot results. The residual tissue on the section after transfer should be limited, and may be difficult to interpret. If complex diagnostic features are to be examined, an adjacent H and E section should be utilized. Spatial resolution is determined by the scanning methodology, as well as transfer conditions and abundance of the protein. “Acinar” resolution on the order of 100–200 μm is obtainable with optimized conditions.
Fig. 1.
A schematic diagram of the multiplex tissue immunoblotting for tissue protein transfer. The tissue section is treated with enzymes and then assembled for heat-facilitated capillary transfer. Assemble the transfer unit carefully and then seal the assembled transfer unit using a Kapak SealPAK pouch. After the sealing, the transfer unit is moved to the heat block and then applied to a multiple-serial heating system (1 h for 55 °C, 0.5 h for 65 °C, and 2 h for 80 °C). The protein leaves the tissue slide and is deposited onto the membrane stack. A weight applied to the top of the transfer assembly unit helps to ensure a tight connection between the layers of material used in the transfer system. Total protein is detected by biotinylation and Cy5 fluorescence (red emission). Individual target proteins are detected with specific antibodies and FITC fluorescence (green emission)
3.1. Deparaffinization and Enzyme Treatment
Deparaffinize the formalin-fixed, paraffin-embedded tissue section in xylene (3 × 5 min) in glass or plastic coplin jars. Transfer to 100 % EtOH (2 × 5 min), 95 % EtOH (2 × 5 min), 70 % EtOH (1 × 5 min), and then to PBS (2 × 5 min) (see Note 6).
Equilibrate the deparaffinized tissue slide for 5 min with 50 mM ammonium bicarbonate buffer (pH 8.2).
Prepare the enzyme cocktail solution by mixing 100 μL of trypsin stock solution, 50 μL of Proteinase-K stock solution, and 20 mL of 50 mM ammonium bicarbonate buffer (pH 8.2).
Place the enzyme cocktail solution in an incubation chamber. After the equilibration is completed, incubate the slide for 30 min at 37 °C with enzyme cocktail solution (see Note 7).
Pour off the enzyme cocktail solution and rinse the surface of the tissue twice with PBS.
Place the slide on a flat surface and then apply immediately 2 mL of proBuffer to the slide for 15 min at RT.
Briefly wash the slide with transfer buffer. The slide is ready for transfer.
3.2. Transferring from the Tissue Slide to Membrane
After deparaffinization and enzyme treatment are completed, equilibrate the tissue slide in 1 mL of transfer buffer for 15 min.
During the equilibration of the tissue slide, prepare a spacer membrane, a stack of membranes (see 8), four absorbent pads, 3MM paper, and a nitrocellulose membrane trap (see Note 9). Equilibrate all membranes for 5 min in transfer buffer.
The pre-equilibrated spacer membrane is laid on the top of the tissue and subsequently the five-membrane stack is laid on the top of the spacer membrane. Gently roll over the sandwich using a disposable pipette to ensure that no air bubbles are present.
The pre-wetted nitrocellulose membrane trap is carefully laid on the top of the stack of the membrane, ensuring that no air bubbles are trapped in the resulting sandwich.
Place the pre-wetted 3MM paper and two absorbent pads on the top of the nitrocellulose membrane trap.
Two additional two absorbent pads are wetted in the transfer buffer and laid on the bottom of the tissue slide. Support the transfer assembly using two slide glasses as shown in Fig. 1.
Place the transfer assembly unit in a Kapak SealPAK pouch and then heat seal by an impulse sealer.
After the assembly of the transfer unit is completed, the transfer unit is transferred under serial conditions for 1 h at 55 °C, for 0.5 h at 65 °C, and for 2 h at 80 °C using heat block (see Note 10).
3.3. Immunoblotting
Once the transfer is completed, the SealPAK pouch is opened and the membrane stack carefully disassembled, with the top absorbent pads and 3MM paper removed.
Remove excess transfer buffer by washing the membranes (3 × 5 min) in PBS.
- Stain the nitrocellulose and the spacer membranes using the Blot FastStain kit (see Note 11).
- Prepare 1:7-fold diluted working solutions of reagent A and B with distilled water.
- Incubate the nitrocellulose and spacer membranes in 7 mL of reagent A for 10 min, followed by incubation of the both membranes in reagent B for 10 min or until spot visualized.
- Move the staining container to 4 °C and then let stand for 10–30 min.
The five-membrane stack is then incubated in 20 mL biotinylation solution for 10 min at RT on a rocking platform (see Note 12).
The biotinylation solution is discarded and then the membrane washed three times for 5 min each with TBST.
Incubate each membrane by adding 0.5 mL of an appropriately diluted primary antibody in the antibody dilution buffer for overnight (16–18 h) at 4 °C on a Kapak SealPAK pouch. Rocking platform is recommended (see Note 13).
The primary antibody is then removed and the membrane washed three times for 5 min each with TBST.
Prepare the mixture of secondary antibodies by mixing 5 μL of streptavidin linked Cy5 and 5 μL of FITC conjugated anti-rabbit IgG or anti-mouse IgG, and 5 mL of the antibody dilution buffer.
Add the mixture of secondary antibodies to the membrane and then incubate for 30 min at RT on a rocking platform. The membrane should be protected form light until scan is acquired.
The secondary antibodies are discarded and the membrane washed five times for 10 min each with PBST.
3.4. Image Scanning and Quantitation
Dry the membranes after the final wash between two sheets of 3MM paper.
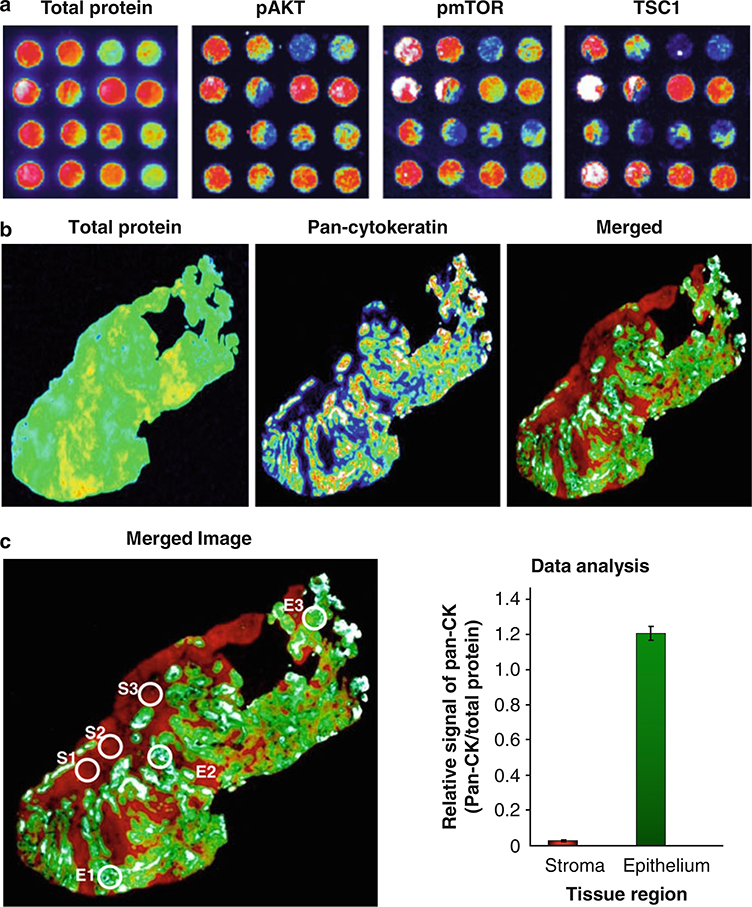
Scan each membrane at appropriate wavelength with GE Typhoon 9410 Imager. Examples of the signals for total protein and pan-cytokeratin are shown in Fig. 2a (see Note 14).
Analyze the scanned images using ImageQuant 5.2 software. Example of quantitation of scanned image is shown in Fig. 2b.
Fig. 2.
Representative images and quantitation of whole tissue section by multiplex tissue immunoblotting. (a) Prior to immunoblotting, lung cancer TMA tissue section (5 μm thickness) was transferred to the five-membrane stack by heat-facilitated capillary transfer system. The transferred proteins are detected on the membranes by conventional immunoblotting methodologies. (b) A formalin-fixed and paraffin-embedded tissue section of a gastrointestinal tumor (5 μm thickness) was transferred to the five-membrane stack by heat-facilitated capillary transfer system. The third membrane was incubated in biotinylation solution and then was reacted with anti-pan-cytokeratin antibodies (1:200-fold diluted). After primary antibody incubation is completed, total protein and specific target signals were detected using streptavidin linked Cy5 and FITC conjugated anti-mouse IgG (1:1,000-fold diluted). Membranes were imaged with a microarray scanner. Fluorescent scans are represented in pseudo-color, where signal intensity is white-red-yellow-green-blue-black from maximum to minimum signal. (c) We selected three different representative stroma and epithelium tissue regions based on the H and E slide. We subsequently quantified those areas using ImageQuant 5.2 software. After normalization with total protein level, relative expressional signals were represented as a ratio. The bar graph shows the average ± SD of three circle areas (S stroma, E epithelium)
Acknowledgment
The authors wish to thank Michael R. Emmert-Buck for his advice and insights into applications for this platform.
Footnotes
We have found that Dako Proteinase-K is best for this method. Substitution with other Proteinase-K can diminish reproducibility of results.
This protocol can be adapted for ethanol-fixed, paraffin-embedded tissue sections. In that case, the enzyme solution should be changed to 0.001 % trypsin only for 15 min at 37 °C. Overall the final condition of enzyme digestion should be optimized depending on tissue type with a minor change of enzyme concentration and time.
When handling membranes, always wear gloves to prevent contamination. The P-Film membrane is very thin and flexible and requires care in handling to avoid bubbles and folds.
The spacer is an uncoated polycarbonate PVPF membrane (pore size: 0.4 μm) and is used for filtrations of inappropriately digested proteins and debris during heat-facilitated capillary transfer procedure.
The primary antibodies which are suggested for western blotting as well as immunohistochemistry are compatible in this platform. We recommend 1:200-fold starting antibody dilution in this protocol. The cytoplasmic markers are in general better targets than nuclear and membrane-bound molecules; however, we have detected proteins in all these locations. For membrane-bound targets, thicker tissue sections (10 μm) may produce better results.
We found aqueous based dewaxers such as AutoDewaxer (Openbiosystems, Huntsville, AL, USA) could be used as a deparaffinizing agent in place of xylene at high temperature with equal results and greater safety (see ref. 16). The temperature of xylene should be controlled under 65 °C. Inappropriate deparaffinization can result in poor protein transfer to the membrane.
The dynamic range of the enzyme condition is relatively narrow. For this reason we recommend avoiding repeated freezing and thawing of the enzyme stock solution. The stock solution can be stored up to 6 months in the freezer (−20 °C).
There are two different features (glossy vs. non-glossy sides) on the membrane. Before pre-soaking the membrane, we marked membrane and case numbers in the margin of nonglossy side for further information such as number of membrane and case using a regular ballpoint pen. The glossy side of the membrane stack should face the tissue on the slide (Fig. 1).
This protocol is used for a regular tissue slide, and can be adapted for irregular slide size with appropriate membrane size. Cut all membrane and pads with size of approximately 2.2 × 4.5 cm. The cover of cover slip box can be used a container for incubation chamber, to prevent excess buffer use.
The use of a multi-serial temperature condition produce an even distribution of proteins across membranes compared to a single temperature, which resulted in uneven transfer or bubble spots. This procedure generated a linear decrease in protein concentration through the membrane stack, with a great correlation coefficient (R2 = 0.985) (see ref. 11).
This step is just a confirmation stage for protein transferred to the membrane stack. The staining kit is very sensitive but is a transient staining. If you want to keep original image you should scan the semi-wet membrane between two transparent films and e save the image.
Do not use a plastic petri dish coated for cell culture. A 20 mL of biotinylation solution can be covered up to 20 membranes (2.2 × 4.5 cm).
It is not necessary to block the membranes. The carrier protein (BSA) in the antibody dilution is sufficient to inhibit nonspecific binding. Do not use dry milk as a carrier protein.
Excitation at 633 nm induces the Cy5 fluorescence (red emission) for total protein, while excitation at 488 nm induces FITC fluorescence (blue emission). This fluorescence labeling system can be adapted for user purpose.
References
- 1.Chuaqui RF, Bonner RF, Best CJ, Gillespie JW, Flaig MJ, Hewitt SM et al. (2002) Post-analysis follow-up and validation of microarray experiments. Nat Genet 32:509–514 [DOI] [PubMed] [Google Scholar]
- 2.Hewitt SM, Dear J, Star RA (2004) Discovery of protein biomarkers for renal diseases. J Am Soc Nephrol 15:1677–1689 [DOI] [PubMed] [Google Scholar]
- 3.Simone NL, Paweletz CP, Charboneau L, Petricoin EF, Liotta LA (2000) Laser capture microdissection: beyond functional genomics to proteomics. Mol Diagn 5:301–307 [DOI] [PubMed] [Google Scholar]
- 4.Ornstein DK, Englert C, Gillespie JW, Paweletz CP, Linehan WM, Emmert-Buck MR et al. (2000) Characterization of intracellular prostate-specific antigen from laser capture microdissected benign and malignant prostatic epithelium. Clin Cancer Res 6:353–356 [PubMed] [Google Scholar]
- 5.Craven RA, Totty N, Harnden P, Selby PJ, Banks RE (2002) Laser capture microdissection and two-dimensional polyacrylamide gel electrophoresis: evaluation of tissue preparation and sample limitations. Am J Pathol 160: 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camp RL, Chung GG, Rimm DL (2002) Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med 11:1323–1327 [DOI] [PubMed] [Google Scholar]
- 7.Liotta LA, Petricoin E (2000) Molecular profiling of human cancer. Nat Rev Genet 1: 48–56 [DOI] [PubMed] [Google Scholar]
- 8.Miyaji T, Hewitt SM, Liotta LA, Star RA (2002) Frozen protein arrays: a new method for arraying and detecting recombinant native tissue proteins. Proteomics 2:1489–1493 [DOI] [PubMed] [Google Scholar]
- 9.Lopez MF (2000) Better approaches to finding the needle in a haystack: optimizing proteome analysis through automation. Electrophoresis 21:1082–1093 [DOI] [PubMed] [Google Scholar]
- 10.Molloy MP, Phadke ND, Maddock JR, Andrews PC (2001) Two-dimensional electrophoresis and peptide mass fingerprinting of bacterial outer membrane proteins. Electrophoresis 22:1686–1696 [DOI] [PubMed] [Google Scholar]
- 11.Chung JY, Braunschweig T, Baibakov G, Galperin M, Ramesh A, Skacel M et al. (2006) Transfer and multiplex immunoblotting of a paraffin embedded tissue. Proteomics 6:767–774 [DOI] [PubMed] [Google Scholar]
- 12.Chung JY, Braunschweig T, Hu N, Roth M, Traicoff JL, Wang QH et al. (2006) Profiling of biomarkers in the normal to tumor transition zone of esophageal squamous cell carcinoma by multiplex tissue immunoblotting. Cancer Epidemiol Biomarkers Prev 15:1403–1408 [DOI] [PubMed] [Google Scholar]
- 13.Chung JY, Braunschweig T, Tuttle K, Hewitt SM (2007) Tissue microarrays as a platform for proteomic investigation. J Mol Histol 38: 123–128 [DOI] [PubMed] [Google Scholar]
- 14.Traicoff JL, Chung JY, Braunschweig T, Mazo I, Shu Y, Ramesh A et al. (2007) Expression of EIF3-p48/INT6, TID1 and Patched in cancer, a profiling of multiple tumor types and correlation of expression. J Biomed Sci 14:395–405 [DOI] [PubMed] [Google Scholar]
- 15.Chung JY, Hong SM, Choi BY, Cho HJ, Yu ES, Hewitt SM (2009) The expression of phospho-AKT, phospho-mTOR, and PTEN in extrahepatic cholangiocarcinoma. Clin Cancer Res 15:660–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung JY, Braunschweig T, Hewitt SM (2006) Optimization of recovery of RNA from formalin-fixed, paraffin-embedded tissue. Diagn Mol Pathol 15:229–236 [DOI] [PubMed] [Google Scholar]