Abstract
Inherited muscle disorders are caused by pathogenic changes in numerous genes. Herein, we aimed to investigate the etiology of muscle disease in 24 consecutive Greek patients with myopathy suspected to be genetic in origin, based on clinical presentation and laboratory and electrophysiological findings and absence of known acquired causes of myopathy. Of these, 16 patients (8 females, median 24 years-old, range 7 to 67 years-old) were diagnosed by Whole Exome Sequencing as suffering from a specific type of inherited muscle disorder. Specifically, we have identified causative variants in 6 limb-girdle muscular dystrophy genes (6 patients; ANO5, CAPN3, DYSF, ISPD, LAMA2, SGCA), 3 metabolic myopathy genes (4 patients; CPT2, ETFDH, GAA), 1 congenital myotonia gene (1 patient; CLCN1), 1 mitochondrial myopathy gene (1 patient; MT-TE) and 3 other myopathy-associated genes (4 patients; CAV3, LMNA, MYOT). In 6 additional family members affected by myopathy, we reached genetic diagnosis following identification of a causative variant in an index patient. In our patients, genetic diagnosis ended a lengthy diagnostic process and, in the case of Multiple acyl-CoA dehydrogenase deficiency and Pompe’s disease, it enabled specific treatment to be initiated. These results further expand the genotypic and phenotypic spectrum of inherited myopathies.
Keywords: Genetics, Inherited myopathy, Limb-girdle muscular dystrophy, Whole exome sequencing, Genetic diagnosis
Highlights
-
•
24 consecutive patients with myopathy were investigated via WES.
-
•
16 patients and 6 family members with causative variants in 14 different genes
-
•
Causative variants in 6 LGMD genes (ANO5, CAPN3, DYSF, ISPD, LAMA2, SGCA)
-
•
Other genes involved were CPT2, ETFDH, GAA, CLCN1, LMNA, MT-TE, MYOT, CAV3.
-
•
These results expand the genotypic and phenotypic spectrum of inherited myopathies.
1. Introduction
Inherited muscle diseases are a heterogeneous group of disorders, that includes congenital, metabolic and mitochondrial myopathies, muscular dystrophies and myotonias [1,2]. Each of these categories of genetic muscle disorders is in turn broad, with muscular dystrophies subdivided to Duchenne/Becker, myotonic, facioscapulohumeral, distal and limb-girdle muscular dystrophies (LGMD) [1]. This predominantly phenotype-based classification has been adequate for several decades; however, it has become obsolete after the delineation of the genetic and molecular basis of most of these disorders in recent years. These advances have led to the realization that defects in a certain gene can cause various myopathic phenotypes, and inversely, the same phenotype can be caused by different pathogenic variants in the same or multiple genes (allelic and locus heterogeneity) [3]. For instance, LGMDs, characterized by weakness affecting predominantly the shoulder and pelvic girdle muscles, are caused by pathogenic changes in more than 30 genes, with the gene altered and the mode of inheritance determining the LGMD subtype[[4], [5], [6], [7], [8], [9], [10], [11], [12]].
Clinical and other characteristics, such as the precise distribution of muscle weakness or the levels of serum creatine phosphokinase (CPK) have been associated with specific inherited myopathy subtypes. However, predicting the genetic defect involved in a myopathy based on the history, clinical features, MRI findings and muscle biopsy results is practically impossible [9]. Furthermore, there is significant clinical and genetic overlap among various inherited muscle disorders. For instance, LGMDs phenotypically and genotypically overlap with other types of muscular dystrophies, such as the Congenital Muscular Dystrophies [13,14].
Next generation sequencing (NGS) techniques, such as Targeted Gene Panels and Whole Exome Sequencing (WES), have revolutionized the diagnostic process of inherited myopathies [4,7,[15], [16], [17], [18], [19]]. Compared with other, gene-panel based, NGS techniques, WES provides a non-biased approach towards the identification of established pathogenic variants and the discovery of novel variants and genes associated with heterogeneous disorders, such as myopathies [20]. This improved diagnostic efficacy through high-throughput genetic testing is a prerequisite for management decisions, prognostication, and genetic counseling for patients with inherited myopathies.
Here, using WES, we expand further the phenotypic and genotypic spectrum of inherited muscle diseases, by establishing the genetic etiology of myopathy in 16 patients from Greece and 6 of their family members. In this cohort of unselected patients, referred to us from neurologists and pediatric neurologists across Greece, we have identified causative variants, including some novel ones, in 14 inherited myopathy genes (ANO5, CAPN3, CAV3, CLCN1, CPT2, DYSF, ETFDH, GAA, ISPD, LAMA2, LMNA, MYOT, MT-TE and SGCA).
2. Patients and methods
2.1. Study subjects
The study group included 24 consecutive patients with suspected genetically determined myopathy, based on the clinical presentation, electrophysiological studies, and absence of known acquired causes of myopathy. Also, patients with a complex phenotype where myopathy was not a prominent feature were excluded. For 16 of the 24 patients included in the study, the genetic cause of myopathy was identified via WES in the Neurology/Neurogenetics Laboratory, Medical School, University of Crete and are described herein. For 8 of the 24 patients (3 males, 5 females), including 5 patients with LGMD type of weakness, 2 patients with congenital myopathy, and 1 patient with prominent rhabdomyolysis and quadriceps weakness, we were not able to reach a specific genetic diagnosis. For these patients, the median age at testing was 19.5 years (range 4-72) and the median age at symptom onset 15 years (0-52). There were no major phenotypic differences between patients for whom a genetic diagnosis was reached and those for whom the cause of their myopathy remained unresolved, with the exception of presence of 2 patients with congenital myopathy in the latter group.
Informed consent for performing clinical WES was obtained from the patients and/or their legal guardians. The study protocol was performed following the ethical guidelines of the World Medical Association Declaration of Helsinki (version 2008) and was also approved by the Institutional Review Board of the University Hospital of Heraklion, Crete, Greece.
2.2. Blood sampling and DNA extraction
Peripheral blood (~5ml) was collected in ethylene diamine tetra-acetic acid (EDTA) tubes and stored at −80 °C until use. DNA was extracted from 1mL of whole blood, using the QIAamp DNA blood midi kit (Qiagen, USA) following the manufacturer’s centrifugation-based protocol. DNA concentration and purity were determined spectrophotometrically by absorbance measurement at 260 and 280 nm and agarose electrophoresis.
2.3. Whole exome sequencing
WES and initial bioinformatics analysis were performed in a CLIA-certified laboratory (Otogenetics Corporation, Norcross, GA, USA). Exome library preparation was performed using the Agilent V5 (51Mb) Sure-Select Target Enrichment System. Exon-enriched DNA libraries were sequenced on a HiSeq 2000/2500 (Illumina, USA) platform using paired end reads of 100-125 bp with an estimated average coverage of 50X. The data were then processed using the DNA Nexus platform, consisting of alignment of reads for each patient to the human reference genome hg19/GRCh37, removal of PCR duplicates using Picard, indel realignment and base quality score recalibration, variant calling and quality evaluation using the Genome Analysis ToolKit version 3.6. On average, the percentage of nucleotides with at least 50x coverage was more than 60%, and the average depth of coverage per interval was over 60. Sequencing results were transmitted through the DNAnexus.com platform (Mountain View, CA, USA).
2.4. WES data analysis
Annotation of called variants was performed at the Neurogenetics Laboratory, University of Crete through a combined approach using the automated Ingenuity Variant Analysis (IVA, Qiagen, USA) and GenomeTrax 2015.1 (Biobase, Wolfenbüttel, Germany) software, followed by semi-automated annotation of variants, integrating information from online databases (i.e. ClinVarTM, HGMD®), other bioinformatics tools and literature sources. The semi-automatic curation of variants was performed using stepwise filtering, to generate a list of possible disease-associated variants. Ascertainment was based on features of both the gene (e.g. inheritance patterns, functional evidence) and the specific variant (e.g. location, type, population frequency, computational predictions of effect). We therefore excluded variants with minor allele frequency > 1% on databases such as the Exome Aggregation Consortium (ExAC) dataset and focused on variants which produced a missense, non-sense, frameshift or splicing change. We also kept all potentially pathogenic genetic variants listed in the Human Gene Mutation Database (HGMD®). The functional consequences of identified variants on encoded proteins were also assessed using the VarSome database (https://varsome.com) that compiles prediction scores from several prediction algorithms including SIFT, PROVEAN, MutationTaster, PhyloP, FATHMM, and MetaSVM [21]. The Combined Annotation Dependent Depletion (CADD) score was also taken into consideration to ascribe pathogenicity to given variants [22]. Final interpretation of the pathogenicity of identified variants was performed manually taking into consideration data available in public databases and published in the literature. The classification of variants followed the recommendations by the American College of Medical Genetics and the Association for Molecular Pathology Laboratory Practice Committee Working Groups [23].
2.5. Sanger sequencing and MLPA analyses
Variants related to patient phenotypes were confirmed by Sanger sequencing for the patients, parents and affected relatives, if applicable, either at the Neurology Laboratory, University of Crete or at Diagenom (Rostock, Germany). MLPA analysis, needed to search for a trans variant in patient #2 with a single variant identified via WES, were performed at Diagenom (Rostock, Germany).
3. Results/case descriptions (Table 1, Table 2)
Table 1.
Demographic, clinical and laboratory features of the inherited muscle disorder cases identified through whole exome sequencing (WES).
| Patient # | Sex | Age at diagnosis (y) | Age at onset | Clinical features | Family history | NCS/EMG features | Muscle biopsy/Labs (CPK) |
|---|---|---|---|---|---|---|---|
| 1 | M | 16 | 14 | Mild lower extremity weakness, easy fatigability | - | - | Elevated CPK |
| 2 | M | 34 | 16 | Central-type muscle weakness, abolished tendon reflexes | Affected brother | - | Elevated CPK |
| 3 | F | 45 | 40 | Myopathy (exercise induced muscle stiffness and myalgias, moderate proximal muscle weakness) No cardiomyopathy |
- | - | - |
| 4 | F | 29 | 10 | Myotonic symptoms | Affected sister | Myotonia | - |
| 5 | F | 14 | 13 | Myalgias | Affected sister, father, paternal siblings | - | Elevated CPK |
| 6 | M | 17 | 14 | Myalgias | Affected sister | - | Elevated CPK |
| 7 | M | 19 | 7 | Pain and cramps in legs | - | Myopathy | Elevated CPK, transaminases |
| 8 | F | 7 | 2 | Lower extremity weakness, easy fatigability | - | Myopathy | Muscle biopsy: lipid myopathy |
| 9 | F | 67 | 57 | Central type muscle weakness, respiratory difficulties | - | Myopathy | Muscle biopsy non-diagnostic / DBS, cultured fibroblasts low GAA activity |
| 10 | F | 19 | 10 | Scapular winging, tibial hypertrophy, scoliosis, waddling gait, central type muscle weakness, respiratory dysfunction, decreased tendon reflexes | Affected sister | Μyopathy | Elevated CPK |
| 11 | M | 38 | 1 | Progressive muscle weakness | - | - | Elevated CPK |
| 12 | F | 39 | Childhood | Myopathy, cardiomyopathy | De novo | - | Elevated CPK |
| 13 | F | 47 | 35 | Myopathy, cardiomyopathy | Possibly affected father and sibling (sudden deaths) | - | - |
| 14 | Μ | 17 | 3 | Progressive muscle weakness, glucose intolerance | Affected mother with myopathy, diabetes in family | - | Elevated CPK |
| 15 | M | 54 | 53 | Proximal weakness, atrophy in the shoulder girdle and the face | Son: cardiac defibrillator at the age of 14 years | Myopathy with face-shoulder-arm distribution | Elevated CPK |
| 16 | Μ | 12 | 5 | Tested for increased CPK values | - | - | Elevated CPK |
Table 2.
Characteristics of the variants identified through whole exome sequencing (WES) in our cohort.
| Patient # |
Gene | Variants | CADD score |
ID |
gnomAD frequency (%) |
rs | Pathogenicity Criteria (ACMG)⁎ |
Phenotype |
|---|---|---|---|---|---|---|---|---|
| 1 | ΑΝΟ5 | p.Phe506fs⁎6 (c.1517delT) / p.Met542fs⁎11 (c.1624dupA) |
23.1 / 35.0 |
NM_01142649.2 |
0.000 / 0.004 |
794727158 / 281865480 |
PVS1, PS4, PM2, PM3 / PVS1, PS4, PM2, PM3 |
LGMD R12 (former 2L) |
| 2 | CAPN3 | p.Tyr537⁎ (c.1611C>A) / deletion of exons 2 to 8 (c.310-?_c.1115+?del) |
38.0 / - |
NM_000070.3 |
0.000 / 0.000 |
886042439 / - |
PVS1, PS3, PS4, PM2, PM3 / PM3, PM4 |
LGMD R1 (former 2A) |
| 3 | CAV3 | p.Thr78Met (c.233C>T) | 23.0 | NM_001234.5 | 0.270 | 72546668 | PS3, PS4, PM1, PM5, PP3, BS1, BP5, BP6 |
Caveolinopathy (former LGMD 1C) |
| 4 | CLCN1 | p.Phe167Leu (c.501C>G) / c.1471+1G>A |
20.3 / 34.0 |
NM_000083.3 |
0.110 / 0.002 |
149729531 / 375596425 |
PS4, PM1, PM2, PM3, PP1, PP5, BP6 / PVS1, PS3, PS4, PM2, PM3, BS2 |
Myotonia Congenita |
| 5 , 6 | CPT2 | p.Ser113Leu (c.338C>T) -Homozygous |
34.0 | NM_000098.3 | 0.140 | 74315294 | PA2, PS3, PS4, PM3, PP3, PP5, BS1, BS2 |
Metabolic Myopathy |
| 7 | DYSF | c.997-1G>A / p.Ala927fs⁎21 (c.2779delG) |
33.0 / 35.0 |
NM_003494.4 |
0.000 / 0.002 |
- / 727503909 |
PVS1, PM2 / PVS1, PS4, PM2, PM3, PP5 |
LGMD R2 (former 2B) |
| 8 | ETFDH | p.Pro483Leu (c.1448C>T) / p.Arg559⁎ (c.1675C>T) |
27.4 / 42.0 |
NM_04453.4 |
0.002 / 0.000 |
377656387 / 186023896 |
PS4, PM2, PM3, PP3, PP5 / PVS1, PS4, PM2 |
Metabolic Myopathy |
| 9 | GAA | c.32-13T>G / p.Tyr292Cys (c.875A>G) |
<10 / 28.5 |
NM_000152.5 |
0.340 / 0.001 |
386834236 / 1057516600 |
PA2, PS3, PS4, PM3, PP5, BS1, BS4, BP2, BP4 / PS3, PS4, PM1, PM2, PM3, PP3, PP5 |
Metabolic Myopathy (former LGMD 2V) |
| 10 | CRPPA (ISPD) | p.Thr238Ala (c.712A> G) -Homozygous |
24.9 |
NM_01101426.4 |
0.000 |
1038301242 |
PS4, PM1, PM2, PM5, PP3 |
LGMD R20 |
| 11 | LAMA2 | c.2208+4_2208+19delAGCTTGCAAGAATGTA / p.Ile2508fs⁎4 (c.7521dupT) |
- / - |
NM_000426.4 |
0.000 / 0.000 |
- / - |
PM2 / PVS1, PM2 |
LGMD R23 |
| 12 | LMNA | p.Arg249Gln (c.746G>A) | 27.1 | NM_170707.4 | 0.000 | 59332535 | PS4, PM1, PM2, PM6, PP3, PP5, BP2 |
Laminopathy (former LGMD 1B) |
| 13 | LMNA | c.1608+2T>C | 29.4 | NM_170707.4 | 0.000 | - | PVS1, PM2 | Laminopathy (former LGMD 1B) |
| 14 | MT-TE | m.14709T>C | <10 | NC_012920.1 | - | 121434453 | BP4 | Mitochondrial Myopathy |
| 15 | MYOT | p.Thr57fs⁎2 (c.170delC) | 33.0 | NM_006790.3 | 0.000 | - | PVS1, PM2 | Myotilinopathy (former LGMD 1A) |
| 16 | SGCA | p.Val247Met (c.739G>A) / p.Arg284Cys (c.850C>T) |
25.8 / 33.0 |
NM_000023.4 |
0.010 / 0.020 |
143570936 / 137852623 |
PS3, PS4, PM1, PM2, PM3, PP1, PP3, PP5 / PS3, PS4, PM1, PM2, PM3, PP3, PP5 |
LGMD R3 (former 2D) |
Criteria according to Richards et al, Genet Med, 2015 (Standards and Guidelines for the Interpretation of Sequence Variants) through the Ingenuity software.
3.1. Patient #1 (ΑΝΟ5)
This 16-year-old male patient presented easy fatigability, mainly of the lower extremities, since about two years and showed increased CPK (up to 1,200 U/L) on several occasions. Via WES, he was found to be heterozygous for the p.Phe506fs*6 (c.1517delT) and p.Met542fs*11 (c.1624dupA) changes in the ANO5 gene, as a cause of LGMD type R12 (formerly 2L). Both variants lead to a shift in the reading frame and a truncated protein product. Moreover, both p.Phe506fs*6 and p.Met542fs*11 are characterized as deleterious according to CADD (scores of 23.1 and 35.0, respectively; Table 2).
3.2. Patient #2 (CAPN3)
This 34-year-old man presented with progressive proximal weakness and atrophies of the upper and lower extremities since the age of 16 years (Fig. 1). On examination, tendon reflexes were abolished, except for the Achilles tendon reflexes. Serum testing revealed repeatedly elevated CPK values. The patient's brother, but not his sister, had a similar clinical picture, with proximal muscle weakness in all four limbs since the age of 10 years. Based on the clinical picture of both brothers and the absence of myopathy in the sister, an X-linked inheritance was initially suspected. However, both the patient and his brother tested negative for DMD gene pathogenic changes. WES in the patient identified the p.Tyr537* (c.1611C>A) change in the CAPN3 gene in a heterozygous state. Further testing of the CAPN3 gene by MLPA showed an additional heterozygous deletion of exons 2 to 8 (c.310-?_c.1115+?del) which leads to the p.Glu104_Arg372delfshX11 amino acid change causing a frameshift and a new stop codon located 11 codons downstream. By testing the father of the patient, who harbored only the heterozygous deletion of exons 2 to 8, we established that the pathogenic variants in the patient were in a trans state. Thus, we reached the diagnosis of LGMD type R1 (previous classification, 2A).
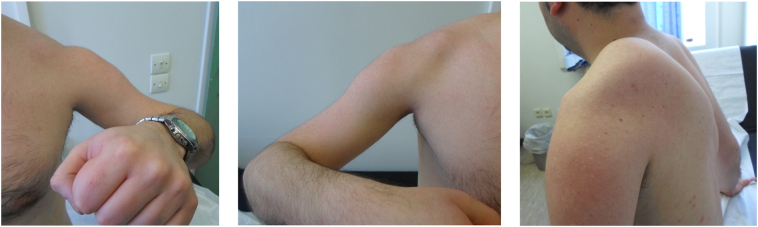
Fig. 1.
Photos of the upper extremities of patient #2, showing marked atrophy of the proximal muscles and scapular winging. This patient and his similarly affected brother were found to harbor both the p.Tyr537* change and the deletion of exons 2 to 8 in the CAPN3 gene in compound heterozygosity.
3.3. Patient #3 (CAV3)
This 44-year-old female patient presented with a history of exercise-induced muscle stiffness and myalgias, symptoms that developed since early adulthood and had been initially misdiagnosed as fibromyalgia. She developed progressive proximal muscle weakness, mainly in the pelvic area and the lower limbs. There was no family history of myopathy. Biochemical testing, CPK and inflammatory markers were normal. The patient underwent muscle biopsy that produced no specific findings, whereas only the latest of a series of EMGs revealed mild myopathic findings. The ECG, heart U/S and investigation for glycogen storage disorders produced normal results. Via WES we found that the patient carried the previously described as pathogenic p.Thr78Met (c.233C>T) CAV3 variant, in the heterozygous state, as a cause of caveolinopathy (formerly known as LGMD-1C). The unaffected father was also heterozygous for the same variant.
3.4. Patient #4 (CLCN1)
This 29-year-old patient has been experiencing myotonic symptoms, especially in the lower extremities, since the age of 10 years. These symptoms appeared at the initiation of movement, as in standing up from a seating position or climbing stairs. EMG studies in the patient and her affected sister, but not in her clinically unaffected mother, showed myotonic activity. Testing by WES established the diagnosis of Myotonia Congenita due to the exonic p.Phe167Leu (c.501C>G) and splice site c.1471+1G>A variants in the CLCN1 gene. These changes were verified by Sanger sequencing and were also found in her similarly affected sister.
3.5. Patient #5 (CPT2)
This 14-year-old patient presented with episodes of myalgia and rhabdomyolysis since at least one year, with increased CPK (up to 7,355 U/L) found on several occasions. Testing for both organic acids and carnitine esters, even when the patient had significantly elevated CPK levels, gave normal results. Her 12-year-old sister had episodes of myalgias and increased CPK since the age of 4 years. Myalgias were also reported in the father of the two sisters, as well as in 2 paternal uncles. Testing by WES followed by Sanger sequencing established the diagnosis of CPT II deficiency due to the homozygous p.Ser113Leu (c.338C>T) CPT2 variant in the patient and her similarly affected sister. The variant has been identified in 0.2% (134/66568) of European chromosomes by the Exome Aggregation Consortium (ExAC, http://exac.broadin stitute.org; dbSNP rs74315294).
3.6. Patient #6 (CPT2)
This 17-year-old patient has presented severe rhabdomyolysis after a viral upper respiratory tract infection. Three years earlier he had been hospitalized because of severe rhabdomyolysis and acute renal failure, necessitating temporary renal dialysis, that had been attributed to salmonellosis. He had repeatedly increased CPK values, up to 200,000 U/L. Testing for organic acids and carnitine esters, on repeated occasions, gave results within normal range. In this patient, WES established the diagnosis of CPT II deficiency due to the homozygous p.Ser113Leu (c.338C>T) CPT2 gene variant. The latter was verified by Sanger sequencing and found in homozygous and heterozygous state in the patient’s sister and brother, respectively. He was apparently unrelated to patient #5, even though they both carried the same p.Ser113Leu CPT2 change. Following genetic diagnosis, both the patient and his similarly affected sister followed special nutrition directions and avoided strenuous exercise.
3.7. Patient #7 (DYSF)
This 18-year-old male presented with progressive weakness, pain and cramps in both lower extremities. At the age of 7 years, testing for growth hormone deficiency, due to mild delay in growth, showed no abnormal findings. At the age of 8 years, laboratory testing revealed increased CPK values, with progressive increase in the ensuing years (peak value of about 23,000 U/L). Testing for Pompe disease was negative. Neurophysiological testing, performed in 2006 and repeated in 2014, showed myopathic features, with recording of spontaneous activity (fibrillation potentials and positive sharp waves) and early recruitment of motor units. A muscle biopsy, performed in 2007, showed no abnormal findings. Genetic testing for muscular dystrophy by sequencing and MLPA analysis of the DMD gene did not reveal any pathogenic changes. By using WES, we found that the patient was a compound heterozygote for the pathogenic c.907-1G>A and p.Ala927fs*21 (c.2779delG) changes in the DYSF gene. The former is a novel variant predicted to affect splicing and characterized by strong computational support for pathogenicity (CADD score = 33.0). The latter is present in population databases (rs745407251, ExAC 0.02%), but is associated mainly with LGMD-R2 pathogenicity and causes frameshift and premature termination of the protein translation.
3.8. Patient #8 (ETFDH)
This 7-year-old female patient presented lower and upper extremity weakness and easy fatigability since the age of 2 years. Muscle biopsy at the time showed findings compatible with metabolic myopathy due to derangement of lipid metabolism (lipid storage myopathy). VLCAD measurements in fibroblasts produced normal results. Testing by WES established the diagnosis of metabolic myopathy due to the p.Pro483Leu (c.1448C>T) and p.Arg559* (c.1675C>T) changes in the ETFDH gene. The latter is highly deleterious (CADD=42.0) and causes premature translation termination resulting in a truncated protein. Sanger sequencing verified the presence of these variants in the patient and testing of the parents and the unaffected brother showed that these two variants were in trans. This allowed the initiation of specific therapy with riboflavin, with significant improvement in muscle strength and exercise endurance already noted in the first few months of treatment.
3.9. Patient #9 (GAA)
This 67-year-old woman presented with a 10-year history of progressive difficulties in climbing stairs, getting up from sitting position and walking, as well as orthopnea. On neurological examination, proximal muscle weakness was noted in all four extremities; the patient was walking with a cane. Pulmonary function studies showed reduced Forced Vital Capacity-FVC (49% of predicted). CPK levels were mildly elevated (160-200 U/L), while EMG showed non-specific myopathic changes. Muscle biopsy was non diagnostic, since it revealed vacuolar myopathy, ragged-red, moth-eaten fibers and absence of glycogen accumulation. WES revealed heterozygosity for two GAA gene pathogenic variants, one in intron 1 (c.-32-13T>G) and one in exon 5 (p.Tyr292Cys, c.875A>G). These were confirmed by Sanger sequencing, while genotyping the patient’s mother confirmed compound heterozygosity in the patient. An initial dried blood spot (DBS) GAA enzyme assay showed normal activity. Repeated testing showed abnormally low GAA activity in DBS (1.13 pmoles/punch/hr; normal range: 5.3-31.5) and fibroblasts (0.23 nmoles/mg/min; normal range: 0.34-4.9). Thus, the diagnosis of Pompe's disease was confirmed and substitution therapy was initiated. This led to an initial improvement in motor scores (mean 14.5m increase on the 6-minute walk test) and pulmonary function studies. However, the patient developed a severe lower respiratory tract infection and passed away 3 years after the initial genetic diagnosis.
3.10. Patient #10 (CRPPA-ISPD)
This 19-year-old female had experienced progressive muscle weakness and walking difficulties in the past 9 years. On clinical examination, scapular winging, calf hypertrophy, scoliosis, wadding gait, muscular weakness in a limb-girdle distribution, impaired respiratory function (FVC at 65% of normal values) and markedly decreased deep tendon reflexes were noted. Serum CPK levels were persistently elevated (8–10X normal values). Repeated EMG studies were suggestive of myopathy. At the age of 14 years, she underwent muscle biopsy, where immune-reactivity to antibodies against dystrophin (DYS-1, DYS-2, DYS-3) and β, γ and δ sarcoglycan, but not against caveolin, adhalin, merosin, dysferlin, merosin, emerin, desmin and collagen VI was found, thus pointing to a sarcoglycanopathy or dystrophinopathy. Heart U/S showed only mild mitral and tricuspid regurgitation and brain MRI did not reveal any abnormal findings. The patient’s 14-year-old sister presented with similar clinical features, albeit milder, and again her brain MRI was unrevealing. Both parents were unaffected by myopathy or other neurological ailment and they denied any possibility of consaguinity. The homozygous c.712A>G change in the CRPPA gene (previously known as ISPD), leading to the p.Thr238Ala amino acid substitution, was found by WES, pointing to the diagnosis of LGMD R20. These results were verified by Sanger sequencing, which also showed the same amino acid change in the similarly affected sister, again in a homozygous state. The p.Thr238Ala variant has an exceptionally low frequency in controls (0.18% ExAC) and is computationally shown to produce a deleterious effect (CADD score = 24.9).
3.11. Patient #11 (LAMA2)
This 39-year-old male had been diagnosed as a “floppy” infant with generalized muscle weakness and hypotonia, receiving a diagnosis of congenital myopathy/muscular dystrophy. He walked at the age of five years but completed elementary and high school. When he was 14 years old, he underwent Achilles tendon elongation for pes equinus. On clinical examination, scoliosis, wadding gait, muscle weakness in a limb-girdle distribution and markedly decreased deep tendon reflexes were evident. Also, mild cognitive deficits (MOCA score of 24/30) were noted. Serum CPK levels were persistently elevated. Repeated EMG studies were suggestive of myopathy. Heart U/S and 24hr ECG recording were normal. Pulmonary function tests showed FVC at 68% of normal values and moderate restrictive defects. Muscle biopsy showed normal immune-reactivity to antibodies against dystrophin (DYS-1, DYS-2, DYS-3), albeit mild deficiency of complex I was observed. Brain MRI was suggestive of leukoencephalopathy. Testing by WES established the diagnosis of LGMD-R23 due to the LAMA2 gene c.2208+4_2208+19delAGCTTGCAAGAATGTA and p.Ile2508fs*4 (c.7521dupT) variants. Both are novel with the former leading to a splice-site loss, while the latter causing a frameshift in the reading frame and a truncated product.
3.12. Patient #12 (LMNA)
This 39-year-old female patient had a long history of slowly progressive muscle weakness and atrophy in a limb girdle distribution since her childhood, initially presenting as difficulty in climbing stairs and running. In the past few years, she was diagnosed with second degree atrioventricular block and dilated cardiomyopathy and underwent implantation of implantable cardioverter defibrillator (ICD). She had mildly increased CPK values in repeated measurements. Her EMG showed a myopathic pattern and a deltoid muscle biopsy revealed dystrophic changes (degeneration / regeneration) with type 1 fiber predominance. Her family's history was negative for neurologic disease. In this patient, we identified the p.Arg249Gln (c.746G>A) heterozygous LMNA gene variant, establishing the diagnosis of laminopathy (formerly LGMD-1B). Sanger sequencing confirmed the presence of the p.Arg249Gln LMNA variant in the patient, and showed absence of this variant in her parents, raising the possibility of a de novo mutation. This variant is not present in population databases (rs59332535, no ExAC frequency)
3.13. Patient #13 (LMNA)
This 47-year-old female patient presented with history of ventricular arrhythmias, second degree atrioventricular block, dilated cardiomyopathy (diagnosed with both MRI and U/S) and mild proximal muscle weakness. Due to her heart problems, an ICD was implanted. Cardiac and skeletal muscle involvement developed gradually during her third decade of life. The EMG revealed distinct myopathic features. Family history was positive for sudden cardiac deaths and dilated cardiomyopathy (Fig. 2), indicating a possible autosomal dominant inheritance pattern. Via WES, we identified a heterozygous novel splice site variant (c.1608+2T>C) in the LMNA gene in this patient, reaching the diagnosis of laminopathy (formerly LGMD-1B). The variant is predicted to result in a decreased splicing efficiently, as shown by a MaxEntScan score decrease of 79% (from 9.82 to 2.06). Also, in silico analysis using different bioinformatic tools supports the pathogenic effect of the c.1608+2T>C variant (CADD score = 29.4).
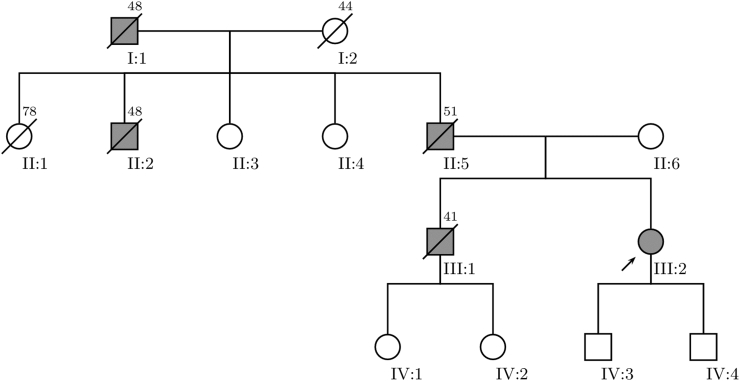
Fig. 2.
Family pedigree of patient #13, harboring a heterozygous novel splice site variant (c.1608+2T>C) in the LMNA gene. The proband is indicated by an arrowhead (III-2) and subjects possibly affected by laminopathy are colored in grey. Subjects II-5, III-1 and III-2 suffered from second degree atrioventricular block and dilated cardiomyopathy and underwent either a permanent pacemaker or an implantable cardioverter-defibrillator insertion. The symbol for deceased subjects is marked with a diagonal bar and age of death is placed in the upper right corner.
3.14. Patient #14 (MT-TE)
This 17-year-old patient has presented difficulty walking and frequent falls since the age of 3 years and later inability to walk unassisted. In recent years, he is wheel-chair dependent, has swallowing difficulties and needs assisted ventilation at nights. The patient also had osteopenia and glucose intolerance. Electrophysiology revealed myopathic features and serum testing persistently elevated CPK values. His mother and maternal grandmother have developed muscle weakness in a more advanced age compared to the patient. Several maternal family members have been diagnosed with diabetes. Muscle biopsy in the patient and his affected mother showed signs of mitochondrial myopathy. As an off-target finding of WES, we identified a heteroplasmic variant in the MT-TE mitochondrial gene (m.14709T>C) in this patient. Sanger sequencing confirmed the presence of this variant in the patient and his affected mother, establishing the diagnosis of mitochondrial myopathy.
3.15. Patient #15 (MYOT)
This 54-year-old man presented with a history of progressive muscle weakness and atrophy in his shoulders and face of about one-year duration. Neurological examination revealed proximal weakness in all four extremities and muscle atrophy in the shoulder girdle and the lower half of the facial musculature (Fig. 3). Tendon reflexes were brisk in all four extremities, but plantar responses were flexor. The patient had a waddling type of gait and could not jump on one leg, perform a squat or walk on his heels. Electrophysiological testing revealed a myopathic disorder with face-shoulder-arm distribution. MRI of cervical and lumbar spine showed cervical and lumbar stenosis due of degenerative changes. Serum testing showed mildly increased CPK values (375 U/L). A heart U/S showed only mild aortic valve deficiency. Pulmonary function testing revealed minor disturbances of expiratory flows and reduced maximum inspiratory and expiratory pressures and diffusion lung capacity. The mother of the patient died at the age of 45 years old, reportedly from acute ischemic stroke, and his son had a cardiac defibrillator implanted at the age of 14 years old. WES in this patient identified the heterozygous p.Thr57fs*2 (c.170delC) change in the MYOT gene, suggesting the diagnosis of myotilinopathy (formerly LGMD-1A). It is a novel frameshift variant that leads to a very premature termination of protein translation and strong computational evidence exists for a damaging effect on the resulting protein (CADD = 33.0), supporting its pathogenicity.
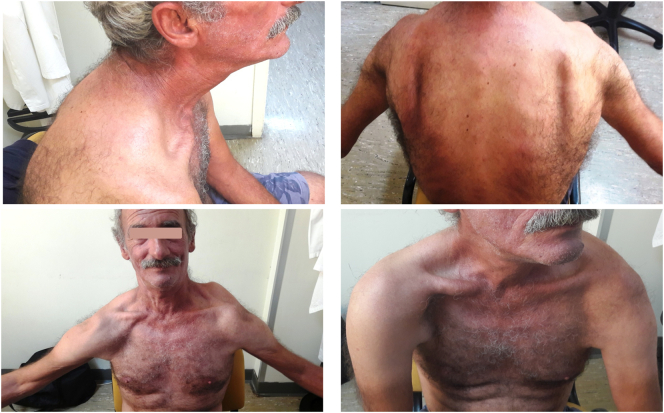
Fig. 3.
Photos of the patient #15 showing muscle atrophy in the shoulder girdle and the lower half of the facial musculature. WES testing identified the p.Thr57fs*2 (c.170delC) change in the MYOT gene, suggesting the diagnosis of myotilinopathy.
3.16. Patient #16 (SGCA)
This 11-year-old patient was referred due to increased CPK values (up to 1,158 U/L) in repeated measurements. He did not show any symptoms of significant muscle weakness and his growth had been normal. Muscle biopsy was non-diagnostic, revealing only mild histopathological changes. Heart U/S did not show abnormal findings. No pathogenic changes were found in the DMD gene. By performing WES, we identified the p.Val247Met (c.739G>A) and p.Arg284Cys (c.850C>T) pathogenic variants in the SGCA gene.
4. Discussion
Here we have tested by WES consecutive unselected Greek patients affected with LGMD and other inherited myopathies and found in 16 of them causative variants in 14 genes (Table 1). Specifically, we found pathogenic variants in 6 LGMD genes (6 patients; ANO5, CAPN3, DYSF, ISPD, LAMA2, SGCA), in 3 metabolic myopathy genes (4 patients; CPT2, ETFDH, GAA), in 1 congenital myotonia gene (1 patient; CLCN1), in 1 mitochondrial myopathy gene, as an off target result of WES (1 patient; MT-TE) and in 3 other myopathy genes (4 patients; CAV3, LMNA, MYOT). In our study, we have identified some novel variants, unique to the Greek population. Also, most of the known pathogenic variants we describe have been associated with multiple phenotypes in the literature, and we present here their phenotypic expression in cohort from Greece.
The diagnostic rate in our cohort (16/24; 66.7%) is considered high compared to other similar cohorts. In a recent US study, the application of NGS targeted gene panels resulted in genetic diagnosis in 27% of 4,656 LGMD patients, with the most common pathogenic variants identified in the CAPN3, DYSF, FKRP and ANO5 genes [6]. A similar NGS panel study from Latin America identified a definite genetic cause in 16% of 2,103 LGMD patients [24]. The higher diagnostic rate in our cohort compared to these LGMD cohorts probably relates to the inclusion of muscle disorders, such as metabolic myopathies and congenital myotonias, that are more prone to genetic diagnosis. Also, WES, that we used in our study, compared to panel-based NGS techniques, allows the identification of pathogenic variants in genes not covered in gene panels, and thus improved diagnostic yield [20].
Patient #1 was found to be compound heterozygote for the p.Met542fs*11 and p.Phe506fs*6 changes in the ANO5 gene. Pathogenic variants in the ANO5 gene are causing autosomal recessive Miyoshi muscular dystrophy type 3, autosomal recessive LGMD type 12 (R12, formerly 2L), asymptomatic hyperCKaimia, exercise associated myalgia/myoglobinuria and autosomal dominant gnathodiaphyseal dysplasia [25,26]. Anoctamin 5, encoded by the ANO5 gene, is a member of the anoctamins, a family of 10 calcium-activated proteins, with diverse cellular functions, including Cl- channeling, intracellular signaling and membrane trafficking and repair [27]. Each of these proteins has a distinct tissue distribution and physiological function, with ANO5 mainly expressed in skeletal and cardiac muscle and bone [25]. The shift in the reading frame caused by both ANO5 variants found in our patient (p.Met542fs*11 and p.Phe506fs*6) leads to a truncated protein product, in a gene where loss of function is not well tolerated. The p.Phe506fs*6 change has been described before in LGMD patients [[27], [28], [29]]. Specifically, it has been described (as c.1520delT), along with the intronic splice site variant c.1899–4A>G, in an Italian patient with onset of proximal upper limb weakness and increased CPK at the age of 50 years [30]. Also, it has been found, concomitantly with another frameshift change (c.191dupA), in a 39-year-old asymptomatic patient with increased CPK and skeletal muscle involvement in MRI [31,32]. About the p.Met542fs*11 change, it has been described, as homozygous c.1627dupA change, in two Italian brothers with LGMD [33].
In patient #2, the p.Tyr537* change and a deletion of exons 2 to 8 in the CAPN3 gene were identified by WES and MLPA, respectively. Pathogenic variants in the CAPN3 gene are the cause of the autosomal recessive LGMD-R1 (formerly known as LGMD-2A) [34] and the less severe autosomal dominant LGMD-D4 [35,36]. Calpain-3, the protein encoded by the CAPN3 gene, is the skeletal muscle specific member of the calpains, a family of Ca2+ dependent cysteine proteases with heterogeneous function and tissue distribution [34]. Calpain-3 has several unique properties, including rapid auto-degradation, Na+ dependence and non-proteolytic functions [37]. Both CAPN3 variants found it our patient have been described before as pathogenic. In specific, the p.Tyr537* variant has been found in homozygosity in LGMD patients from Turkey [38,39] and in compound heterozygosity with 550delA in a family from Croatia [40]. Likewise, the deletion of exons 2-8 (p.Glu104_Arg372delfshX11) has been described before in two Bulgarian patients, again in compound heterozygosity with c.550delA [41].
In patient #3 we found the pathogenic CAV3 variant (c.233C>T, p.Thr78Met) in the heterozygous state. CAV3 pathogenic variants are associated with caveolinopathies, that include skeletal muscle pathologies such as LGMD type 1C (not included in the new LGMD classification), rippling muscle disease, distal myopathy, and isolated hyperCKemia, as well as cardiac abnormalities such as long QT syndrome, sudden infant death syndrome and hypertrophic cardiomyopathy[42]. Caveolinopathies present with heterogeneous features such as exercise intolerance, myalgia, rhabdomyolysis, and rippling and percussion-induced muscle contractions [43]. Caveolin-3, the protein encoded by CAV3, forms flask-shaped invaginations on the cytoplasmic surface of the sarcolemma, termed caveolae. These caveolae have an essential role in maintenance of sarcolemmal integrity, vesicular trafficking and signal transduction [44]. The p.Thr78Met CAV3 variant found in our patient has been repeatedly described in the literature as pathogenic, associated with many different phenotypes [43,[45], [46], [47], [48], [49], [50], [51], [52], [53], [54]]. This variant is part of the central hydrophobic transmembrane domain of the caveolin 3 and has been shown to affect its function. In spite of the association of p.Thr78Met CAV3 with both skeletal and cardiac muscle pathologies, its functional role regarding cardiac diseases has been a matter of controversy. In particular, it has been characterized as a variant of unknown or benign significance after it has been found in healthy individuals with frequencies comparable to cases with long QT syndrome [55,56]. Even though this variant has been found also in the patient’s asymptomatic father, we ascribe this to a possible concerted action with another yet unidentified variant that predisposes to the specific phenotypic characteristics, as it has been previously suggested for the role of p.Thr78Met CAV3 in cardiac diseases [57].
In patient #4 and her similarly affected sister we reached the diagnosis of Myotonia Congenita due to the exonic p.Phe167Leu (c.501C>G) and splice site c.1471+1G>A variants in the CLCN1 gene. Pathogenic variants in the CLCN1 gene are the cause of autosomal dominant (Thomsen) and autosomal recessive (Becker) Myotonia Congenita, an inherited skeletal muscle channelopathy [58,59]. Chloride channels, encoded by the CLCN1 gene, are important for the muscle fiber repolarization and the stability of the sarcollemal resting potential [60]. Both variants found in our patient and her sister have been described before in the literature [[61], [62], [63], [64], [65]].
In patients #5 and #6, as well as the affected sister of patient #5, all presenting with episodes of acute rhabdomyolysis, we found the p.Ser113Leu (c.338C>T) CPT2 gene variant in homozygosity. CPT2 gene variants are the cause of CPT II deficiency, one of the most common mitochondrial fatty acid oxidation disorders [66]. CPT II deficiency manifests as recurrent acute rhabdomyolysis associated with metabolic decompensation and intermittent myopathic symptoms, both usually triggered by exercise or infection [67]. The carnitine palmitoyltransferase (CPT) system mediates long chain fatty acid translocation across the mitochondrial membrane from the cytosol to the mitochondrial matrix, where these fatty acids are used for energy production through the β-oxidation process [66]. The CPT-2 protein, encoded by the CPT2 gene, is embedded in the inner mitochondrial membrane and forms part of the CPT system [68]. Given that the p.Ser113Leu CPT2 gene variant is the most frequently identified CPT II pathogenic variant in large series of Caucasian patients, with most of the patients reported to date being homozygous, and since our patients were apparently unrelated, the possibility of a founder effect seems unlikely [67,[69], [70], [71], [72], [73]]. For both patients 5 and 6, although their clinical picture was high suspicious for beta-oxidation defect, especially CPT II deficiency, repeated testing for organic acids and carnitine esters yielded results within normal range, even at times of significantly elevated CPK levels. Thus, given that other diagnostic modalities proved unsuccessful, genetic diagnosis was important to avoid serious and even life-threatening complications, such as acute renal failure by timely intervening with high-glucose fluids, correction of metabolic acidosis and early dialysis.
In patient #7, an 18-year-old male with progressively deteriorating leg pain and cramps, myopathic EMG features and increased CPK values, we identified the pathogenic c.907-1G>A and p.Ala927fs*21 (c.2779delG) DYSF gene changes in compound heterozygosity. Pathogenic variants in the DYSF gene cause LGMD-R2 (formerly 2B), Miyoshi muscular dystrophy type 1 and distal myopathy with anterior tibial onset, collectively called dysferlinopathies [74,75]. Dysferlin, encoded by the DYSF gene, regulates vesicle fusion contributing patching of sarcolemmal defects [76]. Concerning the two DYSF variants found in our patient, the novel c.907-1G>A change is absent from controls and is predicted to be deleterious probably by affecting splicing, while the p.Ala927fs*21 DYSF change causes frameshift and premature termination of the protein translation. Furthermore, it has been described as pathogenic in myopathy patients in Iran, Turkey, South Spain and more importantly in Israel, in several members of the community of Jews originating from Caucasus, where a founder effect and a 4% carrier frequency has been identified [[77], [78], [79], [80], [81], [82]].
In patient #8, WES established the diagnosis of metabolic myopathy due to the p.Pro483Leu (c.1448C>T) and p.Arg559* (c.1675C>T) variants in the ETFDH gene. Variants in the ETFDH gene are one of the causes of Glutaric acidemia type II (Multiple acyl-CoA dehydrogenase deficiency -MADD), specifically type IIC. The two other causes of MADD are pathogenic variants in the ETFA and ETFB genes. The proteins encoded by the ETFA, ETFB and ETFDH genes, electron transfer flavoprotein (ETF) A and ETF-B and EFT Dehydrogenase, respectively, are involved in the transfer of electrons to the mitochondrial respiratory chain by flavoproteins. MADD’s clinical manifestations range from a neonatal form characterized by acidosis, hypoglycemia, cardiomyopathy and encephalopathy, to an adult-onset form with progressive muscle weakness, exercise intolerance, and episodes of vomiting, hypoglycemia, and metabolic acidosis. MADD is considered one of the curable myopathies, with riboflavin administration having excellent results in a sizable proportion of patients [83], as was the case in our patient. The p.Pro483Leu ETFDH change found in our patient has been described initially in a 13-year-old Turkish girl, born to consanguineous parents and presenting muscle weakness with inability to walk, myalgias and increased CPK. The patient showed full recovery following administration of riboflavin [84]. Subsequently, the p.Pro483Leu variant has been reported as pathogenic, but responsive to riboflavin, several other times [[85], [86], [87], [88]]. Concerning the second ETFDH variant (p.Arg559*) found in our patient, this causes premature translation termination resulting in a truncated protein and has also been described before as pathogenic [89].
In patient #9, we detected trans heterozygosity of the pathogenic variants c.-32-13T>G and p.Tyr292Cys (c.875A>G) in the GAA gene. Variants in the GAA gene are the cause of Pompe’s disease (formerly LGMD-2V), the clinical spectrum of which ranges from severe infantile forms to milder late-onset forms, with the diagnosis of the latter being challenging [90,91]. Our patient with late-onset Pompe disease was finally diagnosed by WES following a 10-year delay, despite inconclusive initial muscle biopsy and biochemical studies. The role of WES in detecting pathogenic GAA variants has been recently emphasized [92], despite initial shortcomings probably due to limitations of earlier WES techniques [93]. Pompe’s disease is characterized by lysosomal glycogen accumulation, due to acid alpha-glucosidase (GAA) deficiency, progressively affecting skeletal, respiratory and heart muscles [94]. About the c.-32-13T>G change of our patient, it has been repeatedly described, being the most common in cohorts of patients with late onset Pompe’s disease, with a frequency up to 87.5% [6,19,91,92,[95], [96], [97], [98]]. The c.-32-13T>G change has been shown to affect splicing, leading to omission of exon 2, where the start codon is located [95,99]. Thus, it is established as pathogenic, despite the low computational evidence supporting its pathogenicity (CADD score <10). About the p.Tyr292Cys change, it has also been described in the literature in cases of juvenile and other forms of glycogenosis [97,[100], [101], [102]]. It is located in a mutational hotspot, in a critical domain of the GAA enzyme, whereas functional studies strongly support its pathogenicity [103,104]. Of note, both variants of our patient have been found in compound heterozygosity in a patient from Spain, with approximately the same age at disease onset as our patient (60 years) [96].
About patient #10 and her similarly affected sister, the p.Thr238Ala (c.712A>G) change in exon 4 of the CRPPA (ISPD) gene was found in homozygous state, leading to the diagnosis of LGMD type R20. CRPPA gene pathogenic variants have been reported to cause a spectrum of phenotypes ranging from congenital muscular dystrophy with cerebral and ocular involvement (Walker-Warburg syndrome), through mild muscle weakness and subtentorial brain involvement, to mild adult onset LGMD phenotype without CNS involvement [[105], [106], [107], [108], [109]]. CNS involvement was unlikely in our patients, since no cognitive deficits were found, and brain MRI did not show any abnormalities. CRPPA variants are a rare cause of LGMD; in an Italian study, they were found in less than 1% of the entire LGMD cohort [110]. The D-ribitol-5-phosphate cytidylyltransferase protein encoded by the CRPPA gene contributes to the glycosylation of dystroglycan in the cytosol [109]. Dystroglycan is a major adhesion complex, composed of one α- and one β-subunit, that connects the subsarcolemmal actin cytoskeleton to the extracellular matrix, thus providing stability to skeletal muscles [111]. CRPPA mutations disrupt α-dystroglycan glycosylation, leading to reduced binding capacity of ECM ligands and thus compromising myocyte integrity and function [18,109]. The p.Thr238Ala (c.712A>G) variant found in homozygosity in our patient and her sister is located in a mutational hot spot and in a well-established CRPPA functional domain (Glyco_tranf_GTA_type) where no benign variation has been found. It is noteworthy that a similar change (c.713C>T, leading also to the p.Thr238Ile amino acid substitution) in compound heterozygosity with a nonsense variant (p.Arg86*) in the CRPPA gene has been reported to result in severe Cobblestone Lissencephaly [112].
In patient #11, we found the c.2208+4_2208+19delAGCTTGCAAGAATGTA and p.Ile2508fs*4 (c.7521dupT) LAMA2 gene variants. Pathogenic variants in the LAMA2 gene are the cause of congenital muscular dystrophy type 1A (MDC1A) and LGMD-R23. Again, as in other genes, all the myopathic phenotypes caused by LAMA2 gene variants have been reclassified under the comprehensive name “LAMA2-related muscular dystrophies” [113]. Brain MRI in our patient showed diffuse cerebral hemispheric white matter abnormalities; this is not uncommon in congenital muscular dystrophies [114].The LAMA2 gene encodes for the laminin a2 chain (also known as merosin) of the heterotrimeric laminin-211 complex that stably connects the sarcolemma to the extracellular matrix. The c.7521dupT (p.I2508fs*4) LAMA2 variant in our patient has not been described before in the literature; however, it is considered pathogenic since it is a frameshift variant that leads to a truncated protein, in a gene where loss of function is a known pathogenetic mechanism. The second LAMA2 variant found in our patient (c.2208+4_2208+19delAGCTTGCAAGAATGTA) leads to a splice-site loss, and as such it is also considered pathogenic.
In two female patients (#12, #13) with cardiomyopathy and skeletal myopathy, we report the p.Arg249Gln (c.746G>A) and the splice-site c.1608+2T>C LMNA gene variants, respectively. LMNA gene pathogenic variants are implicated in laminopathies, a group of disorders manifesting as heart disease, Emery-Dreifuss, congenital and limb-girdle muscular dystrophies, lipodystrophy, metabolic disorders, Charcot-Marie-Tooth disease and premature aging syndromes [115,116]. Heart involvement, characterized by conduction system defects, arrhythmias, dilated cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy, heart failure and sudden cardiac death, is highly prevalent [[117], [118], [119]]. Lamins A and C, encoded by the LMNA gene, are major structural components of the nuclear lamina, a fibrillar network that stabilizes the nuclear envelope [120]. Additionally, lamins are involved in multiple cellular processes, such as chromatin organization, DNA replication, gene and cell-cycle regulation and nucleo-cytoskeletal coupling [118]. Even though the frequency of the p.Arg249Gln LMNA gene variant found in our patient #12 is probably extremely low, it has already been extensively described in the literature as pathogenic, being reported in multiple unrelated individuals affected with Emery-Dreifuss muscular dystrophy and a single individual affected with LGMD [15,16,[121], [122], [123], [124], [125], [126], [127]]. The c.1608+2T>C LMNA variant found in our patient #13 is predicted to affect splicing. The strongest neighboring alternative splice site lies 72 bases upstream from the exon/intron boundary and this is predicted to result in an in-frame deletion. There have been several splice site variants in the LMNA gene that are described as pathogenic, including the c.1608+5G>C change[128], just 3 nucleotides downstream of the c.1608+2T>C variant of our patient. Finally, this variant is extremely rare in public variant databases.
In patient #14, we identified a heteroplasmic variant in the MT-TE mitochondrial gene (m.14709T>C), as an off-target finding of WES. The MT-TE gene encodes for the mitochondrial t-RNA for glutamate, and as such, pathogenic variants in this gene are expected to disrupt intramitochondrial protein synthesis. The m.14709T>C MT-TE gene variant is reported as pathogenic in both the ClinVar and MitoMap databases and has been associated with several syndromes, including 1) mitochondrial myopathy with diabetes mellitus, 2) maternally transmitted diabetes- deafness syndrome, 3) juvenile myopathy, encephalopathy, lactic acidosis and stroke. There have been numerous publications describing large families in which the m.14709T>C MT-TE gene variant segregates with diabetes mellitus and myopathy, independently, concurrently or sequentially [[129], [130], [131], [132], [133], [134], [135], [136], [137], [138], [139]], as was the case in our patient’s family. The presence in our patient #14’s extended family of members with myopathy, diabetes or both is an interesting finding, and indicates that physician should have raised awareness in similar clinical scenarios to consider a mitochondrial disorder in their differential diagnosis. Interestingly, there is still debate about the exact pathophysiological mechanism of the phenotype associated with the m.14709T>C variant [140].
In the case of patient #15, with progressive quadriparesis and proximal limb and lower face atrophy, we found the p.Thr57fs*2 MYOT variant. Variants in the MYOT gene have been associated with various autosomal dominant disease phenotypes (myotilinopathies), that include the former LGMD-1A subtype and a subgroup of the myofibrillar myopathies [[141], [142], [143], [144], [145]]. Myotilin, the 498 amino acid long protein encoded by the MYOT gene, is a sarcomeric Z-disk component that acts together with a-actinin and filamin C to cross link actin and support the integrity of the contracting myocyte [143]. The p.Thr57fs*2 (c.170delC) MYOT variant found in our patient has not been previously described in the literature. However, the predicted effect on protein translation, in a gene where loss of function usually leads to disease, combined with the high CADD score, justifies its pathogenicity. Interestingly, an amino acid substitution at the same residue (p.Thr57Ile) has been found as the cause of LGMD-1A in a large North American pedigree of German descent [146].
In patient #16, with persistent hyperCKemia, by performing WES we identified the p.Val247Met (c.739G>A) and p.Arg284Cys (c.850C>T) pathogenic variants in the SGCA gene. Variants in the SGCA gene have been associated with autosomal recessive LGMD-R3, formerly known as LGMD-2D [147]. The protein encoded by the SGCA gene, α-sarcoglycan (also known as adhalin), is part of the multi-protein Dystrophin Glycoprotein Complex (DGC) that spans the sarcolemma and links cytoskeletal actin to the extracellular matrix [148]. Other components of the DGC include dystrophin, syntrophins and dystroglycans [149]. This complex has an important role in maintaining the structural integrity of myocytes and the stability of the neuromuscular junction [148,150]. The p.Val247Met SGCA change found in our patient is considered the second most common SGCA variant associated with LGMD and hyperCKemia, having been described repeatedly [[151], [152], [153], [154], [155]]. Similarly, there are several reports describing LGMD patients harboring the second SGCA variant (p.Arg284Cys) found in our patient [6,152,[155], [156], [157], [158], [159], [160]].
5. Conclusions
As shown here, WES can aid in the diagnostic investigation across the inherited muscle disease spectrum, including LGMDs, metabolic myopathies, myotonias, and even mitochondrial myopathies, as an off-target effect. Thus, even though WES cannot identify disorders associated with repeat expansions or large-scale deletions/duplications, such as facioscapulohumeral dystrophy and myotonic dystrophies, it has become an indispensable tool in investigating patients with heterogeneous inherited myopathies. The accurate genetic diagnosis achieved by WES or other NGS strategies ends the diagnostic marathon of the patients, assists in family planning and, in some cases, opens treatment prospects, as in the case of Pompe’s disease or MADD. In the near future, this will be even more important with the advent of gene therapies, as for example in the case of LGMD-R3, where SGCA gene delivery has already been accomplished [161].
Overall, our case study underscores the diagnostic value of WES in unselected patients with inherited myopathies and expands further their phenotypic and genotypic heterogeneity, by revealing novel pathogenic variants (such as the p.Thr57fs*2 MYOT change and the two LAMA2 variants) and diverse myopathic phenotypes in the Greek population for known pathogenic variants with multiple phenotypic presentations. Also, our results hint to the nearly impossible task of phenotypically and genotypically categorizing myopathies according to existing classification systems. This failure, in conjunction with the expected availability of gene therapies, calls for a new gene-based approach for classifying inherited myopathies.
Submission of data to a genetic database
All variants described herein have been uploaded to the LOVD database (https://www.lovd.nl/).
Declaration of Competing Interest
None.
References
- 1.Narayanaswami P., Weiss M., Selcen D., David W., Raynor E., Carter G. Evidence-based guideline summary: Diagnosis and treatment of limb-girdle and distal dystrophies. Neurology. 2014;83(16):1453–1463. doi: 10.1212/WNL.0000000000000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lilleker J.B., Keh Y.S., Roncaroli F., Sharma R., Roberts M. Metabolic myopathies: a practical approach. Pract. Neurol. 2018;18(1):14–26. doi: 10.1136/practneurol-2017-001708. [DOI] [PubMed] [Google Scholar]
- 3.González-Jamett A.M., Bevilacqua J.A., Cárdenas Díaz A.M. Hereditary myopathies. In: Sakuma K., editor. Muscle Cell and Tissue - Current Status of Research Field. IntechOpen; 2018. [Google Scholar]
- 4.Angelini C., Giaretta L., Marozzo R. An update on diagnostic options and considerations in limb-girdle dystrophies. Expert. Rev. Neurother. 2018;18(9):693–703. doi: 10.1080/14737175.2018.1508997. [DOI] [PubMed] [Google Scholar]
- 5.Liu W., Pajusalu S., Lake N.J., Zhou G., Ioannidis N., Mittal P. Estimating prevalence for limb-girdle muscular dystrophy basedon public sequencing databases. Genet. Med. 2019;21:2512–2520. doi: 10.1038/s41436-019-0544-8. [DOI] [PubMed] [Google Scholar]
- 6.Nallamilli B.R.R., Chakravorty S., Kesari A., Tanner A., Ankala A., Schneider T. Genetic landscape and novel disease mechanisms from a large LGMD cohort of 4656 patients. Ann. Clin. Transl. Neurol. 2018;5(12):1574–1587. doi: 10.1002/acn3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghaoui R., Cooper S.T., Lek M., Jones K., Corbett A., Reddel S.W. Use of whole-exome sequencing for diagnosis of limb-girdle muscular dystrophy: outcomes and lessons learned. JAMA Neurol. 2015;72(12):1424–1432. doi: 10.1001/jamaneurol.2015.2274. [DOI] [PubMed] [Google Scholar]
- 8.Taghizadeh E., Rezaee M., Barreto G.E., Sahebkar A. Prevalence, pathological mechanisms, and genetic basis of limb-girdle muscular dystrophies: A review. J. Cell. Physiol. 2019;234(6):7874–7884. doi: 10.1002/jcp.27907. [DOI] [PubMed] [Google Scholar]
- 9.Liewluck T., Milone M. Untangling the complexity of limb-girdle muscular dystrophies. Muscle Nerve. 2018;58(2):167–177. doi: 10.1002/mus.26077. [DOI] [PubMed] [Google Scholar]
- 10.Wang L., Zhang V.W., Li S., Li H., Sun Y., Li J. The clinical spectrum and genetic variability of limb-girdle muscular dystrophy in a cohort of Chinese patients. Orphanet J. Rare Dis. 2018;13(1):133. doi: 10.1186/s13023-018-0859-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nigro V., Savarese M. Genetic basis of limb-girdle muscular dystrophies: the 2014 update. Acta Myologica Myopathies Cardiomyopathies. 2014;33(1):1–12. [PMC free article] [PubMed] [Google Scholar]
- 12.Straub V., Murphy A., Udd B., Corrado A., Aymé S., Bönneman C. 229th ENMC international workshop: limb girdle muscular dystrophies: nomenclature and reformed classification. Naarden, the Netherlands, 19 March 2017. Neuromuscul. Disord. 2018;28(8):702–710. doi: 10.1016/j.nmd.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Sframeli M., Sarkozy A., Bertoli M., Astrea G., Hudson J., Scoto M. Congenital muscular dystrophies in the UK population: Clinical and molecular spectrum of a large cohort diagnosed over a 12-year period. Neuromuscul. Disord. 2017;27(9):793–803. doi: 10.1016/j.nmd.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Mercuri E., Muntoni F. Muscular dystrophies. Lancet. 2013;381(9869):845–860. doi: 10.1016/S0140-6736(12)61897-2. [DOI] [PubMed] [Google Scholar]
- 15.Harris E., Topf A., Barresi R., Hudson J., Powell H., Tellez J. Exome sequences versus sequential gene testing in the UK highly specialised Service for Limb Girdle Muscular Dystrophy. Orphanet J. Rare Dis. 2017;12(1):151. doi: 10.1186/s13023-017-0699-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy H.M., Cho K.-A., Lek M., Estrella E., Valkanas E., Jones M.D. The sensitivity of exome sequencing in identifying pathogenic mutations for LGMD in the United States. J. Hum. Genet. 2017;62(2):243–252. doi: 10.1038/jhg.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fichna J.P., Macias A., Piechota M., Korostyński M., Potulska-Chromik A., Redowicz M.J. Whole-exome sequencing identifies novel pathogenic mutations and putative phenotype-influencing variants in Polish limb-girdle muscular dystrophy patients. Hum. Genom. 2018;12(1):34. doi: 10.1186/s40246-018-0167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson K., Bertoli M., Phillips L., Töpf A., Van den Bergh P., Vissing J. Detection of variants in dystroglycanopathy-associated genes through the application of targeted whole-exome sequencing analysis to a large cohort of patients with unexplained limb-girdle muscle weakness. Skelet. Muscle. 2018;8(1):23. doi: 10.1186/s13395-018-0170-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonorazky H.D., Naumenko S., Ramani A.K., Nelakuditi V., Mashouri P., Wang P. Expanding the boundaries of RNA sequencing as a diagnostic tool for rare Mendelian disease. Am. J. Hum. Genet. 2019;104(3):466–483. doi: 10.1016/j.ajhg.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18(7):696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 21.Kopanos C., Tsiolkas V., Kouris A., Chapple C.E., Albarca Aguilera M., Meyer R. VarSome: the human genomic variant search engine. Bioinformatics. 2018;35(11):1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2018;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17(5):405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bevilacqua J.A., Guecaimburu Ehuletche M.d.R., Perna A., Dubrovsky A., Franca M.C., Vargas S. The Latin American experience with a next generation sequencing genetic panel for recessive limb-girdle muscular weakness and Pompe disease. Orphanet J. Rare Dis. 2020;15(1):11. doi: 10.1186/s13023-019-1291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benarroch E.E. 89(7) 2017. Anoctamins (TMEM16 Proteins). Functions and Involvement in Neurologic Disease; pp. 722–729. [DOI] [PubMed] [Google Scholar]
- 26.Bolduc V., Marlow G., Boycott K.M., Saleki K., Inoue H., Kroon J. Recessive mutations in the putative calcium-activated chloride channel anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am. J. Hum. Genet. 2010;86(2):213–221. doi: 10.1016/j.ajhg.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savarese M., Di Fruscio G., Tasca G., Ruggiero L., Janssens S., De Bleecker J. Next generation sequencing on patients with LGMD and nonspecific myopathies: findings associated with ANO5 mutations. Neuromuscul. Disord. 2015;25(7):533–541. doi: 10.1016/j.nmd.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hicks D., Sarkozy A., Muelas N., Köehler K., Huebner A., Hudson G. A founder mutation in Anoctamin 5 is a major cause of limb girdle muscular dystrophy. Brain. 2010;134(1):171–182. doi: 10.1093/brain/awq294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarkozy A., Hicks D., Hudson J., Laval S.H., Barresi R., Hilton-Jones D. ANO5 gene analysis in a large cohort of patients with anoctaminopathy: confirmation of male prevalence and high occurrence of the common exon 5 gene mutation. Hum. Mutat. 2013;34(8):1111–1118. doi: 10.1002/humu.22342. [DOI] [PubMed] [Google Scholar]
- 30.Penttilä S., Palmio J., Suominen T., Raheem O., Evilä A., Muelas Gomez N. Eight new mutations and the expanding phenotype variability in muscular dystrophy caused by ANO5. Neurology. 2012;78(12):897–903. doi: 10.1212/WNL.0b013e31824c4682. [DOI] [PubMed] [Google Scholar]
- 31.Sarkozy A., Deschauer M., Carlier R.-Y., Schrank B., Seeger J., Walter M.C. Muscle MRI findings in limb girdle muscular dystrophy type 2L. Neuromuscul. Disord. 2012;22:S122–S129. doi: 10.1016/j.nmd.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Wahbi K., Béhin A., Bécane H.M., Leturcq F., Cossée M., Laforêt P. Dilated cardiomyopathy in patients with mutations in anoctamin 5. Int. J. Cardiol. 2013;168(1):76–79. doi: 10.1016/j.ijcard.2012.09.070. [DOI] [PubMed] [Google Scholar]
- 33.Magri F., Del Bo R., D’Angelo M.G., Gandossini S., Corti S., Lucchini V. P2.50 LGMD2L in Italian population: New mutations and clinical and morphological aspects. Neuromuscul. Disord. 2011;21(9):675. [Google Scholar]
- 34.Zatz M., Starling A. Calpains and disease. N. Engl. J. Med. 2005;352(23):2413–2423. doi: 10.1056/NEJMra043361. [DOI] [PubMed] [Google Scholar]
- 35.Vissing J., Barresi R., Witting N., Van Ghelue M., Gammelgaard L., Bindoff L.A. A heterozygous 21-bp deletion in CAPN3 causes dominantly inherited limb girdle muscular dystrophy. Brain. 2016;139(8):2154–2163. doi: 10.1093/brain/aww133. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Thompson J.M., Niu Z., Tracy J.A., Moore S.A., Swenson A., Wieben E.D. Autosomal dominant calpainopathy due to heterozygous CAPN3 C.643_663del21. Muscle Nerve. 2018;57(4):679–683. doi: 10.1002/mus.25970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono Y., Ojima K., Shinkai-Ouchi F., Hata S., Sorimachi H. An eccentric calpain, CAPN3/p94/calpain-3. Biochimie. 2016;122:169–187. doi: 10.1016/j.biochi.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Richard I., Brenguier L., Dinçer P., Roudaut C., Bady B., Burgunder J.M. Multiple independent molecular etiology for limb-girdle muscular dystrophy type 2A patients from various geographical origins. Am. J. Hum. Genet. 1997;60(5):1128–1138. [PMC free article] [PubMed] [Google Scholar]
- 39.Balci B., Aurino S., Haliloglu G., Talim B., Erdem S., Akcören Z. Calpain-3 mutations in Turkey. Eur. J. Pediatr. 2006;165(5):293–298. doi: 10.1007/s00431-005-0046-3. [DOI] [PubMed] [Google Scholar]
- 40.Milic A., Canki-Klain N. Calpainopathy (LGMD2A) in Croatia: molecular and haplotype analysis. Croat. Med. J. 2005;46(4):657–663. [PubMed] [Google Scholar]
- 41.Todorova A., Georgieva B., Tournev I., Todorov T., Bogdanova N., Mitev V. A large deletion and novel point mutations in the calpain 3 gene (CAPN3) in Bulgarian LGMD2A patients. Neurogenetics. 2007;8(3):225–229. doi: 10.1007/s10048-007-0083-3. [DOI] [PubMed] [Google Scholar]
- 42.Papadopoulos C., Papadimas G.K., Kekou K., Spengos K., Svigou M., Kitsiou-Tzeli S. Caveolinopathies in Greece. Neurologist. 2015;20(1):8–12. doi: 10.1097/NRL.0000000000000036. [DOI] [PubMed] [Google Scholar]
- 43.Scalco R.S., Gardiner A.R., Pitceathly R.D.S., Hilton-Jones D., Schapira A.H., Turner C. CAV3 mutations causing exercise intolerance, myalgia and rhabdomyolysis: expanding the phenotypic spectrum of caveolinopathies. Neuromuscul. Disord. 2016;26(8):504–510. doi: 10.1016/j.nmd.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 44.Gazzerro E., Sotgia F., Bruno C., Lisanti M.P., Minetti C. Caveolinopathies: from the biology of caveolin-3 to human diseases. Eur. J. Hum. Genet. 2010;18(2):137–145. doi: 10.1038/ejhg.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vatta M., Ackerman M.J., Ye B., Makielski J.C., Ughanze E.E., Taylor E.W. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114(20):2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]
- 46.Andreasen C., Refsgaard L., Nielsen J.B., Sajadieh A., Winkel B.G., Tfelt-Hansen J. Mutations in genes encoding cardiac ion channels previously associated with sudden infant death syndrome (SIDS) are present with high frequency in new exome data. Can. J. Cardiol. 2013;29(9):1104–1109. doi: 10.1016/j.cjca.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Cronk L.B., Ye B., Kaku T., Tester D.J., Vatta M., Makielski J.C. Novel mechanism for sudden infant death syndrome: persistent late sodium current secondary to mutations in caveolin-3. Heart Rhythm. 2007;4(2):161–166. doi: 10.1016/j.hrthm.2006.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donkervoort S., Schindler A., Tesi-Rocha C., Schreiber A., Leach M.E., Dastgir J. ‘Double trouble’: diagnostic challenges in Duchenne muscular dystrophy in patients with an additional hereditary skeletal dysplasia. Neuromuscul. Disord. 2013;23(12):955–961. doi: 10.1016/j.nmd.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hedley P.L., Kanters J.K., Dembic M., Jespersen T., Skibsbye L., Aidt F.H. The role of CAV3 in long QT syndrome. Circ. Cardiovasc. Genet. 2013;6(5):452–461. doi: 10.1161/CIRCGENETICS.113.000137. [DOI] [PubMed] [Google Scholar]
- 50.Refsgaard L., Holst A.G., Sadjadieh G., Haunsø S., Nielsen J.B., Olesen M.S. High prevalence of genetic variants previously associated with LQT syndrome in new exome data. Eur. J. Hum. Genet. 2012;20(8):905–908. doi: 10.1038/ejhg.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ricci G., Scionti I., Alì G., Volpi L., Zampa V., Fanin M. Rippling muscle disease and facioscapulohumeral dystrophy-like phenotype in a patient carrying a heterozygous CAV3 T78M mutation and a D4Z4 partial deletion: further evidence for “double trouble” overlapping syndromes. Neuromuscul. Disord. 2012;22(6):534–540. doi: 10.1016/j.nmd.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Flores J.L., Fakhro K., Hackett N.R., Salit J., Fuller J., Agosto-Perez F. Exome sequencing identifies potential risk variants for Mendelian disorders at high prevalence in Qatar. Hum. Mutat. 2014;35(1):105–116. doi: 10.1002/humu.22460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Traverso M., Gazzerro E., Assereto S., Sotgia F., Biancheri R., Stringara S. Caveolin-3 T78M and T78K missense mutations lead to different phenotypes in vivo and in vitro. Lab. Investig. 2008;88(3):275–283. doi: 10.1038/labinvest.3700713. [DOI] [PubMed] [Google Scholar]
- 54.Vaidyanathan R., Vega A.L., Song C., Zhou Q., Tan B., Berger S. The interaction of caveolin 3 protein with the potassium inward rectifier channel Kir2.1: physiology and pathology related to long QT syndrome 9 (LQT9) J. Biol. Chem. 2013;288(24):17472–17480. doi: 10.1074/jbc.M112.435370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spadafora P., Liguori M., Andreoli V., Quattrone A., Gambardella A. CAV3 T78M mutation as polymorphic variant in South Italy. Neuromuscul. Disord. 2012;22(7):669–670. doi: 10.1016/j.nmd.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Ghouse J., Have C.T., Weeke P., Bille Nielsen J., Ahlberg G., Balslev-Harder M. Rare genetic variants previously associated with congenital forms of long QT syndrome have little or no effect on the QT interval. Eur. Heart J. 2015;36(37):2523–2529. doi: 10.1093/eurheartj/ehv297. [DOI] [PubMed] [Google Scholar]
- 57.Campostrini G., Bonzanni M., Lissoni A., Bazzini C., Milanesi R., Vezzoli E. The expression of the rare caveolin-3 variant T78M alters cardiac ion channels function and membrane excitability. Cardiovasc. Res. 2017;113(10):1256–1265. doi: 10.1093/cvr/cvx122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matthews E., Fialho D., Tan S.V., Venance S.L., Cannon S.C., Sternberg D. The non-dystrophic myotonias: molecular pathogenesis, diagnosis and treatment. Brain. 2009;133(1):9–22. doi: 10.1093/brain/awp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heatwole C.R., Statland J.M., Logigian E.L. The diagnosis and treatment of myotonic disorders. Muscle Nerve. 2013;47(5):632–648. doi: 10.1002/mus.23683. [DOI] [PubMed] [Google Scholar]
- 60.Stunnenberg B., LoRusso S., Arnold W.D., Barohn R.J., Cannon S.C., Fontaine B. Guidelines on clinical presentation and management of non-dystrophic myotonias. Muscle Nerve. 2020;62:430–444. doi: 10.1002/mus.26887. n/a(n/a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.George A., Sloan-Brown K., Fenichel G., Mitchell G., Spiegel R., Pascuzzi R. Nonsense and missense mutations of the muscle chloride channel gene in patients with myotonia congenita. Hum. Mol. Genet. 1994;3(11):2071–2072. [PubMed] [Google Scholar]
- 62.Meyer-Kleine C., Steinmeyer K., Ricker K., Jentsch T.J., Koch M.C. Spectrum of mutations in the major human skeletal muscle chloride channel gene (CLCN1) leading to myotonia. Am. J. Hum. Genet. 1995;57(6):1325–1334. [PMC free article] [PubMed] [Google Scholar]
- 63.Tincheva S., Georgieva B., Todorov T., Savov A., Tsaneva S., Litvinenko I. Myotonia congenita type Becker in Bulgaria: first genetically proven cases and mutation screening of two presumable endemic regions. Neuromuscul. Disord. 2016;26(10):675–680. doi: 10.1016/j.nmd.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 64.Lucchiari S., Ulzi G., Magri F., Bucchia M., Corbetta F., Servida M. Clinical evaluation and cellular electrophysiology of a recessive CLCN1 patient. J. Physiol. Pharmacol. 2013;64(5):669–678. [PubMed] [Google Scholar]
- 65.Vindas-Smith R., Fiore M., Vásquez M., Cuenca P., del Valle G., Lagostena L. Identification and functional characterization of CLCN1 mutations found in nondystrophic myotonia patients. Hum. Mutat. 2016;37(1):74–83. doi: 10.1002/humu.22916. [DOI] [PubMed] [Google Scholar]
- 66.Sigauke E., Rakheja D., Kitson K., Bennett M.J. Carnitine palmitoyltransferase II deficiency: a clinical, biochemical, and molecular review. Lab. Investig. 2003;83(11):1543–1554. doi: 10.1097/01.lab.0000098428.51765.83. [DOI] [PubMed] [Google Scholar]
- 67.Tajima G., Hara K., Yuasa M. Carnitine palmitoyltransferase II deficiency with a focus on newborn screening. J. Hum. Genet. 2019;64(2):87–98. doi: 10.1038/s10038-018-0530-z. [DOI] [PubMed] [Google Scholar]
- 68.Rufer A.C., Thoma R., Hennig M. Structural insight into function and regulation of carnitine palmitoyltransferase. Cell. Mol. Life Sci. 2009;66(15):2489–2501. doi: 10.1007/s00018-009-0035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fanin M., Anichini A., Cassandrini D., Fiorillo C., Scapolan S., Minetti C. Allelic and phenotypic heterogeneity in 49 Italian patients with the muscle form of CPT-II deficiency. Clin. Genet. 2012;82(3):232–239. doi: 10.1111/j.1399-0004.2011.01786.x. [DOI] [PubMed] [Google Scholar]
- 70.Taroni F., Verderio E., Dworzak F., Willems P.J., Cavadini P., DiDonato S. Identification of a common mutation in the carnitine palmitoyltransferase II gene in familial recurrent myoglobinuria patients. Nat. Genet. 1993;4(3):314–320. doi: 10.1038/ng0793-314. [DOI] [PubMed] [Google Scholar]
- 71.Balasubramanian M., Jenkins T.M., Kirk R.J., Nesbitt I.M., Olpin S.E., Hill M. Recurrent rhabdomyolysis caused by carnitine palmitoyltransferase II deficiency, common but under-recognised: Lessons to be learnt. Mol. Genet. Metabol. Rep. 2018;15:69–70. doi: 10.1016/j.ymgmr.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vivante A., Ityel H., Pode-Shakked B., Chen J., Shril S., van der Ven A.T. Exome sequencing in Jewish and Arab patients with rhabdomyolysis reveals single-gene etiology in 43% of cases. Pediatr. Nephrol. 2017;32(12):2273–2282. doi: 10.1007/s00467-017-3755-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joshi P.R., Deschauer M., Zierz S. Clinically symptomatic heterozygous carnitine palmitoyltransferase II (CPT II) deficiency. Wien. Klin. Wochenschr. 2012;124(23):851–854. doi: 10.1007/s00508-012-0296-9. [DOI] [PubMed] [Google Scholar]
- 74.Patel N.J., Van Dyke K.W., Espinoza L.R. Limb-girdle muscular dystrophy 2B and miyoshi presentations of dysferlinopathy. Am J Med Sci. 2017;353(5):484–491. doi: 10.1016/j.amjms.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 75.Cacciottolo M., Numitone G., Aurino S., Caserta I.R., Fanin M., Politano L. Muscular dystrophy with marked Dysferlin deficiency is consistently caused by primary dysferlin gene mutations. Eur. J. Hum. Genet. 2011;19(9):974–980. doi: 10.1038/ejhg.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amato A.A., Brown R.H. Chapter 7 - dysferlinopathies. In: Griggs R.C., Amato A.A., editors. Handbook of Clinical Neurology. Elsevier; 2011. pp. 111–118. [Google Scholar]
- 77.Gallardo E., de Luna N., Diaz-Manera J., Rojas-García R., Gonzalez-Quereda L., Flix B. Comparison of dysferlin expression in human skeletal muscle with that in monocytes for the diagnosis of dysferlin myopathy. PLoS One. 2011;6(12):e29061. doi: 10.1371/journal.pone.0029061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nguyen K., Bassez G., Bernard R., Krahn M., Labelle V., Figarella-Branger D. Dysferlin mutations in LGMD2B, Miyoshi myopathy, and atypical dysferlinopathies. Hum. Mutat. 2005;26(2):165. doi: 10.1002/humu.9355. [DOI] [PubMed] [Google Scholar]
- 79.Nguyen K., Bassez G., Krahn M., Bernard R., Laforêt P., Labelle V. Phenotypic study in 40 patients with dysferlin gene mutations: high frequency of atypical phenotypes. Arch. Neurol. 2007;64(8):1176–1182. doi: 10.1001/archneur.64.8.1176. [DOI] [PubMed] [Google Scholar]
- 80.Paradas C., González-Quereda L., De Luna N., Gallardo E., García-Consuegra I., Gómez H. A new phenotype of dysferlinopathy with congenital onset. Neuromuscul. Disord. 2009;19(1):21–25. doi: 10.1016/j.nmd.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 81.Ceyhan-Birsoy O., Talim B., Swanson L.C., Karakaya M., Graff M.A., Beggs A.H. Whole exome sequencing reveals DYSF, FKTN, and ISPD mutations in congenital muscular dystrophy without brain or eye involvement. J. Neuromusc. Dis. 2015;2:87–92. doi: 10.3233/JND-140038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leshinsky-Silver E., Argov Z., Rozenboim L., Cohen S., Tzofi Z., Cohen Y. Dysferlinopathy in the Jews of the Caucasus: a frequent mutation in the dysferlin gene. Neuromuscul. Disord. 2007;17(11):950–954. doi: 10.1016/j.nmd.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 83.Silva A.M.S., Mendonça R.H., Soares D.C., Callegaro D., Caldas V.M., Perissinotti I.N. Pearls and Oy-sters: a curable myopathy manifesting as exercise intolerance and respiratory failure. Neurology. 2018;91(4):187–190. doi: 10.1212/WNL.0000000000005867. [DOI] [PubMed] [Google Scholar]
- 84.Gempel K., Topaloglu H., Talim B., Schneiderat P., Schoser B.G.H., Hans V.H. The myopathic form of coenzyme Q10 deficiency is caused by mutations in the electron-transferring-flavoprotein dehydrogenase (ETFDH) gene. Brain. 2007;130(8):2037–2044. doi: 10.1093/brain/awm054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sahai I., Garganta C.L., Bailey J., James P., Levy H.L., Martin M. Newborn screening for glutaric aciduria-II: the New England experience. In: Zschocke J., editor. JIMD Reports - Case and Research Reports. Vol. 13. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cornelius N., Frerman F.E., Corydon T.J., Palmfeldt J., Bross P., Gregersen N. Molecular mechanisms of riboflavin responsiveness in patients with ETF-QO variations and multiple acyl-CoA dehydrogenation deficiency. Hum. Mol. Genet. 2012;21(15):3435–3448. doi: 10.1093/hmg/dds175. [DOI] [PubMed] [Google Scholar]
- 87.Olsen R.K.J., Olpin S.E., Andresen B.S., Miedzybrodzka Z.H., Pourfarzam M., Merinero B. ETFDH mutations as a major cause of riboflavin-responsive multiple acyl-CoA dehydrogenation deficiency. Brain. 2007;130(8):2045–2054. doi: 10.1093/brain/awm135. [DOI] [PubMed] [Google Scholar]
- 88.van der Westhuizen F.H., Smuts I., Honey E., Louw R., Schoonen M., Jonck L.-M. A novel mutation in ETFDH manifesting as severe neonatal-onset multiple acyl-CoA dehydrogenase deficiency. J. Neurol. Sci. 2018;384:121–125. doi: 10.1016/j.jns.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 89.Yotsumoto Y., Hasegawa Y., Fukuda S., Kobayashi H., Endo M., Fukao T. Clinical and molecular investigations of Japanese cases of glutaric acidemia type 2. Mol. Genet. Metab. 2008;94(1):61–67. doi: 10.1016/j.ymgme.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 90.Papadimas G.K., Papadimas G.K., Spengos K., Papadopoulos C., Manta P. Late onset glycogen storage disease type II: pitfalls in the diagnosis. Eur. Neurol. 2012;67(2):65–68. doi: 10.1159/000334398. [DOI] [PubMed] [Google Scholar]
- 91.Golsari A., Nasimzadah A., Thomalla G., Keller S., Gerloff C., Magnus T. Prevalence of adult Pompe disease in patients with proximal myopathic syndrome and undiagnosed muscle biopsy. Neuromuscul. Disord. 2018;28(3):257–261. doi: 10.1016/j.nmd.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 92.Johnson K., Töpf A., Bertoli M., Phillips L., Claeys K.G., Stojanovic V.R. Identification of GAA variants through whole exome sequencing targeted to a cohort of 606 patients with unexplained limb-girdle muscle weakness. Orphanet J. Rare Dis. 2017;12(1):173. doi: 10.1186/s13023-017-0722-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mori M., Haskell G., Kazi Z., Zhu X., DeArmey S.M., Goldstein J.L. Sensitivity of whole exome sequencing in detecting infantile- and late-onset Pompe disease. Mol. Genet. Metab. 2017;122(4):189–197. doi: 10.1016/j.ymgme.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boentert M., Prigent H., Várdi K., Jones H.N., Mellies U., Simonds A.K. Practical recommendations for diagnosis and management of respiratory muscle weakness in late-onset Pompe disease. Int. J. Mol. Sci. 2016;17(10):1735. doi: 10.3390/ijms17101735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hule M.L., Chen A.S., Tsujino S., Shanske S., DiMauro S., Engel A.G. Aberrant splicing in adult onset glycogen storage disease type II (GSDII): molecular identification of an IVS1 (– 13T→G) mutation in a majority of patients and a novel IVS10 (+ 1GT → CT) mutation. Hum. Mol. Genet. 1994;3(12):2231–2236. doi: 10.1093/hmg/3.12.2231. [DOI] [PubMed] [Google Scholar]
- 96.Gort L., Coll M.J., Chabás A. Glycogen storage disease type II in Spanish patients: high frequency of c.1076-1G>C mutation. Mol. Genet. Metab. 2007;92(1):183–187. doi: 10.1016/j.ymgme.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 97.Gutiérrez-Rivas E., Bautista J., Vílchez J.J., Muelas N., Díaz-Manera J., Illa I. Targeted screening for the detection of Pompe disease in patients with unclassified limb-girdle muscular dystrophy or asymptomatic hyperCKemia using dried blood: a Spanish cohort. Neuromuscul. Disord. 2015;25(7):548–553. doi: 10.1016/j.nmd.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 98.Löscher W.N., Huemer M., Stulnig T.M., Simschitz P., Iglseder S., Eggers C. Pompe disease in Austria: clinical, genetic and epidemiological aspects. J. Neurol. 2018;265(1):159–164. doi: 10.1007/s00415-017-8686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boerkoel C.F., Exelbert R., Nicastri C., Nichols R.C., Miller F.W., Plotz P.H. Leaky splicing mutation in the acid maltase gene is associated with delayed onset of glycogenosis type II. Am. J. Hum. Genet. 1995;56(4):887–897. [PMC free article] [PubMed] [Google Scholar]
- 100.Castro-Gago M., Eirís-Puñal J., Rodríguez-Núñez A., Pintos-Martínez E., Benlloch-Marín T., Barros-Angueira F. Severe form of juvenile type II glycogenosis in a compound-heterozygous boy (Tyr-292--> Cys/Arg-854-->Stop) Rev. Neurol. 1999;29(1):46–49. [PubMed] [Google Scholar]
- 101.Lee D.-H., Qiu W.-J., Lee J., Chien Y.-H., Hwu W.-L. Hypertrophic cardiomyopathy in pompe disease is not limited to the classic infantile-onset phenotype. In: Zschocke J., editor. JIMD Reports. Vol. 17. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. pp. 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park H., Lee D., Choi T., Lee Y., Lee S., Kim J. Three patients with glycogen storage disease type II and the mutational spectrum of GAA in Korean patients. Ann. Clin. Lab. Sci. 2013;43(3):311–316. [PubMed] [Google Scholar]
- 103.Hermans M.M.P., Leenen D.V., Kroos M.A., Beesley C.E., Van der Ploeg A.T., Sakuraba H. Twenty-two novel mutations in the lysosomal α-glucosidase gene (GAA) underscore the genotype–phenotype correlation in glycogen storage disease type II. Hum. Mutat. 2004;23(1):47–56. doi: 10.1002/humu.10286. [DOI] [PubMed] [Google Scholar]
- 104.Flanagan J.J., Rossi B., Tang K., Wu X., Mascioli K., Donaudy F. The pharmacological chaperone 1-deoxynojirimycin increases the activity and lysosomal trafficking of multiple mutant forms of acid alpha-glucosidase. Hum. Mutat. 2009;30(12):1683–1692. doi: 10.1002/humu.21121. [DOI] [PubMed] [Google Scholar]
- 105.Cirak S., Foley A.R., Herrmann R., Willer T., Yau S., Stevens E. ISPD gene mutations are a common cause of congenital and limb-girdle muscular dystrophies. Brain. 2013;136(1):269–281. doi: 10.1093/brain/aws312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Willer T., Lee H., Lommel M., Yoshida-Moriguchi T., de Bernabe D.B.V., Venzke D. ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. Nat. Genet. 2012;44(5):575–580. doi: 10.1038/ng.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tasca G., Moro F., Aiello C., Cassandrini D., Fiorillo C., Bertini E. Limb-girdle muscular dystrophy with α-dystroglycan deficiency and mutations in the <em>ISPD</em> gene. Neurology. 2013;80(10):963–965. doi: 10.1212/WNL.0b013e3182840cbc. [DOI] [PubMed] [Google Scholar]
- 108.Baranello G., Saredi S., Sansanelli S., Savadori P., Canioni E., Chiapparini L. A novel homozygous ISPD gene mutation causing phenotype variability in a consanguineous family. Neuromuscul. Disord. 2015;25(1):55–59. doi: 10.1016/j.nmd.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 109.Roscioli T., Kamsteeg E.-J., Buysse K., Maystadt I., van Reeuwijk J., van den Elzen C. Mutations in ISPD cause Walker-Warburg syndrome and defective glycosylation of α-dystroglycan. Nat. Genet. 2012;44(5):581–585. doi: 10.1038/ng.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Magri F., Colombo I., Del Bo R., Previtali S., Brusa R., Ciscato P. ISPD mutations account for a small proportion of Italian Limb Girdle Muscular Dystrophy cases. BMC Neurol. 2015;15(1):172. doi: 10.1186/s12883-015-0428-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brancaccio A. A molecular overview of the primary dystroglycanopathies. J. Cell. Mol. Med. 2019;23(5):3058–3062. doi: 10.1111/jcmm.14218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vuillaumier-Barrot S., Bouchet-Séraphin C., Chelbi M., Devisme L., Quentin S., Gazal S. Identification of mutations in TMEM5 and ISPD as a cause of severe cobblestone lissencephaly. Am. J. Hum. Genet. 2012;91(6):1135–1143. doi: 10.1016/j.ajhg.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oliveira J., Gruber A., Cardoso M., Taipa R., Fineza I., Gonçalves A. LAMA2 gene mutation update: Toward a more comprehensive picture of the laminin-α2 variome and its related phenotypes. Hum. Mutat. 2018;39(10):1314–1337. doi: 10.1002/humu.23599. [DOI] [PubMed] [Google Scholar]
- 114.van der Knaap M.S., Smit L.M.E., Barth P.G., Catsman-Berrevoets C.E., Brouwer O.F., Begeer J.H. Magnetic resonance imaging in classification of congenital muscular dystrophies with brain abnormalities. Ann. Neurol. 1997;42(1):50–59. doi: 10.1002/ana.410420110. [DOI] [PubMed] [Google Scholar]
- 115.Kang S.-m., Yoon M.-H., Park B.-J. Laminopathies; mutations on single gene and various human genetic diseases. BMB Rep. 2018;51(7):327–337. doi: 10.5483/BMBRep.2018.51.7.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Madej-Pilarczyk A., Kochański A. Emery-Dreifuss muscular dystrophy: the most recognizable laminopathy. Folia Neuropathol. 2016;54(1):1–8. doi: 10.5114/fn.2016.58910. [DOI] [PubMed] [Google Scholar]
- 117.Peretto G., Di Resta C., Perversi J., Forleo C., Maggi L., Politano L. Cardiac and neuromuscular features of patients with LMNA-related cardiomyopathy. Ann. Intern. Med. 2019;171(7):458–463. doi: 10.7326/M18-2768. [DOI] [PubMed] [Google Scholar]
- 118.Captur G., Arbustini E., Bonne G., Syrris P., Mills K., Wahbi K. Lamin and the heart. Heart. 2018;104(6):468–479. doi: 10.1136/heartjnl-2017-312338. [DOI] [PubMed] [Google Scholar]
- 119.Ditaranto R., Boriani G., Biffi M., Lorenzini M., Graziosi M., Ziacchi M. Differences in cardiac phenotype and natural history of laminopathies with and without neuromuscular onset. Orphanet J. Rare Dis. 2019;14(1):263. doi: 10.1186/s13023-019-1245-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brayson D., Shanahan C.M. Current insights into LMNA cardiomyopathies: existing models and missing LINCs. Nucleus. 2017;8(1):17–33. doi: 10.1080/19491034.2016.1260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Raffaele di Barletta M., Ricci E., Galluzzi G., Tonali P., Mora M., Morandi L. Different mutations in the LMNA gene cause autosomal dominant and autosomal recessive Emery-Dreifuss muscular dystrophy. Am. J. Hum. Genet. 2000;66(4):1407–1412. doi: 10.1086/302869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Coutance G., Labombarda F., Cauderlier E., Belin A., Richard P., Bonne G. Hypoplasia of the aorta in a patient diagnosed with LMNA gene mutation. Congenit. Heart Dis. 2013;8(4):E127–E129. doi: 10.1111/j.1747-0803.2012.00695.x. [DOI] [PubMed] [Google Scholar]
- 123.Emerson L.J., Holt M.R., Wheeler M.A., Wehnert M., Parsons M., Ellis J.A. Defects in cell spreading and ERK1/2 activation in fibroblasts with lamin A/C mutations. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2009;1792(8):810–821. doi: 10.1016/j.bbadis.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 124.Ki C.S., Hong J.S., Jeong G.Y., Ahn K.J., Choi K.M., Kim D.K. Identification of lamin A/C (LMNA) gene mutations in Korean patients with autosomal dominant Emery-Dreifuss muscular dystrophy and limb-girdle muscular dystrophy 1B. J. Hum. Genet. 2002;47(5):225–228. doi: 10.1007/s100380200029. [DOI] [PubMed] [Google Scholar]
- 125.Komaki H., Hayashi Y.K., Tsuburaya R., Sugie K., Kato M., Nagai T. Inflammatory changes in infantile-onset LMNA-associated myopathy. Neuromuscul. Disord. 2011;21(8):563–568. doi: 10.1016/j.nmd.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 126.Mitsuhashi H., Hayashi Y.K., Matsuda C., Noguchi S., Wakatsuki S., Araki T. Specific phosphorylation of Ser458 of A-type lamins in LMNA-associated myopathy patients. J. Cell Sci. 2010;123(22):3893–3900. doi: 10.1242/jcs.072157. [DOI] [PubMed] [Google Scholar]
- 127.Park H.J., Jang H., Kim J.H., Lee J.H., Shin H.Y., Kim S.M. Discovery of pathogenic variants in a large Korean cohort of inherited muscular disorders. Clin. Genet. 2017;91(3):403–410. doi: 10.1111/cge.12826. [DOI] [PubMed] [Google Scholar]
- 128.Muchir A., Bonne G., van der Kooi A.J., van Meegen M., Baas F., Bolhuis P.A. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B) Hum. Mol. Genet. 2000;9(9):1453–1459. doi: 10.1093/hmg/9.9.1453. [DOI] [PubMed] [Google Scholar]
- 129.Hao H., Bonilla E., Manfredi G., DiMauro S., Moraes C.T. Segregation patterns of a novel mutation in the mitochondrial tRNA glutamic acid gene associated with myopathy and diabetes mellitus. Am. J. Hum. Genet. 1995;56(5):1017–1025. [PMC free article] [PubMed] [Google Scholar]
- 130.Mancuso M., Ferraris S., Nishigaki Y., Azan G., Mauro A., Sammarco P. Congenital or late-onset myopathy in patients with the T14709C mtDNA mutation. J. Neurol. Sci. 2005;228(1):93–97. doi: 10.1016/j.jns.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 131.Damore M., Speiser P., Slonim A., New M., Shanske S., Xia W. Early onset of diabetes mellitus associated with the mitochondrial DNA T14709C point mutation: patient report and literature review. J. Pediatr. Endocrinol. Metab. 1999;12(2):207–213. doi: 10.1515/jpem.1999.12.2.207. [DOI] [PubMed] [Google Scholar]
- 132.Hanna M.G., Nelson I., Sweeney M.G., Cooper J.M., Watkins P.J., Morgan-Hughes J.A. Congenital encephalomyopathy and adult-onset myopathy and diabetes mellitus: different phenotypic associations of a new heteroplasmic mtDNA tRNA glutamic acid mutation. Am. J. Hum. Genet. 1995;56(5):1026–1033. [PMC free article] [PubMed] [Google Scholar]
- 133.McFarland R., Schaefer A.M., Gardner J.L., Lynn S., Hayes C.M., Barron M.J. Familial myopathy: new insights into the T14709C mitochondrial tRNA mutation. Ann. Neurol. 2004;55(4):478–484. doi: 10.1002/ana.20004. [DOI] [PubMed] [Google Scholar]
- 134.Vialettes B., Paquis-Flucklinger V., Pelissier J., Bendahan D., Narbonne H., Silvestre-Aillaud P. Phenotypic expression of diabetes secondary to a T14709C mutation of mitochondrial DNA. Comparison with MIDD syndrome (A3243G mutation): a case report. Diabetes Care. 1997;20(11):1731–1737. doi: 10.2337/diacare.20.11.1731. [DOI] [PubMed] [Google Scholar]
- 135.Rigoli L., Prisco F., Caruso R., Lafusco D., Ursomanno G., Zuccarello D. Association of the T14709C mutation of mitochondrial DNA with maternally inherited diabetes mellitus and/or deafness in an Italian family. Diabet. Med. 2001;18(4):334–336. doi: 10.1046/j.1464-5491.2001.00429-2.x. [DOI] [PubMed] [Google Scholar]
- 136.Gorman G.S., Schaefer A.M., Ng Y., Gomez N., Blakely E.L., Alston C.L. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann. Neurol. 2015;77(5):753–759. doi: 10.1002/ana.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lax N.Z., Hepplewhite P.D., Reeve A.K., Nesbitt V., McFarland R., Jaros E. Cerebellar ataxia in patients with mitochondrial DNA disease: a molecular clinicopathological study. J. Neuropathol. Exp. Neurol. 2012;71(2):148–161. doi: 10.1097/NEN.0b013e318244477d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lax N.Z., Pienaar I.S., Reeve A.K., Hepplewhite P.D., Jaros E., Taylor R.W. Microangiopathy in the cerebellum of patients with mitochondrial DNA disease. Brain. 2012;135(6):1736–1750. doi: 10.1093/brain/aws110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mezghani N., Mkaouar-Rebai E., Mnif M., Charfi N., Rekik N., Youssef S. The heteroplasmic m.14709T>C mutation in the tRNAGlu gene in two Tunisian families with mitochondrial diabetes. J. Diabetes Complicat. 2010;24(4):270–277. doi: 10.1016/j.jdiacomp.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 140.Perucca-Lostanlen D., Taylor R.W., Narbonne H., Mousson de Camaret B., Hayes C.M., Saunieres A. Molecular and functional effects of the T14709C point mutation in the mitochondrial DNA of a patient with maternally inherited diabetes and deafness. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2002;1588(3):210–216. doi: 10.1016/s0925-4439(02)00167-9. [DOI] [PubMed] [Google Scholar]
- 141.Fichna J.P., Maruszak A., Żekanowski C. Myofibrillar myopathy in the genomic context. J. Appl. Genet. 2018;59(4):431–439. doi: 10.1007/s13353-018-0463-4. [DOI] [PubMed] [Google Scholar]
- 142.Olivé M., Goldfarb L.G., Shatunov A., Fischer D., Ferrer I. Myotilinopathy: refining the clinical and myopathological phenotype. Brain. 2005;128(10):2315–2326. doi: 10.1093/brain/awh576. [DOI] [PubMed] [Google Scholar]
- 143.Selcen D., Engel A.G. Mutations in myotilin cause myofibrillar myopathy. Neurology. 2004;62(8):1363–1371. doi: 10.1212/01.wnl.0000123576.74801.75. [DOI] [PubMed] [Google Scholar]
- 144.Claeys K.G., Fardeau M., Schröder R., Suominen T., Tolksdorf K., Behin A. Electron microscopy in myofibrillar myopathies reveals clues to the mutated gene. Neuromuscul. Disord. 2008;18(8):656–666. doi: 10.1016/j.nmd.2008.06.367. [DOI] [PubMed] [Google Scholar]
- 145.Ramos-Fransi A., Martínez-Piñeiro A., Almendrote M., Lucente G., Carrato C., Ballester-Lopez A. Myotilinopathy unmasked by statin treatment: a case report. Muscle Nerve. 2018;57(6):E138–E140. doi: 10.1002/mus.26078. [DOI] [PubMed] [Google Scholar]
- 146.Hauser M.A., Horrigan S.K., Salmikangas P., Torian U.M., Kristi D., Viles R. Dancel. Myotilin is mutated in limb girdle muscular dystrophy 1A. Hum. Mol. Genet. 2000;9(14):2141–2147. doi: 10.1093/hmg/9.14.2141. [DOI] [PubMed] [Google Scholar]
- 147.Mojbafan M., Bahmani R., Bagheri S.D., Sharifi Z., Zeinali S. Mutational spectrum of autosomal recessive limb-girdle muscular dystrophies in a cohort of 112 Iranian patients and reporting of a possible founder effect. Orphanet J. Rare Dis. 2020;15(1):14. doi: 10.1186/s13023-020-1296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Belhasan D.C., Akaaboune M. The role of the dystrophin glycoprotein complex on the neuromuscular system. Neurosci. Lett. 2020;722 doi: 10.1016/j.neulet.2020.134833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bhat H.F., Mir S.S., Dar K.B., Bhat Z.F., Shah R.A., Ganai N.A. ABC of multifaceted dystrophin glycoprotein complex (DGC) J. Cell. Physiol. 2018;233(7):5142–5159. doi: 10.1002/jcp.25982. [DOI] [PubMed] [Google Scholar]
- 150.Gawor M., Prószyński T.J. The molecular cross talk of the dystrophin–glycoprotein complex. Ann. N. Y. Acad. Sci. 2018;1412(1):62–72. doi: 10.1111/nyas.13500. [DOI] [PubMed] [Google Scholar]
- 151.Bianchini E., Fanin M., Mamchaoui K., Betto R., Sandonà D. Unveiling the degradative route of the V247M α-sarcoglycan mutant responsible for LGMD-2D. Hum. Mol. Genet. 2014;23(14):3746–3758. doi: 10.1093/hmg/ddu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tarnopolsky M., Hoffman E., Giri M., Shoffner J., Brady L. Alpha-sarcoglycanopathy presenting as exercise intolerance and rhabdomyolysis in two adults. Neuromuscul. Disord. 2015;25(12):952–954. doi: 10.1016/j.nmd.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 153.Piccolo F., Roberds S.L., Jeanpierre M., Leturcq F., Azibi K., Beldjord C. Primary adhalinopathy: a common cause of autosomal recessive muscular dystrophy of variable severity. Nat. Genet. 1995;10(2):243–245. doi: 10.1038/ng0695-243. [DOI] [PubMed] [Google Scholar]
- 154.Walter M., Dekomien G., Schlotter-Weigel B., Reilich P., Pongratz D., Müller-Felber W. Respiratory insufficiency as a presenting symptom of LGMD2D in adulthood. Acta Myol. 2004;23(1):1–5. [PubMed] [Google Scholar]
- 155.Soheili T., Gicquel E., Poupiot J., N'Guyen L., Le Roy F., Bartoli M. Rescue of sarcoglycan mutations by inhibition of endoplasmic reticulum quality control is associated with minimal structural modifications. Hum. Mutat. 2012;33(2):429–439. doi: 10.1002/humu.21659. [DOI] [PubMed] [Google Scholar]
- 156.Duggan D.J., Gorospe J.R., Fanin M., Hoffman E.P., Angelini C., Pegoraro E. Mutations in the sarcoglycan genes in patients with myopathy. N. Engl. J. Med. 1997;336(9):618–625. doi: 10.1056/NEJM199702273360904. [DOI] [PubMed] [Google Scholar]
- 157.Mongini T., Doriguzzi C., Bosone I., Chiadò-Piat L., Hoffman E.P., Palmucci L. Alpha-sarcoglycan deficiency featuring exercise intolerance and myoglobinuria. Neuropediatrics. 2002;33(02):109–111. doi: 10.1055/s-2002-32374. [DOI] [PubMed] [Google Scholar]
- 158.Avila De Salman S., Taratuto A.L., Dekomien G., Carrero-Valenzuela R. Alpha vs. gamma sarcoglycanopathy: DNA tests solve a case from Argentina. Acta Myologica Myopathies Cardiomyopathies. 2007;26(2):115–118. [PMC free article] [PubMed] [Google Scholar]
- 159.Cantero D., Hernández-Laín A., Martínez J.F.G., Pérez M.R., Ruano Y., Lleixà C. Milder forms of α-sarcoglicanopathies diagnosed in adulthood by NGS analysis. J. Neurol. Sci. 2018;394:63–67. doi: 10.1016/j.jns.2018.08.026. [DOI] [PubMed] [Google Scholar]
- 160.Wu L., Brady L., Shoffner J., Tarnopolsky M.A. Next-generation sequencing to diagnose muscular dystrophy, rhabdomyolysis, and HyperCKemia. Can. J. Neurol. Sci. 2018;45(3):262–268. doi: 10.1017/cjn.2017.286. [DOI] [PubMed] [Google Scholar]
- 161.Mendell J.R., Chicoine L.G., Al-Zaidy S.A., Sahenk Z., Lehman K., Lowes L. Gene delivery for limb-girdle muscular dystrophy type 2D by isolated limb infusion. Hum. Gene Ther. 2019;30(7):794–801. doi: 10.1089/hum.2019.006. [DOI] [PMC free article] [PubMed] [Google Scholar]