Abstract
Correlative light and electron microscopy harnesses the best from each of the two modalities of microscopy it utilizes; while light microscopy provides information about the dynamic properties of the cellular structure or fluorescently labeled protein, electron microscopy provides ultrastructural information in an unsurpassed resolution. However, tracing a particular cell and its rare and small structures such as centrosomes throughout numerous steps of the experiment is not a trivial task. In this chapter, we present the experimental workflow for combining live-cell fluorescence microscopy analysis with classical transmission electron microscopy, adapted for the studies of the centrosomes and basal bodies. We describe, in a step-by-step manner, an approach that can be affordably and successfully employed in any typical cell biology laboratory. The article details all key phases of the analysis starting from cell culture, live-cell microscopy, and sample fixation, through the steps of sample preparation for electron microscopy, to the identification of the target cell on the electron microscope.
INTRODUCTION
Centrosome structure is precisely defined and conserved among various eukaryotic organisms (Carvalho-Santos, Azimzadeh, Pereira-Leal, & Bettencourt-Dias, 2011). The centrosome consists of an unduplicated or a duplicated centriole (Alvey, 1985; Rattner & Phillips, 1973; Vorobjev & Chentsov, 1982), which organizes a proteinaceous pericentriolar material (PCM) in a highly hierarchical and ordered fashion (Lawo, Hasegan, Gupta, & Pelletier, 2012; Mennella, Agard, Huang, & Pelletier, 2014; Mennella et al., 2012; Sonnen, Schermelleh, Leonhardt, & Nigg, 2012). Centrosome function and its ultrastructural features are intimately linked, so the findings discerned through biochemistry and light microscopy should be correlated with analysis at the ultrastructural level whenever possible. Centrioles are ninefold symmetrical microtubule-based structures, easily detectable by electron microscopy (EM). The PCM component of the centrosome is, on the other hand, not electron dense. The PCM components can be visualized by fluorescence microscopy, and their structural organization examined using recently developed super-resolution microscopy techniques (Leung & Chou, 2011; Yamanaka, Smith, & Fujita, 2014) predicts the localization of the florescence signals beyond the resolution limit of classical light microscopy (Keller et al., 2014; Lau, Lee, Sahl, Stearns, & Moerner, 2012; Mennella et al., 2012; Sillibourne et al., 2011). A centriole’s ultrastructural features are less reliably predicted by light microscopy due to their small size (centrioles are, depending on the species, ~120–200 nm in diameter and ~200–500 nm in length).
Correlative light and electron microscopy (CLEM) is an imaging approach that combines various modalities of light microscopy andEMin one experiment (Rieder & Bowser, 1985; Rizzo, Parashuraman, & Luini, 2014; Spiegelhalter, Laporte, & Schwab, 2014). It has been used to analyze centrosome-associated processes during cell cycle progression or after various genetically or chemically induced treatments (Kong et al., 2014; Lončarek, Hergert, & Khodjakov, 2010; Loncarek, Hergert, Magidson, & Khodjakov, 2008; Rieder & Bowser, 1985; Tsou et al., 2009). A combination of live- or fixed-cell light microscopy and EM allows the investigator to capitalize on the strengths of both individual techniques. However, it also brings a new layer of complexity to the experiment, as it requires an expertise in both modalities of microscopy.
One of the challenges of CLEM is to trace down the target cell previously analyzed by light microscopy through multiple steps of sample preparation, down to the imaging on the electron microscope. Finding the right cell among hundreds of surrounding cells might seem impossible, but various strategies can be employed to accomplish this task. In this chapter, we describe the strategy utilized in our laboratory for routine study of centrosomes by CLEM. This strategy allows us to first analyze centrosomes by light microscopy, and to reproducibly follow the same cell through embedding, trimming, and sectioning, and to image the same centrosomes on the electron microscope. Due to the complexity of the technique, many researchers interested in centrosome biology may hesitate to employ CLEM and therefore not benefit from the insight provided by ultrastructural analysis. We hope that detailing our approach to CLEM will inspire some to introduce this highly rewarding technique into their daily experimental practice. The strategy we describe can also be employed for the analysis of other cellular organelles with minimal adaptations to the described protocol.
1. EXPERIMENTAL PROCEDURE
1.1. CELL CULTURE AND PREPARATION OF THE CELLS FOR MICROSCOPY
1.1.1. Cell culture
Cells are plated on sterile 25-mm round, 0.17-mm thick coverslips previously washed in deionized H2O, followed by 70% ethanol, then absolute ethanol, and individually dried in a sterile cell culture hood. Most cells will adhere and proliferate on these clean glass coverslips; however, some cell types will require coverslips to be coated with poly-l-lysine for better adherence.
To visualize centrioles by fluorescent light microscopy, we express Centrin1-green fluorescent protein (GFP) (or another fluorescent-tagged centrosomal protein) and select for the population of cells with optimal signal-to-noise ratio for imaging (see Figure 5) using cell sorting.
For easier identification of the target cell after embedding, the cells should be less than 60% confluent at the time of analysis. This allows us to use the shape of neighboring cells as identification landmarks during trimming and sectioning. In confluent cultures, identification of the target cell throughout the experiment is possible but the incidence for misidentification increases.
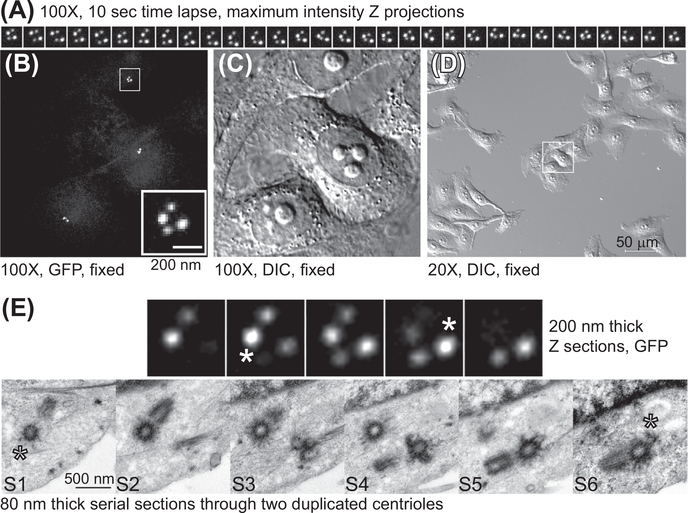
FIGURE 5. CLEM analysis of the centrioles from Centrin1-GFP expressing HeLa cell arrested in G2.
(A) A series of maximal density projections illustrating the movement of the centriole pairs. A Z stack spanning the entire centrosome content was collected using spinning disc confocal and 100× objective lens, every 10 s (B and C) The same cells imaged by time-lapse microscopy were fixed with 2.5% glutaraldehyde and returned to the microscope. The position of the centrioles within the cell was then recorded in fluorescence and DIC. (D) Low-magnification image of the target cell and its neighbor cells in DIC. (E) The centrioles recorded by time-lapse imaged on the electron microscope. Four consecutive immunofluorescence Z sections and six serial EM sections are presented (S1–S6). The asters in S1 and S6 correspond to the immunofluorescence Z section 2 and 4, with maximal intensity for centrin-GFP signal belonging to the mother centrioles (indicating the position of the distal part of the centriole).
1.1.2. Preparation of the cells for microscopy
To image the cells we use homemade reusable Rose chambers (see Figure 1) which are a proven, affordable alternative to glass bottom Petri dishes (Pereira, Matos, Lince-Faria, & Maiato, 2009; Stout, Rizk, & Walczak, 2009). Rose chambers are ideally suited for long-term live-cell imaging at high resolution. Most cell types will grow in a Rose chamber without a change in viability for days. In addition, their square shape allows for an easy orientation and quick repositioning of the sample on the microscope during fixation, marking the target cells, and switching between the high- and low-magnification objectives (as described in the next chapter). Before imaging, assemble a Rose chamber in a sterile hood as follows:
Put an empty coverslip on the flat surface of the lower metal plate.
Place a sterile silicone gasket on the top followed by a coverslip with the cells facing down. For sterilization, wash the gasket in 70% ethanol and dry in a sterile hood, or sterilize the spacers by autoclaving.
Put the upper metal plate on the rubber gasket. Be sure to assemble the chamber fast enough to prevent the cells from drying.
Press lightly on the assembly to hold it in place while sealing the chamber with four bolts.
Perfuse the chamber with complete, warm, CO2-independent medium (which will maintain pH during imaging) using a 21–22 gauge needle and a 3 mL syringe. Punch through the rubber gasket, and slowly inject the medium. Be sure to insert another needle to serve as a vent on the opposite side of the rubber gasket before injecting the medium into the chamber.
Remove all traces of media and cell debris from the coverslip. The Rose chamber is now ready for imaging.
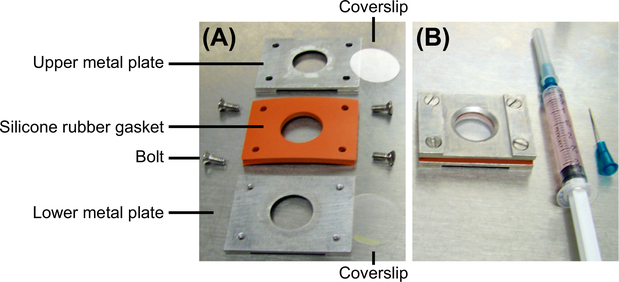
FIGURE 1. Rose chamber for live-cell imaging.
(A) A disassembled Rose chamber to illustrate its components. A Rose chamber contains two metal plates, one 1–3 mm thick silicon rubber gasket, and two coverslips (one that carries the cells and another one that is empty). All components are held together by four bolts. (B) An assembled Rose chamber and a syringe filled with medium. The side of the Rose chamber facing up will be facing the objective during imaging.
Many variations of the original Rose chamber have been developed since its first description in the 1950s (Rose, Pomerat, Shindler, & Trunnell, 1958). We designed a modified version of the chamber to account for our specific microscope setting and objective features. In designing a Rose chamber, be sure that the silicone rubber used for the spacer is made of high quality, FDA approved, nontoxic material as it will be in direct contact with the medium. An alternative to using a Rose chamber is to culture cells in a culture dish with a glass bottom (also available with a gridded bottom, e.g., from MatTek).
1.1.3. Light microscopy
For live-cell microscopy use a research-grade inverted microscope equipped with 20×, 60×, and 100× objectives, a spinning disc confocal, a sensitive CCD camera, bright field illumination, and an environmental enclosure set to 37 °C. To resolve individual centrin-GFP labeled centrioles, cells must be imaged using 60× or 100× objective with a high numerical aperture (NA = 1.4 or higher). The exposure to illuminating fluorescence light should be minimized during imaging by using automated shutters to avoid photo damage. Photo damage may induce a nonphysiological response of the cells during imaging. This is especially important if the cells are imaged for a long period of time. The cell cycle and the centriole cycle can both be perturbed or even completely stalled in cell cultures stressed by excitation light.
1.2. CELL FIXATION AND POSTFIXATION RECORDING OF CELL/CENTRIOLE POSITION
Centrosomes occupy only a small fraction of a cell’s volume and can be easily missed during EM analysis. In addition, centrioles in live cells continuously change their position and orientation with respect to the coverslip. The average thickness of a section for transmission EM analysis is 60–80 nm, meaning the centriole(s) belonging to one centrosome will, depending on their orientation, be present in no more than three to six serial sections (out of ~100 needed to span entire volume of a typical adherent interphase cell). Thus, it is important to record the position of the centrosome/centriole with respect to the coverslip and other cellular landmarks as accurately as possible on the light microscope after fixation. This is crucial for overall success of the experiment.
After and analysis of the target cell by light microscopy, fix the cells by perfusing the Rose chamber with freshly prepared and prewarmed fixative and return the chamber to the microscope in the same position and orientation as prior to fixation.
Within 1–2 min of fixative addition, intracellular movements will cease. A soon as possible after fixation, record a Z-stack (200-nm z step) spanning the entire cell to register the position and orientation of the centrioles.
In parallel, record a Z-stack in differential interference contrast (DIC) (or phase) at the same magnification to facilitate subsequent localization of the centrioles within the cell using morphological clues of the cell (the shape, the position of the nucleus, etc.) while searching on the electron microscope.
The position of the target cell should then be permanently marked on the coverslip. For this purpose, we use an objective diamond scriber placed on the microscope objective turret. If you have such a scriber, scratch a circular mark on the glass surface around the cell of interest (see Figure 2(A)). Alternatively, the position of the target cell can be marked by an objective slide marker (available from Nikon), and subsequently reinforced using a glass writing diamond pen (many versions are available on the market). The bottom of the glass coverslip can also be prescratched using a diamond pen before plating the cells. In addition, cells can be cultured on “grid” glass bottom dishes (MatTek) or on gridded coverslips (which is a more expensive alternative).
Clean immersion oil from the coverslip and use a low-magnification objective (for instance 10× or 20×) to record DIC images of the targeted cell and its surroundings (see Figure 5). Neighboring cells will be helpful landmarks when identifying the cell of interest during further steps, such as trimming of the embedded sample. We recommend taking several images of the area surrounding the target cell.
When the position of the target cell is clearly marked on the surface of the coverslip, disassemble the Rose chamber and place the coverslip (cell side facing up) into 5 mL of the fixative in a 60-mm Petri dish, seal the dish with parafilm, and store it at 4 °C until embedding.
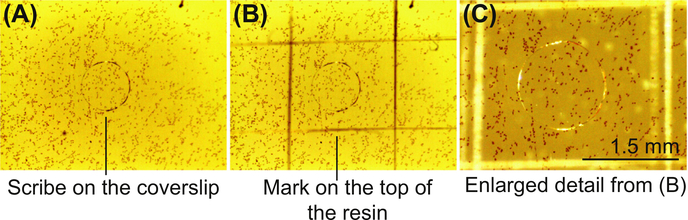
FIGURE 2. Marking the position of the target cell on polymerized resin.
(A) The scribe on the coverslip encircling the target cell is easily visible after embedding. (B) The position of the target cell is marked on the resin with a razor blade. (C) Enlarged detail from (B).
1.3. PRESTAINING, DEHYDRATION, AND EMBEDDING
After fixation and marking the position of the target cell on the light microscope, prestaining, dehydration, and embedding, with many possible variations in the protocol, can be performed in any EM laboratory by a technician skilled in the preparation of EM samples. We will delineate here the protocol used in our laboratory.
Transfer the coverslips with the fixed cells into a 35-mm Petri dish and carry out all the following steps in Petri dish at room temperature. Wash the cells with 1X phosphate buffered saline (PBS) (pH 7.2) three times, 10 min/each wash, to remove the fixative.
Incubate the cells in 0.15% Tannic acid in 1X-PBS for 1 min. Tannic acid allows for better contrasting of microtubule-based structures such as centrioles. Wash the cells with 1X-PBS for 5 min.
Postfix the cells in 2% Osmium tetroxide (OsO4) diluted in distilled water (hereafter dH2O) for 1 h at 4 °C in a Petri dish sealed with parafilm. Wash the cells in dH2O three times, 5 min/each wash.
Prestain the cells with 1% uranyl acetate diluted in dH2O for 1 h at 4 °C in a dish sealed with parafilm. Wash the cells in dH2O two or three times, 5 min/each wash.
Dehydrate the sample by sequential exchange of 20%, 30%, 40%, 60%, 80%, and 95% ethanol solutions, each exchange being 5 min. Finally, dehydrate the sample in three exchanges of 100% ethanol, 10 min each. During this dehydration procedure, be sure that the cells are not allowed to dry out between ethanol exchanges.
Incubate the cells in a mixture of 100% ethanol and Embed 812 resin at 1:1 ratio overnight. In the morning remove the old resin and replace with pure Embed 812 resin, and leave for 1 h.
Tilt the Petri dish for a few seconds to let the resin flow down, and remove it using a pipette. Keep the Petri dish tilted, apply fresh pure Embed 812 resin to the cells from the top of the coverslip and leave for 1 h. Repeat this procedure two times to completely remove all traces of ethanol.
Remove the resin from the Petri dish as described in Step 7. Take the coverslip out of the Petri dish and clean residual resin from the bottom of the coverslip with a Kimwipe tissue. Place the coverslip (with the cells up) on some sort of the holder resistant to high temperature. Apply ~1 mL of fresh Embed 812 resin to the center of the coverslip, and the resin will slowly spread out to cover the whole coverslip. Put the coverslip with resin in an oven at 60 °C for 48 h to polymerize. Be sure that the holder with the coverslips is in horizontal orientation to avoid leaking of the resin from the coverslip. As a holder we use a pipette tips box, with some tips left in the box to support the coverslip from below.
1.4. MARKING THE POSITION OF THE TARGET CELL ON THE POLYMERIZED RESIN
After embedding, the cells will be localized within a thin layer of polymerized resin adjacent to the glass coverslip. The mark on the glass, indicating the position of the target cell, must be transferred to the top of the resin before the glass coverslip is removed. After embedding, the bottom of the embedded sample is usually covered with a thin layer of polymerized resin which makes the mark on the coverslip invisible. To observe the mark, scrape the resin off the glass using a razor blade. Place the sample on the dissecting microscope and transfer the mark from the glass to the top of the resin with a fine needle, diamond pen, or razor blade, as illustrated in Figure 2.
1.5. REMOVAL (DISSOLVING) OF THE GLASS COVERSLIP
This step potentially exposes a researcher to the harmful effects of hydrofluoric acid. Therefore, all steps described in this section should be performed in a vented chemical hood in agreement with required safety procedures.
Place embedded sample in a small Petri dish with the glass coverslip facing upwards. Add enough concentrated hydrofluoric acid to overlay the coverslip. Incubate for 1 h at room temperature. Hydrofluoric acid will gradually dissolve the glass coverslip.
Check if the glass is completely dissolved; if not, continue incubation until it is.
Using tweezers transfer the sample into a large beaker containing glass beads at the bottom and wash the sample under a gentle stream of running water for 1–2h. The resin will appear turbid. Let the resin thoroughly dry until it becomes clear again. Alternatively, fill up the beaker with water, and exchange water in 20 min intervals.
If the cells were cultured on a plastic surface, separate the plastic from the resin using two pliers. Note that this step may damage the tissue close to the Petri dish. Alternatively, immerse the sample into liquid nitrogen to break the plastic.
1.6. TRIMMING, ULTRATHIN SERIAL SECTIONING, AND THE PICKUP OF THE SERIAL SECTIONS
Until this point, all described steps of the sample preparation can be accomplished in a typical laboratory setting after appropriate training. However, trimming, ultrathin serial sectioning, and the pickup of the serial sections is the most delicate part of CLEM, and it is not expected to be routinely preformed in the laboratory. Extensive experience and great skill are required to perform this part of experiment. The assistance of a specialist will usually be required.
1.6.1. Trimming
Roughly cut out the region of the resin where your cell of interest is located using pliers and glue it onto the flat bottom of a premade resin block (see Figure 3(A)). Before trimming, make sure that the sample tightly attaches to the resin block. Note that after removal of the glass or the plastic, the thin layer of the resin that contains the cells is unprotected and vulnerable to damage. It is important to keep this surface of the resin unscratched during cutting and trimming, as the scratches may damage the cells or make it impossible to identify the target cell later on.
Place the resin block into the sample holder of an ultramicrotome and secure it tightly. Place the holder into the trimming adaptor that is locked on the ultramicrotome stage.
Use previously recorded low-magnification DIC images of the target cell and the neighboring cells to identify the cell of interest under the ultramicrotome stereomicroscope. Trim the sample with a sharp razor blade to the shape of a trapezoidal prism. The trapezoid should have one side slightly slanted, and another side straight (see Figure 3(B)).
Center the prism on the target cell, and then carefully continue fine trimming until you form a pyramid with the small trapezoid on the top. The final top surface of the trapezoid should be about 0.5 mm2 containing only a few neighboring cells, with the target cell in the center (see Figure 3(C)). To get a straight ribbon of serial sections during future sectioning, it is important that the bases of the trapezoid are smooth and parallel to each other. To obtain smooth sides of the pyramid during final trimming steps, it is the best to use only the sharpest and previously unused razor blades.
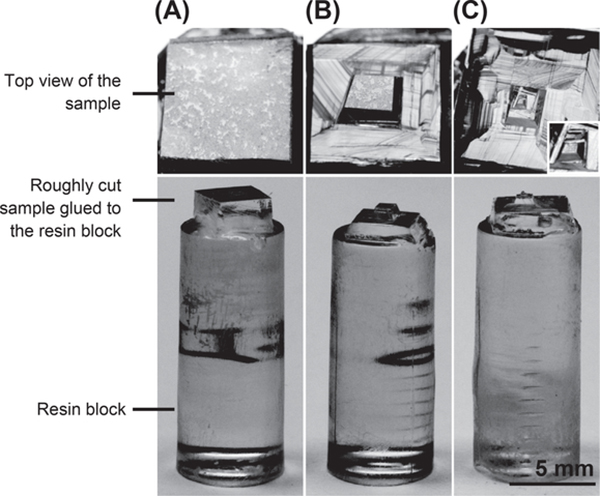
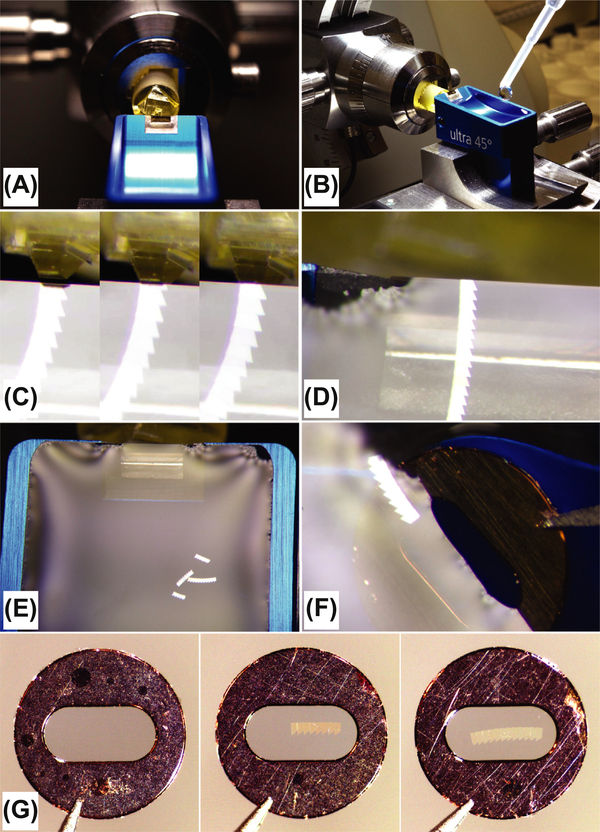
FIGURE 3. Trimming.
(A) Roughly cut the part of the resin containing the cell of interest. A layer of cells is visible under the ultramicrotome stereomicroscope (top view). (B) Shaping of a trapezoidal prism with the target cell in the center of the trapezoid. (C) Final size of the trapezoidal prism ready for sectioning. The inset is showing enlarged prism (top view).
1.6.2. Ultrathin serial sectioning
This section describes the procedure for sectioning using Leica EM UC7 ultramicrotome (Leica Microsystems). This procedure might have to be adapted for the specific features of other instruments.
After fine trimming, place the sample holder into the specimen arm and clamp it with the knurled screw. Set the diamond knife into the knife holder with its clearance angle to 6°, as recommended, and lock the knife in this setting (see Figure 4(A)).
Turn off all the overhead lights to reduce distraction and only leave the bottom light on. Go to the highest magnification of the stereomicroscope and slowly advance the diamond knife to the trapezoid, bringing them as close together as possible (within 1 mm) until you can see a bright narrow gap between them. Align the bottom base of the trapezoid to the diamond knife edge to make them parallel to each other.
Adjust the orientation of the trapezoid to make the gap between its surface and the diamond knife edge identical for both trapezoid bases (until the width of the bright gap does not change during the up and down movement of the trapezoid). This alignment is important to get the section plane parallel to the sample plane.
Retract the diamond knife 1–2 mm from the trapezoid. Take great care not to damage the surface of the sample during the whole procedure.
Turn on all overhead lights and set the cutting window with buttons “START” and “END” on the touch screen control panel following the manufacturer’s instructions.
Fill the boat of the knife with dH2O (see Figure 4(B)). Water should be leveled with the diamond knife edge until you can see a silver reflection of water surface on the surface of the water. Set the cutting speed to 1 mm/s, the section thickness to 70–80 nm, and start sectioning. One long ribbon of silver-to-light yellow sections should appear floating on the surface of the water (see Figure 4(C) and (D), and Video 1). Usually, one can see the cells within the sections through binoculars.
Stop the cutting after the ribbon is 30–40 sections long and gently pull the ribbon toward the water boat to make it float on the surface of the water. Retract the diamond knife ~1 mm away before you are ready to pick up the sections. Separate the long ribbon into shorter segments (7–10 sections) using an eyelash glued to the end of a wooden toothpick (see Figure 4(E)). The eyelash tip should be wet, as a dry eyelash tends to stick to the sections. The eyelash should be precleaned in acetone.
Do not cut too many sections at a time, as this can make it harder to distinguish the correct order of those separated shorter ribbons. If the ribbon breaks into too many segments during sectioning, it could mean that the bases of the trapezoid were poorly made, or that the alignment between the trapezoid and the knife was not correct. Try your best to track the correct order of those segments.
FIGURE 4. Serial sectioning and picking up the sections.
(A) The sample in a position ready for sectioning. (B) A knife boat is filled with water before sectioning. (C) Stills from Video 1 illustrating the movement of the trapezoidal prism during sectioning and the formation of the ribbon of sections. (D) The ribbon floating on the water with the last section still attached to the edge of the knife. (E) The long ribbon is separated in four shorter ribbons before the pickup. The ribbons are free-floating in a knife boat filled with water. (F) The coated grid partially submerged in the water in preparation for the ribbon pickup. (G) Examples of three formvar coated grids; the left one is empty, and the middle and the right one carry a ribbon of serial sections.
1.6.3. Picking up the serial sections
The ribbons of serial sections are picked up on the formvar-coated slot grid which must be prepared at least 24 h in advance prior to sectioning (how to prepare coated slot grids is described in the following section). Great care must be taken not to cause wrinkles in the sections during the picking up procedure. To avoid wrinkles, a ribbon of serial sections should be picked from underneath the water as follows:
Hold the formvar-coated grid at the four o’clock position and inspect it under the binoculars. Use only the best coated grids with no cracks, wrinkles, or any other irregularity on the formvar.
Paddle the water around the ribbon to bring the ribbon to an isolated area in the water boat. Submerge approximately three-fourth of the grid in the water, keeping the top area of the film dry (see Figure 4(F)). Tilt the grid a bit toward the ribbon, slowly advancing it to the ribbon, then waft the ribbon closer to the grid by gently moving the grid back and forth, or by paddling the water around the ribbon toward the grid using the eyelash.
Once one edge of the ribbon attaches to the top, dry area of the film, slowly pull the grid out of the water. The entire ribbon will attach to the formvar film (see Figure 4(G)). Touch the lower edge of the grid with a small piece of filter paper to absorb water, air dry the grid for a few seconds, and put it into a well of the grid storage box. Pick up all ribbons in this way and keep them in the grid storage box in a systematic order.
Continue sectioning and picking up the ribbons.
1.6.4. Staining of the sections
Before they are used for imaging on the transmission electron microscope, the sections have to be stained with uranyl acetate and lead citrate. We stain the grids using a grid staining matrix system, which allows for fast and uniform staining of multiple grids.
Put the grids into the wells of the matrix body in a sequential order during this procedure (for the analysis of serial sections, it is very important to preserve the order of the grids).
Stain the grids in 2% uranyl acetate diluted in dH2O for 20–30 min at room temperature, followed by three washes in dH2O, each for 1–2 min. Dry the grids, or immediately stain them in lead citrate solution for 1 min, followed by three 1–2 min washes in dH2O that has been previously boiled and cooled down to room temperature.
Return the grids to a grid storage box and air dry the grids at room temperature for 24 h (do not close the box). Samples are now ready to be analyzed on the transmission electron microscope.
1.6.5. Electron microscopy
To facilitate the identification of the targeted cell by electron microscope, use both low- and high-magnification recordings of the target cell previously obtained by the light microscope. Use the three-dimensional Z-stacks to assess the position of the centrioles in XY and Z planes. Note that serial sections, if collected by following the above procedure, will be flipped with respect to the images collected on the light microscope. Thus, we recommend printing the flipped versions of the original images in preparation for the search on the electron microscope. Using the recordings in both low and high magnification, and knowing at which region and cell depth the centrioles were at the time of fixation, one can easily find the target cell and the centrioles within. As long as serial sectioning was preformed parallel to the confocal plain, it will be relatively simple to translate the Z position of the centrioles into the sectioning depth.
Start imaging from the first to the last section of the first grid. Once the centrioles are identified, acquire images of them at low magnification (2.5–5K) and then at higher magnification (10–12K). Lower magnifications will assist during the alignment of the images from consecutive serial sections, as images of each section will be slightly rotated and shifted with respect to the previous one. Images in lower magnification contain more cellular landmarks which are often used to assist the alignment of the images obtained on higher magnification.
After images of the centrioles are captured from serial sections, the centrioles can be aligned (see Figure 5) using Photoshop, if no specialized image aligning software is available.
1.7. PREPARATION OF FORMVAR-COATED SLOT GRIDS
Slot grids need to be coated with a thin layer of formvar supporting film before they are used to pick up serial sections. The procedure for preparing the coated grids is illustrated in Figure 6.
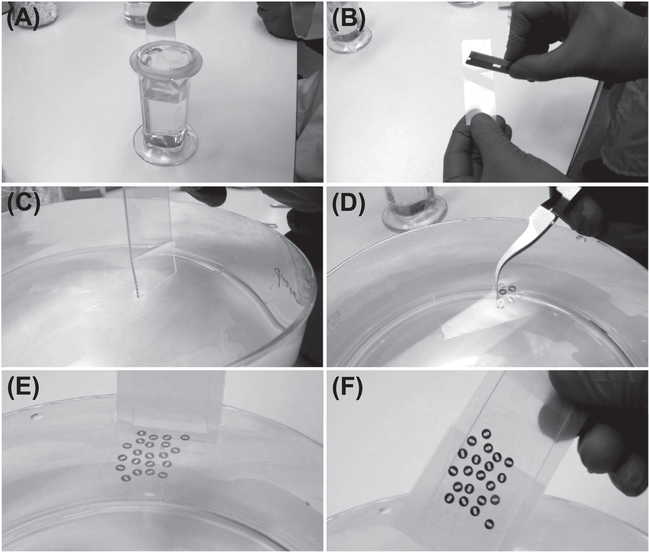
FIGURE 6. Preparation of the formvar-coated slot grids.
Selected frames from Videos 2–5 illustrating the key steps in the coating of the slot grids with formvar. (A) Dipping microscope slide in 0.5% formvar solution. (B) Scraping formvar film off the edges of the slide. (C) Detaching formvar film from the slide. (D) Putting the grids on the top of the floating formvar film. (E and F) Collecting coated grids from the water.
Preclean the slot grids by putting them in a clean glass beaker filled with 10–20 mL of pure ethanol or acetone. Soak the grids for 15 min at room temperature, swirling several times. Pure out the ethanol or acetone and dry the grids well before use.
Clean both sides of a microscopy glass slide with ethanol, air dry, and dip into 0.5% formvar solution. Pull the slide steadily out of the solution, and air dry the slide for several minutes. The thin film of formvar will form on the glass slide (see Video 2).
Fill a big dish with dH2O. To detach formvar from the glass slide, scrape off the formvar film from the edges of the slide with a razor blade (see Video 3).
Slowly dip the glass slide in the water. The formvar film will detach from the slide and stay afloat on the water. Gently pull the slide out of the water (see Video 4).
Place precleaned slot grids one by one on the top of the floating film with their shiny sides facing toward the film. The film will permanently stick to the cooper grid.
To collect coated grids from the water, wrap another microscope slide with parafilm. Submerge one edge of the slide under the water, and establish the contact with the film carrying grids. Slowly push the slide deeper into the water until the film with the grids is attached to the parafilm. Then pull the slide with the grids attached to it from the water (see Video 5).
Dry coated grids in a clean glass dish at room temperature. Protect the grids from dust. Note that the film and the shiny side of the slot grids are now facing up.
1.8. CHEMICALS, BUFFERS, AND MEDIA
CO2-independent medium (18045–088; Life Technologies).
Formvar (15800; Electron Microscopy Sciences (EMS)): 0.5% (m/v) solution in chloroform; can be reused many times.
Fixative: 2.5% Glutaraldehyde in 1X-PBS pH 7.2, or 2.5% glutaraldehyde in 0.1Mcacodylate buffer, pH 7.4. The fixative should be prepared fresh each time.
Glutaraldehyde (G5882; Sigma-Aldrich); once opened, a bottle of glutaraldehyde should be stored at 4 °C and used within 2 months.
Hydrofluoric acid, 48% (339261; Sigma-Aldrich)
Lead citrate (178000; EMS): put 0.01 g (or 0.04 g) lead citrate powder into 9 mL dH2O (boiled and cooled down to room temperature), add 1 mL of freshly made 1 N sodium hydroxide (21160; EMS), and mix for several minutes until lead citrate is completely dissolved. Filter the solution with a 0.22 μm filter before use.
Osmium tetroxide (19100; EMS): 2% (m/v) solution in dH2O.
Poly-l-lysine (P1524; Sigma): Prepare a stock of 1 mg/mL in sterile dH2O. Aliquot and store at −20 °C.
Resin for embedding: Thoroughly mix 25 mL EMbed-812 (14900; EMS), 15 mL Araldite 502 (10900; EMS), 55 mL DDSA (13710; EMS), and 1.5–1.9 mL DMP-30 (13600; EMS) in a 250 mL glass flask on a magnetic stirrer. Transfer the resin into several 50 mL conical tubes, and get rid of the air bubbles in the resin by centrifugation at 1000 rpm for 5 min. The resin is ready for use or the tubes can be sealed with parafilm and stored at −20 °C for several months.
Resin blocks: Fill the flat bottom embedding capsules (70021; EMS) with Embed 812 resin, and put them into the 60 °C oven to polymerize for 48 h. After that, take the resin blocks out of the capsules. Prepare these resin blocks before trimming and sectioning.
Sodium cacodylate (12300; EMS): stock solution: 0.4 M in dH2O, pH 7.4).
Tannic acid (21700; EMS): 0.15% (m/v) solution in 1X-PBS.
Uranyl acetate (22400; EMS): 1 or 2% (m/v) solution in dH2O filtered through a 0.22 μm syringe filter unit (SLGS033GS; Millipore).
1.9. INSTRUMENTATION
Coverslips, 25-mm round, 0.17 mm thick (640715; Warner Instruments). Most high-end objectives are infinity-corrected for a glass coverslip of 0.17 mm thickness. This thickness corresponds to coverslip # 1.5.
Glass slides (48312–003; VWR International)
Grid staining matrix system kit (71179–01; EMS)
Diamond knife with a boat (Ultra 45°, 3 mm, Diatome)
Diamond knife cleaning kit (70600; EMS)
Microscope Objective Marker (MBW10000; Nikon)
Loctite liquid super glue
Objective Diamond Scriber (Leica), a pen with a diamond tip (various vendors)
Glass-grid-bottom Petri dishes, 35 mm (35G-2–14-C-grid; MatTek Corporation)
Razor blades for manual trimming (71980; EMS)
Rose chambers (homemade) of noncorrosive aluminum, with FDA approved silicone rubber spacer. Syringes (3 and 20 mL) and needles (305165; BD) for perfusion
Slot grids (2330P-XA; SPI Supplies)
Tweezers (72864-D; EMS)
Ultramicrotome, Leica EM UC7 (Leica Microsystems)
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Anil Shukla for his enthusiasm and help in acquiring all videos and numerous pictures that we used during image preparation. We also thank Ms Veronica Farmer for reading of the manuscript. JL feels beholden to her former mentor Dr Alexey Khodjakov for sharing his knowledge of correlative light and electron microscopy, and to Dr Tatiana Vinogradova for a generous offer of a diamond scriber. Work in the lab of JL is supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
SUPPLEMENTARY DATA
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/bs.mcb.2015.03.003.
REFERENCES
- Alvey PL (1985). An investigation of the centriole cycle using 3T3 and CHO cells. Journal of Cell Science, 78(1), 147–162. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, & Bettencourt-Dias M (2011). Tracing the origins of centrioles, cilia, and flagella. Journal of Cell Biology, 194(2), 165–175. 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller D, Orpinell M, Olivier N, Wachsmuth M, Mahen R, Wyss R, et al. (2014). Mechanisms of HsSAS-6 assembly promoting centriole formation in human cells. Journal of Cell Biology, 204(5), 697–712. 10.1083/jcb.201307049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Farmer V, Shukla A, James J, Gruskin R, Kiriyama S, et al. (2014). Centriole maturation requires regulated Plk1 activity during two consecutive cell cycles. Journal of Cell Biology, 206(7), 855–865. 10.1083/jcb.201407087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L, Lee YL, Sahl SJ, Stearns T, & Moerner WE (2012). STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophysical Journal, 102(12), 2926–2935. 10.1016/j.bpj.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, & Pelletier L (2012). Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nature Cell Biology, 14(11), 1148–1158. http://www.nature.com/ncb/journal/v14/n11/abs/ncb2591.html-supplementary-information. [DOI] [PubMed] [Google Scholar]
- Leung BO, & Chou KC (2011). Review of super-resolution fluorescence microscopy for biology. Applied Spectroscopy, 65(9), 967–980. 10.1366/11-06398. [DOI] [PubMed] [Google Scholar]
- Lončarek J, Hergert P, & Khodjakov A (2010). Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Current Biology, 20(14), 1277–1282. 10.1016/j.cub.2010.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Magidson V, & Khodjakov A (2008). Control of daughter centriole formation by the pericentriolar material. Nature Cell Biology, 10(3), 322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Agard DA, Huang B, & Pelletier L (2014). Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends in Cell Biology, 24(3), 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, et al. (2012). Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nature Cell Biology, 14(11), 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A, Matos I, Lince-Faria M, & Maiato H (2009). Dissecting mitosis with laser microsurgery and RNAi in Drosophila cells In McAinsh AD (Ed.), Mitosis (Vol. 545, pp. 145–164). Humana Press. [DOI] [PubMed] [Google Scholar]
- Rattner JB, & Phillips SG (1973). Independence of centriole formation and DNA synthesis. Journal of Cell Biology, 57(2), 359–372. 10.1083/jcb.57.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, & Bowser SS (1985). Correlative immunofluorescence and electron microscopy on the same section of epon-embedded material. Journal of Histochemistry & Cytochemistry, 33(2), 165–171. 10.1177/33.2.3881520. [DOI] [PubMed] [Google Scholar]
- Rizzo R, Parashuraman S, & Luini A (2014). Correlative video-light–electron microscopy: development, impact and perspectives. Histochemistry and Cell Biology, 142(2), 133–138. 10.1007/s00418-014-1249-3. [DOI] [PubMed] [Google Scholar]
- Rose GG, Pomerat CM, Shindler TO, & Trunnell JB (1958). A cellophane-strip technique for culturing tissue in multipurpose culture chambers. Journal of Biophysical and Biochemical Cytology, 4(6), 761–764. 10.1083/jcb.4.6.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne JE, Specht CG, Izeddin I, Hurbain I, Tran P, Triller A, et al. (2011). Assessing the localization of centrosomal proteins by PALM/STORM nanoscopy. Cytoskeleton, 68(11), 619–627. 10.1002/cm.20536. [DOI] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, & Nigg EA (2012). 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biology Open, 1(10), 965–976. 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelhalter C, Laporte J, & Schwab Y (2014). Correlative light and electron microscopy: from live cell dynamic to 3D ultrastructure In Kuo J (Ed.), Electron microscopy (Vol. 1117, pp. 485–501). Humana Press. [DOI] [PubMed] [Google Scholar]
- Stout J, Rizk R, & Walczak C (2009). Protein inhibition by microinjection and RNA-mediated interference in tissue culture cells: complementary approaches to study protein function In Carroll DJ (Ed.), Microinjection (Vol. 518, pp. 77–97). Humana Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou M-FB, Wang W-J, George KA, Uryu K, Stearns T, & Jallepalli PV (2009). Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Developmental Cell, 17(3), 344–354. 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, & Chentsov Y (1982). Centrioles in the cell cycle. I. Epithelial cells. Journal of Cell Biology, 93(3), 938–949. 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka M, Smith NI, & Fujita K (2014). Introduction to super-resolution microscopy. Microscopy, 63(3), 177–192. 10.1093/jmicro/dfu007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.