Abstract
Background
Cutaneous melanoma is an aggressive cancer with increasing incidence and mortality rates worldwide. Metastasis is one of the primary elements that influence the prognosis of patients with cutaneous melanoma. This study aims to clarify the potential mechanism underlying the low survival rate of metastatic melanoma and to search for novel target genes to improve the survival rate of patients with metastatic tumors.
Methods
Gene expression dataset and clinical data were downloaded from The Cancer Genome Atlas portal. Differentially expressed genes (DEGs) were identified, and their functions were studied through gene ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses. Survival and multivariate Cox regression analyses were used to screen out candidate genes that could affect the prognosis of patients with metastatic melanoma.
Results
After a series of comprehensive statistical analysis, 464 DEGs were identified between primary tumor tissues and metastatic tissues. Survival and multivariate Cox regression analyses revealed four vital genes, namely, POU2AF1, ITGAL, CXCR2P1, and MZB1, that affect the prognosis of patients with metastatic melanoma.
Conclusion
This study provides a new direction for studying the pathogenesis of metastatic melanoma. The genes related to cutaneous metastatic melanoma that affect the overall survival time of patients were identified.
Keywords: cutaneous melanoma, metastasis, prognosis, gene analysis
1. Introduction
Cutaneous melanoma is a highly malignant and invasive skin cancer, in which melanocytes switch to cancerous cells through variations at molecular and biochemical levels [1]. The incidence rates of melanoma continuously increase [2]. The 5-year overall survival rate for patients in all stages is 92%, whereas that of patients with advanced metastasis (Stage IV) is 23% [2]. Patients with metastatic melanoma tend to have a poor prognosis. Although various treatments, including surgery, chemotherapy, and radiotherapy, are often effective, no certain treatment can improve the overall survival rate due to the consequence of recurrence and severe metastasis relevant to cutaneous melanoma [3]. Thus, potential biomarkers that can evaluate the prognosis of patients with metastatic melanoma and that can serve as potential therapeutic targets for these patients must be explored.
This study aimed to identify the differentially expressed genes (DEGs) between primary and metastatic melanoma and to determine their main functions through a series of comprehensive biostatistical analyses by using the data from The Cancer Genome Atlas (TCGA) public database. Candidate genes that affect the prognosis of patients with metastatic melanoma were further identified by survival and multivariate cox regression analyses.
2. Methods
2.1. Data sources
Gene expression chart and clinical information were obtained from the TCGA portal as of 17 September 2019. All 472 cutaneous melanoma samples were matched with the corresponding dataset of clinical information for this study. Some samples with no records of the clinical data (n = 4), survival data (n = 20), and solid normal tissue (n = 1) were excluded. In total, 447 samples, including 354 cutaneous metastatic melanoma and 93 primary tumors, were included. Ethics approval was not needed because TCGA is an available public database.
2.2. Differential gene expression in cutaneous melanoma
DEGs were determined by using the edgeR package, and P < 0.01 and |log FC| ≥ 1 were considered statistically significant. Volcano maps were applied to illustrate the results clearly by R package. Cluster profile R package was used to conduct the gene ontology (GO) analysis with a cutoff of P < 0.01 and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis with a cutoff of P < 0.05 and to clarify the biological functional category of these DEGs. Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database was used to construct the interaction network of DEGs with regard to the connection between these genes. Cytoscape software was used to reveal the interaction between the DEGs. Finally, the crucial genes in the interaction network were determined according to the number of edges connected by each gene.
2.3. Survival analysis to select the candidate genes
Univariate Cox regression analysis was applied to evaluate the effect on the expression of these DEGs on overall survival time. Multivariate cox regression analysis was then performed on the top 30 genes according to the results of the univariate Cox regression analysis to distinguish the candidate genes, which are the independent risk factors of prognosis for patients with cutaneous metastatic melanoma.
2.4. Analysis of clinical features
Based on the univariate Cox regression analysis, the gene with the biggest impact on the overall survival time of patients with cutaneous metastatic melanoma was selected for further analysis to investigate the correlation between the expression level of a candidate gene and clinical characteristics. The samples were divided into high expression and low expression groups according to the expression level of the genes, and the Chi-square test was then conducted. Clinical characteristics such as age, gender, TNM stage, and tumor status obtained from the TCGA dataset were included.
3. Results
3.1. DEGs in cutaneous metastatic melanoma
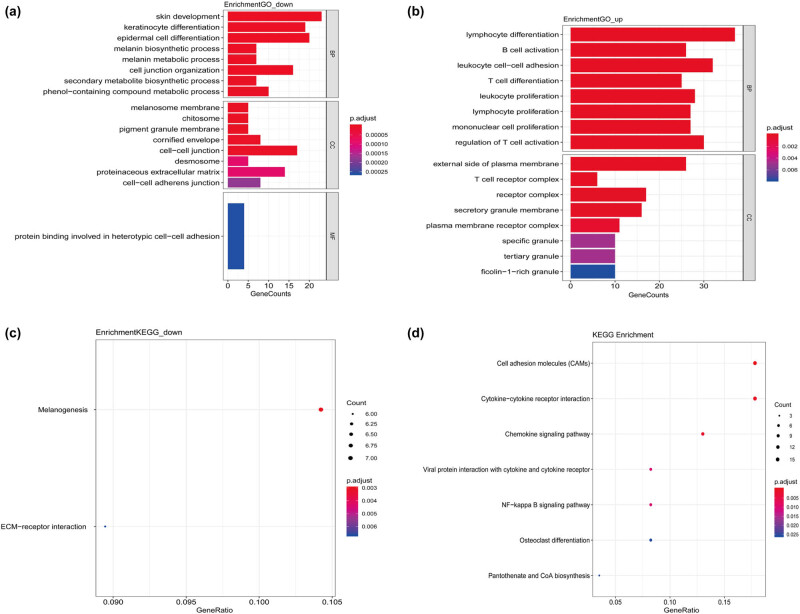
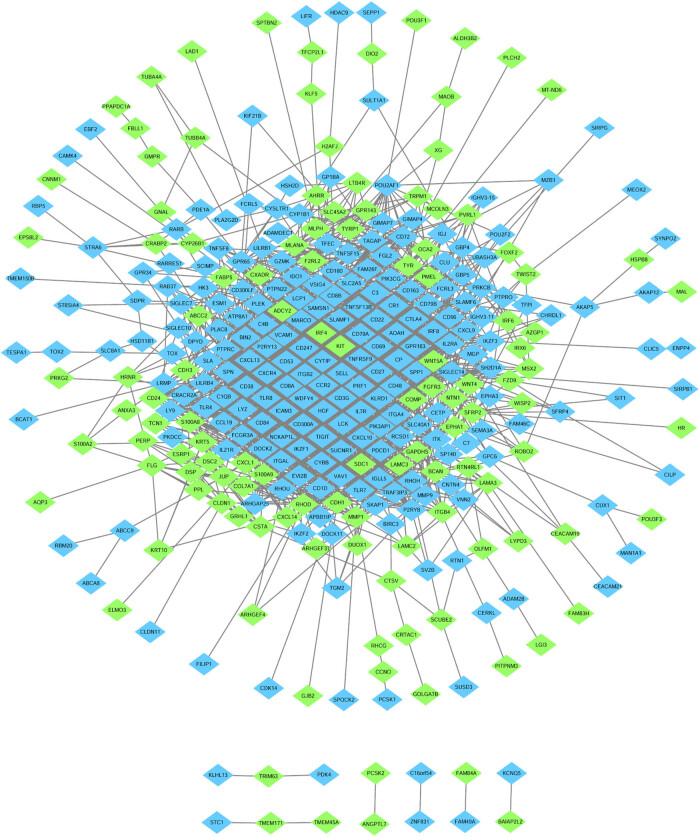
After data screening and processing by R software, standardized data were used to compare the gene expression in primary and metastatic tumors. The edgeR package identified 464 DEGs between primary tumor tissues and cutaneous metastasis tissues. Among these DEGs, 177 were downregulated and 287 were upregulated according to the cutoff of P < 0.01 and |log FC| > 1. Volcano map was used to illustrate the results (Figure 1). GO and KEGG analyses were performed to investigate the functions in which these DEGs were enriched (Figure 2). Downregulated DEGs were enriched in the skin development, keratinocyte differentiation, epidermal cell differentiation, melanin biosynthetic process, and melanin metabolic process. Meanwhile, upregulated DEGs were mainly enriched in lymphocyte differentiation, B-cell activation, leukocyte cell-to-cell adhesion, T cell differentiation, and leukocyte proliferation. KEGG pathway enrichment was also performed to identify the enriched pathways of these genes. The signaling pathways primarily enriched by the downregulated DEGs were the ECM–receptor interaction and tyrosine metabolism signaling way, whereas those primarily enriched in upregulated DEGs were cell adhesion molecules (CAMs), cytokine-to-cytokine receptor interaction, chemokine signaling pathway, viral protein interaction with cytokine and cytokine receptor, and NF-kappa B signaling pathway. STRING (a database of known and predicted protein interactions) was used to predict protein interactions among these DEGs. We then constructed a protein–protein interaction (PPI) network of DEGs, and Cytoscape software was applied to construct a network visualizing the result with 338 nodes and 2,061 edges (Figure 3). The nodes represent the proteins that correspond to the DEGs, and the proteins connected by edges interact with each other. Among them, PTPRC, CTLA4, SELL, ITGB2, TLR4, CXCR4, TLR8, PLEK, CD69, and IKZF1 with high degrees are considered as hub-genes, which suggests they may play important roles in the development of metastasis of melanoma (Figure 4).
Figure 1.
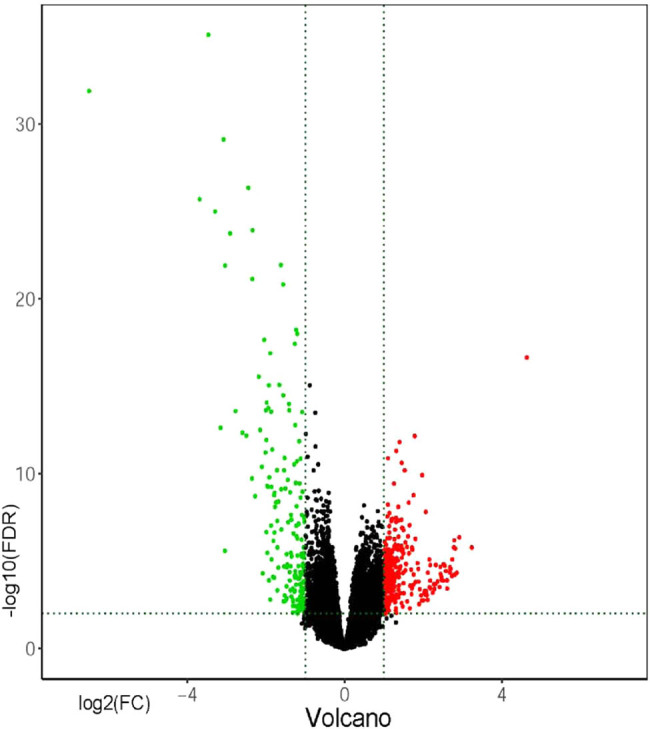
The DEGs of metastasizes and primary tumors. The y-axis value is log FC, and the x-axis value is −log 10(FDR). The red plots represent the upregulated DEGs, while green plots represent the downregulated DEGs.
Figure 2.
GO and KEGG enrichment analyses of differentially expressed genes. (a) GO analysis results of the downregulated DEGs. (b) GO analysis results of the upregulated DEGs. (c) KEGG analysis results of the downregulated DEGs. (d) KEGG analysis results of the upregulated DEGs.
Figure 3.
The protein–protein interaction network of DEGs. The nodes represent proteins that correspond to the DEGs. Upregulated DEGs are represented in blue, while downregulated DEGs are represented in green. The proteins connected by edges interact with each other.
Figure 4.
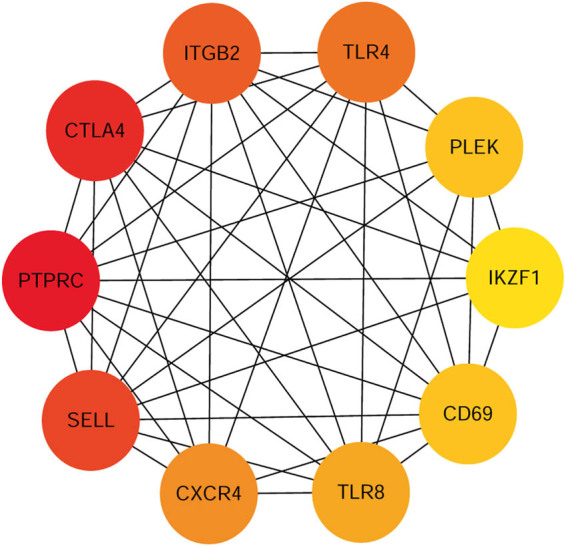
Hub genes in protein–protein interaction network. The top10 hub genes ranked by the degree in the protein–protein interaction network make up a subnetwork.
3.2. Survival analysis
Cutaneous melanoma is a highly invasive disease, and the appearance of metastatic tumor indicates its further deterioration. The survival analysis indicated that 147 DEGs have a significant effect on the overall survival time with a cutoff of P < 0.01. The top 30 DEGs, including IGLV2-14, MZB1, CD27, POU2AF1, CD72, PDCD1, HSH2D, PRF1, SIT1, LY9, PLA2G2D, IGLV2-8, RNF39, CXCL9, CD38, CXCR2P1, SIRPG, IGKV4-1, FCRL5, RAB37, ADAMDEC1, LCK, IGLV3-21, IGHV3-15, IGHM, IGLC2, ITGAL, RHOH, IGLC3, and ICAM3, were chosen as the potential risk factors for the prognosis of patients with metastatic tumors (Table 1). In addition, the multivariate Cox regression analysis was performed on these genes to determine the candidate genes that can exhibit a significant prognostic value. Finally, POU2AF1 (P = 0.00660, HR = 1.36, B = 0.31, CI [1.09–1.70]), ITGAL (P = 0.02181, HR = 1.72, B = 0.54, CI [1.08–2.75]), CXCR2P1 (P = 0.02379, HR = 1.19, B = 0.17, CI [1.02–1.37]), and MZB1 (P = 0.03646, HR = 1.30, B = 0.26, CI [1.02–1.66]) were discovered to be the independent risk factors with P < 0.05 and hazard rate (HR) > 1 (Table 2).
Table 1.
Top30 genes significantly affect the overall survival time of patients by survival analysis
| Symbol | HR | Lower 95 | Upper 95 | P-value | |
|---|---|---|---|---|---|
| ENSG00000211666 | IGLV2-14 | 1.064586836 | 1.033802118 | 1.096288266 | 2.91 × 10−5 |
| ENSG00000170476 | MZB1 | 1.070874267 | 1.036688679 | 1.106187151 | 3.52 × 10−5 |
| ENSG00000139193 | CD27 | 1.104194794 | 1.053468406 | 1.157363747 | 3.62 × 10−5 |
| ENSG00000110777 | POU2AF1 | 1.076582536 | 1.039061559 | 1.115458411 | 4.56 × 10−5 |
| ENSG00000137101 | CD72 | 1.142400427 | 1.07114116 | 1.218400324 | 5.09 × 10−5 |
| ENSG00000188389 | PDCD1 | 1.088919949 | 1.044023795 | 1.135746773 | 7.33 × 10−5 |
| ENSG00000196684 | HSH2D | 1.093259058 | 1.046041572 | 1.142607904 | 7.55 × 10−5 |
| ENSG00000180644 | PRF1 | 1.100839397 | 1.049524241 | 1.154663542 | 7.99 × 10−5 |
| ENSG00000137078 | SIT1 | 1.097245862 | 1.047646805 | 1.149193103 | 8.42 × 10−5 |
| ENSG00000122224 | LY9 | 1.09801634 | 1.047826183 | 1.150610572 | 8.97 × 10−5 |
| ENSG00000117215 | PLA2G2D | 1.062038864 | 1.029489807 | 1.095617015 | 0.000150662 |
| ENSG00000278196 | IGLV2-8 | 1.060655117 | 1.028816517 | 1.09347902 | 0.000152534 |
| ENSG00000204618 | RNF39 | 0.858613102 | 0.792757339 | 0.92993962 | 0.00018116 |
| ENSG00000138755 | CXCL9 | 1.075199152 | 1.035120947 | 1.116829121 | 0.000183342 |
| ENSG00000004468 | CD38 | 1.092412444 | 1.042907907 | 1.144266851 | 0.000187308 |
| ENSG00000229754 | CXCR2P1 | 1.069877224 | 1.032359785 | 1.108758101 | 0.000208432 |
| ENSG00000089012 | SIRPG | 1.083532648 | 1.038542691 | 1.130471581 | 0.000209063 |
| ENSG00000211598 | IGKV4-1 | 1.055884647 | 1.025779669 | 1.086873157 | 0.000229057 |
| ENSG00000143297 | FCRL5 | 1.062535038 | 1.028736881 | 1.097443602 | 0.000235294 |
| ENSG00000172794 | RAB37 | 1.105656553 | 1.047967049 | 1.166521805 | 0.000239164 |
| ENSG00000134028 | ADAMDEC1 | 1.075612496 | 1.03447626 | 1.118384525 | 0.000248689 |
| ENSG00000182866 | LCK | 1.090707281 | 1.041152223 | 1.14262098 | 0.000252373 |
| ENSG00000211662 | IGLV3-21 | 1.055848619 | 1.025517144 | 1.087077202 | 0.000257917 |
| ENSG00000211943 | IGHV3-15 | 1.055896242 | 1.025467865 | 1.08722751 | 0.000266718 |
| ENSG00000211899 | IGHM | 1.057699963 | 1.026263913 | 1.090098947 | 0.000268378 |
| ENSG00000211677 | IGLC2 | 1.060772 | 1.027622014 | 1.094991369 | 0.000270526 |
| ENSG00000005844 | ITGAL | 1.098116455 | 1.044162118 | 1.154858741 | 0.000271455 |
| ENSG00000168421 | RHOH | 1.09005463 | 1.040546325 | 1.141918499 | 0.000277017 |
| ENSG00000211679 | IGLC3 | 1.060198803 | 1.027263638 | 1.094189905 | 0.00028281 |
| ENSG00000076662 | ICAM3 | 1.096557746 | 1.042964911 | 1.152904453 | 0.000311657 |
Table 2.
Results of multivariate Cox regression analysis of top30 genes
| Symbol | Coef. | HR | Lower 95 | Upper 95 | Pr (>|z|) | |
|---|---|---|---|---|---|---|
| ENSG00000211666 | IGLV2–14 | 0.019054 | 1.019237 | 0.8858 | 1.1728 | 0.79017 |
| ENSG00000170476 | MZB1 | 0.262876 | 1.300665 | 1.0167 | 1.6639 | 0.03646* |
| ENSG00000139193 | CD27 | 0.103152 | 1.10866 | 0.7666 | 1.6034 | 0.58371 |
| ENSG00000110777 | POU2AF1 | 0.30893 | 1.361966 | 1.0898 | 1.7021 | 0.0066* |
| ENSG00000137101 | CD72 | −0.276222 | 0.758644 | 0.5625 | 1.0232 | 0.07033 |
| ENSG00000188389 | PDCD1 | −0.129255 | 0.878749 | 0.6704 | 1.1519 | 0.34929 |
| ENSG00000196684 | HSH2D | −0.268995 | 0.764147 | 0.589 | 0.9913 | 0.04279* |
| ENSG00000180644 | PRF1 | −0.017232 | 0.982916 | 0.7773 | 1.2429 | 0.88557 |
| ENSG00000137078 | SIT1 | −0.186718 | 0.829678 | 0.5577 | 1.2342 | 0.35679 |
| ENSG00000122224 | LY9 | −0.086585 | 0.917057 | 0.6824 | 1.2324 | 0.56587 |
| ENSG00000117215 | PLA2G2D | −0.227453 | 0.79656 | 0.6802 | 0.9328 | 0.00476* |
| ENSG00000278196 | IGLV2–8 | 0.003758 | 1.003765 | 0.8987 | 1.1211 | 0.9469 |
| ENSG00000204618 | RNF39 | 0.081054 | 1.084429 | 0.9514 | 1.2361 | 0.22498 |
| ENSG00000138755 | CXCL9 | 0.024433 | 1.024733 | 0.8602 | 1.2208 | 0.78441 |
| ENSG00000004468 | CD38 | −0.248007 | 0.780355 | 0.6448 | 0.9444 | 0.01084* |
| ENSG00000229754 | CXCR2P1 | 0.170242 | 1.185592 | 1.0229 | 1.3742 | 0.02379* |
| ENSG00000089012 | SIRPG | 0.085592 | 1.089361 | 0.7577 | 1.5662 | 0.64404 |
| ENSG00000211598 | IGKV4-1 | 0.068116 | 1.07049 | 0.9404 | 1.2186 | 0.30297 |
| ENSG00000143297 | FCRL5 | −0.123312 | 0.883988 | 0.6869 | 1.1376 | 0.33804 |
| ENSG00000172794 | RAB37 | 0.024174 | 1.024469 | 0.8445 | 1.2428 | 0.80625 |
| ENSG00000134028 | ADAMDEC1 | −0.032565 | 0.96796 | 0.8507 | 1.1013 | 0.62103 |
| ENSG00000182866 | LCK | −0.004812 | 0.9952 | 0.6297 | 1.5728 | 0.98356 |
| ENSG00000211662 | IGLV3–21 | 0.030352 | 1.030817 | 0.9201 | 1.1549 | 0.60058 |
| ENSG00000211943 | IGHV3–15 | −0.028086 | 0.972304 | 0.8631 | 1.0954 | 0.64412 |
| ENSG00000211899 | IGHM | 0.053087 | 1.054521 | 0.9358 | 1.1882 | 0.3835 |
| ENSG00000211677 | IGLC2 | −0.164464 | 0.848348 | 0.6886 | 1.0451 | 0.12231 |
| ENSG00000005844 | ITGAL | 0.544648 | 1.724001 | 1.0825 | 2.7457 | 0.02181* |
| ENSG00000168421 | RHOH | −0.040537 | 0.960274 | 0.7026 | 1.3125 | 0.7993 |
| ENSG00000211679 | IGLC3 | −0.088797 | 0.915031 | 0.7835 | 1.0687 | 0.26214 |
| ENSG00000076662 | ICAM3 | 0.0512 | 1.052534 | 0.7176 | 1.5438 | 0.79332 |
Significant codes: 0 “***”, 0.001 “**”, 0.01 “*”, 0.05 “.”, 0.1 “” 1.
3.3. Clinicopathological parameters with regard to the expression of MZB1
Compared with the other three candidate genes, MZB1 has the biggest impact on the overall survival time according to the survival analysis. The Chi-square analysis was performed to evaluate the connection between the expression of MZB1 and clinical characteristics such as gender, age, clinical stage, pathologic-T, pathologic-M, pathologic-N, and neoplasm status. According to the analysis, the expression level of MZB1 was significantly associated with pathologic-N (P = 0.046) and tumor status (P = 0.002) in patients with cutaneous melanoma (Table 3). By contrast, characteristics such as clinical stage, gender, age, pathologic-T, and pathologic-M were not connected with the expression of MZB1 (all P > 0.05).
Table 3.
Clinicopathological parameters of patients with cutaneous melanoma with regard to the expression of MZB1
| Characteristic | Expression of MZB1 | P-value | |
|---|---|---|---|
| Low | High | ||
| Age, years | 0.566 | ||
| <60 | 46 | 51 | |
| ≥60 | 176 | 171 | |
| Gender | 0.105 | ||
| Female | 76 | 93 | |
| Male | 147 | 131 | |
| Tumor_status | 0.002a | ||
| Tumor free | 86 | 123 | |
| With tumor | 119 | 93 | |
| Pathologic-T | 0.166 | ||
| Tis + T1 + T2 | 60 | 64 | |
| T3 + T4 | 129 | 101 | |
| Pathologic-M | 0.505 | ||
| M0 | 197 | 202 | |
| M1 | 13 | 10 | |
| Pathologic-N | 0.046a | ||
| N0 | 120 | 102 | |
| N1 + N2 + N3 | 76 | 97 | |
a P < 0.05. T, tumor; M, metastasis; N, node.
4. Discussion
Cutaneous melanoma is an invasive disease with high recurrence and metastasis rates. The overall survival time of patients with cutaneous metastatic tumors was remarkably shorter than that of patients without metastases, suggesting that the presence of metastases always means a poor prognosis. Identifying vital molecules that participate in the pathogenesis of metastatic melanoma is valuable for the potential development of therapeutic targets. Various biomarkers of cutaneous metastatic melanoma have been previously identified. However, the results are always different due to the variations in the samples and the focus of the analysis. Glypican 6 expression is higher in metastatic melanoma than in primary melanoma and is higher in primary melanoma than in normal melanocytes; therefore, this gene may be a biomarker for the metastatic progression of melanoma [4]. Glypican 6 is used to distinguish between the primary tumors of melanoma and regional cutaneous/subcutaneous metastases during early metastasis. The expression of nestin, an intermediate filament that can be a biomarker for stem cells, is also different between primary tumors and cutaneous metastatic melanoma [5]. Dong Wei identified that ATF2, SOX2, and RAC1 are involved in the metastasis of melanoma [6]. This article focused on the functional analysis of DEGs and did not explore the impact of DEGs on prognosis.
In our study, 464 DEGs were identified by the edgeR package, in which 177 were downregulated and 287 were upregulated. GO and KEGG enrichment analyses were conducted in these DEGs. The univariate Cox regression analysis was applied for the survival analysis based on the diverse expression of these DEGs. The top 30 DEGs with the most significant influence on the overall survival time according to the P-value were selected for further multivariate Cox regression analysis. Four candidate genes, namely, POU2AF1, ITGAL, CXCR2P1, and MZB1, were considered to be the substantial independent risk factors for metastatic cutaneous melanoma.
POU2AF1, which was upregulated in the cutaneous metastatic melanoma, is a B cell-specific coactivator that can stimulate the gene transcription. Its expression is regulated by the B-cell receptor (BCR) and CD40-L; a continuous stimulation may lead to an overexpression of this gene on B cells [7]. POU2AF1 plays a role in the pre-B1-to-pre-B2 cell transition and affects the pre-BCR and BCR signaling at multiple stages of B-cell development [8].
POU2AF1 is closely related to the immune system and various lymphopoietic system diseases. Kan Zhai’s study revealed that the gene mutation in 3′-UTR regulates POU2AF1 expression and subsequently gives rise to lymphoma [9]. The co-expression of POU2AF1 and Oct-2 can be a helpful prognosis for patients with acute myeloid leukemia (AML) [10]. POU2AF1 helps in the progression of multiple myeloma (MM) when activated by amplification or other mechanisms [11]. In an analysis of gastrointestinal stromal tumors, POU2AF1 was found to be one of the four genes that act as biomarkers for the prognosis of the high-risk gastrointestinal stromal tumors [12].
ITGAL, also known as CD11a, is upregulated in metastatic melanoma, highly expressed in most immune cell populations, and encodes a subunit of LFA-1 integrin [13]. LFA-1 interacts with its ligand, ICAMs 1–3, which acts as a rolling and signaling molecule that plays a crucial role in intercellular adhesion between white blood cells and lymphocyte co-stimulation signaling [14,15]. LFA-1 also mediates lymphocyte, monocyte, natural killer cell, and granulocyte interaction with other cells in immunity and inflammation [16].
ITGAL is closely linked to the pathogenesis of diverse immune-related diseases. In addition, ITGAL or LFA-1 encoded by ITGAL plays a role in various tumors. In the research of prostate cancer, the expression of ITGAL, along with four other genes, is connected with a number of positive lymph nodes [17]. ITGAL is also involved in immune response, inflammatory response, and formation of the tumor microenvironment, thus contributing to the pathogenesis of head and neck squamous cell carcinoma [18]. ITGAL is one of the prognostic factors for the survival and the risk of death of men with castration-resistant prostate cancer [19]. This finding is consistent with the results of the present study, in which the high expression of ITGAL is a prognostic risk factor for metastatic melanoma. By contrast, LFA-1 and ICAM-1 upregulated by IL-18 can facilitate the eosinophil-mediated tumoricidal activity against a colon carcinoma cell line [20].
MZB1 (or pERp1) is upregulated in the metastatic melanoma and is a B cell-specific and endoplasmic reticulum (ER)-localized protein that is abundantly expressed in marginal zone B and B1 cells [21]. MZB1 regulates calcium signaling, antibody secretion, integrin-mediated adhesion, and lymphocyte adhesion and migration [22,23]. Herold et al. found that the expression of MZB1 can be a valuable prognostic factor for different lymphoma subtypes [24]. MZB1 is also a biomarker of favorable prognosis in pancreatic cancer resected after the neoadjuvant chemoradiotherapy [25]. By reviewing the studies, we found that the impact of B lymphocytes on tumors has two sides. On the one hand, B lymphocytes can produce tumor antigen-specific immunoglobulin G antibodies [26]. On the other hand, B lymphocytes scattered in the tumor stroma suppress the antitumor immunity [27]. Hence, we speculated that MZB1 can be a favorable or a poor prognosis in different tumors.
CXCR2P1 upregulated in the metastatic melanoma is also known as CXCR2P and IL8RBP. The high expression level of CXCR2 is always relevant to metastasis and poor prognosis of tumors. CXCR2 can facilitate breast cancer metastasis and chemoresistance [28] and gastric cancer metastasis [29]. Interleukin-8 promotes cell migration via CXCR1 and CXCR2 in liver cancer [30]. In malignant melanoma, the expression of CXCL8 and CXCR2 promotes aggressive growth and metastasis [31]. Singh et al. found that the host’s CXCR2 contributes to the melanoma growth, angiogenesis, and experimental lung metastasis in mice [32]. Small molecule antagonists targeting CXCR2 inhibit the proliferation, migration, and invasion of melanoma cells, such as SCH-527123 [33]. Although CXCR2 participates in melanoma metastasis, only a few studies for CXCR2P are available. Whether CXCR2P participates in this process and the extent or mechanism of its participation are still unknown. Thus, further study is needed in the future.
The functions of the candidate genes are relevant to the immune system and inflammation and are consistent with the primary enrichment functions of upregulated DEGs. Melanoma microenvironment is composed of diverse immune cells and stromal cells that regulate the initiation and the development of disease and cellular response to therapies [34]. Innate immune cells in cutaneous melanoma, such as macrophages, NK cells, and dendritic cells, have strong plasticity and play a protumor or antitumor role through cell-to-cell and cell-to-tumor interactions and soluble molecules in the microenvironment [35]. This finding could explain why some genes are bidirectional in regulating tumors. Hence, this bidirectional action should be considered and explored from multiple perspectives when discussing the relationship between genes and melanoma. Tumor-related inflammation caused by the accumulation of white blood cells, which secrete cytokines and chemokines, can actively remodel tissues and angiogenesis, leading to conditions conducive to tumor growth, invasion, and metastasis [36]. However, the relationship between candidate genes and cutaneous metastatic melanoma is still unclear and warrants additional research.
In the clinical analysis, MZB1 has the biggest effect on the overall survival time. Furthermore, the rate of lymph node metastasis is higher in the MZB1 high expression group than in the low expression group. This result coincides with other studies that MZB1 regulates lymphocyte adhesion and migration. The number of patients exhibiting a tumor-free status was higher in the MZB1 high expression group than in the low expression group. However, the patients’ economic status, basic diseases, surgical approach, or other treatments, all of which can influence clinical outcomes, were not considered.
Cutaneous metastatic melanoma always has a worse prognosis than primary melanoma. Patients with advance-staged melanoma do not respond well to the treatment due to primary or acquired resistance. In addition, only a few studies on the genes related to the prognosis of metastatic melanoma have been performed. In the present study, we determined four candidate genes associated with metastatic melanoma that could be potentially prognostic risk factors for patients with cutaneous metastatic melanoma. We found that all of these genes are related to immunity and inflammation, but the specific processes of how these genes participated in metastatic melanoma have not been proven. This study provides a new direction for further research on metastatic melanoma and may provide a new target for the treatment or prevention of metastatic melanoma.
Acknowledgments
This research did not receive any specific grant from funding agencies in the public, commercial, or for-profit sectors.
Footnotes
Conflict of interest: All authors declare no conflicts of interest.
Statement of originality: This article has not been published in other journals.
References
- [1].Coricovac D, Dehelean C, Moaca E-A, Pinzaru I, Bratu T, Navolan D, et al. Cutaneous melanoma—a long road from experimental models to clinical outcome: a review. Int J Mol Sci. 2018;19:1566. [DOI] [PMC free article] [PubMed]; Coricovac D, Dehelean C, Moaca E-A, Pinzaru I, Bratu T, Navolan D. et al. Cutaneous melanoma—a long road from experimental models to clinical outcome: a review. Int J Mol Sci. 2018;19:1566. doi: 10.3390/ijms19061566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: Cancer J Clinic. 2019;69:7–34. [DOI] [PubMed]; Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: Cancer J Clinic. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- [3].Zhang X, Ding C, Tian H, Dong X, Meng X, Zhu W, et al. ZNF23 suppresses cutaneous melanoma cell malignancy via mitochondria-dependent pathway. Cell Physiol Biochem. 2017;43:147–57. [DOI] [PubMed]; Zhang X, Ding C, Tian H, Dong X, Meng X, Zhu W. et al. ZNF23 suppresses cutaneous melanoma cell malignancy via mitochondria-dependent pathway. Cell Physiol Biochem. 2017;43:147–57. doi: 10.1159/000480333. [DOI] [PubMed] [Google Scholar]
- [4].Ahmad A, Li Y, Li M, Shats I, Krahn JM, Flake GP, et al. Glypican 6 is a putative biomarker for metastatic progression of cutaneous melanoma. PLoS One. 2019;14:e0218067. [DOI] [PMC free article] [PubMed]; Ahmad A, Li Y, Li M, Shats I, Krahn JM, Flake GP. et al. Glypican 6 is a putative biomarker for metastatic progression of cutaneous melanoma. PLoS One. 2019;14:e0218067. doi: 10.1371/journal.pone.0218067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Klein WM, Wu BP, Zhao S, Wu H, Klein-Szanto AJ, Tahan SR. Increased expression of stem cell markers in malignant melanoma. Mod Pathol. 2007;20:102–7. [DOI] [PubMed]; Klein WM, Wu BP, Zhao S, Wu H, Klein-Szanto AJ, Tahan SR. Increased expression of stem cell markers in malignant melanoma. Mod Pathol. 2007;20:102–7. doi: 10.1038/modpathol.3800720. [DOI] [PubMed] [Google Scholar]
- [6].Wei D. A multigene support vector machine predictor for metastasis of cutaneous melanoma. Mol Med Rep. 2018;17:2907–14. [DOI] [PMC free article] [PubMed]; Wei D. A multigene support vector machine predictor for metastasis of cutaneous melanoma. Mol Med Rep. 2018;17:2907–14. doi: 10.3892/mmr.2017.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Teitell MA. OCA-B regulation of B-cell development and function. Trends Immunol. 2003;24:546–53. [DOI] [PubMed]; Teitell MA. OCA-B regulation of B-cell development and function. Trends Immunol. 2003;24:546–53. doi: 10.1016/j.it.2003.08.002. [DOI] [PubMed] [Google Scholar]
- [8].Siegel R, Kim U, Patke A, Yu X, Ren X, Tarakhovsky A, et al. Nontranscriptional regulation of SYK by the coactivator OCA-B is required at multiple stages of B cell development. Cell. 2006;125:761–74. [DOI] [PubMed]; Siegel R, Kim U, Patke A, Yu X, Ren X, Tarakhovsky A. et al. Nontranscriptional regulation of SYK by the coactivator OCA-B is required at multiple stages of B cell development. Cell. 2006;125:761–74. doi: 10.1016/j.cell.2006.03.036. [DOI] [PubMed] [Google Scholar]
- [9].Zhai K, Chang J, Hu J, Wu C, Lin D. Germline variation in the 3′-untranslated region of the POU2AF1 gene is associated with susceptibility to lymphoma. Mol Carcinogen. 2017;56:1945–52. [DOI] [PubMed]; Zhai K, Chang J, Hu J, Wu C, Lin D. Germline variation in the 3′-untranslated region of the POU2AF1 gene is associated with susceptibility to lymphoma. Mol Carcinogen. 2017;56:1945–52. doi: 10.1002/mc.22652. [DOI] [PubMed] [Google Scholar]
- [10].Advani AS, Lim K, Gibson S, Shadman M, Jin T, Copelan E, et al. OCT-2 expression and OCT-2/BOB.1 co-expression predict prognosis in patients with newly diagnosed acute myeloid leukemia. Leukemia Lymphoma. 2010;51:606–12. [DOI] [PubMed]; Advani AS, Lim K, Gibson S, Shadman M, Jin T, Copelan E. et al. OCT-2 expression and OCT-2/BOB.1 co-expression predict prognosis in patients with newly diagnosed acute myeloid leukemia. Leukemia Lymphoma. 2010;51:606–12. doi: 10.3109/10428191003592735. [DOI] [PubMed] [Google Scholar]
- [11].Zhao C, Inoue J, Imoto I, Otsuki T, Iida S, Ueda R, et al. POU2AF1, an amplification target at 11q23, promotes growth of multiple myeloma cells by directly regulating expression of a B-cell maturation factor, TNFRSF17. Oncogene. 2008;27:63–75. [DOI] [PubMed]; Zhao C, Inoue J, Imoto I, Otsuki T, Iida S, Ueda R. et al. POU2AF1, an amplification target at 11q23, promotes growth of multiple myeloma cells by directly regulating expression of a B-cell maturation factor, TNFRSF17. Oncogene. 2008;27:63–75. doi: 10.1038/sj.onc.1210637. [DOI] [PubMed] [Google Scholar]
- [12].Jin S, Zhu W, Li J. Identification of key genes related to high-risk gastrointestinal stromal tumors using bioinformatics analysis. J Cancer Res Therapeut. 2018;14:S243–S7. [DOI] [PubMed]; Jin S, Zhu W, Li J. Identification of key genes related to high-risk gastrointestinal stromal tumors using bioinformatics analysis. J Cancer Res Therapeut. 2018;14:S243–S7. doi: 10.4103/0973-1482.207068. [DOI] [PubMed] [Google Scholar]
- [13].Keum S, Lee HK, Chu PL, Kan MJ, Huang MN, Gallione CJ, et al. Natural genetic variation of integrin alpha L (Itgal) modulates ischemic brain injury in stroke. PLoS Genet. 2013;9:e1003807. [DOI] [PMC free article] [PubMed]; Keum S, Lee HK, Chu PL, Kan MJ, Huang MN, Gallione CJ. et al. Natural genetic variation of integrin alpha L (Itgal) modulates ischemic brain injury in stroke. PLoS Genet. 2013;9:e1003807. doi: 10.1371/journal.pgen.1003807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Samatov TR, Tonevitsky AG, Schumacher U. Epithelial-mesenchymal transition: focus on metastatic cascade, alternative splicing, non-coding RNAs and modulating compounds. Mol Cancer. 2013;12:107. [DOI] [PMC free article] [PubMed]; Samatov TR, Tonevitsky AG, Schumacher U. Epithelial-mesenchymal transition: focus on metastatic cascade, alternative splicing, non-coding RNAs and modulating compounds. Mol Cancer. 2013;12:107. doi: 10.1186/1476-4598-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–89. [DOI] [PubMed]; Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–89. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- [16].Heidenkummer HP, Kampik A. Intercellular adhesion molecule-1 (ICAM-1) and leukocyte function-associated antigen-1 (LFA-1) expression in human epiretinal membranes. Graefe’s Arch Clin Exp Ophthalmol. 1992;230:483–7. [DOI] [PubMed]; Heidenkummer HP, Kampik A. Intercellular adhesion molecule-1 (ICAM-1) and leukocyte function-associated antigen-1 (LFA-1) expression in human epiretinal membranes. Graefe’s Arch Clin Exp Ophthalmol. 1992;230:483–7. doi: 10.1007/BF00175938. [DOI] [PubMed] [Google Scholar]
- [17].Zhao X, Lei Y, Li G, Cheng Y, Yang H, Xie L, et al. Integrative analysis of cancer driver genes in prostate adenocarcinoma. Mol Med Rep. 2019;19:2707–15. [DOI] [PMC free article] [PubMed]; Zhao X, Lei Y, Li G, Cheng Y, Yang H, Xie L. et al. Integrative analysis of cancer driver genes in prostate adenocarcinoma. Mol Med Rep. 2019;19:2707–15. doi: 10.3892/mmr.2019.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Song Y, Pan Y, Liu J. The relevance between the immune response-related gene module and clinical traits in head and neck squamous cell carcinoma. Cancer Manage Res. 2019;11:7455–72. [DOI] [PMC free article] [PubMed]; Song Y, Pan Y, Liu J. The relevance between the immune response-related gene module and clinical traits in head and neck squamous cell carcinoma. Cancer Manage Res. 2019;11:7455–72. doi: 10.2147/CMAR.S201177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xie W, Stopsack KH, Drouin SJ, Fu H, Pomerantz MM, Mucci LA, et al. Association of genetic variation of the six gene prognostic model for castration-resistant prostate cancer with survival. Prostate. 2019;79:73–80. [DOI] [PMC free article] [PubMed]; Xie W, Stopsack KH, Drouin SJ, Fu H, Pomerantz MM, Mucci LA. et al. Association of genetic variation of the six gene prognostic model for castration-resistant prostate cancer with survival. Prostate. 2019;79:73–80. doi: 10.1002/pros.23712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gatault S, Delbeke M, Driss V, Sarazin A, Dendooven A, Kahn JE, et al. IL-18 is involved in eosinophil-mediated tumoricidal activity against a colon carcinoma cell line by upregulating LFA-1 and ICAM-1. J Immunol. 2015;195:2483–92. [DOI] [PubMed]; Gatault S, Delbeke M, Driss V, Sarazin A, Dendooven A, Kahn JE. et al. IL-18 is involved in eosinophil-mediated tumoricidal activity against a colon carcinoma cell line by upregulating LFA-1 and ICAM-1. J Immunol. 2015;195:2483–92. doi: 10.4049/jimmunol.1402914. [DOI] [PubMed] [Google Scholar]
- [21].Rosenbaum M, Andreani V, Kapoor T, Herp S, Flach H, Duchniewicz M, et al. MZB1 is a GRP94 cochaperone that enables proper immunoglobulin heavy chain biosynthesis upon ER stress. Genes Dev. 2014;28:1165–78. [DOI] [PMC free article] [PubMed]; Rosenbaum M, Andreani V, Kapoor T, Herp S, Flach H, Duchniewicz M. et al. MZB1 is a GRP94 cochaperone that enables proper immunoglobulin heavy chain biosynthesis upon ER stress. Genes Dev. 2014;28:1165–78. doi: 10.1101/gad.240762.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Flach H, Rosenbaum M, Duchniewicz M, Zhang SL, Cahalan MD, Mittler G, et al. Mzb1 protein regulates calcium homeostasis, antibody secretion, and integrin activation in innate-like B cells. Immunity. 2010;33:723–35. [DOI] [PMC free article] [PubMed]; Flach H, Rosenbaum M, Duchniewicz M, Zhang SL, Cahalan MD, Mittler G. et al. Mzb1 protein regulates calcium homeostasis, antibody secretion, and integrin activation in innate-like B cells. Immunity. 2010;33:723–35. doi: 10.1016/j.immuni.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Andreani V, Ramamoorthy S, Pandey A, Lupar E, Nutt SL, Lammermann T, et al. Cochaperone Mzb1 is a key effector of Blimp1 in plasma cell differentiation and beta1-integrin function. Proc Natl Acad Sci U S A. 2018;115:E9630–9. [DOI] [PMC free article] [PubMed]; Andreani V, Ramamoorthy S, Pandey A, Lupar E, Nutt SL, Lammermann T. et al. Cochaperone Mzb1 is a key effector of Blimp1 in plasma cell differentiation and beta1-integrin function. Proc Natl Acad Sci U S A. 2018;115:E9630–9. doi: 10.1073/pnas.1809739115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Herold T, Mulaw MA, Jurinovic V, Seiler T, Metzeler KH, Dufour A, et al. High expression of MZB1 predicts adverse prognosis in chronic lymphocytic leukemia, follicular lymphoma and diffuse large B-cell lymphoma and is associated with a unique gene expression signature. Leuk Lymphoma. 2013;54:1652–7. [DOI] [PubMed]; Herold T, Mulaw MA, Jurinovic V, Seiler T, Metzeler KH, Dufour A. et al. High expression of MZB1 predicts adverse prognosis in chronic lymphocytic leukemia, follicular lymphoma and diffuse large B-cell lymphoma and is associated with a unique gene expression signature. Leuk Lymphoma. 2013;54:1652–7. doi: 10.3109/10428194.2012.753445. [DOI] [PubMed] [Google Scholar]
- [25].Miyake K, Mori R, Homma Y, Matsuyama R, Okayama A, Murakami T, et al. MZB1 in borderline resectable pancreatic cancer resected after neoadjuvant chemoradiotherapy. J Surg Res. 2017;220:391–401. [DOI] [PubMed]; Miyake K, Mori R, Homma Y, Matsuyama R, Okayama A, Murakami T. et al. MZB1 in borderline resectable pancreatic cancer resected after neoadjuvant chemoradiotherapy. J Surg Res. 2017;220:391–401. doi: 10.1016/j.jss.2017.07.003. [DOI] [PubMed] [Google Scholar]
- [26].Carmi YC, Spitzer MH, Linde IL, Burt BM, Prestwood TR, Perlman N, et al. Allogeneic IgG combined with dendritic cell stimuli induce antitumour T-cell immunity. Nature. 2015;521:99–104. [DOI] [PMC free article] [PubMed]; Carmi YC, Spitzer MH, Linde IL, Burt BM, Prestwood TR, Perlman N. et al. Allogeneic IgG combined with dendritic cell stimuli induce antitumour T-cell immunity. Nature. 2015;521:99–104. doi: 10.1038/nature14424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Inoue S, Leitner WW, Golding B, Scott D. Inhibitory effects of B cells on antitumor immunity. Cancer Res. 2006;66:7741–7. [DOI] [PubMed]; Inoue S, Leitner WW, Golding B, Scott D. Inhibitory effects of B cells on antitumor immunity. Cancer Res. 2006;66:7741–7. doi: 10.1158/0008-5472.CAN-05-3766. [DOI] [PubMed] [Google Scholar]
- [28].Xu H, Lin F, Wang Z, Yang L, Meng J, Ou Z, et al. CXCR2 promotes breast cancer metastasis and chemoresistance via suppression of AKT1 and activation of COX2. Cancer Lett. 2018;412:69–80. [DOI] [PubMed]; Xu H, Lin F, Wang Z, Yang L, Meng J, Ou Z. et al. CXCR2 promotes breast cancer metastasis and chemoresistance via suppression of AKT1 and activation of COX2. Cancer Lett. 2018;412:69–80. doi: 10.1016/j.canlet.2017.09.030. [DOI] [PubMed] [Google Scholar]
- [29].Xiang Z, Zhou ZJ, Xia GK, Zhang XH, Wei ZW, Zhu JT, et al. A positive crosstalk between CXCR4 and CXCR2 promotes gastric cancer metastasis. Oncogene. 2017;36:5122–33. [DOI] [PubMed]; Xiang Z, Zhou ZJ, Xia GK, Zhang XH, Wei ZW, Zhu JT. et al. A positive crosstalk between CXCR4 and CXCR2 promotes gastric cancer metastasis. Oncogene. 2017;36:5122–33. doi: 10.1038/onc.2017.108. [DOI] [PubMed] [Google Scholar]
- [30].Bi H, Zhang Y, Wang S, Fang W, He W, Yin L, et al. Interleukin-8 promotes cell migration via CXCR1 and CXCR2 in liver cancer. Oncol Lett. 2019;18:4176–84. [DOI] [PMC free article] [PubMed]; Bi H, Zhang Y, Wang S, Fang W, He W, Yin L. et al. Interleukin-8 promotes cell migration via CXCR1 and CXCR2 in liver cancer. Oncol Lett. 2019;18:4176–84. doi: 10.3892/ol.2019.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Varney ML, Johansson SL, Singh RK. Distinct expression of CXCL8 and its receptors CXCR1 and CXCR2 and their association with vessel density and aggressiveness in malignant melanoma. Am J Clin Pathol. 2006;125:209–16. [DOI] [PubMed]; Varney ML, Johansson SL, Singh RK. Distinct expression of CXCL8 and its receptors CXCR1 and CXCR2 and their association with vessel density and aggressiveness in malignant melanoma. Am J Clin Pathol. 2006;125:209–16. doi: 10.1309/VPL5-R3JR-7F1D-6V03. [DOI] [PubMed] [Google Scholar]
- [32].Singh S, Varney M, Singh RK. Host CXCR2-dependent regulation of melanoma growth, angiogenesis, and experimental lung metastasis. Cancer Res. 2009;69:411–5. [DOI] [PMC free article] [PubMed]; Singh S, Varney M, Singh RK. Host CXCR2-dependent regulation of melanoma growth, angiogenesis, and experimental lung metastasis. Cancer Res. 2009;69:411–5. doi: 10.1158/0008-5472.CAN-08-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shang FM, Li J. A small-molecule antagonist of CXCR1 and CXCR2 inhibits cell proliferation, migration and invasion in melanoma via PI3K/AKT pathway. Med Clin. 2019;152:425–30. [DOI] [PubMed]; Shang FM, Li J. A small-molecule antagonist of CXCR1 and CXCR2 inhibits cell proliferation, migration and invasion in melanoma via PI3K/AKT pathway. Med Clin. 2019;152:425–30. doi: 10.1016/j.medcli.2018.08.006. [DOI] [PubMed] [Google Scholar]
- [34].Yang S, Liu T, Nan H, Wang Y, Chen H, Zhang X, et al. Comprehensive analysis of prognostic immune-related genes in the tumor microenvironment of cutaneous melanoma. J Cell Physiol. 2019;1–11. [DOI] [PubMed]; Yang S, Liu T, Nan H, Wang Y, Chen H, Zhang X. et al. Comprehensive analysis of prognostic immune-related genes in the tumor microenvironment of cutaneous melanoma. J Cell Physiol. 2019:1–11. doi: 10.1002/jcp.29018. [DOI] [PubMed] [Google Scholar]
- [35].Marzagalli M, Ebelt ND, Manuel ER. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment. Semin Cancer Biol. 2019;59:236–50. [DOI] [PubMed]; Marzagalli M, Ebelt ND, Manuel ER. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment. Semin Cancer Biol. 2019;59:236–50. doi: 10.1016/j.semcancer.2019.08.002. [DOI] [PubMed] [Google Scholar]
- [36].Ladanyi A. Prognostic and predictive significance of immune cells infiltrating cutaneous melanoma. Pigm Cell Melanoma Res. 2015;28:490–500. [DOI] [PubMed]; Ladanyi A. Prognostic and predictive significance of immune cells infiltrating cutaneous melanoma. Pigm Cell Melanoma Res. 2015;28:490–500. doi: 10.1111/pcmr.12371. [DOI] [PubMed] [Google Scholar]