Abstract
Lung cancer remains the leading cause of cancer related death world-wide despite advances in treatment. This largely relates to the fact that many of these patients already have advanced diseases at the time of initial diagnosis. As most lung cancers present as nodules initially, an accurate classification of pulmonary nodules as early lung cancers is critical to reducing lung cancer morbidity and mortality. There have been significant recent advances in artificial intelligence (AI) for lung nodule evaluation. Deep learning (DL) and convolutional neural networks (CNNs) have shown promising results in pulmonary nodule detection and have also excelled in segmentation and classification of pulmonary nodules. This review aims to provide an overview of progress that has been made in AI recently for pulmonary nodule detection and characterization with the ultimate goal of lung cancer prediction and classification while outlining some of the pitfalls and challenges that remain to bring such advancements to routine clinical use.
Keywords: Artificial intelligence (AI), machine learning (ML), pulmonary nodule
Introduction
Lung cancer remains the leading cause of cancer related death among males and females in the United States (1). Over the past few decades, while the one-year survival rate for lung cancer has increased, the overall 5-year survival rate remains dismally low at 19% (1). A major factor for this poor prognosis relates to the fact that many patients already have advanced disease at the time of initial diagnosis. Previous literature reported that up to 55% of lung cancer patients already have distant disease when they are initially diagnosed (2), with a current predicted 5-year survival rate of 5% (1). An accurate classification of pulmonary nodules as early stage lung cancers is critical to help reduce lung cancer morbidity and mortality.
Chest radiography remains one of the most common imaging tests being ordered (3,4) and is often the first examination in the clinician’s arsenal when working up the first symptoms of lung cancer such as cough. Interpretations of chest radiographs by human readers remains neither specific nor sensitive in the diagnosis of lung cancers with reported sensitivity ranging between 36–84% depending on the study population and tumor size (3,5-8). In fact, data from the literature has demonstrated that 19–26% of pulmonary neoplasms seen on chest radiographs were not detected during their first interpretation (8,9).
Such shortcomings have led to increasing reliance on computed tomography (CT) as a diagnostic and screening tool. Additionally, there is an expanding role of CT in the use of lung cancer screening after the National Lung Cancer Screening Trial (NLST) demonstrated an improvement in mortality in high risk patients when screened with low-dose CT (LDCT) (10). However, although sensitivity for pulmonary nodule detection is improved with CT, specificity for lung cancer diagnosis remains somewhat low (7). Moreover, human interpretation of CT remains subjective with high reader variability for lung cancer detection and diagnosis (11).
One approach to improve the aforementioned shortcomings of radiography and CT is the use of artificial intelligence (AI). Over the last few years, there has been a surge in research and development of AI and much has been published on the use of AI in the detection and characterization of lung nodules on both radiography and CT. This review is not meant to be all-comprehensive, however, our aim is to provide an overview of the progress in AI for pulmonary nodule evaluation while outlining some of the pitfalls and challenges that remain to bring such advancement to routine clinical use.
Terminology
Although applying AI to medicine has been conceptualized since the late 1950s (12), the extent of its use was limited. However, significant advances in both hardware and software have facilitated the recent explosive growth of this field. It is important to first define AI and other commonly used terms in the field of AI including machine learning (ML) and deep learning (DL).
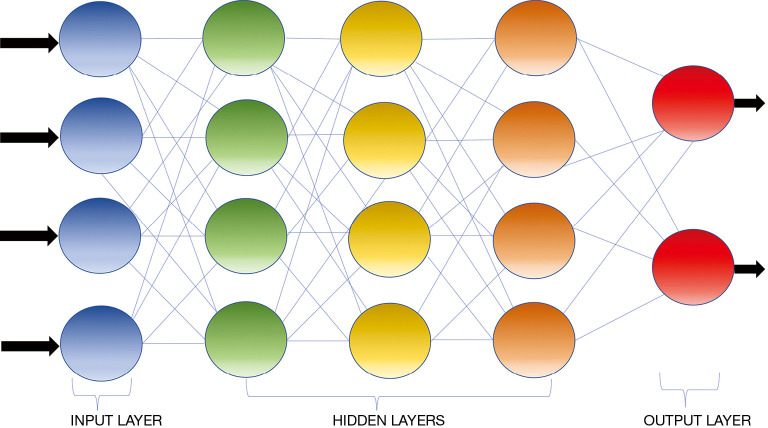
AI is defined as a discipline of computer science which focuses on the creation of machines that are able to perceive the world and perform similar to humans (13). The initial AI algorithms, meant for simple data analysis, were hard-coded by programmers and did not recognize patterns not specially programmed (14). ML is a subfield of AI where algorithms can recognize and learn patterns within complex data sets to produce intellectual predictions rather than by explicit programming (14,15). However, most traditional ML algorithms still required human input and the patterns such algorithms are capable of evaluating are still fairly simple. DL can be conceptualized as a class of ML where algorithms are organized into many processing layers based on artificial neural networks, similar to the human brain. The most commonly used DL model for medical imaging is the convolutional neural network (CNN) (16) (Figure 1) which was originally described by Fukushima in 1980 (17). LeCun et al. first described the use of backpropagation to train CNNs for image recognition in 1989 (18). In 2012, Krizhevsky et al. were the first to use a graphics processing unit (GPU) to train a CNN to classify objects and as result won the ImageNet Large Scale Visual Recognition Challenge (19). CNN does not require human intervention for complex data analysis (20). Modeled off the human brain with neurons organized into multiple layers (21), CNN contains input and output layers and the computational strength of the networks lies in the integration of multiple “neurons” within the multiple deep hidden layers between the input and output layers, where the outputs of one layer serve as the input of the next layer (Figure 2) (22).
Figure 1.
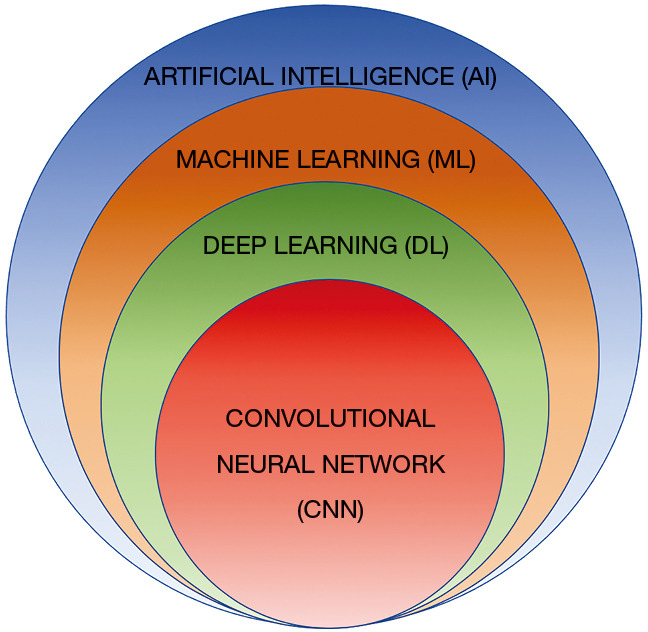
Euler diagram demonstrating the AI hierarchy. ML is a subfield of AI where algorithms can recognize and learn patterns using complex data sets to produce without explicit programming. DL can be conceptualized as a class of ML where algorithms are organized into many processing layers based on artificial neural networks, similar to the human brain. CNN is the type of DL model most commonly used for medical imaging presently. AI, artificial intelligence; ML, machine learning; DL, deep learning; CNN, convolutional neural network.
Figure 2.
Schematic representation of a convolutional neural network containing an input layer, three hidden connected layers, and an output layer. The computational strength of such network lies in the integration of multiple “neurons” (represented by the circles) within the deep hidden layers between the input and output layers. Typically the outputs of one layer serve as the input of the next layer.
DL has been applied to multiple facets of imaging, including thoracic imaging, where it is used for quantification of diffuse lung diseases (23-26) detection of tuberculosis (27) or pneumonia (28), and evaluation of lung nodules (29) among other things. The most common utility of DL in thoracic imaging at this time is pulmonary nodule assessment. AI can help detect, segment and characterize pulmonary nodules on chest radiography, CT and positron emission tomography (PET).
Applications
Nodule segmentation
In imaging, segmentation is utilized to isolate an object from its surroundings for analysis, such as for nodule size evaluation. It is well known that nodule size is a strong predictor of malignancy (30,31). Volumetry is currently being promoted as the preferred method of nodule size measurement and growth determination, given its superior reproducibility, potential for three-dimensional analysis of the entire nodule and sensitivity in detecting nodule growth compared to linear measurements (32-36). Volumetry was utilized in the Dutch-Belgian Randomized Lung Cancer Screening Trial (Dutch acronym: NELSON) (37) and is now incorporated into the United States Lung CT Screening Reporting and Data System (Lung-RADS 1.1).
There have been multiple attempts to develop CAD algorithms for nodule segmentation since the 1980s. Rules for many segmentation algorithms are programed explicitly into the software. For example, on CT, the criteria used for segmentation could be pixel attenuation (14). However, this approach was limited, as the anatomy to be segmented is often too complex to yield optimal results; for instance when a nodule is adjacent to a structure, such as a vessel, with similar attenuation (38). Moreover, most segmentation approaches are semi-automated, requiring user input (such as drawing a region of interest to initiate segmentation) which can be labor intensive and can introduce intra- and inter-observer variability.
Such shortcomings can be obviated with DL automated nodule segmentation. DL semantic segmentation refers to the process of linking each pixel in an image to a class label to determine the boundary conditions which delineate a specific object. The fully convolutional network is a significant breakthrough in DL semantic segmentation. Convolutions with large receptive fields replace the fully connected layers that are present in the standard CNN (39). U-Net is one of the most recognized segmentation CNN used in biomedical imaging and combines a uniform amount of up- and down-sampling layers with skip connections between opposing convolution and deconvolution layers (40). A receptive field is the region in the input layer that a corresponding CNN feature looks at. Object detection and segmentation mask production for each occurrence can be achieved simultaneously with mask region-based CNN (R-CNN) (41).
Nodule detection
Since the development of graphical processing units and CNN, there has been a significant boost in the performance of computer-aided detection (CAD). The goal of CAD is to have high sensitivity while having a low number of false positives.
Radiography
Multiple strategies have been developed using CAD to detect pulmonary nodules on chest radiographs, ranging from CAD alone to CAD with concomitant bone suppression using dual-energy. These approaches report variable success, with nodule detection sensitivity ranging from 51.6% to 87% (4,42-46).
More recently, CAD systems using DL algorithms have shown increased accuracy for nodule detection on chest radiographs compared to conventional ML. Hwang et al. demonstrated that a DL algorithm trained on a dataset of 54,221 normal and 35,613 abnormal chest radiographs was able to distinguish normal from neoplasm, active tuberculosis, pneumothorax, and pneumonia with a median (range) area under the curve (AUC) of 0.979 (0.973–1.000) for image-wise classification and 0.972 (0.923–0.985) for lesion-wise identification (47). The algorithm also demonstrated significantly better performance than thoracic radiologists, general board-certified radiologists, and non-radiology physicians. Additionally, all three categories of human readers improved when they used this algorithm as a second reader.
Pesce et al. using CNN with a visual attention network demonstrated an accuracy of 0.76 for nodule detection and 0.65 for nodule localization on chest radiographs (48). In 2018, Nam et al. developed a DL algorithm for malignant pulmonary nodule detection on chest radiographs and compared its performance with that of 18 physicians (half were radiologists) (49). In the data set they used, there were a total of 43,292 chest radiographs with a normal to diseased ratio of 3.67. Using an external validation data set, they demonstrated that the AUC of the algorithm was higher than that of 17 of the 18 human interpreters. Additionally, as with previous studies, the human interpreters also demonstrated improved nodule detection when the algorithm was used as a second reader.
CT
The use of AI for chest CTs is more complicated than for radiographs given the large number of images and 3D nature of each CT exam. Detection of small pulmonary nodules on CT can be challenging given the fact that a volumetric CT has over 9 million voxels and a 5-mm nodule only occupies approximately 130 voxels (38). Multiple studies have shown that there is significant variability in nodule detection sensitivity amongst radiologists (50-52), with as many as 8.9% of cancers missed on the National Lung Screening Trial (NLST) (53). Although concurrent reading of scans by two radiologists improves detection sensitivity, it is impractical in daily practice given its time-consuming and inefficient nature (54). For this reason, it is extremely useful to use AI to aid radiologists in nodule identification and to act as a “second radiologist” to concurrently read the study. Additionally, low dose CT (LDCT) has been widely accepted after the NLST demonstrated improvement in mortality in high risk patients when screened with LDCT (10). This has caused an increase in CT utilization and an even greater need for a CAD system to aid in the radiologist’s workflow by maintaining interpretation accuracy in the face of increased work volume.
CAD for lung nodule detection began in the early 2000s using traditional ML approaches such as support vector machines (SVM) (55). Traditional CAD systems have provided good results on CT, though, often involving complex pipelines of algorithms that rely deeply on manual input which limited their performance (55). Traditional CAD algorithms for pulmonary nodules include nodule segmentation, feature extraction, classification of lesions as non-nodules as opposed to true nodules. The number of selected features and the type of ML model used for classification (Fisher linear discriminant, massive training artificial neural network, random forest, distance weighted nearest neighbor support vector among others) is dependent on the type of CAD system being utilized (21,56). Although conventional ML CAD successfully assisted nodule detection, one pitfall of conventional ML is overfitting where there is apparent high algorithm performance for a particular training data set that cannot be replicated on other independent datasets (57).
Algorithms using DL can possibly eliminate the innate obstacles in traditional CAD systems by eliminating the need for complex human-led pipelines and their ability to self-learn previously unknown features with limited direct supervision (38,39). In 2015, Hua et al. were the first to publish results of a DL pulmonary nodule detection system on CT, reporting a sensitivity of 73% and a specificity of 80%, which was superior to the conventional CAD systems available at that time (58). Since that time, there have been multiple studies showing the superiority of CAD with DL compared to conventional CAD, including a DL system studied by Setio et al. in 2016 which reached 85.4% sensitivity for nodule detection with only one false positive lesion per scan (59). In 2018, Huang et al. developed a DL network for detection of pulmonary nodules using the LUNA 16 and Ali Tianchi databases and evaluated its performance on the LUNA 16 dataset. They noted false positive rates of 0.125 and 0.25 per scan with sensitivities reaching as high as 81.7% and 85.1%, respectively (60). There have also been studies that exhibit sensitivity of nodule detection as high as 95%, however, they have a wide variety of false positive rates (1.17 to 22.4) (61-63). In 2020, Schwyzer et al. studied the diagnostic performance of DL for small 18F-fluorodeoxyglucose (FDG) avid pulmonary nodules in PET scans and examined whether different image reconstruction [block sequential regularized expectation maximization (BSREM) and ordered subset expectation maximization (OSEM)] affected nodule detection accuracy (64). They found that the DL algorithm they implemented may aid detection of small FDG avid pulmonary nodules and is affected by image reconstruction. On a per-slice analysis, the sensitivity and specificity were 66.7% and 79% for OSEM, respectively. For BSREM, the sensitivity and specificity were 69.2% and 84.5%, respectively.
Nodule classification and cancer prediction
After nodules are detected, the classification of these nodules as benign or malignant is critical as it guides management. Distinguishing cancerous from noncancerous nodules is particularly important during early stages of the disease, given a 5-year survival rate of 61% for localized non-small cell lung cancer (NSCLC) compared to a dismal 6% for metastasized NSCLC (65).
Unfortunately, imaging appearance of benign and malignant nodules can have considerable overlap, resulting in significant inter-observer variability among radiologist which can lead to missed malignancies, unwarranted interventional procedures, such as biopsies and/or resections with attendant potential complications, and/or unnecessary imaging surveillance which can be costly (66). For this reason, classification of nodules and prediction of malignancy is an area generating a great deal of interest.
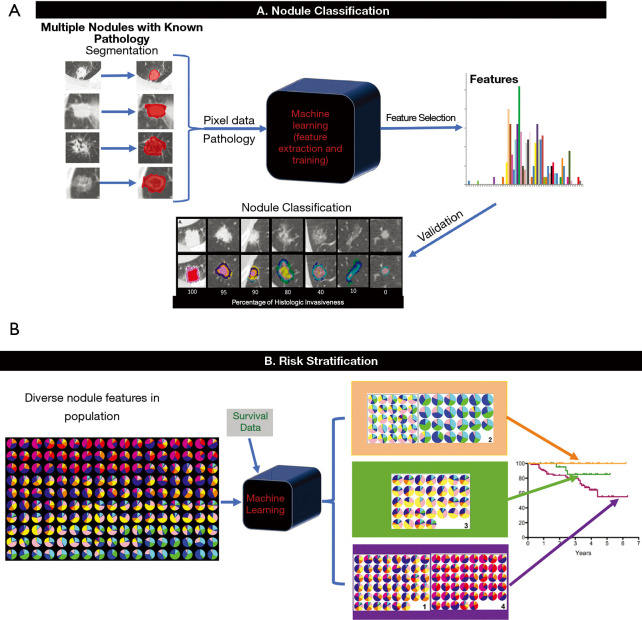
Pulmonary nodules are low-contrast tissues that are not easily distinguished from its surroundings. However, each nodule contains characteristics that can be represented by “features” in ML. For medical imaging, these features are typically numeric. “Radiomics” is a term created by Lambin and colleagues to describe the automatic extraction of characteristics from diagnostic imaging by turning image voxels into a collection of numbers that cannot only help determine lesion malignancy or tumor grade but also monitor treatment response (67). A set of numeric features can be conveniently described by a feature vector. Feature vectors typically represent a lesion’s textures, density, intensity value, shape, and/or geometry. A classifier is the ML model that can differentiate feature vectors between different pulmonary nodule types by application of a training algorithm and labeled data (Figure 3) (68).
Figure 3.
General illustration of a feature based machine learning nodule classification and risk stratification model [modified from Computer-Aided Nodule Assessment and Risk Yield (CANARY), Mayo Clinic, Rochester, MN, USA]. (A) Nodule Classification. After nodule segmentation, radiomic features are extracted from the images. Following feature-pathology correlation and feature selection, the machine learning model is trained to classify pulmonary nodules. There is typically a validation step with a separate set of data to further refine the algorithm before final testing prior to use. (B) Risk stratification. Multiple nodules with features representative of the population and the corresponding survival data are used to train the algorithm, which in turn classifies the features into three main groups in the case of CANARY and performs survival analysis of each group of nodules (the orange group has good, the green group has intermediate and the purple group has poor prognosis).
There is a large volume of articles devoted to multiple kinds of classifiers used such as SVM and random forest to classify pulmonary nodules or determine risk of malignancy (69). Using the SPIE-AAPM Lung CT Challenge dataset of 70 thoracic CTs, Rendon-Gonzalez et al. used SVM trained textural and shape features for lung nodule classification, reporting 78.08% accuracy, 84.93% sensitivity, and 80.92% specificity (70). Lynch et al. applied various ML techniques to the Surveillance, Epidemiology and End Results (SEER) program database to help predict survival in lung cancer patients and concluded that the performance of these techniques, when applied to this particular database, may be on par with classical methods (71).
Newer ML algorithms can also identify spatial complexity, intensity pattern, and a range of other texture features that are beyond human capability to perceive (38). Pathology literature has shown that malignant lung nodules have increased heterogeneity which are not appreciable with the human eye but can be quantified with radiomics (72).
In addition to being applied in CT, there has been recent work using AI for detecting and classifying pulmonary nodules on PET/CT. In 2019, Teramoto et al. developed an automated scheme using a ML technique (random forest) for the classification of nodules using conventional CT in combination with early and delayed phase PET/CT (73). They reported that 94.4% of malignant nodules were identified accurately. The accuracy rate of benign nodule detection using CT in addition to two-phase PET images was 44.4% higher than that obtained by CT images alone and 11.1% higher than CT plus early PET images.
There have also been several studies using ML in examining the radiomic features extracted from the parenchyma surrounding lung nodules (perinodular) to help distinguish benign and malignant nodules. Overall, these studies have found that the perinodular radiomic features can risk stratify lung cancers. For example, in 2019, Uthoff et al. reported that the inclusion of parenchymal imaging features improved the performance of the ML tool over exclusively nodular features (P<0.01) (74).
A key advantage of DL over traditional ML systems is that they are able to maximize classification with limited direct supervision because of their ability to self-learn previously unknown features (38). Studies comparing the use of DL in pulmonary nodule classification has shown superiority over standard ML techniques (43,56,75,76). Ciompi et al. introduced a DL system based on Lung-RADS in 2017 that surpasses classical ML in nodule classification performance while keeping inter-observer variability comparable to four experienced human observers (77). In 2018, Shaffie et al. described a generalized DL system with the potential to be a valuable tool for lung cancer detection because it achieved an accuracy of 91.2% for distinguishing malignant from benign nodules (78).
In 2019, Ardila et al. developed a DL network that used the patient’s prior and current CT volumes to help predict the risk of lung cancer (79). The model demonstrated an excellent performance (94.4% AUC) on 6,716 National Lung Cancer Screening Trial cases and demonstrated similar performance on an independent clinical validation set of 1,139 cases. In this project, they conducted two reader studies. When prior CT imaging was available, the model performance was on-par with the radiologists. The model, however, outperformed all six radiologists when prior CT imaging was not available, with an absolute reduction of 11% in false positives and 5% in false negatives.
Zhou et al. have reported that particular nodule imaging characteristics correlate with specific metagene groups (80) and there has been significant interest in trying to define a radiogenomic signature for specific gene mutations (such as ALK, EGFR, and K-RAS) in order to assess a targeted inhibitory agent treatment response (81-83). Rios Velazquez et al. obtained a radiomic signature that was able to successfully discriminate between positive and negative EGFR cases with an AUC of 0.69 (84).
Pitfalls, challenges, and potential solutions
Although AI offers many potential opportunities for improved patient care, pitfalls and multiple challenges must be overcome prior to routine clinical adoption. Most algorithms, especially DL algorithms, require large, well-labeled anonymized data sets, which are challenging to curate and the process of generating such data sets can be highly labor intensive. Several methods are being adopted to overcome the challenge of limited data, including the development and release of publicly available databases. For example, the National Institutes of Health released 100,000 labeled chest radiographs (85) in 2017. The labels in this database were prepared using natural language processing to derive disease classification data from radiology reports. This allows implementation of larger databases so that the labeling step can be omitted.
Another strategy to tackle the lack of large datasets is to artificially generate data sets with features similar to a given training dataset using CNN such as the generative adversarial network (GAN) (86). Such GANs could be trained to learn representative features in a completely unsupervised manner. Since the features are generated and not selected from pre-existing images, the labeling step can be obviated. GANs can be incorporated with supervised strategies or used independently.
Transfer learning is another strategy that is being employed to help with the need of large annotated databases. This method consists of training large non-medical imaging data sets and then transferring the learned parameters onto the smaller medical imaging datasets (21). For example, in 2016, Bush et al. pretrained a CNN model on a subset of an ImageNet dataset containing millions of labeled real-word images and retrained it to detect the presence or absence of pulmonary nodules on chest radiographs with a sensitivity of 92% and specificity of 86% (87).
An additional hurdle stems from the fact that pathologically proven datasets are typically needed for nodule classification; however, there is potential interpretation variation between pathologists. More sophisticated ML models might need to account for molecular markers given their emerging significance, however, such markers are not uniformly acquired by all institutions. Moreover, uniformed labeling of training cases might be problematic as not all physicians utilize the same terminologies for characterizing lung nodules (e.g., confusion between subsolid versus nonsolid or ground glass) or classifying the actual pathology of the nodules (e.g., minimal invasive adenocarcinoma or adenocarcinoma in situ as opposed to low grade adenocarcinoma). One potential solution is for leading radiology and pathology societies to work on standardized, clearly defined terminologies.
Another challenge that arises in the age of big data are the ethical and legal aspects of data sharing and patient privacy. In the US, the Health Insurance Portability and Accountability Act imposes severe monetary fines for privacy breach. It is therefore extremely important that the data sets used in training are fully anonymized and comply with the laws. Internationally, data protection laws vary. Some geographic locations prevent data from leaving physical locations legally which limits research. Furthermore, implementation of AI into clinical practice requires an interconnected network of patient datasets so that AI is both robust and generalized across different patient diseases, demographics, and regions around the world (66).
To many, ML remains a “black box”, rendering output features not apparent to the human eyes questionable. It is uncertain if correlation with pathology will decrease this skepticism. Education of fellow physicians might mitigate anxiety toward the unknown. In real world clinical settings, physicians typically synthesize clinical as well as imaging features to derive differential diagnoses which can add complexity to any given prediction model. Algorithms incorporating non-imaging and multi-modality imaging features are emerging (88).
Lastly, the most challenging task at present is how best to rigorously validate any algorithm prior to clinical application, because many ML algorithms suffer from overfitting and perform very well for a specific set of data but lack generalizability. Overfitting describes the situation where a model has seemingly high performance for a given data set but fails on unseen independent data. This can be seen when there are insufficient number of samples in a training set or the training set is not well balanced. This problem can be detected using model validation techniques such as cross validation, which estimates how accurately a predictive model will perform in practice. Overfitting is common with DL algorithms that contains many layers generating numerous variables to learn from small training sets. This problem would be alleviated by using larger and/or more varied training sets (89).
A related issue to overfitting is the lack of generalizability (63). Kim et al. studied whether the diagnostic performance of ML based radiomic models for differentiating subsolid nodules and invasive pulmonary adenocarcinomas (IPAs) was affected by image reconstruction. Specifically, the group compared images reconstruction with the recently popularized model-based iterative reconstruction (MBIR) to the traditional filtered back projection (90) and found that inclusion of a CT scan reconstructed using MBIR significantly decreased diagnostic performance for the identification of IPAs. One potential solution would be to use diverse data sets that encompass different image reconstructions to train ML algorithms.
Underfitting is another problem that can be seen during training of algorithms. This happens when there is too much regularization, resulting in an inflexible model that fails to learn from the data set or when the model is too simple, with too few features. For example, models with an insufficient number of features in the presence of multiple subpopulations in the training set will fail to encompass the entire population (89). Therefore, one remedy for this issue is to ensure an adequate number of features during training and to use just enough regularization to prevent overfitting but not so much that generates underfitting.
Conclusions
There has been much progress in AI assisted nodule segmentation, detection and classification in the recent years. However, ML for nodule evaluation is at its infancy and there are still many limitations to overcome, including general acceptance of such disruptive innovation. Nonetheless, ML is here to stay and has demonstrated many promising potentials for pulmonary nodule evaluation and management. It is imperative for both radiologists and clinicians to be cognizant of these algorithms’ capabilities and limitations and play an active role in introducing these tools clinically to improve patient care.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors acknowledge the assistance of Lucy Bahn, PhD, in preparation of this manuscript.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editor (Chi Wan Koo) for the series “Contemporary Practice in Thoracic Neoplasm Diagnosis, Evaluation and Treatment” published in Journal of Thoracic Disease. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at: http://dx.doi.org/10.21037/jtd-2019-cptn-03). The series “Contemporary Practice in Thoracic Neoplasm Diagnosis, Evaluation and Treatment” was commissioned by the editorial office without any funding or sponsorship. CWK served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of Journal of Thoracic Disease from Dec 2018 to Nov 2020. Dr. BJB reports personal fees from Promedior, LLC, other royalties from Imbio, LLC, outside the submitted work. In addition, Dr. BJB has a patent SYSTEMS AND METHODS FOR ANALYZING IN VIVO TISSUE VOLUMES USING MEDICAL IMAGING pending and intellectual property rights to CANARY software but no financial relationships from that software. The authors have no other conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Kligerman S, Cai L, White CS. The effect of computer-aided detection on radiologist performance in the detection of lung cancers previously missed on a chest radiograph. J Thorac Imaging 2013;28:244-52. 10.1097/RTI.0b013e31826c29ec [DOI] [PubMed] [Google Scholar]
- 3.de Hoop B, De Boo DW, Gietema HA, et al. Computer-aided detection of lung cancer on chest radiographs: effect on observer performance. Radiology 2010;257:532-40. 10.1148/radiol.10092437 [DOI] [PubMed] [Google Scholar]
- 4.Schalekamp S, van Ginneken B, Koedam E, et al. Computer-aided detection improves detection of pulmonary nodules in chest radiographs beyond the support by bone-suppressed images. Radiology 2014;272:252-61. 10.1148/radiol.14131315 [DOI] [PubMed] [Google Scholar]
- 5.Li F, Arimura H, Suzuki K, et al. Computer-aided detection of peripheral lung cancers missed at CT: ROC analyses without and with localization. Radiology 2005;237:684-90. 10.1148/radiol.2372041555 [DOI] [PubMed] [Google Scholar]
- 6.Potchen EJ, Cooper TG, Sierra AE, et al. Measuring performance in chest radiography. Radiology 2000;217:456-9. 10.1148/radiology.217.2.r00nv14456 [DOI] [PubMed] [Google Scholar]
- 7.Toyoda Y, Nakayama T, Kusunoki Y, et al. Sensitivity and specificity of lung cancer screening using chest low-dose computed tomography. Br J Cancer 2008;98:1602-7. 10.1038/sj.bjc.6604351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gavelli G, Giampalma E. Sensitivity and specificity of chest X-ray screening for lung cancer: review article. Cancer 2000;89:2453-6. [DOI] [PubMed] [Google Scholar]
- 9.Austin JH, Romney BM, Goldsmith LS. Missed bronchogenic carcinoma: radiographic findings in 27 patients with a potentially resectable lesion evident in retrospect. Radiology 1992;182:115-22. 10.1148/radiology.182.1.1727272 [DOI] [PubMed] [Google Scholar]
- 10.National Lung Screening Trial Research T , Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. 10.1056/NEJMoa1102873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh S, Pinsky P, Fineberg NS, et al. Evaluation of reader variability in the interpretation of follow-up CT scans at lung cancer screening. Radiology 2011;259:263-70. 10.1148/radiol.10101254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ledley RS, Lusted LB. Reasoning foundations of medical diagnosis; symbolic logic, probability, and value theory aid our understanding of how physicians reason. Science 1959;130:9-21. 10.1126/science.130.3366.9 [DOI] [PubMed] [Google Scholar]
- 13.Russell S, Norvig P. Artificial Intelligence: A Modern Approach. 3rd ed: Pearson, 2009. [Google Scholar]
- 14.Auffermann WF, Gozansky EK, Tridandapani S. Artificial Intelligence in Cardiothoracic Radiology. AJR Am J Roentgenol 2019. doi:. 10.2214/AJR.18.20771 [DOI] [PubMed] [Google Scholar]
- 15.Chartrand G, Cheng PM, Vorontsov E, et al. Deep Learning: A Primer for Radiologists. Radiographics 2017;37:2113-31. 10.1148/rg.2017170077 [DOI] [PubMed] [Google Scholar]
- 16.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 17.Fukushima K. Neocognitron: a self organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol Cybern 1980;36:193-202. 10.1007/BF00344251 [DOI] [PubMed] [Google Scholar]
- 18.LeCun Y, Boser B, Denker JS, et al. Backpropagation applied to handwritten zip code recognition. Neural Computation 1989;1:541-51. 10.1162/neco.1989.1.4.541 [DOI] [Google Scholar]
- 19.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep conventional neural networks. 25th International Conference of Neural Information Processing Systems, 2012;1:1097-105. [Google Scholar]
- 20.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 21.Chassagnon G, Vakalopoulou M, Paragios N, et al. Artificial intelligence applications for thoracic imaging. Eur J Radiol 2020;123:108774. 10.1016/j.ejrad.2019.108774 [DOI] [PubMed] [Google Scholar]
- 22.Soffer S, Ben-Cohen A, Shimon O, et al. Convolutional Neural Networks for Radiologic Images: A Radiologist's Guide. Radiology 2019;290:590-606. 10.1148/radiol.2018180547 [DOI] [PubMed] [Google Scholar]
- 23.Walsh SLF, Calandriello L, Silva M, et al. Deep Learning for Classifying Fibrotic Lung Disease on High-Resolution Computed Tomography: A Case-Cohort Study. Lancet Respir Med 2018;6:837-45. 10.1016/S2213-2600(18)30286-8 [DOI] [PubMed] [Google Scholar]
- 24.Kim GB, Jung KH, Lee Y, et al. Comparison of Shallow and Deep Learning Methods on Classifying the Regional Pattern of Diffuse Lung Disease. J Digit Imaging 2018;31:415-24. 10.1007/s10278-017-0028-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park B, Park H, Lee SM, et al. Lung Segmentation on HRCT and Volumetric CT for Diffuse Interstitial Lung Disease Using Deep Convolutional Neural Networks. J Digit Imaging 2019;32:1019-26. 10.1007/s10278-019-00254-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardoso I, Almeida E, Allende-Cid H, et al. Analysis of Machine Learning Algorithms for Diagnosis of Diffuse Lung Diseases. Methods Inf Med 2018;57:272-9. 10.1055/s-0039-1681086 [DOI] [PubMed] [Google Scholar]
- 27.Xiong Y, Ba X, Hou A, et al. Automatic detection of mycobacterium tuberculosis using artificial intelligence. J Thorac Dis 2018;10:1936-40. 10.21037/jtd.2018.01.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephen O, Sain M, Maduh UJ, et al. An Efficient Deep Learning Approach to Pneumonia Classification in Healthcare. J Healthc Eng 2019;2019:4180949. 10.1155/2019/4180949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Causey JL, Zhang J, Ma S, et al. Highly accurate model for prediction of lung nodule malignancy with CT scans. Sci Rep 2018;8:9286. 10.1038/s41598-018-27569-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McWilliams A, Tammemagi MC, Mayo JR, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med 2013;369:910-9. 10.1056/NEJMoa1214726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horeweg N, Scholten ET, de Jong PA, et al. Detection of lung cancer through low-dose CT screening (NELSON): a prespecified analysis of screening test performance and interval cancers. Lancet Oncol 2014;15:1342-50. 10.1016/S1470-2045(14)70387-0 [DOI] [PubMed] [Google Scholar]
- 32.Revel MP, Bissery A, Bienvenu M, et al. Are two-dimensional CT measurements of small noncalcified pulmonary nodules reliable? Radiology 2004;231:453-8. 10.1148/radiol.2312030167 [DOI] [PubMed] [Google Scholar]
- 33.Ko JP, Berman EJ, Kaur M, et al. Pulmonary Nodules: growth rate assessment in patients by using serial CT and three-dimensional volumetry. Radiology 2012;262:662-71. 10.1148/radiol.11100878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devaraj A, van Ginneken B, Nair A, et al. Use of Volumetry for Lung Nodule Management: Theory and Practice. Radiology 2017;284:630-44. 10.1148/radiol.2017151022 [DOI] [PubMed] [Google Scholar]
- 35.Han D, Heuvelmans MA, Vliegenthart R, et al. Influence of lung nodule margin on volume- and diameter-based reader variability in CT lung cancer screening. Br J Radiol 2018;91:20170405. 10.1259/bjr.20170405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yankelevitz DF, Reeves AP, Kostis WJ, et al. Small pulmonary nodules: volumetrically determined growth rates based on CT evaluation. Radiology 2000;217:251-6. 10.1148/radiology.217.1.r00oc33251 [DOI] [PubMed] [Google Scholar]
- 37.Ru Zhao Y, Xie X, de Koning HJ, et al. NELSON lung cancer screening study. Cancer Imaging 2011;11 Spec No A:S79-84. [DOI] [PMC free article] [PubMed]
- 38.Ather S, Kadir T, Gleeson F. Artificial intelligence and radiomics in pulmonary nodule management: current status and future applications. Clin Radiol 2020;75:13-9. 10.1016/j.crad.2019.04.017 [DOI] [PubMed] [Google Scholar]
- 39.Lee SM, Seo JB, Yun J, et al. Deep Learning Applications in Chest Radiography and Computed Tomography: Current State of the Art. J Thorac Imaging 2019;34:75-85. 10.1097/RTI.0000000000000387 [DOI] [PubMed] [Google Scholar]
- 40.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical segmentation. International Conference on Medical image computing and computer-assisted intervention, Springer, 2015:3431-40. [Google Scholar]
- 41.He K, Gkioxari G, Dollar P, et al. Mask r-cnn. Computer Vision (ICCV), 2017 IEEE International Conference 2017:2980-8. [Google Scholar]
- 42.Li F, Engelmann R, Armato SG, 3rd, et al. Computer-aided nodule detection system: results in an unselected series of consecutive chest radiographs. Acad Radiol 2015;22:475-80. 10.1016/j.acra.2014.11.008 [DOI] [PubMed] [Google Scholar]
- 43.Lee KH, Goo JM, Park CM, et al. Computer-aided detection of malignant lung nodules on chest radiographs: effect on observers' performance. Korean J Radiol 2012;13:564-71. 10.3348/kjr.2012.13.5.564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szucs-Farkas Z, Schick A, Cullmann JL, et al. Comparison of dual-energy subtraction and electronic bone suppression combined with computer-aided detection on chest radiographs: effect on human observers' performance in nodule detection. AJR Am J Roentgenol 2013;200:1006-13. 10.2214/AJR.12.8877 [DOI] [PubMed] [Google Scholar]
- 45.Dellios N, Teichgraeber U, Chelaru R, et al. Computer-aided Detection Fidelity of Pulmonary Nodules in Chest Radiograph. J Clin Imaging Sci 2017;7:8. 10.4103/jcis.JCIS_75_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kakeda S, Moriya J, Sato H, et al. Improved detection of lung nodules on chest radiographs using a commercial computer-aided diagnosis system. AJR Am J Roentgenol 2004;182:505-10. 10.2214/ajr.182.2.1820505 [DOI] [PubMed] [Google Scholar]
- 47.Hwang EJ, Park S, Jin KN, et al. Development and Validation of a Deep Learning-Based Automated Detection Algorithm for Major Thoracic Diseases on Chest Radiographs. JAMA Netw Open 2019;2:e191095. 10.1001/jamanetworkopen.2019.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pesce E, Ypsilantis PP, Withey S, et al. Learning to detect chest radiographs containing lung nodules using visual attention networks. ArXiv 2017;1712.00996:1-23. [DOI] [PubMed] [Google Scholar]
- 49.Nam JG, Park S, Hwang EJ, et al. Development and Validation of Deep Learning-based Automatic Detection Algorithm for Malignant Pulmonary Nodules on Chest Radiographs. Radiology 2019;290:218-28. 10.1148/radiol.2018180237 [DOI] [PubMed] [Google Scholar]
- 50.Rubin GD. Lung nodule and cancer detection in computed tomography screening. J Thorac Imaging 2015;30:130-8. 10.1097/RTI.0000000000000140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubin GD, Roos JE, Tall M, et al. Characterizing search, recognition, and decision in the detection of lung nodules on CT scans: elucidation with eye tracking. Radiology 2015;274:276-86. 10.1148/radiol.14132918 [DOI] [PubMed] [Google Scholar]
- 52.Pinsky PF, Gierada DS, Nath PH, et al. National lung screening trial: variability in nodule detection rates in chest CT studies. Radiology 2013;268:865-73. 10.1148/radiol.13121530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scholten ET, Horeweg N, de Koning HJ, et al. Computed tomographic characteristics of interval and post screen carcinomas in lung cancer screening. Eur Radiol 2015;25:81-8. 10.1007/s00330-014-3394-4 [DOI] [PubMed] [Google Scholar]
- 54.Nair A, Screaton NJ, Holemans JA, et al. The impact of trained radiographers as concurrent readers on performance and reading time of experienced radiologists in the UK Lung Cancer Screening (UKLS) trial. Eur Radiol 2018;28:226-34. 10.1007/s00330-017-4903-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goo JM. Computer-aided detection of lung nodules on chest CT: issues to be solved before clinical use. Korean J Radiol 2005;6:62-3. 10.3348/kjr.2005.6.2.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu S, Xie Y, Jirapatnakul A, et al. Pulmonary nodule classification in lung cancer screening with three-dimensional convolutional neural networks. J Med Imaging (Bellingham) 2017;4:041308. 10.1117/1.JMI.4.4.041308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chalkidou A, O'Doherty MJ, Marsden PK. False Discovery Rates in PET and CT Studies with Texture Features: A Systematic Review. PLoS One 2015;10:e0124165. 10.1371/journal.pone.0124165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hua KL, Hsu CH, Hidayati SC, et al. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Onco Targets Ther 2015;8:2015-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Setio AAA, Ciompi F, Litjens G, et al. Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks. IEEE Trans Med Imaging 2016;35:1160-9. 10.1109/TMI.2016.2536809 [DOI] [PubMed] [Google Scholar]
- 60.Huang W, Xue Y, Wu Y. A CAD system for pulmonary nodule prediction based on deep three-dimensional convolutional neural networks and ensemble learning. PLoS One 2019;14:e0219369. 10.1371/journal.pone.0219369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamidian S, Sahiner B, Petrick N, et al. 3D Convolutional Neural Network for Automatic Detection of Lung Nodules in Chest CT. Proc SPIE Int Soc Opt Eng 2017;10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang H, Ma H, Qian W, et al. An Automatic Detection System of Lung Nodule Based on Multigroup Patch-Based Deep Learning Network. IEEE J Biomed Health Inform 2018;22:1227-37. 10.1109/JBHI.2017.2725903 [DOI] [PubMed] [Google Scholar]
- 63.Masood A, Sheng B, Li P, et al. Computer-Assisted Decision Support System in Pulmonary Cancer detection and stage classification on CT images. J Biomed Inform 2018;79:117-28. 10.1016/j.jbi.2018.01.005 [DOI] [PubMed] [Google Scholar]
- 64.Schwyzer M, Martini K, Benz DC, et al. Artificial intelligence for detecting small FDG-positive lung nodules in digital PET/CT: impact of image reconstructions on diagnostic performance. Eur Radiol 2020;30:2031-40. 10.1007/s00330-019-06498-w [DOI] [PubMed] [Google Scholar]
- 65.Society AC. Available online: https://www.cancer.org/cancer/lung-cancer/detection-diagnosis-staging/survival-rates.html [Accessed March 3, 2020.
- 66.van Riel SJ, Sanchez CI, Bankier AA, et al. Observer Variability for Classification of Pulmonary Nodules on Low-Dose CT Images and Its Effect on Nodule Management. Radiology 2015;277:863-71. 10.1148/radiol.2015142700 [DOI] [PubMed] [Google Scholar]
- 67.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Foley F, Rajagopalan S, Raghunath SM, et al. Computer-Aided Nodule Assessment and Risk Yield Risk Management of Adenocarcinoma: The Future of Imaging? Semin Thorac Cardiovasc Surg 2016;28:120-6. 10.1053/j.semtcvs.2015.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X, Mao K, Wang L, et al. An Appraisal of Lung Nodules Automatic Classification Algorithms for CT Images. Sensors (Basel) 2019;19:E194. 10.3390/s19010194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rendon-Gonzalez E, Ponomaryov V. editors. Automatic Lung nodule segmentation and classification in CT images based on SVM. 9th International Kharkiv Symposium on Physics and Engineering of Microwaves, Millimeter and Submillimeter Waves (MSMW); 2016 June 2016; Kharkiv, Ukraine. [Google Scholar]
- 71.Lynch CM, Abdollahi B, Fuqua JD, et al. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int J Med Inform 2017;108:1-8. 10.1016/j.ijmedinf.2017.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mitra S, Shankar BU. Integrating Radio Imaging With Gene Expressions Toward a Personalized Management of Cancer. IEEE Transactions on Human-Machine Systems 2014;44:664-77. 10.1109/THMS.2014.2325744 [DOI] [Google Scholar]
- 73.Teramoto A, Tsujimoto M, Inoue T, et al. Automated Classification of Pulmonary Nodules through a Retrospective Analysis of Conventional CT and Two-phase PET Images in Patients Undergoing Biopsy. Asia Ocean J Nucl Med Biol 2019;7:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uthoff J, Stephens MJ, Newell JD, et al. Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT. Med Phys 2019;46:3207-16. 10.1002/mp.13592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kang G, Liu K, Hou B, et al. 3D multi-view convolutional neural networks for lung nodule classification. PLoS One 2017;12:e0188290. 10.1371/journal.pone.0188290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lyu J, Ling SH. Using Multi-level Convolutional Neural Network for Classification of Lung Nodules on CT images. Annu Int Conf IEEE Eng Med Biol Soc 2018;2018:686-9. 10.1109/EMBC.2018.8512376 [DOI] [PubMed] [Google Scholar]
- 77.Ciompi F, Chung K, van Riel SJ, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep 2017;7:46479. 10.1038/srep46479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shaffie A, Soliman A, Fraiwan L, et al. A Generalized Deep Learning-Based Diagnostic System for Early Diagnosis of Various Types of Pulmonary Nodules. Technol Cancer Res Treat 2018;17:1533033818798800. 10.1177/1533033818798800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ardila D, Kiraly AP, Bharadwaj S, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 2019;25:954-61. 10.1038/s41591-019-0447-x [DOI] [PubMed] [Google Scholar]
- 80.Zhou M, Leung A, Echegaray S, et al. Non-Small Cell Lung Cancer Radiogenomics Map Identifies Relationships between Molecular and Imaging Phenotypes with Prognostic Implications. Radiology 2018;286:307-15. 10.1148/radiol.2017161845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamamoto S, Korn RL, Oklu R, et al. ALK molecular phenotype in non-small cell lung cancer: CT radiogenomic characterization. Radiology 2014;272:568-76. 10.1148/radiol.14140789 [DOI] [PubMed] [Google Scholar]
- 82.Aerts HJ, Grossmann P, Tan Y, et al. Defining a Radiomic Response Phenotype: A Pilot Study using targeted therapy in NSCLC. Sci Rep 2016;6:33860. 10.1038/srep33860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rizzo S, Petrella F, Buscarino V, et al. CT Radiogenomic Characterization of EGFR, K-RAS, and ALK Mutations in Non-Small Cell Lung Cancer. Eur Radiol 2016;26:32-42. 10.1007/s00330-015-3814-0 [DOI] [PubMed] [Google Scholar]
- 84.Rios Velazquez E, Parmar C, Liu Y, et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res 2017;77:3922-30. 10.1158/0008-5472.CAN-17-0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, Peng Y, Lu L, et al. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases, 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, 2017:3462-71. [Google Scholar]
- 86.Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative Adversarial Networks 2014. Available online: https://ui.adsabs.harvard.edu/abs/2014arXiv1406.2661G/abstract. Accessed March 2, 2020.
- 87.Bush I. Lung Nodule Detection and Classification 2016;20:196-209. [Google Scholar]
- 88.Suk HI, Shen D. Deep learning-based feature representation for AD/MCI classification. Med Image Comput Comput Assist Interv 2013;16:583-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chassagnon G, Vakalopolou M, Paragios N, et al. Deep learning: definition and perspectives for thoracic imaging. Eur Radiol 2020;30:2021-30. 10.1007/s00330-019-06564-3 [DOI] [PubMed] [Google Scholar]
- 90.Kim H, Park CM, Gwak J, et al. Effect of CT Reconstruction Algorithm on the Diagnostic Performance of Radiomics Models: A Task-Based Approach for Pulmonary Subsolid Nodules. AJR Am J Roentgenol 2019;212:505-12. 10.2214/AJR.18.20018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as