Abstract
Colorectal cancer (CRC) remains one of the leading causes of cancer-related death due to its high metastatic potential. This study aimed to investigate the detection and heterogeneity of circulating tumor cells (CTCs) and the microsatellite instability (MSI) status in advanced CRC patients prior to any systemic front-line treatment. Peripheral whole blood was obtained from 198 patients. CTCs were detected using double immunofluorescence and a real time-polymerase chain reaction assay; whereas MSI status was evaluated using fragment analysis. Median age of the patients was 66 years, 63.1% were males, 65.2% had a colon/sigmoid tumor location and 90.4% had a good performance status (PS). MSI-High status was detected in 4.9% of the patients; 33.3%, 56.1% and 8.6% patients had at least one detectable CEACAM5+/EpCAM+, CEACAM5+/EpCAM− and CEACAM5−/EpCAM+ CTC, respectively, and 9.1% of the patients had CEACAM5mRNA-positive CTCs. Following multivariate analysis, age, PS and MSI were confirmed as independent prognostic factors for decreased time to progression, whereas age, PS and CTC presence were confirmed as independent prognostic factors for decreased overall survival. In conclusion, our data support the use of CEACAM5 as a dynamic adverse prognostic CTC biomarker in patients with metastatic CRC and MSI-High is considered an unfavorable prognostic factor in metastatic CRC patient tumors.
Keywords: colorectal cancer, metastatic, CTCs, CEACAM5, microsatellite instability
1. Introduction
Colorectal cancer (CRC) is still one of the main causes of cancer-related death [1,2]. Its high metastatic potential is due to cancer cell dissemination through hematogenous or lymphatic vasculature.
Circulating tumor cell (CTC) presence in the blood of patients experiencing cancer has been widely described and its impact on decreased progression-free and overall survival in various cancer types has been documented with different technologies (fluorescent microscopy, molecular methods, isolation by size, CellSearch, etc.) [3,4,5,6,7,8,9,10,11,12,13]. The clinically validated CellSearch assay is based on the immunomagnetic epithelial adhesion cellular molecule (EpCAM)-positive CTC enrichment [5]. However, during their dissemination, CTCs undergo epithelial-to-mesenchymal transition and down-regulate their epithelial markers, including EpCAM [14,15]. Therefore, it is obvious that CellSearch fails to detect CTCs that do not express EpCAM.
In CRC, the carcinoembryonic antigen (CEA) is the most frequently analyzed marker, and we have previously developed a reliable and reproducible molecular method for carcino-embryonic antigen-like cellular adhesion molecule 5 (CEACAM5) mRNA-positive CTC detection in CRC patients [11,13]. The presence of CEACAM5mRNA-positive CTCs has been associated with the worst clinical outcome, both in the metastatic and the non-metastatic setting [11,13].
An important molecular pathway in carcinogenesis is implicated in DNA repair mechanisms [16]. Errors are recognized by changes (instability) in hereditary patterns of single nucleotide sequences (microsatellites) that occur throughout the genome. Usually, the mutations refer to the hMLH1 (human mutL homolog 1) and hMSH2 (human mutS homolog 2) genes [17,18,19]. Mutations in these genes cause a phenomenon known as microsatellite instability (MSI), which occurs in most of the 10–15% of sporadic CRC cases [20,21,22]. MSI-High tumors have been shown to be associated with higher survival rates in relation to MSI-Low or MSI-Stable (MSS) tumors [23,24,25]; however, the underlying mechanism is not well understood.
The current study aimed to investigate the CTCs’ heterogeneity, based on the expression of CEACAM5 and EpCAM markers, in advanced CRC patients prior to any systemic first-line therapy. Moreover, the study aimed to investigate the detection of CTCs expressing CEACAM5mRNA using the reverse transcription quantitative polymerase chain reaction (RT-qPCR) technology and the MSI status of the patients. Finally, to correlate the detection of CTCs and MSI status with known clinico-pathological characteristics and patients’ outcomes.
2. Materials and Methods
2.1. Patients’ Population
One hundred and ninety-eight patients enrolled in the study. All these patients had a newly diagnosed metastatic CRC and histologically documented disease. All 198 patients were treated at the Department of Medical Oncology, University Hospital of Heraklion (Greece). The Ethics Committee/Institutional Review Board of the University Hospital of Heraklion (Greece) has approved the current study (Number: 7302/19-8-2009). All enrolled patients have agreed their enrolment and signed their written informed consent. All procedures followed in the current study were in accordance with the ethical standards of the institutional and/or national research committee and are in agreement with the 1964 Helsinki declaration and its later amendments.
2.2. Blood and Tissue Samples
An amount of 15 mL in EDTA of peripheral blood was obtained at the middle-of-vein puncture. Prior collection, the first 5 mL was discarded to exclude skin epithelial cells. Isolation of peripheral blood mononuclear cells (PBMCs) was performed by Ficoll-Hypaque density (d = 1077 g/mL; Sigma-Aldrich, GmbH, Taufkirchen, Germany) gradient centrifugation at 1800 rpm, for 30 min. Centrifugation and cytospin preparation were described previously [26,27].
Total RNA extraction from both patients’ PBMCs and cell lines was carried out under RNAse-free conditions using Trizol (Thermo Fisher Scientific, Fremont, CA, USA) in a laminar flow hood. Following RNA isolation, it was dissolved in RNA storage buffer (Ambion, Fremont, CA, USA) and until used, it was stored at −80 °C. β-actin was used as house-keeping gene amplification to verify the RNA integrity, as previously described [28,29].
Formalin-fixed, paraffin-embedded (FFPE) tumor sections were evaluated by an experienced pathologist. The diagnosis was confirmed, and the tumor-enriched areas prior dissection were defined. Ten serial sections (5 µm thickness) were stained with nuclear fast red (Sigma-Aldrich, St. Louis, MO, USA). Samples with ≥80% malignant cells were scraped under the microscope. When <80% tumor cells, the piezoelectric Eppendorf microdissector (Eppendorf, Hamburg, Germany) was used for microdissection. The Epicentre® Biotechnologies MasterPure™ Complete DNA and RNA Purification Kit was used for DNA extraction, according to the manufacturer’s instructions (Epicentre, Madison, WI, USA).
2.3. Quantitative RT-PCR (RT-qPCR) and Double Immunofluorescence
The NanoDrop (Thermo Scientific, Fremont, CA, USA) equipment was used to determine RNA concentration. The reverse transcription and qPCR conditions as well as the method’s analytical sensitivity and specificity evaluation have been described previously [11,13]. The ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Waltham, MA, USA) was used to quantify gene expression (in triplicates). An external calibration curve using external standard cDNAs was used for quantification [11,13,30]. In addition, cDNA synthesis of serial dilutions of RNA isolated from the Lovo cells, corresponding to 1–105 cells, was also included in each run. The number of CEACAM5mRNA-positive cells for all of the samples tested was expressed as Lovo cell equivalents/5 μg total RNA. The pair of primers and the probe used were as follows: CEACAM5-forward: AATTCCATAGTCAAGAGCATCACAGT, CEACAM5-reverse: CAGTGG CCCCAGCTGAGA, and probe: CTGCATCTGGAACTTCTCCTGGT. The results were analyzed using the SDS 2.3 software (Applied Biosystems, Waltham, MA, USA). Finally, as no RNA transcripts were detected in the absence of reverse transcriptase, genomic DNA contamination was excluded.
Moreover, immediately following their isolation, 5 × 105 PBMCs were cyto-centrifuged on glass microscope slides, at 2000 rpm for 2 min. Cytospins were dried at room temperature and stored at −80 °C, until use. A number of 10 × 105 PBMCs in total (corresponding to 2 microscope slides) from each patient were screened for CEACAM5 (Acris Antibodies, GmbH, Germany) and EpCAM (Acris Antibodies, GmbH, Germany) expression. Briefly, ice cold aceton:methanol 9:1 (v/v) was used for cell-cytospin fixation, for 20 min. All antibodies (primary and secondary) were incubated for a period of 1 h. CEACAM5 was detected using the Zenon technology (FITC-conjugated IGg1 antibody; Molecular Probes, Invitrogen, Fremont, CA, USA); whereas EpCAM was labelled with Alexa 555 (Molecular Probes). Additionally, cytospins were double stained with the anti-CEACAM5 (mouse) or anti-EpCAM (mouse) and anti-CD45 (anti-rabbit Common Leukocyte Antigen; Santa Cruz, CA, USA) antibodies, whereas Alexa 488 (Molecular Probes) and Alexa 555 (Molecular Probes) were used for labelling, respectively. Finally, DAPI anti-fade reagent (Molecular Probes) was used for cell nuclear staining. The omission of the primary antibody was used in control experiment. The detection of CTCs was performed using a fluorescent microscope (Leica DM 2500, Heidelberg, Germany). Results are expressed as number of CTCs/106 PBMCs.
2.4. Microsatellite Instability and Gene Mutational Analysis
Microsatellite instability (MSI) analysis was examined by PCR fragment analysis. For this assay, the five microsatellite loci (BAT-25, BAT-26, D2S123, D5S346 and D1S250), according to the Bethesda panel assay recommended by the 1997 National Cancer Institute-sponsored MSI workshop, were amplified using a multiplex PCR reaction for the BAT-25, BAT-26, D2S123, D5S346 and D1S250 loci using the MSI Analysis System, v1.2, according to the manufacturer instructions (Promega Corp., Madison, WI, USA). A 10 μL reaction containing 40–60 ng of DNA template was prepared for each tumor and normal tissue of every patient. The PCR cycling conditions included an initial denaturation step at 95 °C (11 min) and at 96 °C (1 min), 30 cycles of denaturation at 94 °C (30 s), annealing at 60 °C (1 min) and extension at 72 °C (90 s). A final extension step at 60 °C (45 min) was added. PCR products were analyzed by capillary electrophoresis using an ABI Prism 3130 Genetic Analyzer (Thermo Fisher Scientific). In brief, deionized formamide was combined with GeneScan-500 (LIZ) size standard (Thermo Fisher Scientific) and 1.2 μL of PCR amplified product. Then, samples were denatured at 95 °C for 3 min and ice-chilled prior analysis. The GeneMapper software v4.1 (Thermo Fisher Scientific, Fremont, CA, USA) was used for data analysis. Only fragments in which both tumor and paired normal tissue DNA showed signals above background levels, were considered assessable. For interpretation purposes, MSI at ≥2 loci was defined as MSI-High, MSI at a single locus was defined as MSI-Low, whereas a locus was called stable (MSS) if no instabilities were observed in the tumor sample in comparison to the paired normal DNA of the same patient.
Sanger sequencing following PCR amplification was used for KRAS exon 2 mutational analysis, and an allelic discrimination assay based on real-time PCR for the BRAFV600E mutational analysis was performed as a standard clinical practice [31,32,33]. It has to be mentioned that a retrospective NRAS mutation analysis was not performed due to the fact that this study was initiated prior the description of the NRAS predictive value in treatment efficacy [34]; a retrospective mutation analysis of this gene was not performed.
2.5. Study Design and Statistics
The aim of the current study was to investigate the clinical relevance of CEACAM5-positive detection and CEACAM5mRNA-positive CTCs on diagnosis and to correlate them with MSI and genetic mutational status. The current study has an observational nature of the study; thus, no formal sample size calculation was performed. The cut-off value of ≥1.92 Lovo cell equivalents/5 μg of RNA was used as has been previously suggested [11]. Summary tables (descriptive statistics and/or frequency tables) are provided for all timepoint variables, as appropriate. Continuous variables are demonstrated with descriptive statistics (number, median, standard deviation and range). Ninety-five percent confidence intervals (95% CI) are also mentioned. Progression-free survival (PFS) was interpreted as the time from first cycle of treatment until clinical or radiological disease relapse or any cause of death. Overall survival (OS) was defined from the date of first cycle of treatment until any cause of death or the date of last follow-up. Whenever appropriate, either Pearson’s Chi-square or Fisher’s exact test was used to compare qualitative factors. The Kruskal Wallis test was used to assess differences in continuous variables. The Kaplan-Meier analysis and log-rank test was performed to estimate PFS and OS. Cox proportional hazards regression models were used to observe associations among prognostic factors and PFS/OS. All statistical tests were two-sided. Any p-value < 0.05 was considered statistically significant. The SPSS statistical software, version 22.0 (SPSS Inc., Chicago, IL, USA), was used for data analysis.
3. Results
3.1. Patients’ Clinico-Pathological Characteristics
From 6/2007 to 12/2013, 198 patients with metastatic CRC were enrolled in the study. The patients’ clinico-pathological characteristics are listed in Table 1. The median age was 66 years, 125 (63.1%) patients were males, 129 (65.2%) had a colon/sigmoid tumor location, 179 (90.4%) had a good performance status (PS ECOG—Eastern Cooperative Oncology Group scale: 0–1), and 76 (39.6%) had high-grade tumors. Moreover, 148 (74.7%) patients underwent surgery and 39 (19.7%) underwent metastasectomy. KRAS (Kirsten rat sarcoma) exon 2 and BRAFV600E mutations were detected in 45.0% and 7.8%, respectively; whereas, MSI-High status was detected in 4.9% of the patients (Table 1 and Table S1).
Table 1.
Patients’ demographics and characteristics.
| Total (n = 198) | ||
|---|---|---|
| Frequency (N = 198) | % | |
| Gender | ||
| Male | 125 | 63.1 |
| Female | 73 | 36.9 |
| Median Age | 66.3 years (20–88) | |
| ≤70 years | 121 | 61.1 |
| >70 years | 77 | 38.9 |
| Tumor Location | ||
| Colon/Sigmoid | 129 | 65.2 |
| Rectum | 69 | 34.8 |
| Grade | ||
| I-II | 116 | 60.4 |
| III | 76 | 39.6 |
| Unknown | 6 | |
| Surgery | ||
| Yes | 148 | 74.7 |
| No | 50 | 25.3 |
| Metastasectomy | ||
| Yes | 39 | 19.7 |
| No | 159 | 80.3 |
| Performance Status (ECOG *) | ||
| 0–1 | 179 | 90.4 |
| 2 | 19 | 9.6 |
| CEACAM5 **+/EpCAM ***+/CD45 #- | ||
| Positive | 66 | 33.3 |
| Negative | 132 | 66.7 |
| CEACAM5+/EpCAM-/CD45- | ||
| Positive | 111 | 56.1 |
| Negative | 87 | 43.9 |
| CEACAM5-/EpCAM+/CD45- | ||
| Positive | 17 | 8.6 |
| Negative | 181 | 91.4 |
| CEACAM5mRNA (cutoff >0.91 Lovo cell equivalents) | ||
| Positive | 18 | 90.9 |
| Negative | 18 | 9.1 |
| KRAS ## exon 2 mutations | ||
| Mutated | 85 | 45 |
| Wild type | 104 | 55 |
| Not Determined | 9 | |
| BRAFV600E mutations | ||
| Mutated | 15 | 7.8 |
| Wild type | 177 | 92.2 |
| Not Determined | 6 | |
| Microsatellite Instability (MSI) | ||
| MSI-High | 8 | 4.9 |
| MSI-Stable (MSS) | 155 | 95.1 |
| Not Determined | 35 | |
* ECOG: Eastern Cooperative Oncology Group; ** CEACAM5: carcino-embryonic antigen-like cellular adhesion molecule 5; *** EpCAM: epithelial cellular adhesion molecule; # CD45: cluster of differentiation 45; ## KRAS: Kirsten rat sarcoma.
3.2. Double Immunofluorescence for the Detection of CEACAM5+ and EpCAM+ CTCs
Following double immunofluorescence staining with anti-CEACAM5 and anti-EpCAM antibodies, 66 (33.3%), 111 (56.1%) and 17 (8.6%) patients had at least one detectable CEACAM5+/EpCAM+, CEACAM5+/EpCAM− and CEACAM5-/EpCAM+ CTC, respectively (Table 2 and Table S1). The median number of CEACAM5+/EpCAM+, CEACAM5+/EpCAM− and CEACAM5−/EpCAM+ CTCs were 2 (range, 1–20), 2 (range, 1–22) and 1 (range, 1–6), respectively (Table S1). There was no correlation between the patients’ characteristics and the detection of CEACAM5 and/or EpCAM positivity. Additional double immunofluorescence staining with anti-CD45 and anti-CEACAM5 or anti-EpCAM revealed no CEACAM5+/CD45+ or EpCAM+/CD45+ cells.
Table 2.
Heterogeneity of circulating tumor cell (CTC) sub-populations and correlation with CEACAM5mRNA expression.
| No of Patients (N = 198) | % | Median | Range | CEACAM5mRNA | |
|---|---|---|---|---|---|
| CEACAM5+/EpCAM+ | 66 | 33.3 | 2 | 1–20 | p < 0.001 |
| CEACAM5+/EpCAM- | 111 | 56.1 | 2 | 1–22 | p < 0.001 |
| CEACAM5-/EpCAM+ | 17 | 8.6 | 1 | 1–6 | p < 0.011 |
| CEACAM5mRNA | 18 | 9.1 | 3.1 | 1.92–71.33 |
3.3. Detection of CEACAM5mRNA-Positive CTCs
According to the cut-off point (>1.91 Lovo cell equivalents/5 μg total RNA), CEACAM5mRNA-positivity was detected in 18 (9.1%) of the enrolled patients (Table 2 and Table S1). The median number of CEACAM5mRNA-positive CTCs was 3.1 (range, 1.92–71.33) (Table S1). CEACAM5mRNA-positivity was significantly associated with the detection of CEACAM5+/EpCAM+, CEACAM5+/EpCAM− and CEACAM5−/EpCAM+ CTCs (p < 0.001; p < 0.001 and p < 0.011, respectively). In contrast, there was no correlation between the patients’ clinico-pathologic characteristics and the CEACAM5mRNA-positivity.
3.4. Patients’ Clinical Outcome According to the Genetic Mutational Profiling, MSI Status and CTC Detection
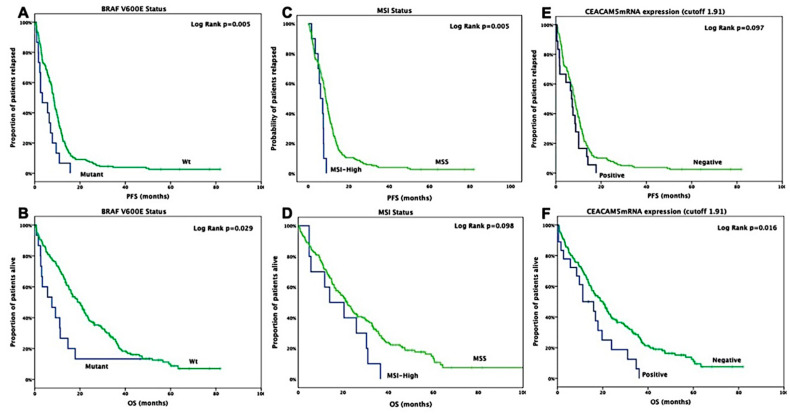
BRAFV600E mutated patients presented with significantly lower PFS (3.4 months, 95% CI: 0.2–6.6 months) compared to those with BRAFV600E wild-type tumors (8.4 months, 95% CI: 7.6–9.2 months; p = 0.005) (Table S1 and Figure 1A); moreover, these patients had a significantly lower OS (7.6 months, 95% CI: 0.3–14.9 months) compared to those with BRAFV600E wild-type status tumors (19.7 months, 95% CI: 16.0–23.4 months; p = 0.029) (Table S1 and Figure 1B). In addition, patients with MSI-High status had a statistically significant shorter PFS (6.1 months, 95% CI: 3.5–8.7 months) compared to MSS patients (8.8 months, 95% CI: 7.9–9.7 months) (p = 0.005); moreover, these patients had a lower OS (14.0 months, 95% CI: 0.7–27.3 months) compared to MSS patients (21.8 months, 95% CI: 18.2–25.5 months), however, such difference was not statistically significant (p = 0.098) (Table S1 and Figure 1C,D). Furthermore, no significant difference was shown in patients with CEACAM5mRNA-positive CTCs and PFS (7.0 months, 95% CI: 5.3–8.7 months) compared to those with CEACAM5mRNA-negative CTCs (8.3 months, 95% CI: 7.5–9.2 months) (p = 0.097) (Table S1 and Figure 1E); whereas, these patients had significantly shorter OS (11.2 months, 95% CI: 2.8–19.6 months) compared to those with CEACAM5mRNA-negative CTCs (19.6 months, 95% CI: 15.7–23.5 months; p = 0.016) (Table S1 and Figure 1F). Finally, no significant difference was shown in patients with KRAS mutational status and PFS (p = 0.610) or OS (p = 0.969) (Table S1).
Figure 1.
Kaplan Meier curves for progression-free and overall survival according to BRAFV600E (A,B), microsatellite instability (C,D) and CEACAM5mRNA (E,F) status, respectively, in stage IV patients.
3.5. Univariate and Multivariate Analysis
Univariate analysis revealed that age, performance status (PS), BRAFV600E and MSI status were associated with decreased PFS (Table 3). Multivariate analysis confirmed that age, PS and MSI were independent prognostic factors for decreased PFS (HR = 1.6; 95% CI: 1.1–2.2; p = 0.008; HR = 3.3; 95% CI: 1.8–6.0; p < 0.001 and HR = 2.5; 95% CI: 1.8–5.3; p < 0.001 (Table 3). Similarly, patients with positive CEACAM5mRNA CTCs had increased risk of death than those with no detectable CEACAM5mRNA-positive cells (HR = 1.8; 95% CI: 1.1–3.1; p = 0.018; Table 3). Age, PS and BRAFV600E were also significantly associated with OS. The multivariate analysis revealed that age, PS and the presence of CEACAM5mRNA-positive CTCs (marginally significant) were independent prognostic factors associated with decreased overall survival (HR = 1.4; 95% CI: 1.0–1.9; p = 0.046; HR = 3.3; 95% CI: 2.0–5.6; p < 0.001 and HR = 1.6; 95% CI: 1.0–2.8; p = 0.056; Table 3).
Table 3.
Univariate and multivariate analysis for progression-free survival (PFS) and overall survival (OS).
| Univariate | Multivariate | |||||||
|---|---|---|---|---|---|---|---|---|
| PFS | OS | PFS | OS | |||||
| Feature | HR (95%CI) | p-Value | HR (95%CI) | p-Value | HR (95%CI) | p-Value | HR (95%CI) | p-Value |
| Age ( >70 y vs. ≤70 y) | 1.4 (1.0–1.8) | 0.039 | 1.4 (1.0–1.9) | 0.043 | 1.6 (1.1–2.2) | 0.008 | 1.4 (1.0–1.9) | 0.046 |
| Performance status (≥2 vs. 0–1) | 4.0 (2.4–6.6) | <0.001 | 4.1 (2.5–6.8) | <0.001 | 3.3 (1.8–6.0) | <0.001 | 3.3 (2.0–5.6) | <0.001 |
| KRAS exon 2 | 1.1 (0.8–1.4) | 0.613 | 1.0 (0.7–1.4) | 0.969 | - | - | - | - |
| BRAFV600E (mutant vs. wild type) | 2.1 (1.2–3.6) | 0.007 | 1.9 (1.1–3.3) | 0.03 | - | - | - | - |
| Microsatellite Instability (MSI) (High vs. Stable) | 2.6 (1.2–5.4) | 0.011 | 1.9 (0.9–4.0) | 0.072 | 2.5 (1.8–5.3) | 0.017 | - | - |
| CEACAM5 **+/EpCAM ***+ | 1.1 (0.8–1.5) | 0.394 | 1.2 (0.8–1.6) | 0.366 | - | - | - | - |
| CEACAM5+/EpCAM- | 1.1 (0.8–1.5) | 0.499 | 1.1 (0.8–1.4) | 0.732 | - | - | - | - |
| CEACAM5-/EpCAM+ | 1.2 (0.8–2.1) | 0.395 | 1.5 (0.9–2.5) | 0.15 | - | - | - | - |
| CEACAM5mRNA (pos vs. neg) | 1.5 (0.9–2.5) | 0.101 | 1.8 (1.1–3.1) | 0.018 | - | - | 1.6 (1.0–2.8) | 0.056 |
** CEACAM5: carcino embryonic antigen cellular adhesion molecule 5; *** EpCAM: epithelial cellular adhesion molecule.
4. Discussion
Several studies have shown that CEA has a metastatic role, which it performs through various ways [35,36,37]. CEA protects tumor cells from the effects of anoikis [38], changes the microenvironment of sinusoids, promotes the adhesion molecules’ expression and malignant cell survival, and this affords a selective advantage for tumor cell survival in circulation [36]. Furthermore, Blumenthal et al. showed that anti-CEACAM6/CEACAM5 mAb fragments targeting the N and A1B1 domains of these antigens lead to block migration, endothelial cells adhesion and increased survival of mice with intrapulmonary micrometastases of human colon cancer [39]. CEA has also been explored, as a target for CRC treatment and diagnosis approaches [35]. Indeed, previous studies from our group report that high CEACAM5mRNA levels are specifically associated with CRC progression, both in patients with operable and metastatic CRC [11,13].
In the present study, we aimed to evaluate the CEACAM5-positive CTCs expression in the blood of metastatic CRC patients, using both immunofluorescence and RT-qPCR technologies. Our results clearly revealed that positive detection of CEACAM5-mRNA was significantly correlated with the detection of CEACAM5 and/or EpCAM positivity by immunofluorescence assay, on initial diagnosis. In line with our previous results, presented data demonstrate the prognostic significance of CEACAM-positive CTCs and the BRAFV600E mutations in metastatic CRC patients. Moreover, positive expression of CEACAM5mRNA was presented as an independent prognostic factor for reduced OS.
A systematic meta-analysis investigated that EpCAM expression might be associated with CRC, while its reduced expression is correlated with the progression, metastasis, and poor prognosis in CRC [40,41]. Interestingly, 56.1% of the enrolled patients had CEACAM5+/EpCAM- detectable CTCs. These results highlight the limitation of the EpCAM-based technologies such as the Food and Drug Administration-approved CellSearch, thus failing to capture these CTC subpopulations. An explanation is that epithelial cell adhesion molecules undergoing EMT lose their epithelial and other characteristics thus acquiring a mesenchymal phenotype [42]. In addition, Kim et al. presented that any metastasizing malignant cell derived from EpCAM loss in CRC might present loss or decreased expression of EpCAM, and in these cases, CTCs cannot be detected despite being present [43]. Therefore, since epithelial markers are not always detectable [44], it might be useful to include additional biomarkers, such as the CEACAM5, thus increasing the detection sensitivity of CTCs.
MSI-High status in adjuvant CRC is associated with improved stage-specific survival; however, the prognostic role of MSI-High status in the metastatic setting remains controversial [45,46]. Several studies have shown that MSI-High metastatic CRC patients have shorter overall survival compared with MSS patients [45,47]. This phenomenon could suggest that MSI-High cancers metastasize earlier, once cancer cells evade the host immune surveillance and are about to metastasize [47]. Similarly, the current study demonstrated that patients with deficiency in the DNA mismatch repair system (MMR) have a significantly more decreased PFS than those with proficient MMR status. Moreover, our observation suggested that MSI-High is considered an unfavorable prognostic factor in metastatic CRC patient tumors.
5. Conclusions
In conclusion, CTCs are a promising alternative for liquid biopsy for use in precision medicine approaches [48]. Our valuable data support the use of CEACAM5 as a dynamic adverse prognostic CTC biomarker in patients with metastatic CRC; the findings should be interpreted with caution due to the limitation of the study (retrospective analysis, non-randomized cases, different treatment regimens, etc.). To our knowledge, this is the first study for the evaluation of the expression among CEACAM5, EpCAM and the MSI status of patients with metastatic CRC. Based on previous findings in a small subset of Lynch syndrome-related CRCs carrying germline EpCAM deletions [49,50], no association was found between loss of EpCAM expression and MMR system [40,43]. Accordingly, there was no significant correlation between MSI status and the expression of EpCAM or CEACAM5 in our results. Prospective studies, preferable as part of randomized trials, may further evaluate the significance of the presented biomarkers. Moreover, significant impact would be added upon evaluation of the presented biomarkers according to survival rates, both in primary tumors and metastasis.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4426/10/4/235/s1, Table S1: Research Data.
Author Contributions
Conceptualization, I.M. and J.S.; methodology, I.M., M.S., K.V., C.K.; validation, I.M., M.S., M.T.; data curation, I.M., M.S., A.K., N.G., J.T., D.M., M.T., J.S.; formal analysis, I.M.; supervision, I.M. and J.S.; writing—original draft preparation, I.M. and M.S.; writing—review and editing, I.M. and J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was fully supported by research grants from the Hellenic Society of Medical Oncology (HeSMO; https://www.hesmo.gr/en/) and the Gastrointestinal Cancer Study Group (GIC-SG; https://www.emkapes.gr/?lang=en).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jemal A., Siegel R., Xu J., Ward E. Cancer statistics, 2010. CA Cancer J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Ries L.A.G., Wingo P.A., Miller D.S., Howe H.L., Weir H.K., Rosenberg H.M., Vernon S.W., Cronin K., Edwards B.K. The annual report to the nation on the status of cancer, 1973-1997, with a special section on colorectal cancer. Cancer. 2000;88:2398–2424. doi: 10.1002/(SICI)1097-0142(20000515)88:10<2398::AID-CNCR26>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 3.Androulakis N., Agelaki S., Perraki M., Apostolaki S., Bozionelou V., Pallis A., Kalbakis K., Xyrafas A., Mavroudis D., Georgoulias V. Clinical relevance of circulating CK-19mRNA-positive tumour cells before front-line treatment in patients with metastatic breast cancer. Br. J. Cancer. 2012;106:1917–1925. doi: 10.1038/bjc.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen S.J., Alpaugh R.K., Gross S., O’Hara S.M., Smirnov D.A., Terstappen L.W., Allard W.J., Bilbee M., Cheng J.D., Hoffman J.P., et al. Isolation and Characterization of Circulating Tumor Cells in Patients with Metastatic Colorectal Cancer. Clin. Color. Cancer. 2006;6:125–132. doi: 10.3816/CCC.2006.n.029. [DOI] [PubMed] [Google Scholar]
- 5.Cristofanilli M., Budd G.T., Ellis M.J., Stopeck A., Matera J., Miller M.C., Reuben J.M., Doyle G.V., Allard W.J., Terstappen L.W., et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 6.Hou J.-M., Krebs M.G., Lancashire L., Sloane R., Backen A., Swain R.K., Priest L.J., Greystoke A., Zhou C., Morris K., et al. Clinical Significance and Molecular Characteristics of Circulating Tumor Cells and Circulating Tumor Microemboli in Patients with Small-Cell Lung Cancer. J. Clin. Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 7.Naito T., Tanaka F., Ono A., Yoneda K., Takahashi T., Murakami H., Nakamura Y., Tsuya A., Kenmotsu H., Shukuya T., et al. Prognostic Impact of Circulating Tumor Cells in Patients with Small Cell Lung Cancer. J. Thorac. Oncol. 2012;7:512–519. doi: 10.1097/JTO.0b013e31823f125d. [DOI] [PubMed] [Google Scholar]
- 8.Okegawa T., Nutahara K., Higashihara E. Prognostic Significance of Circulating Tumor Cells in Patients with Hormone Refractory Prostate Cancer. J. Urol. 2009;181:1091–1097. doi: 10.1016/j.juro.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Messaritakis I., Politaki E., Koinis F., Stoltidis D., Apostolaki S., Plataki M., Dermitzaki E.-K., Georgoulias V., Kotsakis A. Dynamic changes of phenotypically different circulating tumor cells sub-populations in patients with recurrent/refractory small cell lung cancer treated with pazopanib. Sci. Rep. 2018;8:2238. doi: 10.1038/s41598-018-20502-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Messaritakis I., Politaki E., Kotsakis A., Dermitzaki E.-K., Koinis F., Lagoudaki E., Koutsopoulos A., Kallergi G., Souglakos J., Georgoulias V. Phenotypic characterization of circulating tumor cells in the peripheral blood of patients with small cell lung cancer. PLoS ONE. 2017;12:e0181211. doi: 10.1371/journal.pone.0181211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messaritakis I., Sfakianaki M., Papadaki C., Koulouridi A., Vardakis N., Koinis F., Hatzidaki D., Georgoulia N., Kladi A., Kotsakis A., et al. Prognostic significance of CEACAM5mRNA-positive circulating tumor cells in patients with metastatic colorectal cancer. Cancer Chemother. Pharmacol. 2018;82:767–775. doi: 10.1007/s00280-018-3666-9. [DOI] [PubMed] [Google Scholar]
- 12.Milaki G., Messaritakis I., Koinis F., Kotsakis A., Apostolaki S., Dermitzaki E.K., Perraki M., Hatzidaki D., Georgoulias V. Prognostic value of chemotherapy-resistant CK19mRNA-positive circulating tumor cells in patients with advanced/metastatic non-small cell lung cancer. Cancer Chemother. Pharmacol. 2017;80:101–108. doi: 10.1007/s00280-017-3339-0. [DOI] [PubMed] [Google Scholar]
- 13.Vardakis N., Messaritakis I., Papadaki C., Agoglossakis G., Sfakianaki M., Saridaki Z., Apostolaki S., Koutroubakis I.E., Perraki M., Hatzidaki D., et al. Prognostic Significance of the Detection of Peripheral Blood CEACAM5mRNA-Positive Cells by Real-Time Polymerase Chain Reaction in Operable Colorectal Cancer. Clin. Cancer Res. 2010;17:165–173. doi: 10.1158/1078-0432.CCR-10-0565. [DOI] [PubMed] [Google Scholar]
- 14.Polyak K., Weinberg R.A. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Rev. Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 15.Willipinski-Stapelfeldt B., Riethdorf S., Assmann V., Woelfle U., Rau T., Sauter G., Heukeshoven J., Pantel K. Changes in Cytoskeletal Protein Composition Indicative of an Epithelial-Mesenchymal Transition in Human Micrometastatic and Primary Breast Carcinoma Cells. Clin. Cancer Res. 2005;11:8006–8014. doi: 10.1158/1078-0432.CCR-05-0632. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh P., Yamane K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech. Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kheirelseid E.A.H., Miller N., Chang K.H., Curran C., Hennessey E., Sheehan M., Kerin M.J. Mismatch repair protein expression in colorectal cancer. J. Gastrointest. Oncol. 2013;4:397–408. doi: 10.3978/j.issn.2078-6891.2013.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vieira M.L.C., Santini L., Diniz A.L., Munhoz C.D.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016;39:312–328. doi: 10.1590/1678-4685-GMB-2016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pikor L.A., Thu K., Vucic E.A., Lam W. The detection and implication of genome instability in cancer. Cancer Metastasis Rev. 2013;32:341–352. doi: 10.1007/s10555-013-9429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michael-Robinson J.M., Reid L.E., Purdie D.M., Biemer-Hüttmann A.E., Walsh M.D., Pandeya N., Simms L.A., Young J.P., Leggett B.A., Jass J.R., et al. Proliferation, apoptosis, and survival in high-level microsatellite instability sporadic colorectal cancer. Clin. Cancer Res. 2001;7:2347–2356. [PubMed] [Google Scholar]
- 22.Gatalica Z., Vranic S., Xiu J., Swensen J., Reddy S. High microsatellite instability (MSI-H) colorectal carcinoma: A brief review of predictive biomarkers in the era of personalized medicine. Fam. Cancer. 2016;15:405–412. doi: 10.1007/s10689-016-9884-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang S., Na Y., Joung S.Y., Lee S.I., Oh S.C., Min B.W. The significance of microsatellite instability in colorectal cancer after controlling for clinicopathological factors. Medicine (Baltimore) 2018;97:e0019. doi: 10.1097/MD.0000000000010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halpern N., Goldberg Y., Kadouri L., Duvdevani M., Hamburger T., Peretz T., Hubert A. Clinical course and outcome of patients with high-level microsatellite instability cancers in a real-life setting: A retrospective analysis. OncoTargets Ther. 2017;10:1889–1896. doi: 10.2147/OTT.S126905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Copija A., Waniczek D., Witkoś A., Walkiewicz K., Nowakowska-Zajdel E. Clinical Significance and Prognostic Relevance of Microsatellite Instability in Sporadic Colorectal Cancer Patients. Int. J. Mol. Sci. 2017;18:107. doi: 10.3390/ijms18010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kallergi G., Mavroudis D., Politaki E., Mavroudis D., Kouroussis C., Agelaki S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011;13:R59. doi: 10.1186/bcr2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papadaki M.A., Kallergi G., Zafeiriou Z., Manouras L., Theodoropoulos P.A., Mavroudis D., Georgoulias V., Agelaki S. Co-expression of putative stemness and epithelial-to-mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer. 2014;14:651. doi: 10.1186/1471-2407-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stathopoulou A., Gizi A., Perraki M., Apostolaki S., Malamos N., Mavroudis D., Georgoulias V., Lianidou E.S. Real-time quantification of CK-19 mRNA-positive cells in peripheral blood of breast cancer patients using the lightcycler system. Clin. Cancer Res. 2003;9:5145–5151. [PubMed] [Google Scholar]
- 29.Stathopoulou A., Vlachonikolis I., Mavroudis D., Perraki M., Kouroussis C., Apostolaki S., Malamos N., Kakolyris S., Kotsakis A., Xenidis N., et al. Molecular Detection of Cytokeratin-19–Positive Cells in the Peripheral Blood of Patients with Operable Breast Cancer: Evaluation of Their Prognostic Significance. J. Clin. Oncol. 2002;20:3404–3412. doi: 10.1200/JCO.2002.08.135. [DOI] [PubMed] [Google Scholar]
- 30.Xenidis N., Perraki M., Kafousi M., Apostolaki S., Bolonaki I., Stathopoulou A., Kalbakis K., Androulakis N., Kouroussis C., Pallis T., et al. Predictive and Prognostic Value of Peripheral Blood Cytokeratin-19 mRNA-Positive Cells Detected by Real-Time Polymerase Chain Reaction in Node-Negative Breast Cancer Patients. J. Clin. Oncol. 2006;24:3756–3762. doi: 10.1200/JCO.2005.04.5948. [DOI] [PubMed] [Google Scholar]
- 31.Saridaki Z., Papadatos-Pastos D., Tzardi M., Mavroudis D., Bairaktari E., Arvanity H., Stathopoulos E., Georgoulias V., Souglakos J. BRAF mutations, microsatellite instability status and cyclin D1 expression predict metastatic colorectal patients’ outcome. Br. J. Cancer. 2010;102:1762–1768. doi: 10.1038/sj.bjc.6605694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saridaki Z., Tzardi M., Papadaki C., Sfakianaki M., Pega F., Kalikaki A., Tsakalaki E., Trypaki M., Messaritakis I., Stathopoulos E., et al. Impact of KRAS, BRAF, PIK3CA mutations, PTEN, AREG, EREG expression and skin rash in >/= 2 line cetuximab-based therapy of colorectal cancer patients. PLoS ONE. 2011;6:e15980. doi: 10.1371/journal.pone.0015980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benlloch S., Payá A., Alenda C., Bessa X., Andreu M., Jover R., Castells A., Llor X., Aranda F., Massutí B. Detection of BRAF V600E Mutation in Colorectal Cancer. J. Mol. Diagn. 2006;8:540–543. doi: 10.2353/jmoldx.2006.060070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douillard J.-Y., Oliner K.S., Siena S., Tabernero J., Burkes R., Barugel M., Humblet Y., Bodoky G., Cunningham D., Jassem J., et al. Panitumumab–FOLFOX4 Treatment and RAS Mutations in Colorectal Cancer. N. Engl. J. Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 35.Campos-Da-Paz M., Dórea J.G., Galdino A.S., Lacava Z.G.M., Santos M. Carcinoembryonic Antigen (CEA) and Hepatic Metastasis in Colorectal Cancer: Update on Biomarker for Clinical and Biotechnological Approaches. Recent Patents Biotechnol. 2018;12:269–279. doi: 10.2174/1872208312666180731104244. [DOI] [PubMed] [Google Scholar]
- 36.Thomas P., Forse R.A., Bajenova O. Carcinoembryonic antigen (CEA) and its receptor hnRNP M are mediators of metastasis and the inflammatory response in the liver. Clin. Exp. Metastasis. 2011;28:923–932. doi: 10.1007/s10585-011-9419-3. [DOI] [PubMed] [Google Scholar]
- 37.Beauchemin N., Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32:643–671. doi: 10.1007/s10555-013-9444-6. [DOI] [PubMed] [Google Scholar]
- 38.Ordoñez C., Screaton R.A., Ilantzis C., Stanners C.P. Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Cancer Res. 2000;60:3419–3424. [PubMed] [Google Scholar]
- 39.Blumenthal R.D. Inhibition of Adhesion, Invasion, and Metastasis by Antibodies Targeting CEACAM6 (NCA-90) and CEACAM5 (Carcinoembryonic Antigen) Cancer Res. 2005;65:8809–8817. doi: 10.1158/0008-5472.CAN-05-0420. [DOI] [PubMed] [Google Scholar]
- 40.Han S., Zong S., Shi Q., Li H., Liu S., Yang W., Li W., Hou F. Is Ep-CAM Expression a Diagnostic and Prognostic Biomarker for Colorectal Cancer? A Systematic Meta-Analysis. EBioMedicine. 2017;20:61–69. doi: 10.1016/j.ebiom.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munz M., Baeuerle P.A., Gires O. The Emerging Role of EpCAM in Cancer and Stem Cell Signaling. Cancer Res. 2009;69:5627–5629. doi: 10.1158/0008-5472.CAN-09-0654. [DOI] [PubMed] [Google Scholar]
- 42.Serrano M.J., Ortega F.G., Alvarez-Cubero M.J., Nadal R., Sanchez-Rovira P., Salido M., Rodríguez M., García-Puche J.L., Delgado-Rodriguez M., Solé F., et al. EMT and EGFR in CTCs cytokeratin negative non-metastatic breast cancer. Oncotarget. 2014;5:7486–7497. doi: 10.18632/oncotarget.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J.H., Bae J.M., Song Y.S., Cho N.-Y., Lee H.S., Kang G.H. Clinicopathologic, molecular, and prognostic implications of the loss of EPCAM expression in colorectal carcinoma. Oncotarget. 2015;7:13372–13387. doi: 10.18632/oncotarget.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Wit S., Zeune L.L., Hiltermann T.J.N., Groen H.J.M., Van Dalum G., Terstappen L.W. Classification of Cells in CTC-Enriched Samples by Advanced Image Analysis. Cancers. 2018;10:377. doi: 10.3390/cancers10100377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldstein J., Tran B., Ensor J., Gibbs P., Wong H.L., Wong S.F., Vilar E., Tie J., Broaddus R., Kopetz S., et al. Multicenter retrospective analysis of metastatic colorectal cancer (CRC) with high-level microsatellite instability (MSI-H) Ann. Oncol. 2014;25:1032–1038. doi: 10.1093/annonc/mdu100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taieb J., Shi Q., Pederson L., Alberts S., Wolmark N., Van Cutsem E., De Gramont A., Kerr R., Grothey A., Lonardi S., et al. Prognosis of microsatellite instability and/or mismatch repair deficiency stage III colon cancer patients after disease recurrence following adjuvant treatment: Results of an ACCENT pooled analysis of seven studies. Ann. Oncol. 2019;30:1466–1471. doi: 10.1093/annonc/mdz208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin Z., Sanhueza C.T., Johnson B., Nagorney D.M., Larson D.W., Mara K.C., Harmsen W.C., Smyrk T.C., Grothey A., Hubbard J.M. Outcome of Mismatch Repair-Deficient Metastatic Colorectal Cancer: The Mayo Clinic Experience. Oncologist. 2018;23:1083–1091. doi: 10.1634/theoncologist.2017-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onstenk W., Sieuwerts A.M., Mostert B., Lalmahomed Z., Vries J.B.B.-D., Van Galen A., Smid M., Kraan J., Van M., De Weerd V., et al. Molecular characteristics of circulating tumor cells resemble the liver metastasis more closely than the primary tumor in metastatic colorectal cancer. Oncotarget. 2016;7:59058–59069. doi: 10.18632/oncotarget.10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J.H., Bae J.M., Kim K.-J., Rhee Y.-Y., Kim Y., Cho N.-Y., Lee H.S., Chang M.S., Kang G.H. Differential Features of Microsatellite-Unstable Colorectal Carcinomas Depending on EPCAM Expression Status. Korean J. Pathol. 2014;48:276–282. doi: 10.4132/KoreanJPathol.2014.48.4.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kloor M., Voigt A.Y., Schackert H.K., Schirmacher P., von Knebel Doeberitz M., Bläker H. Analysis of EPCAM protein expression in diagnostics of Lynch syndrome. J. Clin. Oncol. 2011;29:223–227. doi: 10.1200/JCO.2010.32.0820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.