Abstract
NKX3.1 (NK3 homeobox 1) is a prostate tumour suppressor protein with a number of activities that are critical for its role in tumour suppression. NKX3.1 mediates the cellular response to DNA damage by interacting with ATM (ataxia telangiectasia mutated) and by activation of topoisomerase I. In the present study we characterized the interaction between NKX3.1 and topoisomerase I. The NKX3.1 homeodomain binds to a region of topoisomerase I spanning the junction between the core and linker domains. Loss of the topoisomerase I N-terminal domain, a region for frequent protein interactions, did not affect binding to NKX3.1 as was shown by the activation of Topo70 (N-terminal truncated topoisomerase I) in vitro. In contrast, NKX3.1 interacts with the enzyme reconstituted from peptide fragments of the core and linker active site domains, but inhibits the DNA-resolving activity of there constituted enzyme in vitro. The effect of NKX3.1 on both Topo70 and the reconstituted enzyme was seen in the presence and absence of camptothecin. Neither NKX3.1 nor CPT (camptothecin) had an effect on the interaction of the other with topoisomerase I. Therefore the interactions of NKX3.1 and CPT with the linker domain of topoisomerase I are mutually exclusive. However, in cells the effect of NKX3.1 on topoisomerase binding to DNA sensitized the cells to cellular toxicity and the induction of apoptosis by low doses of CPT. Lastly, topoisomerase I is important for the effect of NKX3.1 on cell survival after DNA damage as topoisomerase knockdown blocked the effect of NKX3.1 on clonogenicity after DNA damage. Therefore NKX3.1 and topoisomerase I interact in vitro and in cells to affect the CPT sensitivity and DNA-repair functions of NKX3.1.
Keywords: camptothecin (CPT), DNA repair, homeodomain, NK3 homeobox 1 (NKX3.1), prostate cancer, topoisomerase I
INTRODUCTION
NKX3.1 (NK3 homeobox 1) is a homeodomain protein that in the adult the expression of which is restricted almost exclusively to prostate epithelial cells [1,2]. NKX3.1 is a tumour suppressor whose down-regulation in premalignant epithelial cells and non-invasive prostate intraepithelial neoplasia presages the development of invasive prostate cancer in a substantial fraction of cases [3,4]. NKX3.1 suppresses the tumour phenotype by a broad range of effects. The protein is antiproliferative in vitro and in vivo [1,5]. In addition, NKX3.1 plays a significant role in the DNA-damage response of prostate epithelial cells such that the loss of NKX3.1 decreases cell survival after DNA damage as measured by colony-forming ability [6]. The effect on the DNA-damage response is mediated by the interaction of NKX3.1 with at least two proteins involved in DNA-damage response recognition and repair. NKX3.1 enhances the activation of ATM (ataxia telangiectasia mutated) kinase by binding to the N-terminal domain of ATM and accelerating kinase activation within minutes of DNA damage (C. Bowen, J. Ju, J.-H. Lee, T. Paull and E.P. Gelmann, unpublished work). NKX3.l also activates topoisomerase I’s resolving activity and co-localizes with topoisomerase I in the nucleus in response to DNA damage [7].
Topoisomerase I is a DNA-resolving enzyme highly conserved through evolution that relaxes super-coiled DNA by cleaving a single (scissile) strand and allowing the stochastic unwinding of the intact strand around the cleavage complex [8,9]. Topoisomerase I has been implicated in DNA replication and transcription as well as in DNA repair [8]. However, topoisomerase I binding is enhanced at the sites of DNA damage and its activity is affected by the relative location of damaged site to the cleavage site (for a review see [10]).
Topoisomerase I can relax positively or negatively super-coiled DNA by binding and cleaving a single strand via a covalent linkage between Tyr723 and a 3′-end of DNA generated by single-strand DNA cleavage at the site where topoisomerase I binds to DNA. Topoisomerase I nicks DNA preferentially, but not exclusively, at a combination of nucleotides that extends from positions −4 to −1 on the scissile strand that read 5′-(A/T)(G/C)(A/T)T-3′. The enzyme attaches covalently to the −1 T base. The very general function of topoisomerase I and the fact that only a single isoform is widely present throughout evolution suggests that additional proteins control its engagement with DNA. A paradigm for such an interaction is provided by the interaction of topoisomerase I with SV40 (simian virus 40) T antigen which forms a hexameric complex that binds to DNA at the SV40 origin of replication. The T antigen complex with DNA includes topoisomerase I [11]. Moreover, SV40 T antigen hexameric complexes bind topoisomerase I in the absence of DNA [12]. NKX3.1 also binds topoisomerase I in the absence of DNA via its homeodomain. In fact, the two proteins bind with sufficiently high affinity to displace NKX3.1 from its cognate DNA-binding domain [7].
Topoisomerase I is the target for the CPTs (camptothecins), a class of chemotherapeutic agents that bind to the topoisomerase I–DNA complex and prevent DNA re-ligation and release of the enzyme from the DNA-cleavage complex. This interaction activates apoptosis and is toxic to the cell [13]. Acquired resistant to CPTs results in the loss of topoisomerase I expression or mutation that abrogates interaction with the drug [14–17]. CPT binding to the topoisomerase I–DNA complex depends on the linker domain that lies between the core and active sites of the enzyme. As described below, we determined that NKX3.1 binds topoisomerase I in part at the linker domain and further studied the effects of NKX3.1 on topoisomerase I activity and on CPT resistance in vitro and in cells.
EXPERIMENTAL
Cell culture and transfection
The human prostate cell lines LNCaP and PC3 and HEK (human embryonic kidney)-293T cells were cultured in modified IMEM (improved minimum essential medium; Invitrogen) supplemented with 5% FBS (fetal bovine serum) at 5% CO2 and 37°C. Derivative lines of LNCaP, LNCaP (siLuc) and LNCaP (si471) stably expressing siRNAs (short interfering RNAs) direct against luciferase and NKX3.1 respectively, have been described previously [6]. For transient transfection, cells were plated on to six-well plates or 100-mm dishes and transfected using Lipofectamine™ (Invitrogen) as recommended by the manufacturer. Cell lysates were prepared 48 h after the initial transfection and stored in −80°C until use.
Recombinant proteins
GST (glutathione transferase)–Topo70 (N-terminally truncated topoisomerase I) was provided by Dr Daniel T. Simmons (University of Delaware, Newark, DE, U.S.A.). Other polypeptides corresponding to topoisomerase I fragments were overexpressed in Escherichia coli cells and purified as GST fusion proteins. All the peptide fragments were generated by PCR from the wild-type full-length topoisomerase I cDNA. PCR products were purified, digested with BamHI and EcoRI, and inserted into the expression vector pGEX-6P-1 (Amersham). The construction of each plasmid was verified by nucleotide sequencing. To generate the GST peptide E. coli strain BL21 (DE3) cells (Invitrogen) were transformed with the appropriate constructs as well as the pGEX-6p-1 vector. Bacteria were grown at 37°C until D600 = 0.8. Expression of the GST fusion proteins was induced by treatment of the bacteria with 0.5 mM IPTG (isopropyl β-D-thiogalactopyranoside) for 1–3 h at 30°C. Solubilization of the GST fusion proteins was performed essentially as described previously [18]. Briefly, cells were washed once with ice-cold STE buffer [sodium chloride/Tris/EDTA; 10 mM Tris (pH 8.0), 150 mM NaCl and 1 mM EDTA], resuspended in STE buffer containing 100 μg/ml of lysozyme (added prior to resuspension) and incubated on ice for 15 min. DTT (dithiothreitol) was then added to a final concentration of 5 mM. Bacteria were lysed by the addition of N-laurylsarcosine to a final concentration of 1.5%. Cell lysates were vortex mixed, sonicated and the lysates were clarified by centrifuging at 10 000 g for 20 min at 4°C. The supernatants were adjusted to 2% Triton X-100.
To purify the GST fusion proteins, the clarified cell lysates were incubated with glutathione–agarose beads for 30 min at 4°C. The beads were washed six times with ice-cold PBS [8.4 mM Na2HPO4, 1.9 nM NaH2PO4 (pH 7.4) and 150 mM NaCl] and resuspended in ice-cold elution buffer [75 mM Hepes (pH 7.4), 150 mM NaCl, 10 mM reduced glutathione, 5 mM DTT and 0.1% Triton X-100]. The beads were vortex mixed and incubated at 4°C for 30 min. The eluted proteins were collected by centrifugation at 500 g for 1 min. Beads were resuspended with elution buffer, the procedure was repeated as described above and the eluates were pooled. If necessary, the pooled eluted proteins were concentrated with Centrifugal Filter Units (Ultracel YM-10, Millipore). When required, GST fusion proteins were treated with GE Healthcare’s PreScission Protease to remove GST tags according to the manufacturer’s protocol.
GST pull-down assay
The GST pull-down assays were performed as described previously [7]. Briefly, sonicated bacterial lysates containing overexpressed GST or GST fusion proteins (Figure 1) were linked to glutathione–Sepharose beads. Immobilized GST or GST fusion proteins were then incubated overnight at 4°C with 500 μg of cell lysates. After extensive washing, the immobilized proteins were removed from the beads by heating at 90°C for 5 min in 30 μl of 2× SDS loading buffer [7]. The proteins were then separated by SDS/PAGE (4–20% gel), transferred on to nitrocellulose membranes and immunoblotted with NKX3.1 antiserum.
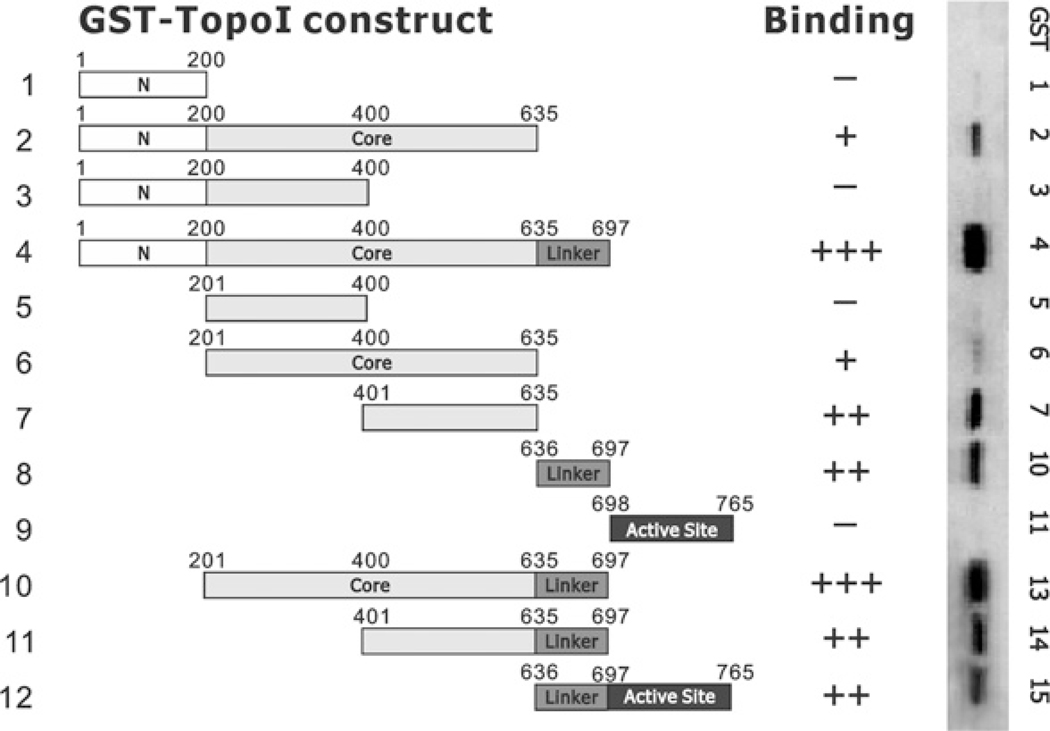
Figure 1. Binding of NKX3.1 to GST fusion peptides of topoisomerase I.
GST–topoisomerase I (Top1) fusion constructs (peptides 1–12) were made as described in the Experimental section. Fusion proteins were expressed in BL21 cells and purified with glutathione beads. For the GST pull-down assay, GST-fusion proteins were bound to glutathione beads, washed and incubated with 500 μg of LNCaP cell lysates overnight. After washing five times with PBS containing 0.2% Triton-X 100, beads were eluted with 2× SDS loading buffer at 99°C for 5 min. Eluted proteins were fractionated by SDS/PAGE (4–20% gels) and transferred on to nitrocellulose membranes. NKX3.1 was Western blotted with an anti-NKX3.1 antibody. the Western blot results are shown in the right-hand panel with the intensity of the signals graded as − to +++.
Affinity chromatography assay for association of the topoisomerase I core domain with the C-terminal domain
GST or GST–topoisomerase I(201–635) was loaded on to glutathione–Sepharose 4B beads by incubation at room temperature (25°C) for 30 min in reconstitution buffer [100 mM potassium phosphate (pH 7.4), 1 mM DTT, 1 mM EDTA and 0.2% Triton-X 100]. Beads were washed four times with reconstitution buffer and incubated for 30 min at room temperature with purified GST-tag removed topoisomerase I peptide 636–765 or peptide 698–765, in the absence or presence of 0, 10 or 20 μg of purified NKX3.1. Beads were washed five times with reconstitution buffer and eluted by boiling for 5 min in SDS loading buffer. Samples were fractionated by SDS/PAGE (20% gel) and the proteins were visualized by Coomassie Blue staining.
DNA-relaxation assay
Topoisomerase I preparations used in the DNA-relaxation assay were either Topo70 or reconstituted topoisomerase I prepared as described previously [19]. Reconstitution was carried out for 30 min at room temperature after mixing the different topoisomerase I peptide fragments. The reaction was performed as described previously [7] based on the protocol from Topogen. For the serial-dilution assays, protein samples were serially diluted and reactions were initiated by the addition of diluted enzyme to a reaction buffer containing 12.5 ng/μl of super-coiled pHOT plasmid substrate DNA. All preparations before the last step were performed on ice and the reaction was carried out at 37°C in a PCR machine. For the time-course assays, the appropriate amount of enzyme was used and the reactions were initiated by the addition of enzyme to the reaction buffer containing 12.5 ng/μl of super-coiled plasmid DNA. Reactions were incubated at 37°C and, at the indicated time points, 10 μl aliquots were removed and added to 2.5 μl of 5× stop buffer/gel loading buffer (2.5% SDS, 0.125% Bromophenol Blue and 25% glycerol) to terminate the reaction. When purified NKX3.1 was included in the reaction, the reaction products were subjected to proteolytic digestion with 0.5 μg/ml proteinase K at 37°C for 30 min. The reaction products were electrophoresed in 1% agarose gels and the DNA bands were visualized by ethidium bromide staining and photographed with UV illumination.
For the graphical representations of the DNA-relaxation assays, the relative DNA amount was quantified using the ImageJ software (http://rsb.info.nih.gov/ij/) and the relaxation activity calculated as 1−(the amount of super-coiled substrate left after incubation)/(total DNA amount). In the assays shown in Figures 4(B) and 4(D) only the major relaxed forms at the lowest molecular mass were quantified and the relative activity was simply calculated as the fold increase in the major relaxed form of DNA.
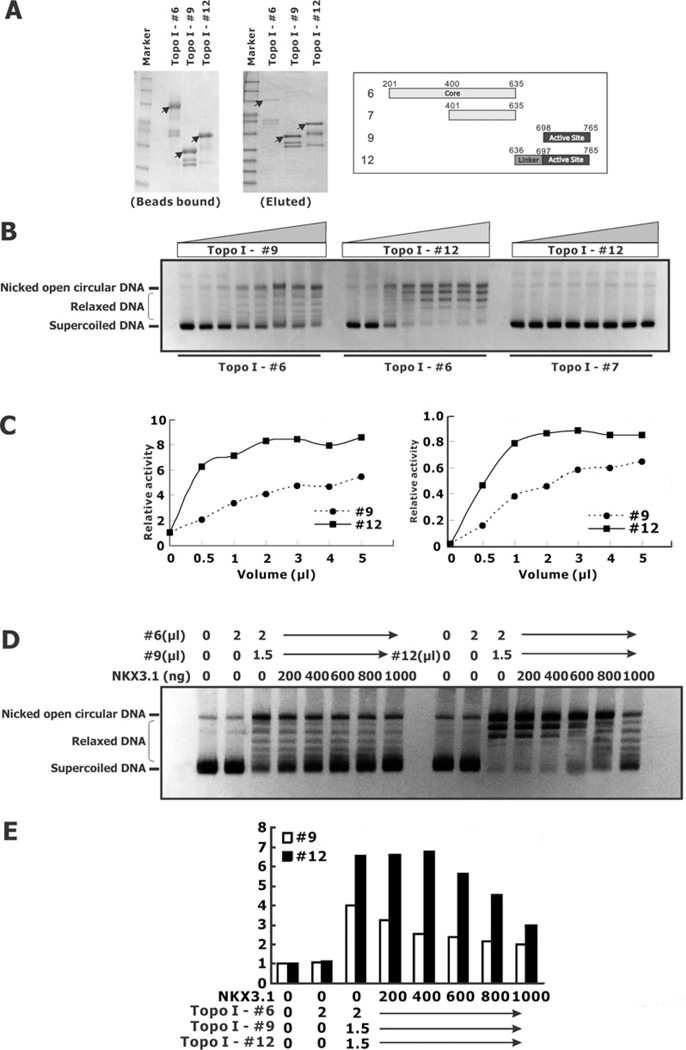
Figure 4. Effect of NKX3.1 on reconstituted topoisomerase I activity.
(A) The topoisomerase I core domain, peptide 6, the C-terminal activity domain, peptide 9, and the linker domain and C-terminal activity domain, peptide 12, were purified with GST beads and eluted with reductive glutathione. (B) Peptide 6 (2 μl) was combined with increasing amount of either peptide 9 or peptide 12 (left-hand and centre panels). Similarly, 2 μl of peptide 7 was combined with increasing amounts of peptide 12 (right-hand panel). (C) The relative relaxation activities of the reconstituted topoisomerase I peptides were quantified and calculated with two different methods, as described in the Experimental section, with similar results. (D) Reconstitution of topoisomerase I activity in the present of NKX3.1. (E) Graphical representation of the relaxation activity from the gel in (D).
Co-immunoprecipitation
HEK-293T cells were engineered to express Myc–NKX3.1 and the cell lysates were prepared 48 h after transfection. Cell lysates (500 μg) were used to perform the co-immunoprecipitation assays with antibodies against either topoisomerase I or Myc (Santa Cruz Biotechnology). After extensive washing, the pellets were fractionated by SDS/PAGE (4–20% gel) and subjected to immunoblotting with anti-(topoisomerase I) (BD Pharmingen) or anti-Myc antibodies.
Detection of PARP [poly(ADP-ribose) polymerase] 1 cleavage
Cells were cultured in six-well plates and were treated with increasing concentrations of CPT for different times. The cells were then harvested and the lysates were prepared as described previously [7]. Samples containing 30 μg of protein were separated by SDS/PAGE (4–20% gel). Immunoblot assays were conducted with primary anti-PARP C2–10 antibody (Enzyme Systems Products) overnight at 4°C. Membranes were washed five times with PBS containing 0.05% Tween 20 and then incubated with horseradish peroxidase-conjugated secondary antibody for 45 min. Membranes were washed again as described above and developed with SuperSignal West Pico Chemiluminescent substrate (Thermo Scientific) and exposed to X-ray film. The membranes were stripped with Thermo Scientific Restore Western Blot Stripping Buffer and reprobed with other antibodies, including antibodies against p53 (Santa Cruz Biotechnology), p21 (Santa Cruz Biotechnology) and β-actin (Sigma).
Clonogenic assay
Long-term survival of LNCaP cells in response to low concentrations of CPT was determined by a colony-forming assay. LNCaP cells (3000) were plated on to six-well plates. After overnight incubation the cells were treated with 20 nM CPT. DMSO was used as the vehicle control. Cells were cultured for 2 weeks and stained with Crystal Violet for colony counting. Colony formation by PC-3 cells was determined as described above except that the cells were co-transfected with a GFP (green fluorescent protein)-expression marker as shown in Figure 8(A). Cells were sorted for GFP expression and plated on to 6-well plates at a cell density of 2000 cells/well and subjected to UV treatment at the indicated doses. Colony formation was counted 7–10 days later and the surviving fraction at each data point was normalized by comparison with colony formation without any treatment.
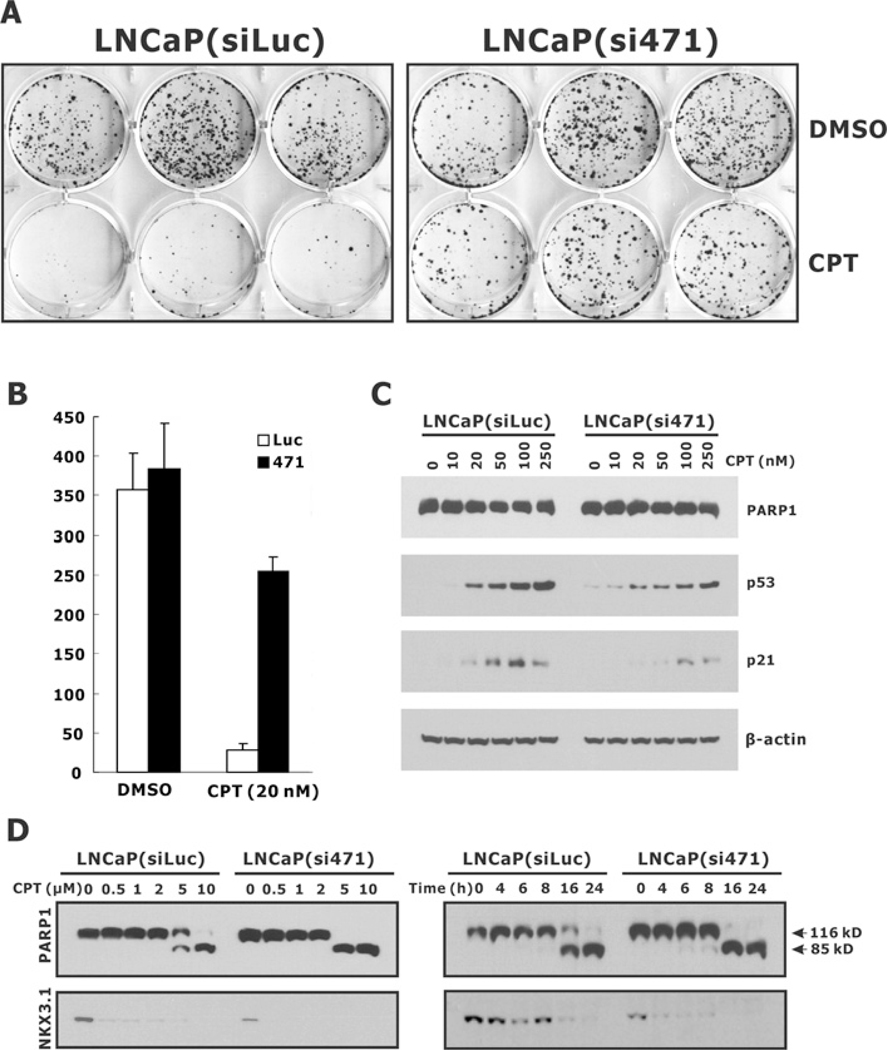
Figure 8. Effect of NKX3.1 knockdown on CPT-induced cytotoxicity in LNCaP cells.
(A) LNCaP(siLuc) and LNcAP(si471) cells were plated into six-well plates at a density of 3000 cells/well and treated with 20 nM CPT. DMSO treatment was used as the vehicle control. Cells were cultured for 2 weeks, stained with Crystal Violet and the colony numbers were counted and plotted (B). (C) LNCaP(siLuc) and LNcAP(si471) cells were treated with increasing amount of CPT for 72 h and the expression of PARP1, p53 and p21 was determined by immunoblotting. (D) Cells were treated with CPT for 24 h (left-hand panel) or treated with 5 μM CPT for the indicated time periods (right-hand panel). The cell lysates were then analysed for PARP1. The intact (116 kD) and cleaved (85 kD) forms of PARP1 are indicated.
RESULTS
Topoisomerase I binds to NKX3.1 via multiple domains
Topoisomerase I is a 765-amino-acid protein that is composed of a non-essential N-terminal domain that is the frequent site for binding by other proteins, a core domain that is essential for resolving activity and DNA binding and contains the catalytic domain, a linker domain that is not essential for activity and is the site for interaction with the inhibitor CPT, and a C-terminal active-site domain that contains the tyrosine residue that forms a covalent bond with DNA. To characterize the domains in topoisomerase I required for NKX3.1 association, we made a series of GST fusion peptides (Figure 1, left-hand panel). GST pull-down assays with the LNCaP cell lysates identified several domains that associated with NKX3.1 (Figure 1). The results suggested that the C-terminal segment of the core domain (amino acids 201–635) and the adjacent linker domain (amino acids 636–697) are two major regions required for interaction with NKX3.1. Among our fusion peptides most fragments that included the core domain C-terminus (constructs 2, 4, 6, 7, 10 and 11) bound strongly to NKX3.1 as did fragments containing the linker domain (constructs 4, 8, 10, 11 and 12).
The NKX3.1 homeodomain is required for its interaction with topoisomerase I
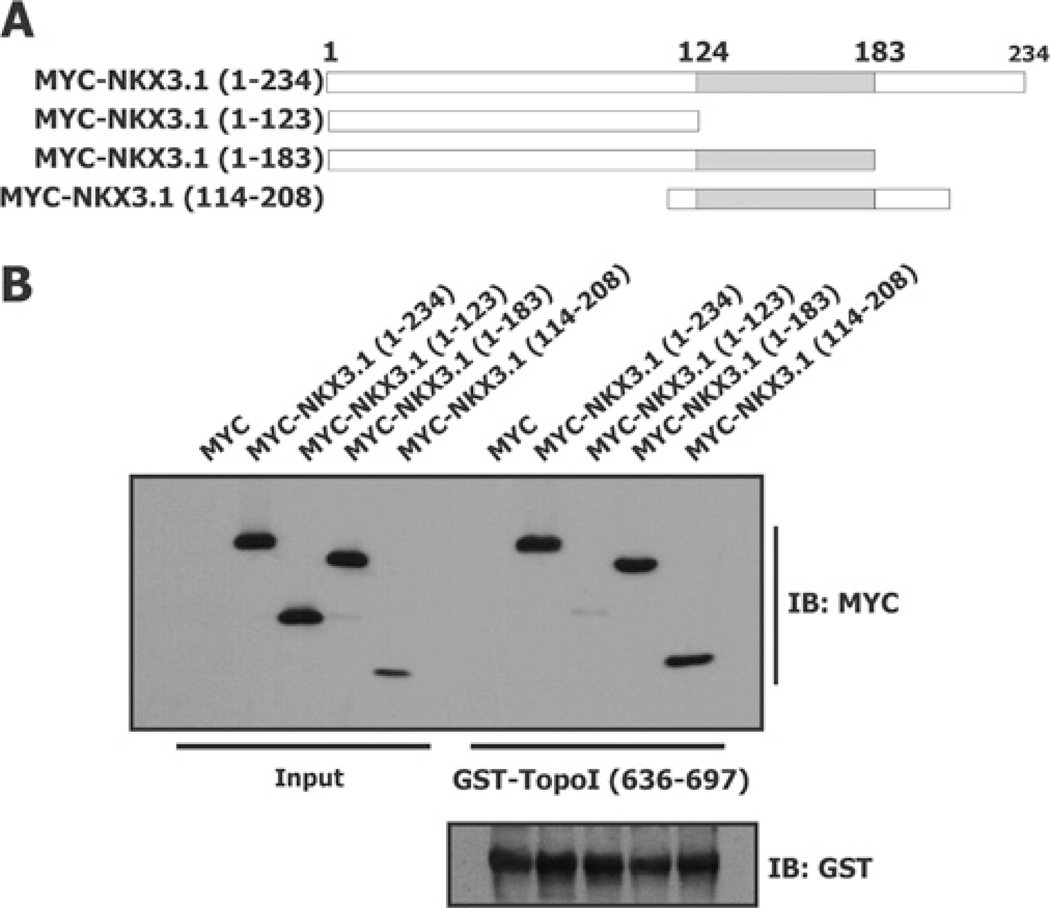
The homeodomain of NKX3.1 is the probable site of binding to topoisomerase I since topoisomerase I can compete with a DNA oligomer that contains the cognate NKX3.1-binding sequence [7]. To demonstrate direct binding of NKX3.1 peptides to topoisomerase I GST pull-down assays were performed with GST–topoisomerase I(636–697) and HEK-293T cell extracts from cells expressing Myc-tagged NKX3.1 peptides (Figure 2A). Binding of the topoisomerase I linker domain to NKX3.1 was highly dependent on the presence of the homeodomain (Figure 2B). To elucidate further the relationship between binding of NKX3.1 to DNA and to topoisomerase I we assayed the binding of several NKX3.1 missense mutant proteins with compromized DNA binding, NKX3.1(T164A) [20], NKX3.1(T179A) [21] and NKX3.1(N174Q). The latter construct contains a residue that forms a hydrogen bond with the major groove of DNA and whose loss diminishes DNA binding by 3 logs (results not shown and [22]). All three missense mutations that affected DNA binding did not affect binding to either the topoisomerase I linker domain or to the topoisomerase I core domain. This binding was compared with GST–topoisomerase I(201–635) (fragment 6) binding (results not shown).
Figure 2. NKX3.1 homeodomain is required for binding to the topoisomerase I linker domain.
HEK-293T cells were transfected with Myc–NKX3.1 fusion constructs (A) and GST pull-down assays were performed with GST–topoisomerase I(636–697) (B). Equal amounts of GST–topoisomerase I(636–697) were used in each reaction as indicated by immunoblot (IB) analysis with an anti-GST antibody (B, lower panel).
Effect of NKX3.1 on topoisomerase I reconstitution in vitro
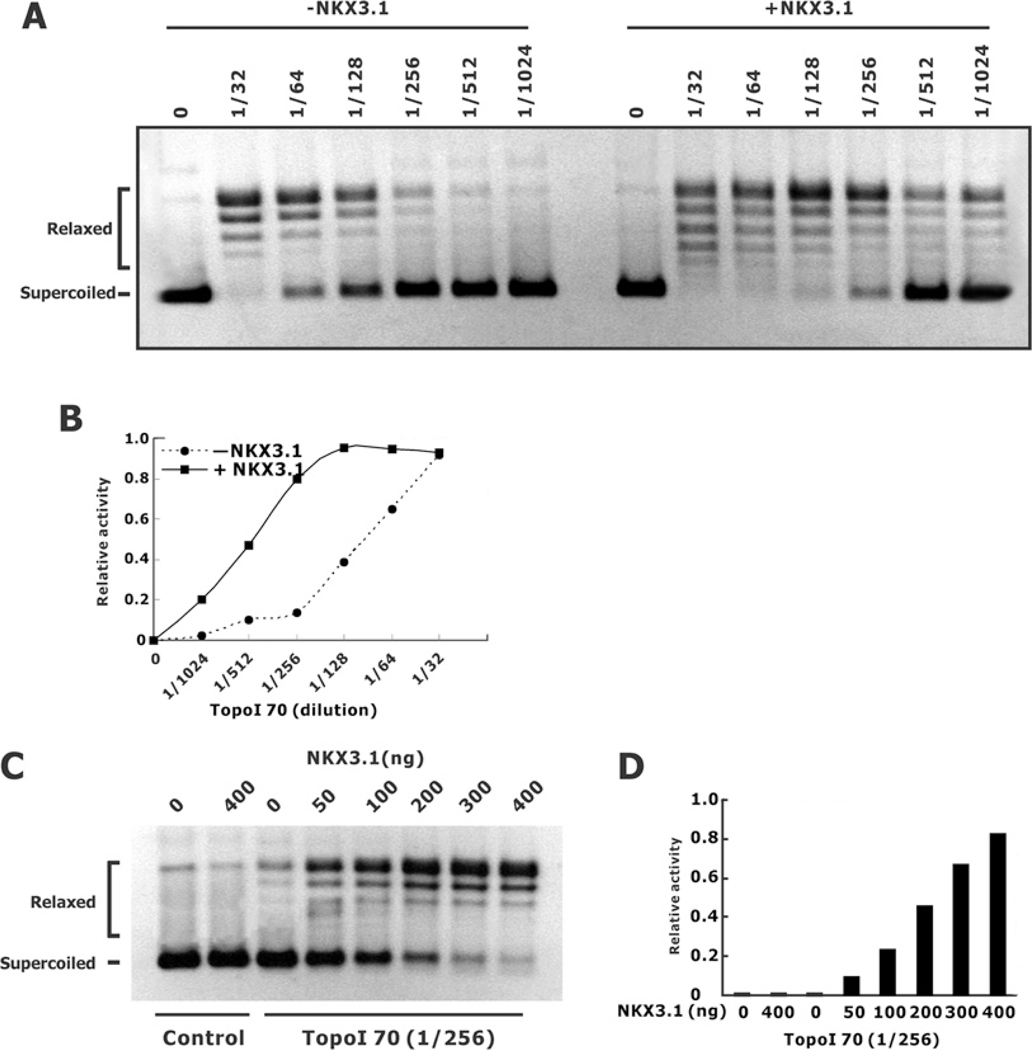
The DNA-relaxation activity of topoisomerase I can be reconstituted in vitro with purified core domain and active site peptides [19,23]. Crystal structures of the reconstituted topoisomerase I core and C-terminal domains revealed that the reconstituted topoisomerase I generated four domains that clamp around the DNA to replicate the activity of the holoenzyme [24]. To elucidate further the interaction between NKX3.1 and the active components of topoisomerase I we determined whether NKX3.1 was able to affect the DNA relaxation activity of the reconstituted core domain and active site. We first determined the effect of purified NKX3.1 on DNA relaxation by Topo70, the enzyme fragment lacking the N-terminal domain. In the presence of NKX3.1 Topo70 relaxation of super-coiled DNA was augmented 4-fold (Figures 3A and 3B). Moreover, increasing amounts of NKX3.1 potentiated the effect of Topo70 in a DNA-relaxation assay (Figures 3C and 3D).
Figure 3. Effect of NKX3.1 on Topo70-induced DNA relaxation.
(A) Purified Topo70 (1:32–1:1024 dilution) was used in DNA-relaxation assays in the absence or presence of 400 ng of purified NKX3.1 per reaction. (B) Graphical depiction of the DNA-relaxation activity from (A). (C) Topo70 (1:256 diluted) was used in DNA-relaxation assays in the absence or presence of increasing amount of purified NKX3.1 (0–400 ng/reaction). (D) The relative relaxation activities from the results shown in (C).
To reconstitute topoisomerase I activity by combining separate components of the enzyme we used GST fusion peptides of the core domain (amino acids 201–635, peptide 6) and one of two C-terminal domain peptides, either amino acids 698–765, peptide 9 containing the active site domain, or amino acids 636–765, peptide 12 containing the linker and active-site domains (Figure 4A). When the fusion peptides were combined in vitro they reconstituted the topoisomerase I relaxation activity (Figure 4B). We found that activity of the core domain and active site peptide 9 (amino acids 698–765) was as much as one tenth the activity of core domain with linker and active site peptide 12 (amino acids 636–765) (Figures 4B and 4C). Thus using the GST fusion peptides we were able to achieve in vitro reconstitution similar to what had been reported previously [24]. The in vitro reconstitution of topoisomerase activity was not seen with the negative control C-terminal peptide (amino acids 401–635, peptide 7) that lacks subdomains I and II, which are important for the formation of the enzyme clamp on DNA. In vitro, NKX3.1 had a marked inhibitory effect on the topoisomerase activity reconstituted from peptide fragments of the enzyme (Figures 4D and 4E). Thus NKX3.1 interacted with the reconstituted enzyme fragments, but has the opposite effect on the resolving activity compared with the native enzyme or Topo70.
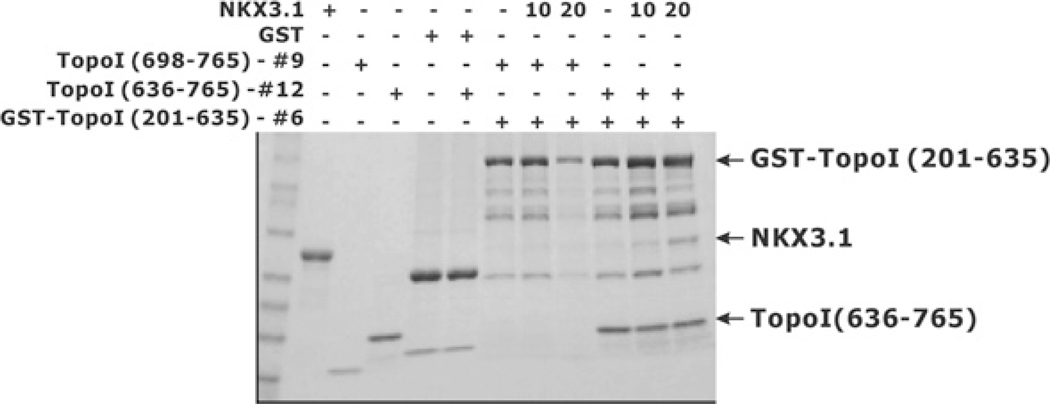
The effect of NKX3.1 on the resolving activity of the reconstituted enzyme fragments was probably owing to direct physical interaction between the peptides. To demonstrate this interaction we performed pull-down experiments with GST-bound topoisomerase I fragments and purified NKX3.1. GST-bound topoisomerase I core domain (amino acids 201–635, peptide 6) was immobilized on glutathione beads and used to retain peptides of topoisomerase I and NKX3.1. When the active site of topoisomerase I (amino acids 698–765, peptide 9) was added to the beads neither that fragment nor the NKX3.1 peptide was retained on the column (Figure 5). However, when the linker and active site of topoisomerase I (amino acids 636–765, peptide 12) was added to the column both it and NKX3.1 were retained. Thus the linker region was not essential for reconstituting the topoisomerase I resolving activity, but markedly enhanced the association of topoisomerase I component peptides and mediated the binding of NKX3.1.
Figure 5. Binding of NKX3.1 to reconstituted topoisomerase I (TopoI) peptides.
GST or GST–topoisomerase I(201–635) was first loaded on to glutathione–Sepharose 4B beads at room temperature for 30 min and the unbound proteins were removed by washing. Beads were then incubated with purified topoisomerase I(698–765) or topoisomerase I(636–765), in the absence or presence of increasing amounts of NKX3.1. The beads were then washed extensively, subjected to SDS/PAGE (4–20% gel) analysis and the proteins visualized by Coomassie Blue staining.
NKX3.1 does not block the effect of CPT in vitro
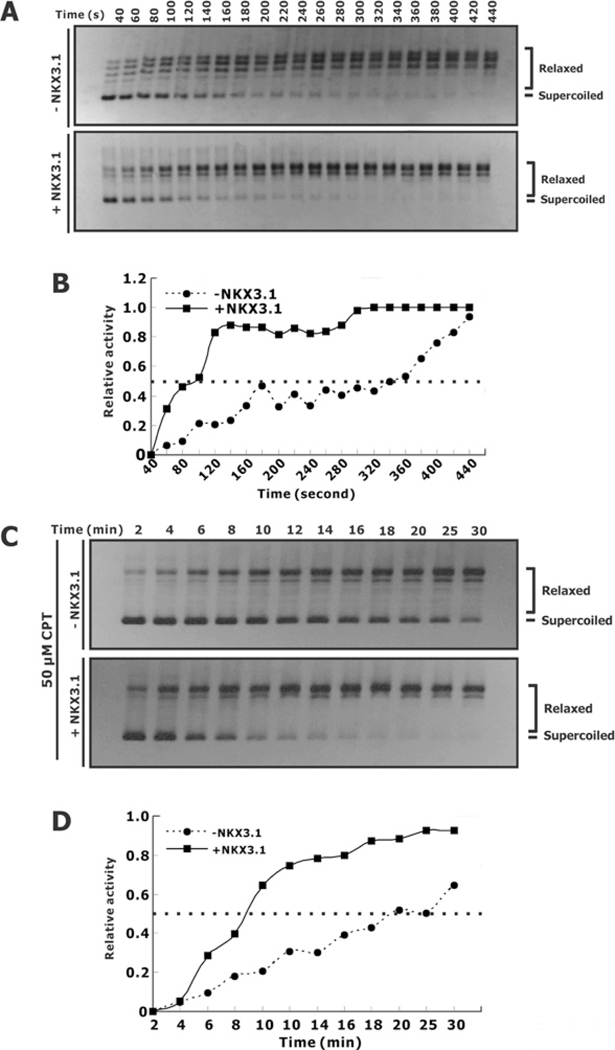
CPT is a plant alkaloid that is used as a chemotherapeutic agent. CPT binds to the topoisomerase I–DNA complex and blocks DNA re-ligation, thus trapping topoisomerase I on DNA and maintaining a single-strand DNA break [25]. CPT is a weak inhibitor of the DNA relaxation activity of the reconstituted topoisomerase I core domain plus the C-terminal domain. Because NKX3.1 binds, in part, to the linker domain, a key element in determining CPT sensitivity, we asked whether NKX3.1 affected the interaction of CPT and topoisomerase I. We first demonstrated the effect of NKX3.1 on DNA relaxation activity mediated by Topo70. NKX3.1 enhanced the DNA relaxation activity by shortening the time needed to complete the relaxation. This result further substantiated the results shown in Figure 4. In the absence of NKX3.1, CPT alone dramatically inhibited the DNA relaxation activity (compare Figures 6A and 6C, upper panels), reflected by a much delayed relaxation of super-coiled DNA. In the absence of CPT Topo70 achieved 50% of the maximal relaxation activity in approximately 3 min. In the presence of 50 μM CPT 50% maximal activity was achieved in 20 min. NKX3.1 affected the rate of the DNA relaxation reaction despite the presence of CPT and can be observed when comparing the 50% maximal activity at 1.5 min in the presence of NKX3.1 with the value at 8.5 min in the presence of NKX3.1 and 50 μM CPT. Similar results were obtained with 100 μM CPT as with 50 μM CPT (results not shown). These results suggested that CPT and NKX3.1 appear to interact with Topo70 independently and, although each binds to Topo70 near the linker domain, we do not see evidence that one interferes with the action of the other, but that their effects are most likely to be additive.
Figure 6. Effect of NKX3.1 and CPT on Topo70 DNA relaxation.
(A) Topo70 with or without NKX3.1 were used in a DNA-relaxation assay as in Figure 3. Aliquots (10 μl) were added to 2.5 μl of 5× stop buffer/gel loading buffer (2.5% SDS, 0.125% Bromophenol Blue and 25% glycerol) to terminate the reaction. DNA was visualized by ethidium bromide staining and photographed with UV illumination. (B) The relative relaxation activities of the reactions from the gel shown in (A) plotted as a function of incubation time. (C) DNA-relaxation assays were performed as described in (A) with the addition of 50 μM CPT. (D) Graphical depiction of the DNA-relaxation activity shown in (C).
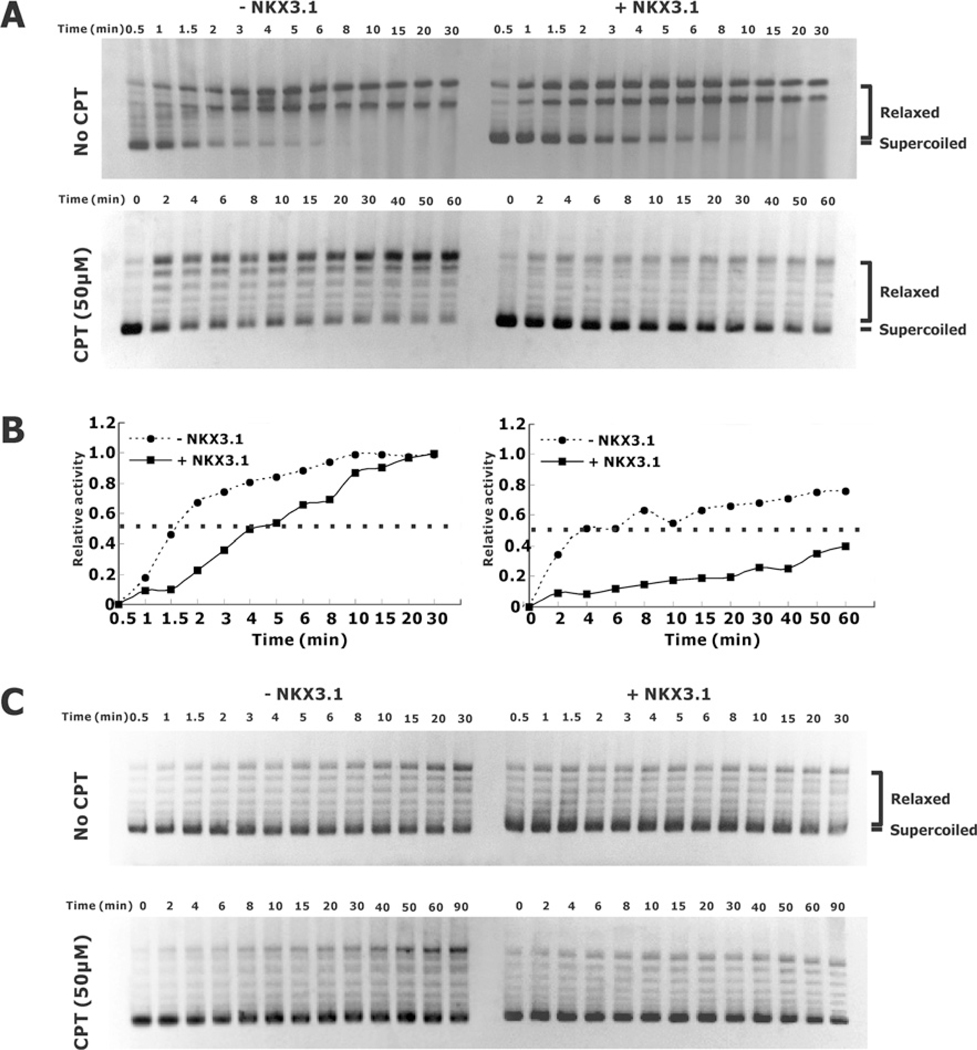
CPT also affects the enzymatic activity of reconstituted topoisomerase I fragments 6 and 9 in vitro. In this reaction, which demonstrates DNA cleavage activity in the absence of a linker domain, we tested for an interaction between CPT and NKX3.1. As shown in Figures 7(A) and 7(B) 50% of the maximal activity of the reconstituted enzyme was achieved in 1.5 min in the absence of NKX3.1 and 4 min in the presence of NKX3.1. When 50 μM CPT was added to the reactions, the time to achieve 50% of the maximal activity increased to 4 min and >60 min respectively. As a control, reconstitution of topoisomerase I activity with fragments 6 and 12 showed an inhibitory effect of NKX3.1, but no effect of CPT owing to the absence of the linker domain. Thus topoisomerase reconstitution was inhibited to a greater degree by the combination of NKX3.1 and CPT more than expected by the sum of their effects. However, it is clear that neither compound interfered with the effect of the other in the reconstitution assay.
Figure 7. Effect of NKX3.1 and CPT on the relaxation activity of reconstituted topoisomerase I peptides.
DNA-relaxation assays were performed as described in Figure 6 except that reconstituted topoisomerase I was used to induce DNA relaxation. The experiments were performed in the absence or presence of NKX3.1 and with or without CPT as indicated. (A) Characterization of the DNA-relaxation activity of the reconstituted topoisomerase I peptides 6 and 12 without (upper panel) or with (lower panel) 50 μM CPT in the absence or presence of NKX3.1 as indicated. Relaxation activities were assayed by incubation at 37°C for the indicated time intervals. (B) Graphical representation of the relaxation activity quantified and calculated from the gels shown in (A), as described in the Experimental section, and plotted as a function of incubation time. (C) Characterization of the DNA relaxation activity of reconstituted topoisomerase I peptides 6 and 9 with or without 50 μM CPT and in the absence or presence of NKX3.1 as indicated.
NKX3.1 affects CPT-induced cytotoxicity
Because NKX3.1 has a substantial effect on the binding of topoisomerase I to DNA and on DNA cleavage, its enhancement of topoisomerase I activity could affect cellular sensitivity to CPT. We predicted, from the results shown in Figure 6, that NKX3.1 enhanced the activity of topoisomerase I in the presence of CPT and thus that NKX3.1 would sensitize cells to the cytotoxic effects of the drug. Indeed, when NKX3.1 was targeted by shRNA (short hairpin RNA)-containing lentivirus the cells became more resistant to low concentrations of CPT (Figures 8A and 8B). The cytotoxic effects of CPT resulted in a decreased cell-cycle arrest as shown by decreased p53 and p21 expression after NKX3.1 knockdown (Figure 8C). When cells were treated with low concentrations of CPT (20 nM), no PARP1 cleavage was detected (Figure 8C); however, when cells were treated with high concentrations of CPT significant apoptosis was seen, reflected by dramatic morphological changes (results not shown) and the cleavage of PARP1. Knockdown of endogenous NKX3.1 expression led to increased apoptosis as shown by the cleavage of PARP1 (Figure 8D).
NKX3.1 and topoisomerase I interact in the DNA-damage response
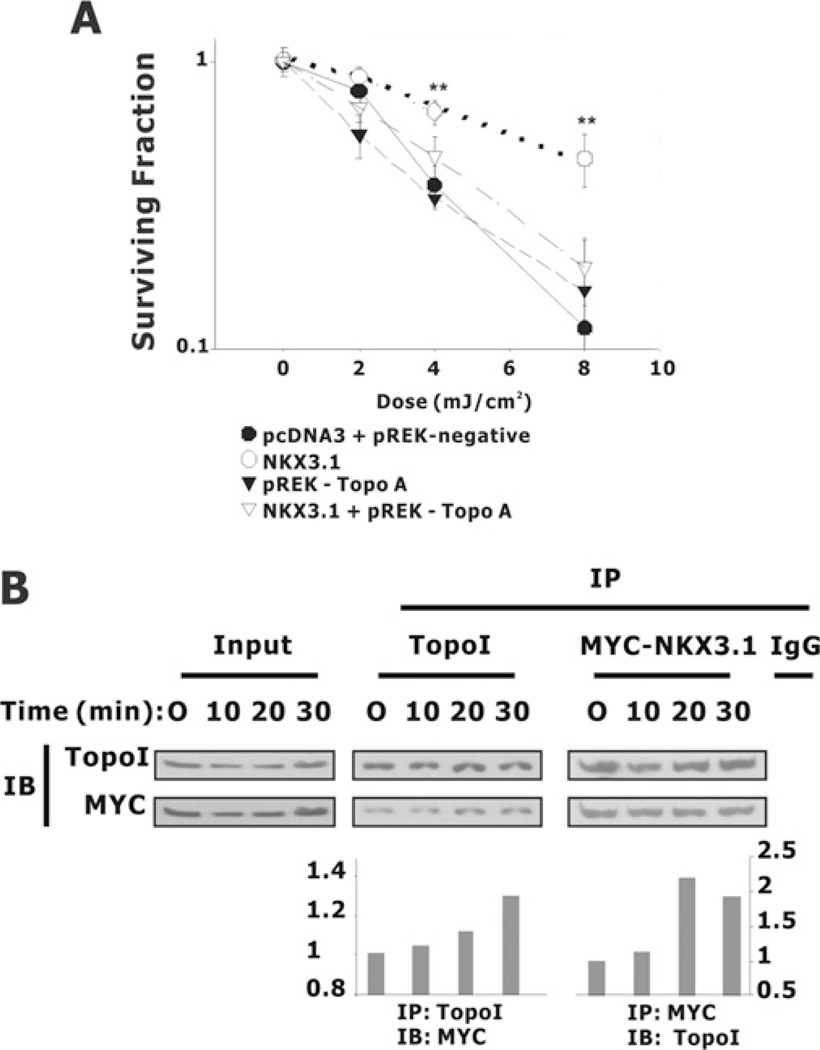
Lastly, we wanted to demonstrate that topoisomerase was important for the role of NKX3.1 in clonogenic survival after DNA damage [6]. After DNA damage, NKX3.1 and topoisomerase I respond in a co-ordinated fashion and remain co-localized in the nucleus [7]. To investigate the role for topoisomerase I in the effect of NKX3.1 on DNA repair we tested topoisomerase I knockdown on clonogenicity after DNA damage. Topoisomerase I knockdown was accomplished with the same siRNA vector that was used to demonstrate the effect of topoisomerase I on chromosomal stability by Pommier and colleagues [26]. Enhanced clonogenicity and cell survival after UV exposure was mediated by NKX3.1 and reversed by topoisomerase I knockdown (Figure 9A). This does not prove that the entire effect of NKX3.1 on DNA repair is via enhancement of topoisomerase I activity. However, the data are consistent with the notion that topoisomerase I and NKX3.1 are involved in related processes of DNA repair. In fact, NKX3.1 and topoisomerase I demonstrate enhanced interaction shortly after the induction of DNA damage. The physical association of topoisomerase I and an epitope-tagged NKX3.1 was seen within minutes of DNA damage as shown by immunoprecipitation and Western blotting (Figure 9B).
Figure 9. NKX3.1 affects colony formation after the exposure of PC-3 cells to UV irradiation.
(A) PC-3 cells were co-transfected with GFP, pcDNA3 or pcDNA3-NKX3.1, together with the topoisomerase I siRNA vector pREK-Topo A or a control vector pREK-negative. At a density of 2000 cells/well cells were plated into six-well plates and subjected to UV irradiation. Colony formation was counted 7–10 days later and the surviving fraction at each data point was normalized by comparison with colony formation without any treatment. (B) Topoisomerase I (TopoI) association with NKX3.1 in response to UV treatment. HEK-293T cells were transiently transfected with the Myc-NKX3.1 expression vector. Cell lysates were prepared and co-immunoprecipitation (IP) was performed with antibodies against topoisomerase I or Myc. After extensive washing, the immunoprecipitated pellets were fractionated by SDS/PAGE (4–20% gel) and subjected to immunoblotting (IB) with antibodies against topoisomerase I and Myc. The amounts of NKX3.1 (left-hand histogram) or topoisomerase I (right-hand histogram) pulled down were determined by densitometry.
DISCUSSION
We have demonstrated a functional and physical association between the prostate tumour suppressor NKX3.1 and topoisomerase I. Topoisomerase I is an essential DNA-resolving enzyme with roles in a broad range of processes including DNA replication [27], transcription [28] and repair [29] (for a review see [30]). Therefore the roles of Nkx3.1 in the development [31,32] and terminal differentiation [1] of prostate epithelial cells could conceivably be mediated, in part, by an effect on topoisomerase I. Although it is apparent that Nkx3.1 affects topoisomerase I activity in an organ-specific manner in the adult, we do not know the physiological purpose for the interaction [7]. To improve our understanding of the interaction between these proteins with the possible goal of targeting their interaction genetically or for therapeutic purposes, we analysed further the molecular determinants of their binding.
NKX3.1 bound to the C-terminal region of the topoisomerase I core domain and the adjacent linker domain. In cell-free preparations NKX3.1 caused the enzymatic activation of an N-terminal truncated Topo70 peptide. Topo70 lacks the N-terminal non-essential domain that is the binding region for a number of other proteins including p53 [33] and ARF (ADP-ribosylation factor) [34]. The interaction of NKX3.1 with a key functional domain of topoisomerase I activated the truncated enzyme in vitro. In contrast, when topoisomerase I peptide fragments were mixed in an assay to investigate possible reconstituted resolving activity, NKX3.1 interfered with the reconstitution and inhibited activity. It may be that NKX3.1 bound to the core domain and interfered with the reconstitution of the core and activation domains. Clearly the results of the present study reflect a close functional interaction of NKX3.1 and topoisomerase I.
We have shown by immunohistochemistry that NKX3.1 and topoisomerase I respond in a co-ordinated way to DNA damage [7]. In the present study we provide further evidence that NKX3.1 and topoisomerase I interact in response to DNA damage. Our data demonstrating that the knockdown of topoisomerase I abrogated the effect of NKX3.l on cell survival after DNA damage (Figure 9A) does not prove that topoisomerase I is the sole mediator of the effect of NKX3.1 on the DNA-damage response. Topoisomerase I affects DNA repair and genomic integrity in cells that do not express NKX3.1 [26]. Therefore knockdown of topoisomerase I in LNCaP cells probably affects the actions of the enzyme both dependent on and independently of NKX3.1. Topoisomerase I has been implicated to have a role in DNA repair [29,35] and has been shown to bind at or near sites of DNA damage [36,37]. Topoisomerase I knockdown has also been associated with chromosomal instability in a colon cancer cell line, further suggesting that topoisomerase I plays a role in DNA repair [26]. Thus knockdown of topoisomerase I very likely had an effect on the DNA-damage response in PC-3 cells that was separate from the effect of NKX3.1.
NKX3.1 is a homeodomain protein in the NK family, many members of which affect organ-specific differentiation. We demonstrated that by binding directly to DNA NKX3.1 could suppress transcription, but could not assemble a transcription-initiation complex and activate gene expression by itself [38]. We presume that NKX3.1 can act as a transcriptional cofactor for other transcription factors, such as serum response factor. In its interaction with serum response factor NKX3.1 binds tightly to the protein [39] and weakly to DNA adjacent to the serum response element [40]. It appears that NKX3.1 binds strongly to other proteins. In two well-characterized examples protein–protein interaction with NKX3.1 is mediated by the homeodomain. NKX3.l binds to and activates ATM in a carefully controlled interaction triggered by DNA damage (Bowen et al, submitted for publication). We have shown previously that NKX3.1 binds to topoisomerase I via the homeodomain with an affinity that exceeds that of the binding of the homeodomain to the NKX3.1 cognate DNA-recognition sequence [7]. Thus we have provided further evidence of a functional interaction between NKX3.1 and topoisomerase I that is likely to reflect a physiological interaction.
Lastly, we examined the effect of NKX3.1 expression on the cytotoxicity of CPT. CPTs bind to the topoisomerase I–DNA cleavage complex and trap the enzyme on the DNA leaving the enzyme-induced DNA single-strand break unrepaired. At nanomolar concentrations of CPT NKX3.1 surprisingly decreased CPT-induced cytotoxicity and protected cells from apoptosis. We interpret these findings as resulting from NKX3.1 accelerating the topoisomerase I reaction and therefore decreasing the likelihood of arresting the cleavage complex in the presence of low CPT concentrations. High CPT concentrations, on the other hand, overwhelmed the reaction and trapped topoisomerase I and DNA in the complex regardless of the presence of NKX3.1.
In conclusion we have further shown the structural domains for a functional interaction between NKX3.1 and topoisomerase I and provided evidence that one role of NKX3.1 is to enhance the activity of topoisomerase I in the DNA-damage response.
Acknowledgments
FUNDING
This work was supported, in part, by the National Institutes for Health via CCSG (Cancer Center Support Grant) [grant number P30 CA013696–36] via Microscopy and Radiation Shared Resources.
Abbreviations used:
- ATM
ataxia telangiectasia mutated
- CPT
camptothecin
- DTT
dithiothreitol
- GFP
green fluorescent protein
- GST
glutathione transferase
- HEK
human embryonic kidney
- NKX3.1
NK3 homeobox 1
- PARP
poly(ADP-ribose) polymerase
- siRNA
small interfering RNA
- SV40
simian virus 40
- Topo70
N-terminally truncated topoisomerase I
REFERENCES
- 1.Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Norton CR, Gridley T, Cardiff RD, Cunha GR, Abate-Shen C and Shen MM (1999) Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 13, 966–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen C, Bubendorf L, Voeller HJ, Slack R, Willi N, Sauter G, Gasser TC, Koivisto P, Lack EE, Kononen J et al. (2000) Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 60, 6111–6115 [PubMed] [Google Scholar]
- 3.Asatiani E, Huang WX, Wang A, Rodriguez OE, Cavalli LR, Haddad BR and Gelmann EP (2005) Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res. 65, 1164–1173 [DOI] [PubMed] [Google Scholar]
- 4.Bethel CR, Faith D, Li X, Guan B, Hicks JL, Lan F, Jenkins RB, Bieberich CJ and De Marzo AM (2006) Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: association with gleason score and chromosome 8p deletion. Cancer Res. 66, 10683–10690 [DOI] [PubMed] [Google Scholar]
- 5.Kim MJ, Bhatia-Gaur R, Banach-Petrosky WA, Desai N, Wang Y, Hayward SW, Cunha GR, Cardiff RD, Shen MM and Abate-Shen C. (2002) Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 62, 2999–3004 [PubMed] [Google Scholar]
- 6.Bowen C and Gelmann EP (2010) NKX3.1 activates cellular response to DNA damage. Cancer Res. 70, 3089–3097 [DOI] [PubMed] [Google Scholar]
- 7.Bowen C, Stuart A, Ju JH, Tuan J, Blonder J, Conrads TP, Veenstra TD and Gelmann EP (2007) NKX3.1 homeodomain protein binds to topoisomerase I and enhances its activity. Cancer Res. 67, 455–464 [DOI] [PubMed] [Google Scholar]
- 8.Champoux JJ (2001) DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 9.Koster DA, Croquette V, Dekker C, Shuman S and Dekker NH (2005) Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 434, 671–674 [DOI] [PubMed] [Google Scholar]
- 10.Kingma PS and Osheroff N. (1998) The response of eukaryotic topoisomerases to DNA damage. Biochim. Biophys. Acta 1400, 223–232 [DOI] [PubMed] [Google Scholar]
- 11.Simmons DT, Roy R, Chen L, Gai D and Trowbridge PW (1998) The activity of topoisomerase I is modulated by large T antigen during unwinding of the SV40 origin. J. Biol. Chem 273, 20390–20396 [DOI] [PubMed] [Google Scholar]
- 12.Gai D, Roy R, Wu C and Simmons DT (2000) Topoisomerase I associates specifically with simian virus 40 large-T-antigen double hexamer-origin complexes. J. Virol. 74, 5224–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pommier Y. (2009) DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 109, 2894–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andoh T, Ishii K, Suzuki Y, Ikegami Y, Kusunoki Y, Takemoto Y and Okada K. (1987) Characterization of a mammalian mutant with a camptothecin-resistant DNA topoisomerase I. Proc. Natl. Acad. Sci. U.S.A 84, 5565–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beidler DR and Cheng YC (1995) Camptothecin induction of a time- and concentration-dependent decrease of topoisomerase I and its implication in camptothecin activity. Mol. Pharmacol. 47, 907–914 [PubMed] [Google Scholar]
- 16.Fiorani P, Bruselles A, Falconi M, Chillemi G, Desideri A and Benedetti P. (2003) Single mutation in the linker domain confers protein flexibility and camptothecin resistance to human topoisomerase I. J. Biol. Chem 278, 43268–43275 [DOI] [PubMed] [Google Scholar]
- 17.Chillemi G, D’Annessa I, Fiorani P, Losasso C, Benedetti P and Desideri A. (2008) Thr729 in human topoisomerase I modulates anti-cancer drug resistance by altering protein domain communications as suggested by molecular dynamics simulations. Nucleic Acids Res. 36, 5645–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frangioni JV and Neel BG (1993) Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210, 179–187 [DOI] [PubMed] [Google Scholar]
- 19.Stewart L, Ireton GC and Champoux JJ (1997) Reconstitution of human topoisomerase I by fragment complementation. J. Mol. Biol 269, 355–372 [DOI] [PubMed] [Google Scholar]
- 20.Zheng SL, Ju JH, Chang BL, Ortner E, Sun J, Isaacs SD, Sun J, Wiley KE, Liu W, Zemedkun M et al. (2006) Germ-line mutation of NKX3.1 cosegregates with hereditary prostate cancer and alters the homeodomain structure and function. Cancer Res. 66, 69–77 [DOI] [PubMed] [Google Scholar]
- 21.Gelmann EP, Steadman DJ, Ma J, Ahronovitz N, Voeller HJ, Swope S, Abbaszadegan M, Brown KM, Strand K, Hayes RB and Stampfer MJ (2002) Occurrence of NKX3.1 C154T polymorphism in men with and without prostate cancer and studies of its effect on protein function. Cancer Res. 62, 2654–2659 [PubMed] [Google Scholar]
- 22.Weiler S, Gruschus JM, Tsao DH, Yu L, Wang LH, Nirenberg M and Ferretti JA (1998) Site-directed mutations in the vnd/NK-2 homeodomain. Basis of variations in structure and sequence-specific DNA binding. J. Biol. Chem 273, 10994–11000 [DOI] [PubMed] [Google Scholar]
- 23.Yang Z and Champoux JJ (2002) Reconstitution of enzymatic activity by the association of the cap and catalytic domains of human topoisomerase I. J. Biol. Chem 277, 30815–30823 [DOI] [PubMed] [Google Scholar]
- 24.Redinbo MR, Stewart L, Kuhn P, Champoux JJ and Hol WG (1998) Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 279, 1504–1513 [DOI] [PubMed] [Google Scholar]
- 25.Laco GS, Collins JR, Luke BT, Kroth H, Sayer JM, Jerina DM and Pommier Y. (2002) Human topoisomerase I inhibition: docking camptothecin and derivatives into a structure-based active site model. Biochemistry 41, 1428–1435 [DOI] [PubMed] [Google Scholar]
- 26.Miao ZH, Player A, Shankavaram U, Wang YH, Zimonjic DB, Lorenzi PL, Liao ZY, Liu H, Shimura T, Zhang HL et al. (2007) Nonclassic functions of human topoisomerase I: genome-wide and pharmacologic analyses. Cancer Res. 67, 8752–8761 [DOI] [PubMed] [Google Scholar]
- 27.Pommier Y, Pourquier P, Fan Y and Strumberg D. (1998) Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta 1400, 83–105 [DOI] [PubMed] [Google Scholar]
- 28.Gilmour DS, Pflugfelder G, Wang JC and Lis JT (1986) Topoisomerase I interacts with transcribed regions in Drosophila cells. Cell 44, 401–407 [DOI] [PubMed] [Google Scholar]
- 29.Subramanian D, Rosenstein BS and Muller MT (1998) Ultraviolet-induced DNA damage stimulates topoisomerase I–DNA complex formation in vivo: possible relationship with DNA repair. Cancer Res. 58, 976–984 [PubMed] [Google Scholar]
- 30.Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 31.Bieberich CJ, Fujita K, He WW and Jay G. (1996) Prostate-specific and androgen-dependent expression of a novel homeobox gene. J. Biol. Chem 271, 31779–31782 [DOI] [PubMed] [Google Scholar]
- 32.Kos L, Chiang C and Mahon KA (1998) Mediolateral patterning of somites: multiple axial signals, including Sonic hedgehog, regulate Nkx-3.1 expression. Mech. Dev. 70, 25–34 [DOI] [PubMed] [Google Scholar]
- 33.Mao Y, Okada S, Chang LS and Muller MT (2000) p53 dependence of topoisomerase I recruitment in vivo. Cancer Res. 60, 4538–4543 [PubMed] [Google Scholar]
- 34.Karayan L, Riou JF, Seite P, Migeon J, Cantereau A and Larsen CJ (2001) Human ARF protein interacts with topoisomerase I and stimulates its activity. Oncogene 20, 836–848 [DOI] [PubMed] [Google Scholar]
- 35.Boothman DA, Trask DK and Pardee AB (1989) Inhibition of potentially lethal DNA damage repair in human tumor cells by β-lapachone, an activator of topoisomerase I. Cancer Res. 49, 605–612 [PubMed] [Google Scholar]
- 36.Mao Y and Muller MT (2003) Down modulation of topoisomerase I affects DNA repair efficiency. DNA Repair 2, 1115–1126 [DOI] [PubMed] [Google Scholar]
- 37.Lebedeva NA, Rechkunova NI, Agama K, Pommier Y and Lavrik OI (2009) Interaction of DNA topoisomerase 1 with DNA intermediates and proteins of base excision repair. Biochemistry 74, 1278–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steadman DJ, Giuffrida D and Gelmann EP (2000) DNA-binding sequence of the human prostate-specific homeodomain protein NKX3.1. Nucleic Acids Res. 28, 2389–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ju JH, Maeng JS, Zemedkun M, Ahronovitz N, Mack JW, Ferretti JA, Gelmann EP and Gruschus JM (2006) Physical and functional interactions between the prostate suppressor homeoprotein NKX3.1 and serum response factor. J. Mol. Biol 360, 989–999 [DOI] [PubMed] [Google Scholar]
- 40.Carson JA, Fillmore RA, Schwartz RJ and Zimmer WE (2000) The smooth muscle γ -actin gene promoter is a molecular target for the mouse bagpipe homologue, mNkx3–1, and serum response factor. J. Biol. Chem 275, 39061–39072 [DOI] [PubMed] [Google Scholar]