Abstract
Though several Diaporthe species have been reported in China, little is known about the species associated with nature reserves in Guizhou province. During a survey of fungi in six nature reserves in Guizhou province of China, thirty-one Diaporthe isolates were collected from different woody hosts. Based on morphology, culture characteristics and molecular phylogenetic analysis, these isolates were characterized and identified. Phylogenetic analysis of internal transcribed spacer region (ITS), combined with translation elongation factor 1-alpha (tef), β-tubulin (tub), calmodulin (cal) and histone H3 (his) gene regions identified five known Diaporthe species and seven distinct lineages representing novel Diaporthe species. The details of five known species: Diaporthe cercidis, D. cinnamomi, D. conica, D. nobilis and D. sackstonii are given and the seven new species D. constrictospora, D. ellipsospora, D. guttulata, D. irregularis, D. lenispora, D. minima, and D. minusculata are introduced with detailed descriptions and illustrations. This study revealed a high diversity of previously undescribed Diaporthe species associated with woody hosts in various nature reserves of Guizhou province, indicating that there is a potential of Diaporthe species remains to be discovered in this unique landform (Karst formations) in China. Interestingly, the five known Diaporthe species have been reported as pathogens of various hosts, and this could indicate that those newly introduced species in this study could be potentially pathogenic pending further studies to confirm.
Keywords: seven new taxa, asexual morph, Diaporthaceae, phylogeny, taxonomy
1. Introduction
Diaporthe Nitschke (including the Phomopsis asexual morph) belongs to family Diaporthaceae, order Diaporthales and class Sordariomycetes [1,2,3] and its species are found worldwide on a diverse range of host plants as endophytes, pathogens and saprobes [4]. Rossman et al. [5] proposed the name Diaporthe over Phomopsis, as both names are well known amongst plant pathologists and subsequent studies have adopted the latter generic name [4,6,7,8,9,10]. More than 1100 epithets for Diaporthe and 986 for Phomopsis are listed in Index Fungorum (2020) (http://www.indexfungorum.org/, accessed August 2020) with names often based on host association. Many Diaporthe species that are morphologically similar have proven to be genetically distinct [11,12], and several isolates formerly identified based on their hosts were shown to represent different taxa [1]. Diaporthe represents a highly complex genus containing numerous cryptic species. In recent studies, Diaporthe species have been distinguished mainly by their molecular phylogenies, and the best five gene regions to conduct a multi-gene phylogenetic analysis are ITS, tef, tub, cal and his [4,13,14,15,16,17,18,19].
A nature reserve is a protected area of importance for flora, fauna or landscapes of geological or other special interest, which is reserved and managed for purposes of conservation and to provide special opportunities for study or research [20]. The Karst region of Guizhou province is comprised of abundant nature reserves that provide a wide range of ecosystem services such as water supply, soil fertility, ecotourism, recreation, biodiversity conservation and carbon sequestration [20]. However, there are few scientific evaluations made for fungi in national nature reserves and national forest parks in Guizhou province, China [21,22]. During the investigation carried out in 2017 to 2019, several isolates of Diaporthe species were collected from six nature reserves in Guizhou province including Fanjing mountain, Guiyang Huaxi wetland park, Guiyang Xiaochehe wetland park, Maolan nature reserve, Suiyang broad water nature reserve and Xingyi Wanfenglin. Fungi isolated from forest trees in China were recorded in old fungal literature, however, most of them lack living culture and molecular data [23,24]. Although several species of Diaporthe have been previously recorded from Guizhou province with details of culture and molecular data [25,26], little is known to associate these with hosts in nature reserves. Thus, the aim of this study is to describe and illustrate Diaporthe taxa from nature reserves in the Karst region of Guizhou province based on morphological characters and phylogenies derived from combined ITS, tef, tub, cal and his gene sequences.
2. Materials and Methods
2.1. Isolation of Fungal Material, Morphology and Culture Characteristics
From 2017 to 2019, thirty-one Diaporthe specimens were collected in field surveys of decaying saprobic woody hosts in different nature reserves including Fanjing mountain, Guiyang Huaxi wetland park, Guiyang Xiaochehe wetland park, Maolan nature reserve, Suiyang broad water nature reserve and Xingyi Wanfenglin in Karst region of Guizhou province (Table 1). Collected samples were taken to the laboratory for isolation and photographed, documented and then kept at 4 °C for further study.
Table 1.
Diaporthe species studied in this study (Figure 1). Details of ex-type species introduced in this study are in bold.
| Species | Isolate | Locality | ITS | tef | tub | cal | his |
|---|---|---|---|---|---|---|---|
| Diaporthe cercidis | GZCC 19-0079 | Guiyang Xiaochehe Wetland Park | MT385942 | MT424677 | MT424698 | MW022466 | MW022482 |
| D. cercidis | GZCC 19-0124 | Maolan Nature Reserve | MT385943 | MT424678 | MT424699 | MW022467 | MW022483 |
| D. cercidis | GZCC 19-0217 | Xingyi Wanfenglin | MT385944 | MT424679 | MT424700 | MW022468 | MW022484 |
| D. cinnamomi | GZCC 19-0274 | Maolan Nature Reserve | MT385945 | MT424680 | N/A | MT424717 | MW022485 |
| D. conica | GZCC 19-0242 | Maolan NatureReserve | MT385946 | MT424681 | MT424701 | MW022469 | MW022486 |
| D. constrictospora | CGMCC 3.20096 | Maolan Nature Reserve | MT385947 | MT424682 | MT424702 | MT424718 | MW022487 |
| D. constrictospora | GZCC 19-0065 | Guiyang Huaxi Wetland Park | MT385948 | MT424683 | MT424703 | MT424719 | N/A |
| D. ellipsospora | CGMCC 3.20099 | Xingyi Wanfenglin | MT385949 | MT424684 | MT424704 | MT424720 | MW022488 |
| D. ellipsospora | GZCC 19-0342 | Xingyi Wanfenglin | MT797176 | MT793019 | MT793030 | MT786247 | MW022489 |
| D. ellipsospora | GZCC 19-0357 | Maolan Nature Reserve | MT797177 | MT793020 | MT793031 | MT786248 | MW022490 |
| D. guttulata | CGMCC 3.20100 | Maolan Nature Reserve | MT385950 | MT424685 | MT424705 | MW022470 | MW022491 |
| D. guttulata | GZCC 19-0371 | Suiyang water nature reserve | MT797178 | MT793021 | MT793032 | MW022471 | MW022492 |
| D. irregularis | CGMCC 3.20092 | Suiyang water nature reserve | MT385951 | MT424686 | MT424706 | MT424721 | N/A |
| D. irregularis | GZCC 19-0344 | Suiyang water nature reserve | MT797179 | MT793022 | MT793033 | MT786249 | N/A |
| D. irregularis | GZCC 19-0362 | Suiyang water nature reserve | MT797180 | MT793023 | MT793034 | MT786250 | N/A |
| D. irregularis | GZCC 19-0352 | Suiyang water nature reserve | MT797181 | MT793024 | MT793035 | MT786251 | N/A |
| D. lenispora | CGMCC 3.20101 | Suiyang water nature reserve | MT385952 | MT424687 | MT424707 | MW022472 | MW022493 |
| D. lenispora | GZCC 19-0343 | Xingyi Wanfenglin | MT797182 | MT793025 | MT793036 | MW022473 | MW022494 |
| D. lenispora | GZCC 19-0351 | Xingyi Wanfenglin | MT797183 | MT793026 | MT793037 | MW022474 | MW022495 |
| D. minima | CGMCC 3.20097 | Guiyang Huaxi Wetland Park | MT385953 | MT424688 | MT424708 | MT424722 | MW022496 |
| D. minima | GZCC 19-0070 | Guiyang Huaxi Wetland Park | MT385954 | MT424689 | MT424709 | MT424723 | MW022497 |
| D. minima | GZCC 19-0061 | Guiyang Huaxi Wetland Park | MT385955 | MT424690 | MT424710 | MT424724 | MW022498 |
| D. minima | GZCC 19-0207 | Guiyang Huaxi Wetland Park | MT385956 | MT424691 | MT424711 | MT424725 | N/A |
| D. minusculata | CGMCC 3.20098 | Xingyi Wanfenglin | MT385957 | MT424692 | MT424712 | MW022475 | MW022499 |
| D. minusculata | GZCC 19-0345 | Suiyang water nature reserve | MT797184 | MT793027 | MT793038 | MW022476 | MW022500 |
| D. minusculata | GZCC 19-0366 | Suiyang water nature reserve | MT797185 | MT793028 | MT793039 | MW022477 | MW022501 |
| D. minusculata | GZCC 19-0372 | Xingyi Wanfenglin | MT797186 | MT793029 | MT793040 | MW022478 | MW022502 |
| D. nobilis | GZCC 19-0213 | Fanjing mountain | MT385958 | MT424693 | MT424713 | MT424726 | MW022503 |
| D. nobilis | GZCC 19-0220 | Xingyi Wanfenglin | MT385959 | MT424694 | MT424714 | MW022479 | MW022504 |
| D. nobilis | GZCC 19-0214 | Fanjing mountain | MT385960 | MT424695 | MT424715 | MW022480 | MW022505 |
| D. sackstonii | GZCC 19-0129 | Maolan Nature Reserve | MT385962 | MT424697 | MT424716 | MT424727 | MW022507 |
Species identification was primarily based on morphological observation of the conidiomata or ascomata from host materials and micromorphology supplemented by culture characteristics. Morphological observations were made using a Motic SMZ (Stereoscopic Zoom Microscope) 168 series stereomicroscope and photographed by a Nikon E80i microscope-camera system. Measurements were made with the Tarosoft (R) Image FrameWork [27] and images used for figures were processed with Adobe Photoshop CS v. 5. Single spore isolations were prepared following the method of Chomnunti et al. [28]. Spore germination on 2% water agar (WA) was examined after 24 h and germinating spores were transferred to potato dextrose agar (PDA) media. Cultures were incubated at 25 °C in the dark and colony morphology and conidial characteristics were examined for a total of 31 isolates. Colony color was determined according to Rayner [29] after 5 d to 10 d on PDA at 25 °C in the dark. More than 20 conidiomata/ascomata, 30 asci, and 50 conidia/ascospores were measured to calculate the mean size/length and respective standard deviations (SD). Conidial shape, color and guttulation were also recorded.
Herbarium specimens were deposited at the Herbarium of Cryptogams, Kunming Institute of Botany Academia Sinica (KUN-HKAS), Kunming, China and herbaria of Guizhou Academy of Agricultural Sciences (GZAAS), Guiyang, China. The living cultures were deposited in the China General Microbiological Culture Collection Center (CGMCC) in Beijing, China and Guizhou Culture Collection (GZCC) in Guiyang, China and (Table 1).
2.2. Molecular Based Amplification
Fungal mycelium of 7 d old cultures was scraped for the extraction of genomic DNA using Biospin Fungus Genomic DNA Extraction Kit (BioFlux®) following the manufacturer’s protocol (Hangzhou, China). For the identification of Diaporthe specimens, the internal transcribed spacer region (ITS) was sequenced for all isolates and BLAST search (basic local alignment search tool) at GenBank was used to reveal the closest matching taxa. Besides ITS gene sequence data, translation elongation factor 1-alpha (tef), β-tubulin (tub), calmodulin (cal) and histone H3 (his) gene regions were also employed to support the species identification. The ITS region was amplified using universal primers ITS1 and ITS4 [30]. The target region of the tef gene was amplified using primer pairs EF-728F and EF-986R [31]. A portion of the tub gene was amplified using the primers BT2a and BT2b [32], while the primer pair CAL228F and CAL737R was used to amplify the cal gene region [31]. The primers CYLH3F [33] and H3-1b [32] were used to amplify part of the his gene. The PCR reactions were accomplished in a Bio Rad C1000 thermal cycler. The amplification procedure was performed in a 50 μL reaction volume containing 5–10 ng DNA, 0.8 units Taq polymerase, 1X PCR buffer, 0.2 mM dNTP, 0.3 μm of each primer with 1.5 mM MgCl2. Following the PCR amplification, products were visualized on 1% agarose gel under UV light using a Gel DocTM XR Molecular Imager following ethidium bromide staining. PCR products were purified using minicolumns, purification resin and buffer according to the manufacturer’s protocols (Amersham product code: 27–9602–01). Sequence analysis was carried out by Shanghai Sangon Biological Engineering Technology and Services Co., Ltd. (Shanghai, China).
2.3. Sequence Alignment and Phylogenetic Analyses
To assure the sequence quality, the resulting sequence chromatograms were checked using BioEdit v.5 [34]. An overview phylogenetic tree (provided as a Supplementary Figure S1) for the genus Diaporthe was constructed from ITS sequence data of all type/ex-type/neo-type Diaporthe species from previous studies [10,13,14,15,33,35,36,37,38,39,40,41,42,43,44,45]. Considering this ITS tree, another phylogenetic analysis was conducted including all the isolates obtained in this study (Table 1) together with several closely associated Diaporthe species (Table 2). Diaporthella corylina (CBS 121124) was selected as the outgroup taxon. The sequences were retrieved from GenBank and aligned with the sequences obtained in this study using MAFFT [46] (http://www.ebi.ac.uk/Tools/msa/mafft/) and manually edited with BioEdit [34] for a maximum alignment. Phylogenetic analysis was performed by using PAUP (Phylogenetic Analysis Using Parsimony) v.4.0b10 for maximum parsimony (MP) method [47], RAxML for maximum likelihood (ML) method [48] and MrBayes v.3.1.2 for Bayesian Inference (BI) method [49]. The best model of evolution was determined by MrModeltest v. 2.3 [50]. Maximum likelihood analyses was accomplished using RAxML GUI v. 0.9b2 [51] with 1000 non-parametric bootstrapping iterations, using the general time-reversible model (GTR) with a discrete gamma distribution. The best scoring trees were chosen with final likelihood values.
Table 2.
GenBank accession numbers of species included in the phylogenetic analysis (Figure 1). Ex-type/ex-epitype/ex-isotype/ex-neotype isolates are in bold.
| Species Name | Isolate Number | ITS | tub | tef | cal | his | Reference |
|---|---|---|---|---|---|---|---|
| Diaporthella corylina | CBS 121124 | KC343004 | KC343972 | KC343730 | KC343246 | KC343488 | Vasilyeva et al. [54] |
| Diaporthe acaciarum | CBS 138862 | KP004460 | KP004509 | N/A | N/A | KP004504 | Crous et al. [55] |
| Diaporthe acuta | PSCG 047 | MK626957 | MK691225 | MK654802 | MK691125 | MK726161 | Guo et al. [15] |
| Diaporthe acuta | PSCG 046 | MK626958 | MK691224 | MK654803 | MK691124 | MK726162 | Guo et al. [15] |
| Diaporthe albosinensis | CFCC 53066 | MK432659 | MK578059 | MK578133 | MK442979 | MK443004 | Yang et al. [14] |
| Diaporthe albosinensis | CFCC 53067 | MK432660 | MK578060 | MK578134 | MK442980 | MK443005 | Yang et al. [14] |
| Diaporthe ampelina | CBS 114016 | AF230751 | JX275452 | AY745056 | AY230751 | N/A | Mostert et al. [56] |
| Diaporthe ampelina | CBS 267.80 | KC343018 | KC343986 | KC343744 | KC343260 | KC343502 | Mostert et al. [56] |
| Diaporthe angelicae | CBS 111592 | KC343027 | KC343995 | KC343753 | KC343269 | KC343511 | Castlebury et al. [57] |
| Diaporthe angelicae | CBS 100871 | KC343025 | KC343993 | KC343751 | KC343267 | KC343509 | Castlebury et al. [57] |
| Diaporthe aquatica | IFRDCC 3015 | JQ797438 | N/A | N/A | N/A | N/A | Hu et al. [58] |
| Diaporthe aquatica | IFRDCC 3051 | JQ797437 | N/A | N/A | N/A | N/A | Hu et al. [58] |
| Diaporthe araucanorum | CBS 145285 | MN509711 | MN509722 | MN509733 | N/A | N/A | Zapata et al. [45] |
| Diaporthe araucanorum | CBS 145284 | MN509710 | MN509721 | MN509732 | N/A | N/A | Zapata et al. [45] |
| Diaporthe asheicola | CBS 136967 | KJ160562 | KJ160518 | KJ160594 | KJ160542 | N/A | Lombard et al. [59] |
| Diaporthe asheicola | CBS 136968 | KJ160563 | KJ160519 | KJ160595 | KJ160543 | N/A | Lombard et al. [59] |
| Diaporthe aspalathi | CBS 117168 | KC343035 | KC344003 | KC343761 | KC343277 | KC343519 | van Rensburg et al. [11] |
| Diaporthe aspalathi | CBS 117169 | KC343036 | KC344004 | KC343762 | KC343278 | KC343520 | van Rensburg et al. [11] |
| Diaporthe australafricana | CBS 111886 | KC343038 | KC344006 | KC343764 | KC343280 | KC343522 | Mostert et al. [56] |
| Diaporthe australafricana | CBS 113487 | KC343039 | KC344007 | KC343765 | KC343281 | KC343523 | Mostert et al. [56] |
| Diaporthe biconispora | ZJUD61 | KJ490596 | KJ490417 | KJ490475 | N/A | KJ490538 | Huang et al. [60] |
| Diaporthe biconispora | ZJUD62 | KJ490597 | KJ490418 | KJ490476 | KJ490539 | KJ490539 | Huang et al. [60] |
| Diaporthe bohemiae | CBS 143347 | MG281015 | MG281188 | MG281536 | MG281710 | MG281361 | Guarnaccia et al. [19] |
| Diaporthe caryae | CFCC 52563 | MH121498 | MH121580 | MH121540 | MH121422 | MH121458 | Yang et al. [13] |
| Diaporthe caryae | CFCC 52564 | MH121499 | MH121581 | MH121541 | MH121423 | MH121459 | Yang et al. [13] |
| Diaporthe cercidis | CFCC 52565 | MH121500 | MH121582 | MH121542 | MH121424 | MH121460 | Yang et al. [13] |
| Diaporthe cercidis | CFCC 52566 | MH121501 | MH121583 | MH121543 | MH121425 | MH121461 | Yang et al. [13] |
| Diaporthe chongqingensis | PSCG 435 | MK626916 | MK691321 | MK654866 | MK691209 | MK726257 | Guo et al. [15] |
| Diaporthe chongqingensis | PSCG 436 | MK626917 | MK691322 | MK654867 | MK691208 | MK726256 | Guo et al. [15] |
| Diaporthe cichorii | MFLUCC 17-1023 | KY964220 | KY964104 | KY964176 | KY964133 | N/A | Dissanayake et al. [9] |
| Diaporthe cinnamomi | CFCC 52569 | MH121504 | MH121586 | MH121546 | N/A | MH121464 | Yang et al. [13] |
| Diaporthe cinnamomi | CFCC 52570 | MH121505 | MH121587 | MH121547 | N/A | MH121465 | Yang et al. [13] |
| Diaporthe cissampeli | CPC 27302 | KX228273 | KX228384 | N/A | N/A | KX228366 | Crous et al. [61] |
| Diaporthe citri | CBS 135422 | KC843311 | KC843187 | KC843071 | KC843157 | MF418281 | Udayanga et al. [6] |
| Diaporthe citri | AR 4469 | KC843321 | KC843197 | KC843081 | KC843167 | N/A | Udayanga et al. [6] |
| Diaporthe conica | CFCC 52571 | MH121506 | MH121588 | MH121548 | MH121428 | MH121466 | Yang et al. [13] |
| Diaporthe conica | CFCC 52572 | MH121507 | MH121589 | MH121549 | MH121429 | MH121467 | Yang et al. [13] |
| Diaporthe coryli | CFCC 53083 | MK432661 | MK578061 | MK578135 | MK442981 | MK443006 | Yang et al. [14] |
| Diaporthe coryli | CFCC 53084 | MK432662 | MK578062 | MK578136 | MK442982 | MK443007 | Yang et al. [14] |
| Diaporthe discoidispora | ZJUD89 | KJ490624 | KJ490445 | KJ490503 | N/A | KJ490566 | Huang et al. [60] |
| Diaporthe discoidispora | ZJUD87 | KJ490622 | KJ490443 | KJ490501 | N/A | KJ490564 | Huang et al. [60] |
| Diaporthe eres | AR 5193 | KJ210529 | KJ420799 | KJ210550 | KJ434999 | KJ420850 | Udayanga et al. [7] |
| Diaporthe eres | CBS 138598 | KJ210521 | KJ420787 | KJ210545 | KJ435027 | KJ420837 | Udayanga et al. [7] |
| Diaporthe foikelawen | CBS 145289 | MN509714 | MN509725 | MN509736 | N/A | N/A | Zapata et al. [45] |
| Diaporthe foikelawen | CBS 145287 | MN509713 | MN509724 | MN509735 | N/A | N/A | Zapata et al. [45] |
| Diaporthe fulvicolor | PSCG 051 | MK626859 | MK691236 | MK654806 | MK691132 | MK726163 | Guo et al. [15] |
| Diaporthe fulvicolor | PSCG 057 | MK626858 | MK691233 | MK654810 | MK691131 | MK726164 | Guo et al. [15] |
| Diaporthe gulyae | BRIP 53158 | JF431284 | KJ197271 | N645799 | N/A | N/A | Thompson et al. [62] |
| Diaporthe gulyae | BRIP 54025 | JF431299 | KJ197272 | JN645803 | N/A | N/A | Thompson et al. [62] |
| Diaporthe helicis | AR 5211 | KJ210538 | KJ420828 | KJ210559 | KJ435043 | KJ420875 | Udayanga et al. [7] |
| Diaporthe hungariae | CBS 143353 | MG281126 | MG281299 | MG281647 | MG281823 | MG281474 | Guarnaccia et al. [19] |
| Diaporthe hungariae | CBS 143354 | MG281127 | MG281300 | MG281648 | MG281824 | MG281475 | Guarnaccia et al. [19] |
| Diaporthe juglandicola | CFCC 51134 | KU985101 | KX024634 | KX024628 | KX024616 | KX024622 | Yang et al. [18] |
| Diaporthe mahothocarpus | CGMCC 3.15181 | KC153096 | KF576312 | KC153087 | N/A | N/A | Gao et al. [63] |
| Diaporthe mahothocarpus | CGMCC 3.15182 | KC153097 | N/A | KC153088 | N/A | N/A | Gao et al. [63] |
| Diaporthe malorum | CAA734 | KY435638 | KY435668 | KY435627 | KY435658 | KY435648 | Santos et al. [16] |
| Diaporthe millettia | GUCC 9167 | MK398674 | MK502089 | MK480609 | MK502086 | N/A | Long et al. [26] |
| Diaporthe nobilis | CBS 587.79 | KC343153 | KC344121 | KC343879 | KC343395 | KC343637 | Li et al. [25] |
| Diaporthe novem | CBS 127270 | KC343155 | KC344123 | KC343881 | KC343397 | KC343640 | Santos et al. [64] |
| Diaporthe novem | CBS 127271 | KC343157 | KC344125 | KC343883 | KC343399 | KC343641 | Santos et al. [64] |
| Diaporthe oraccinii | LC 3166 | KP267863 | KP293443 | KP267937 | N/A | KP293517 | Gao et al. [63] |
| Diaporthe osmanthusis | GUCC 9165 | MK398675 | MK502090 | MK480610 | MK502087 | N/A | Long et al. [26] |
| Diaporthe paranensis | CBS 133184 | KC343171 | KC344139 | KC343897 | KC343413 | KC343655 | Gomes et al. [4] |
| Diaporthe parvae | PSCG 034 | MK626919 | MK691248 | MK654858 | N/A | MK726210 | Guo et al. [15] |
| Diaporthe parvae | PSCG 035 | MK626920 | MK691249 | MK654859 | MK691169 | MK726211 | Guo et al. [15] |
| Diaporthe pascoei | BRIP 54847 | JX862532 | KF170924 | JX862538 | N/A | N/A | Tan et al. [65] |
| Diaporthe passiflorae | CPC 19183 | JX069860 | KY435674 | KY435633 | KY435664 | KY435654 | Crous et al. [66] |
| Diaporthe patagonica | CBS 145291 | MN509717 | MN509728 | MN509739 | N/A | N/A | Zapata et al. [45] |
| Diaporthe patagonica | CBS 145755 | MN509718 | MN509729 | MN509740 | N/A | N/A | Zapata et al. [45] |
| Diaporthe perjuncta | CBS 109745 | KC343172 | KC344140 | KC343898 | KC343414 | KC343656 | van Niekerk et al. [67] |
| Diaporthe phragmitis | CBS 138897 | KP004445 | KP004507 | N/A | N/A | KP004503 | Crous et al. [55] |
| Diaporthe psoraleae | CBS 136412 | KF777158 | KF777251 | KF777245 | N/A | N/A | Crous et al. [68] |
| Diaporthe psoraleae-pinnatae | CBS 136413 | KF777159 | KF777252 | N/A | N/A | N/A | Crous et al. [68] |
| Diaporthe pterocarpicola | MFLUCC 10-0580a | JQ619887 | JX275441 | JX275403 | JX197433 | N/A | Udayanga et al. [69] |
| Diaporthe pterocarpicola | MFLUCC 10-0580b | JQ619887 | JX275441 | JX275403 | JX197433 | N/A | Udayanga et al. [69] |
| Diaporthe pterocarpi | MFLUCC 10-0571 | JX197433 | JX275460 | JX275416 | JX197451 | N/A | Udayanga et al. [69] |
| Diaporthe pterocarpi | MFLUCC 10-0575 | JQ619901 | JX275462 | JX275418 | JX197453 | N/A | Udayanga et al. [69] |
| Diaporthe rostrata | CFCC 50062 | KP208847 | KP208855 | KP208853 | KP208849 | KP208851 | Fan et al. [17] |
| Diaporthe rostrata | CFCC 50063 | KP208848 | KP208856 | KP208854 | KP208850 | KP208852 | Fan et al. [17] |
| Diaporthe rudis | AR 3422 | KC843331 | KC843177 | KC843090 | KC843146 | N/A | Udayanga et al. [6] |
| Diaporthe rudis | AR 3654 | KC843338 | KC843184 | KC843097 | KC843153 | N/A | Udayanga et al. [6] |
| Diaporthe sackstonii | BRIP 54669b | KJ197287 | KJ197267 | KJ197249 | N/A | N/A | Thompson et al. [70] |
| Diaporthe sennae | CFCC 51636 | KY203724 | KY228891 | KY228885 | KY228875 | N/A | Yang et al. [18] |
| Diaporthe sennae | CFCC 51637 | KY203725 | KY228892 | KY228886 | KY228876 | N/A | Yang et al. [18] |
| Diaporthe sennicola | CFCC 51634 | KY203722 | KY228889 | KY228883 | KY228873 | KY228879 | Yang et al. [18] |
| Diaporthe sennicola | CFCC 51635 | KY203723 | KY228890 | KY228884 | KY228874 | KY228880 | Yang et al. [18] |
| Diaporthe shaanxiensis | CFCC 53106 | MK432654 | N/A | MK578130 | MK442976 | MK443001 | Yang et al. [14] |
| Diaporthe shaanxiensis | CFCC 53107 | MK432655 | N/A | MK578131 | MK442977 | MK443002 | Yang et al. [14] |
| Diaporthe sojae | BRIP 54033 | JF431295 | N/A | KC343901 | N/A | N/A | Udayanga et al. [71] |
| Diaporthe sojae | CBS 116019 | KC343175 | KC344143 | KC343901 | KC343417 | KC343659 | Udayanga et al. [71] |
| Diaporthe sojae | FAU 455 | KJ590712 | KJ610868 | KJ590755 | KJ612109 | KJ659201 | Udayanga et al. [71] |
| Diaporthe sojae | FAU 635 | KJ590719 | KJ610875 | KJ590762 | KJ612116 | KJ659208 | Udayanga et al. [71] |
| Diaporthe spartinicola | CPC 24951 | KR611879 | KR857695 | N/A | N/A | KR857696 | Crous et al. [71] |
| Diaporthe spinosa | PSCG 383 | MK626849 | MK691234 | MK654811 | MK691129 | MK726156 | Guo et al. [15] |
| Diaporthe spinosa | PSCG 279 | MK626925 | MK691235 | MK654801 | MK691126 | MK726155 | Guo et al. [15] |
| Diaporthe subordinaria | CBS 464.90 | KC343214 | KC344182 | KC343940 | KC343456 | KC343698 | Gomes et al. [4] |
| Diaporthe subordinaria | CBS 101711 | KC343213 | KC344182 | KC343939 | KC343455 | KC343697 | Gomes et al. [4] |
| Diaporthe taoicola | MFLUCC 16-0117 | KU557567 | KU557591 | KU557635 | N/A | N/A | Dissanayake et al. [9] |
| Diaporthe torilicola | MFLUCC 17-1051 | KY964212 | KY964096 | KY964168 | KY964127 | N/A | Dissanayake et al. [9] |
| Diaporthe toxica | CBS 534.93 | KC343220 | KC344188 | KC343946 | KC343462 | KC343704 | Williamson et al. [72] |
| Diaporthe toxica | CBS 546.93 | KC343222 | KC344190 | KC343948 | KC343464 | KC343706 | Williamson et al. [72] |
| Diaporthe vangueriae | CPC 22703 | KJ869137 | KJ869247 | N/A | N/A | N/A | Crous et al. [55] |
| Diaporthe vawdreyi | BRIP 57887a | KR936126 | KR936128 | KR936129 | N/A | N/A | Crous et al. [73] |
| Diaporthe zaobaisu | PSCG 031 | MK626922 | MK691245 | MK654855 | N/A | MK726207 | Guo et al. [15] |
| Diaporthe zaobaisu | PSCG 032 | MK626923 | MK691246 | MK654856 | N/A | MK726208 | Guo et al. [15] |
AR: Collection of A.Y. Rossman; BRIP: Queensland Plant Pathology herbarium/culture collection, Australia; CBS: Culture collection of the Centraalbureau voor Schimmelcultures, Fungal Biodiversity Centre, Utrecht, The Netherlands; CFCC: China Forestry Culture Collection Center, China; CGMCC: China General Microbiological Culture Collection; CPC: Collection Pedro Crous, housed at CBS; FAU: Isolates in culture collection of Systematic Mycology and Microbiology Laboratory, USDA-ARS, Beltsville, MD, USA; GUCC: Guizhou culture collection, Guizhou, China; IFRDCC: International Fungal Research and Development Centre Culture Collection, Chinese Academy of Forestry, Kunming, China; LC: Corresponding author’s personal collection (deposited in laboratory State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences); MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; ZJUD: Zhejiang University. ITS, internal transcribed spacers 1 and 2 together with 5.8S nrDNA; tub, partial beta-tubulin gene; cal, partial calmodulin gene and tef, partial translation elongation factor 1-a gene, his, histone H3 gene.
Ambiguous regions in the MP alignment were excluded, and gaps were treated as missing data. The stability of the trees was evaluated by 1000 bootstrap replications. Branches of zero length were collapsed, and all multiple parsimonious trees saved. Statistics including tree length (TL), consistency index (CI), retention index (RI), relative consistency index (RC) and homoplasy index (HI) were calculated. Differences between the trees inferred under different optimality criteria were evaluated using Kishino–Hasegawa tests (KHT) [52].
Bayesian analyses were performed in MrBayes v.3.0b4 [49] and posterior probabilities (PP) were determined by Markov Chain Monte Carlo sampling (MCMC). MrModeltest v. 2.3 [50] was used for the statistical selection of the best-fit model of nucleotide substitutions and was integrated into the analysis. Six simultaneous Markov chains were run for 106 generations; sampling the trees at every 100th generation. From the 10,000 trees obtained, the first 2000 representing the burn-in phase were discarded. The remaining 8000 trees were used for calculating posterior probabilities in the majority rule consensus tree.
The details of the fungal strains obtained in this study are listed in Table 1 with information of the type cultures and sequence data. Sequences generated in this study were deposited in GenBank (Table 1); alignments and trees were deposited in TreeBASE (www.treebase.org, study ID S27013). Reviewer access URL: http://purl.org/phylo/treebase/phylows/study/TB2:S27013?x-access-code=1369710211c386567d8b43ba36f49adf&format=html. Taxonomic novelties were submitted to the Faces of Fungi database [53], Index Fungorum (Index Fungorum 2020) and MycoBank (www.mycobank.org) [33].
3. Results
3.1. Phylogenetic Analyses
Saprobic specimens sampled from numerous woody hosts in six nature reserves in the Karst region of Guizhou province, China resulted in the isolation of thirty-one isolates of Diaporthe (Table 1, Figure 1). The ITS gene was employed for the identification of all isolates to the genus level. The ITS, tef, tub, cal and his alignments (including the gaps) were determined to be approximately 570, 470, 450, 610 and 500 bp (base pair) in size, respectively. The combined ITS, tef, tub, cal and his sequences of Diaporthe contained data for 136 isolates, including the outgroup taxon Diaporthella corylina (CBS 121124). The analyses consisted of 31 isolates from this study (Table 1) and 105 sequences (62 type species) originating from GenBank (Table 2). Out of a total of 2594 characters in the MP analyses, 1079 were constant, and 269 were variable and parsimony uninformative. Ten most parsimonious trees resulted from the remaining 1246 parsimony-informative characters (TL = 7439, CI = 0.384, RI = 0.804, RC = 0.309, HI = 0.616). In the ML analyses, the best scoring RAxML tree (Figure 1) with a final likelihood value of −37549.830874 is presented. The matrix had 1675 distinct alignment patterns, with 31.29% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.212875, C = 0.328600, G = 0.237362, T = 0.221162; substitution rates AC = 1.075393, AG = 2.704248, AT = 1.155000, CG = 0.851430, CT = 3.774119, GT = 1.000000; gamma distribution shape parameter alpha = 0.464825. The Maximum likelihood (ML) and Bayesian methods (BI) for phylogenetic analyses performed trees with similar topologies.
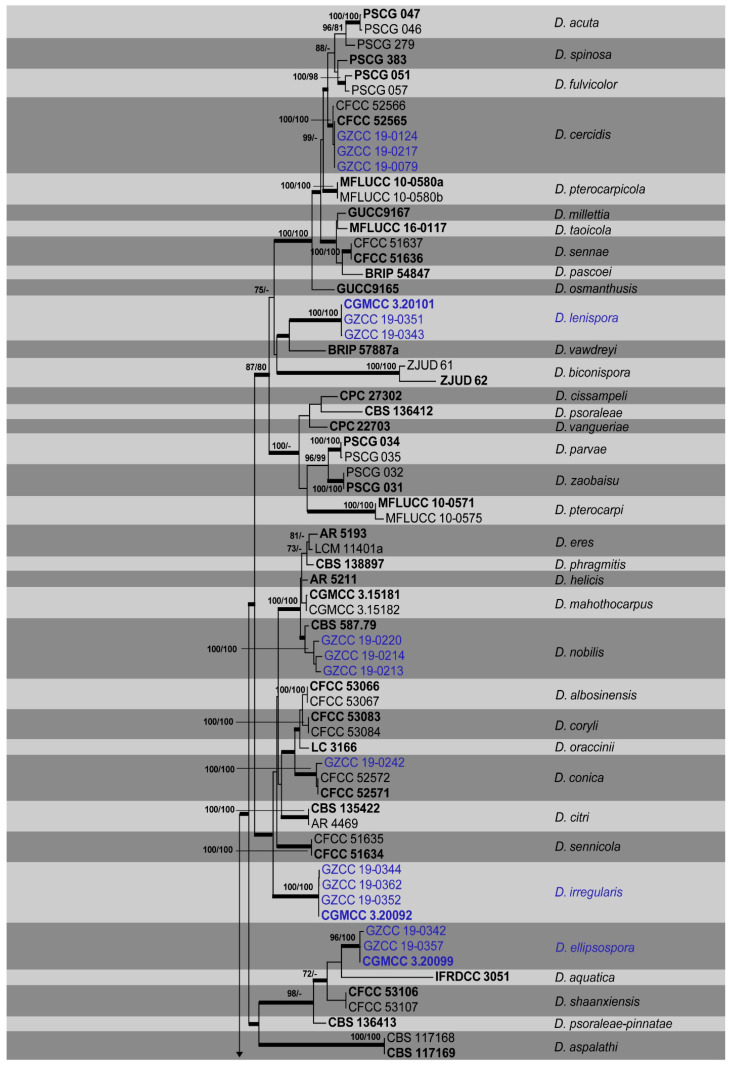
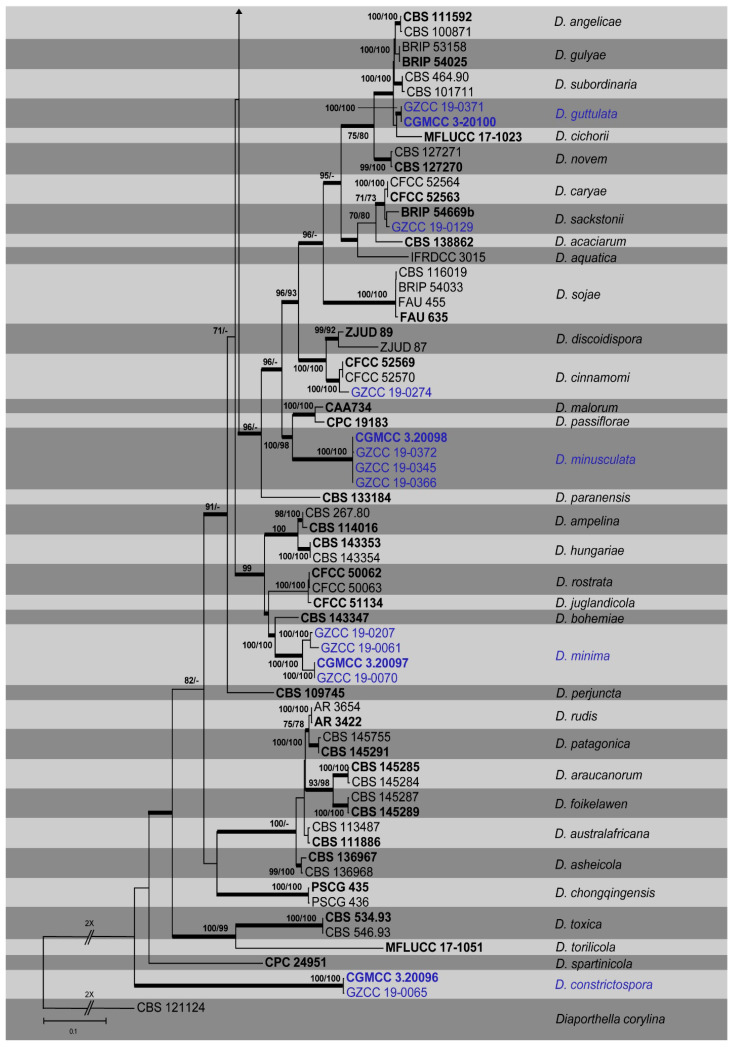
Figure 1.
Phylogram generated from maximum likelihood analysis of Diaporthe species isolated in this study and their phylogenetically closely related species based on combined internal transcribed spacer region (ITS), tef, tub, cal and his sequence data. Bootstrap support values for ML ≥ 70%, MP ≥ 70%, are indicated above the nodes and the branches are in bold indicate Bayesian posterior probabilities ≥0.9. The tree is rooted with Diaporthella corylina (CBS 121124). Isolate numbers of ex-types and reference strains are in bold. Taxa isolated in this study are in blue.
The isolates obtained in this study were grouped into twelve clades. Three isolates were grouped with the ex-type of Diaporthe cercidis (CFCC 52565) while another three isolates were clustered with the ex-type of D. nobilis (CBS 587.79). In addition one isolate with D. cinnamomi (CFCC 52569), D. conica (CFCC 52571) and D. sackstonii (BRIP 54669b) respectively. Twenty-two isolates did not cluster with any known Diaporthe species; thus, seven novel species, Diaporthe constrictospora (2 isolates, Figure 2), Diaporthe ellipsospora (3 isolates, Figure 3), Diaporthe guttulata (2 isolates, Figure 4), Diaporthe irregularis (4 isolates, Figure 5), Diaporthe lenispora (3 isolates, Figure 6), Diaporthe minima (4 isolates, Figure 7) and Diaporthe minusculata (4 isolates, Figure 8) are determined to be new species based on the morphological and phylogenetic evidence (Figure 1).
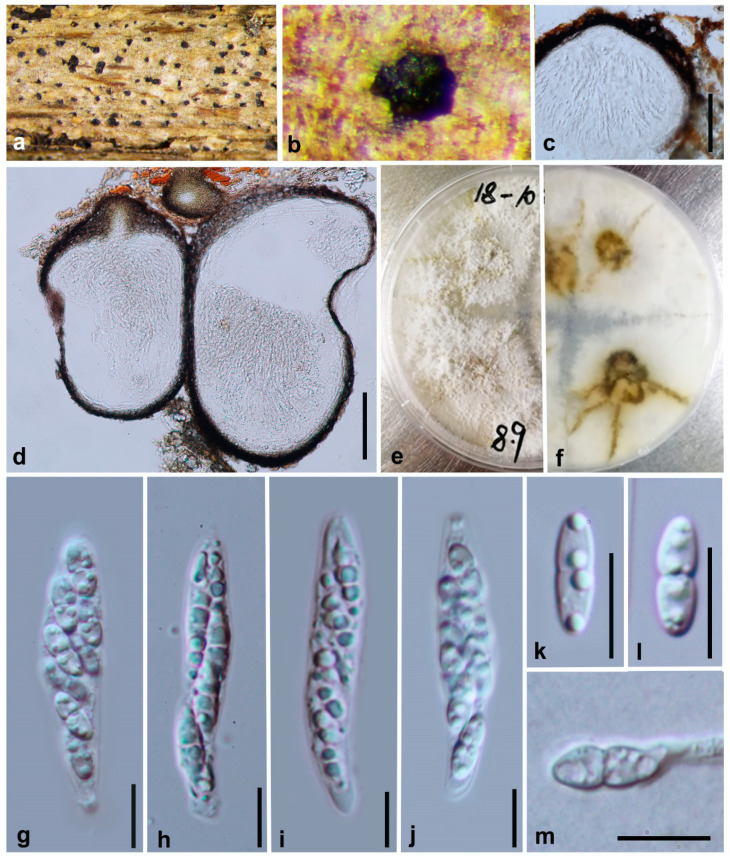
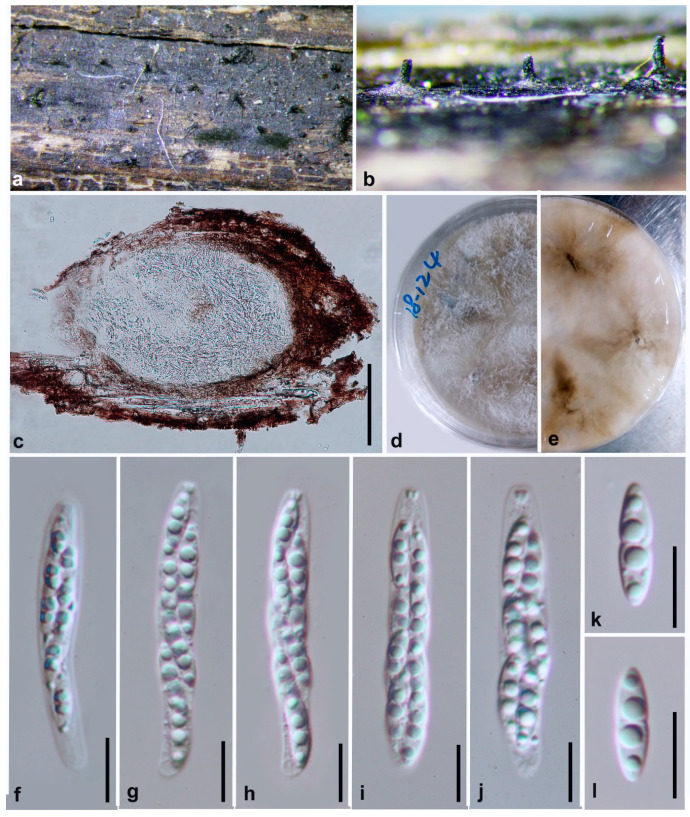
Figure 2.
Diaporthe constrictospora (HKAS 107534, holotype). (a,b) Ascomata on host surface. (c,d) Section ofascomata (e) 10 days old culture on potato dextrose agar (-) from above. (f) 10 days old culture on PDA from reverse. (g–j) Asci. (k,l) Ascospores. (m) Germinating ascospore. Scale bars: (c,d) = 100 μm, (g–m) = 10 μm.
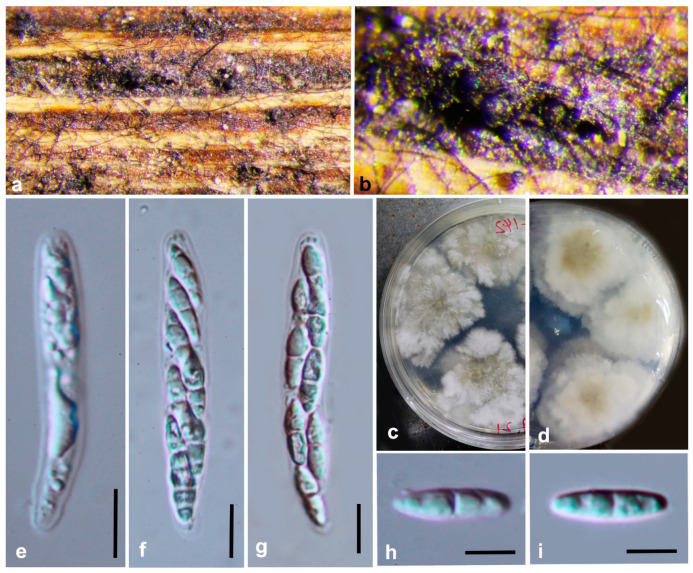
Figure 3.
Diaporthe ellipsospora (HKAS 107535, holotype). (a,b) Ascomata on host surface. (c) Section of an ascoma. (d) 8 days old culture on PDA from above. (e) 8 days old culture on PDA from reverse. (f) Immature ascus. (g–k) Mature asci. (l–p) Ascospores. (q) Paraphyses. Scale bars: (c) = 100 μm, (f–k) = 10 μm, (l–q) = 5 μm.
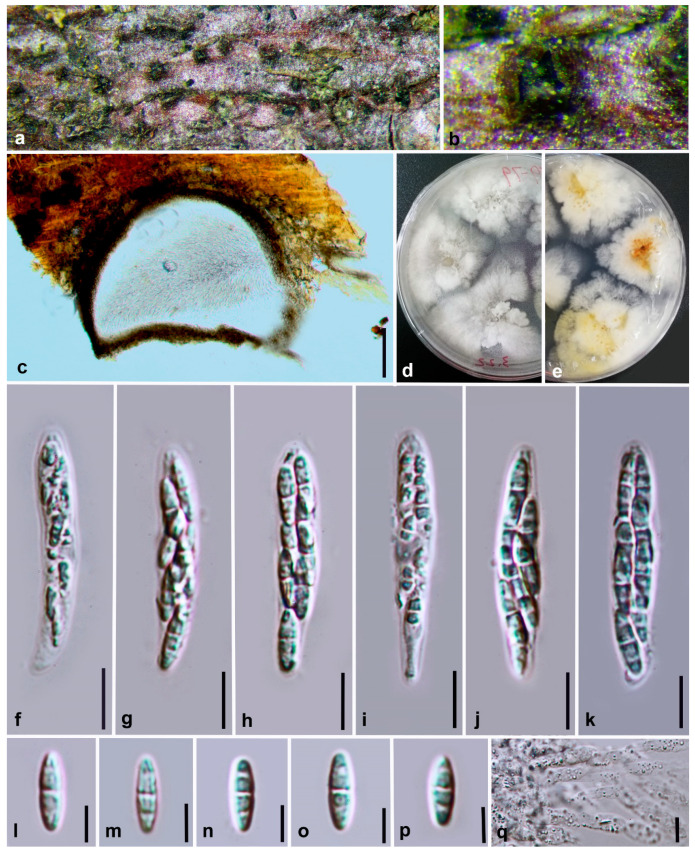
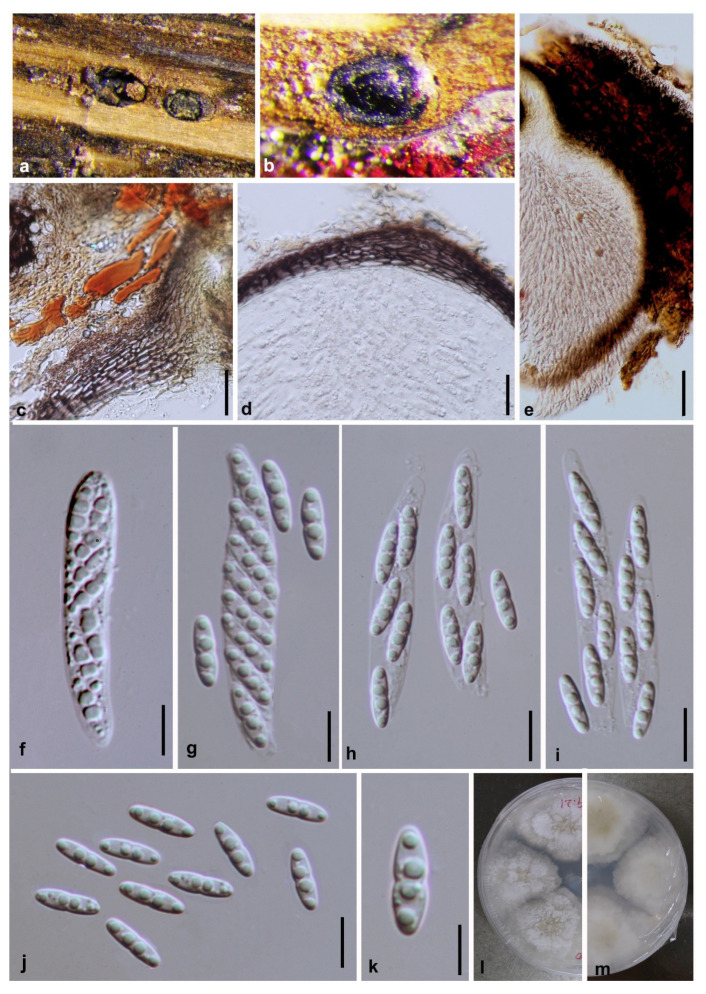
Figure 4.
Diaporthe guttulata (HKAS 107536, holotype). (a,b) Ascomata on host surface. (c) Section of an ascoma. (d) 7 days old culture on PDA from above. (e) 7 days old culture on PDA from reverse. (f) Immature ascus. (g–j) Mature asci. (k,l) Ascospores. Scale bars: (c) = 100 μm, (f–l) = 10 μm.
Figure 5.
Diaporthe irregularis (HKAS 107537, holotype). (a,b) Ascomata on host surface. (c) 10 days old culture on PDA from above. (d) 10 days old culture on PDA from reverse. (e) Immature ascus. (f,g) Mature asci. (h,i) Ascospores. Scale bars: (e–g) = 10 μm, (h,i) = 5 μm.
Figure 6.
Diaporthe lenispora (HKAS 107538, holotype). (a,b) Ascomata on host surface. (c) Ostiole. (d,e) Section of ascomata. (f) Immature ascus. (g–i) Mature asci. (j,k) Ascospores. (l) 10 days old culture on PDA from above. (m) 10 days old culture on PDA from reverse. Scale bars: (c) = 100 μm, (d,e) = 50 μm, (f–j) = 10 μm, (k) = 5 μm.
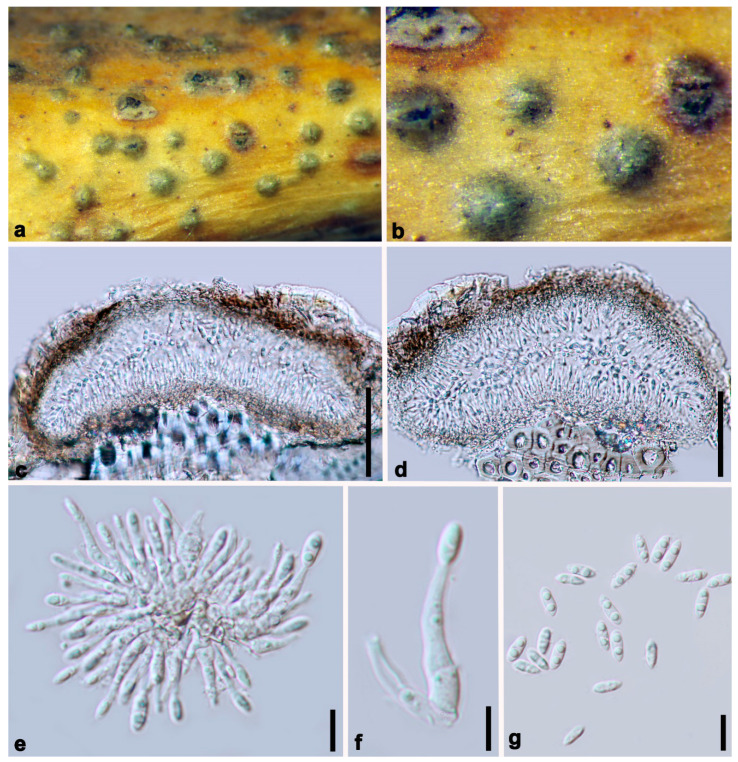
Figure 7.
Diaporthe minima (HKAS 107539, holotype). (a,b) Conidiomata on host surface. (c,d) Section of conidiomata. (e,f) Alpha conidia attached to conidiogenous cells. (g) Alpha conidia. Scale bars: (c,d) = 50 μm, (e–g) = 10 μm.
Figure 8.
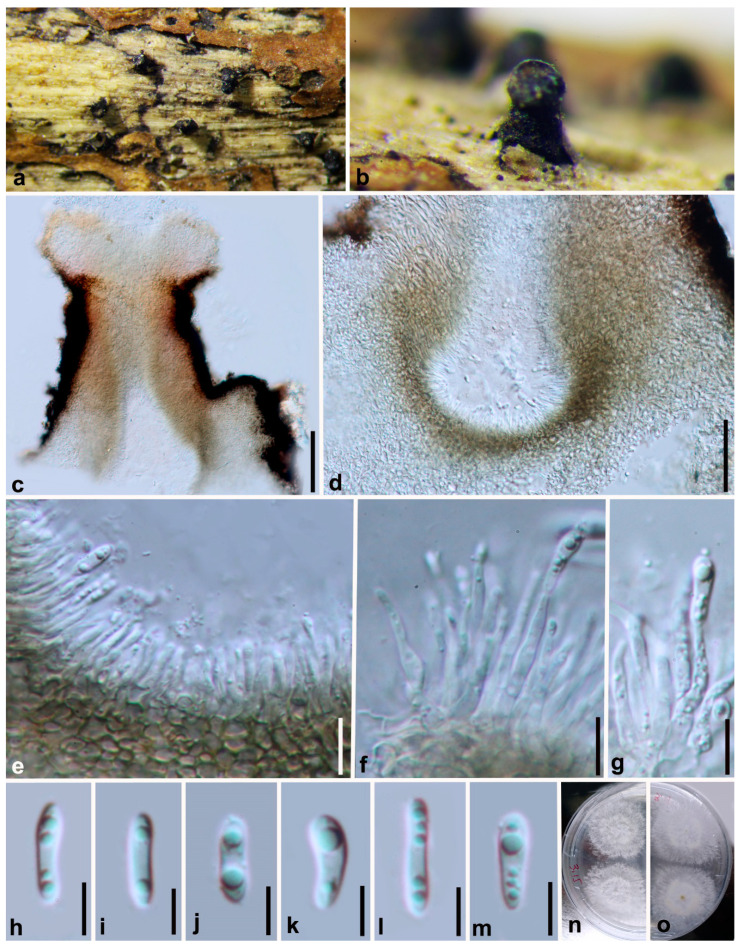
Diaporthe minusculata (HKAS 107540, holotype). (a,b) Conidiomata on host surface. (c–e) Section of conidiomata. (f,g) Alpha conidia attached to conidiogenous cells. (h–m) Alpha conidia. (n) 5 days old culture on PDA from above. (o) 5 days old culture on PDA from reverse. Scale bars: (c) = 100 μm, (d) = 50 μm, (e–g) = 10 μm, (h–m) = 5 μm.
3.2. Morphology and Culture Characteristics
In this study, thirty-one Diaporthe isolates were obtained from decaying woody hosts from six nature reserves in Guizhou province, China (Table 1). The Diaporthe isolates obtained in this study were further categorized based on morphological characteristics. Growth was rapid for all isolates grown on PDA, with mycelia covering the whole surface of the Petri dishes. Aerial mycelium was initially white and turned greyish after incubation in the dark at 25 °C for several days. All species exhibited phenotypic characteristics typical of the genus. The seven new species of Diaporthe described here are phylogenetically distinct from all previously described species for which sequence data are available.
Taxonomy
Diaporthe constrictospora Y.Y. Chen, A.J. Dissanayake and Jian K. Liu sp. nov. (Figure 2)
Index Fungorum number: IF557388; Facesoffungi Number: FoF07853; MycoBank Number: MB836211
Etymology: The epithet from the Latin words constrictus and spora, refers to the slight central constriction often present in the ascospores.
Holotype: HKAS 107534
Saprobic on decaying wood. Sexual morph: Ascomata 190–240 μm diam, black, globose to subglobose or irregular, clustered or solitary, deeply immersed in host tissue. Asci 40–48 μm × 9–11 μm ( = 43 × 8, n = 30), 8-spored, unitunicate, sessile, elongate to clavate. Ascospores 10–12 × 3–4 μm ( = 11 × 4, n = 50), hyaline, elongated to elliptical, two-celled, often 4-guttulate, with larger guttules at center and smaller ones at the ends. Asexual morph: Not observed.
Culture characteristics: Colonies covering entire PDA Petri dishes after 10 d at 25 °C producing abundant white aerial mycelium, reverse fuscous black.
Material examined: China, Guizhou Province, Maolan Nature Reserve, saprobic on decaying woody host, April 2017, Y. Y. Chen (HKAS 107534, holotype); ex-type living culture CGMCC 3.20096 = GZCC19-0084; ibid, Guiyang District, Huaxi Wetland Park, saprobic on decaying woody host, July 2017, Y. Y. Chen (GZAAS 19-1784, paratype), living culture GZCC 19-0065.
Notes: Two strains representing Diaporthe constrictospora cluster in a well-supported basal clade (ML/MP/BI = 100/100/1.0) and appear to be distinct from other Diaporthe species, and can be easily recognized by their distinctive phylogenetic placement (Figure 1). Since this species is not closely related to any Diaporthe species and we were unable to compare the nucleotide differences in the alignment. Diaporthe constrictospora is introduced as a phylogenetically distinct species (Figure 1).
Diaporthe ellipsospora Y.Y. Chen, A.J. Dissanayake and Jian K. Liu sp. nov. (Figure 3)
Index Fungorum number: IF557389; Facesoffungi Number: FoF07854; MycoBank Number: MB836175
Etymology: The specific epithet ellipsospora refers to the shape of the ascospores.
Holotype: HKAS 107535
Saprobic on decaying branch. Sexual morph: Ascomata 380–430 μm diam, black, globose to irregular, scattered on dead twigs, immersed in host tissue, protruding through substrata. Paraphyses up to 100 μm long, rarely present, hyaline, smooth, 1–3-septate, cylindrical with obtuse ends, extending above conidiophores. Asci 40–47 ×7–8.5 μm ( = 43 × 7.5, n = 30), 8-spored, unitunicate, sessile, elongate to clavate. Ascospores 8.5–13 × 2.5–3.5 μm ( = 10.1 × 3.3, n = 50), hyaline, elongated to elliptical, two-celled, often 4-guttulate, with larger guttules at center and smaller ones at ends. Asexual morph: Not observed.
Culture characteristics: Colonies on PDA fast growing, covering entire PDA Petri dish after 8 d at 25 °C. White, aerial mycelium turning grey at edges of plate, reverse yellowish pigmentation developing in centre.
Material examined: China, Guizhou Province, Xingyi Wanfenglin, Saprobic on decaying branch, June 2019, Y.Y. Chen (HKAS 107535, holotype), ex-type living culture CGMCC 3.20099 = GZCC 19-0231; ibid, (GZAAS 19-2061, paratype), living culture GZCC 19-0342; ibid, Maolan Nature Reserve, saprobic on decaying woody host, June 2019, Y. Y. Chen, GZAAS 19-2080, living culture GZCC 19-0357.
Notes:Diaporthe ellipsospora formed an independent clade (Figure 1) and is phylogenetically distinct from D. aquatica in a well-supported clade (ML/MP/BI = 96/100/1.0). Diaporthe ellipsospora can be distinguished from D. aquatica (IFRDCC 3051) only based on ITS locus (17/539) since other gene sequences (tef, tub, cal and his) are unavailable for D. aquatica. Diaporthe ellipsospora can be morphologically differentiated from D. aquatica as the latter has long necks up to 2250 μm [58].
Diaporthe guttulata Y.Y. Chen, A.J. Dissanayake and Jian K. Liu sp. nov. (Figure 4)
Index Fungorum number: IF557390; Facesoffungi Number: FoF07855; MycoBank Number: MB836212
Etymology: Referring to the ascospores with large guttules.
Holotype: HKAS 107536
Saprobic on decaying branch. Sexual morph: Ascomata 560–630 μm diam, black, globose to conical, scattered irregularly, immersed in host tissue with elongated, 300–400 μm long necks protruding through substrata. Asci 45–57 μm × 7–9 μm ( = 50 × 8, n = 30), unitunicate, 8-spored, sessile, elongate to clavate. Ascospores 12–15 × 3–4 μm ( = 13 × 3.1, n = 50), elongated to elliptical, hyaline, two-celled, often 4-guttulate, with larger guttules at centre and smaller one at ends. Asexual morph: Not observed.
Culture characteristics: Colonies covering entire PDA Petri dishes after 7 d at 25 °C producing abundant white aerial mycelium. Reverse white, turning to grey in centre and no conidia produced.
Material examined: China, Guizhou Province, Maolan Nature Reserve, saprobic on decaying branch, July 2017, Y.Y. Chen (HKAS 107536, holotype), ex-type living culture CGMCC 3.20100 = GZCC 19-0140; ibid, Guiyang District, Suiyang broad water nature reserve, saprobic on decaying woody host, June 2018, Y. Y. Chen (GZAAS 19-2067, paratype), living culture GZCC 19-0371.
Notes:Diaporthe guttulata formed a distinct clade with high support (ML/BI = 88/1.0), and differed with the closely related species D. angelicae, D. cichorii, D. gulyae and D. subordinaria. Diaporthe guttulata can be distinguished from D. angelicae (7/539 in ITS, 8/467 in tef, 7/453 in tub, 9/606 in cal and 10/513 in his); D. cichorii (8/539 in ITS, 13/467 in tef and 7/453 in tub and 21/606 in cal); from D. gulyae (11/539 in ITS, 8/467 in tef and 13/453 in tub) and from D. subordinaria (6/539 in ITS, 5/467 in tef, 15/453 in tub, 13/606 in cal and 11/513 in his). Morphologically, D. guttulata differs from D. cichorii in having larger asci (50–8 vs. 45–6 μm) and ascospores (13–3 vs. 10–3 μm) [57]. The morphological characters of Diaporthe guttulata cannot be compared with D. gulyae and D. subordinaria as these two species have no reported sexual morphs.
Diaporthe irregularis Y.Y. Chen, A.J. Dissanayake and Jian K. Liu sp. nov. (Figure 5)
Index Fungorum number: IF557391; Facesoffungi Number: FoF07856; MycoBank Number: MB836213
Etymology: Refers to the irregular shape of the ascomata.
Holotype: HKAS 107537
Saprobic on decaying woody branch. Sexual morph: Ascomata 390–460 μm diam, black, globose to irregular, scattered evenly on dead branches, immersed in host tissue. Asci 52–66 × 7–9 μm ( = 58 × 8, n = 30), 8-spored, unitunicate, sessile, elongate to clavate. Ascospores (10–12 × 3–4 μm ( = 11 × 3.5, n = 50), hyaline, two-celled, often 4-guttulate, with larger guttules at center and smaller ones at ends, elongated to elliptical. Asexual morph: Not observed.
Culture characteristics: Colonies covering entire PDA Petri dishes after 10 d at 25 °C producing abundant white aerial mycelium, reverse fuscous black.
Material examined: China, Guizhou Province, Suiyang broad water nature reserve, saprobic on decaying branch, April 2018. Y.Y. Chen (HKAS 107537, holotype), ex-type living culture CGMCC 3.20092 = GZCC 19-0147; ibid., (GZAAS 19-2064, paratype), living culture GZCC 19-0344; ibid., GZAAS 19-2069, living culture GZCC 19-0362; ibid., GZAAS19-2077, living culture GZCC 19-0352.
Notes: Four isolates, representing Diaporthe irregularis, are retrieved in a well-supported clade (ML/MP/BI = 100/100/1.0) and appear to be distinct from other Diaporthe species phylogenetically (Figure 1). Since this species does not closely related to any particular Diaporthe species, we were unable to compare the nucleotide differences in the concatenated alignment. In addition, Diaporthe irregularis can be morphologically distinguished from other Diaporthe species based on the shape and the position of the ascomata.
Diaporthe lenispora Y.Y. Chen, A.J. Dissanayake and Jian K. Liu sp. nov. (Figure 6)
Index Fungorum number: IF IF557392; Facesoffungi Number: FoF07857; MycoBank Number: MB836214
Etymology: Name reflects the ascospores being smooth-walled, from the Latin lenis referring to smooth and spora.
Holotype: HKAS 107538
Saprobic on decaying woody branch. Sexual morph: Ascomata 435–510 μm diam, black, globose to conical, scattered irregularly, immersed in host tissue with elongated, long necks protruding through substrata. Asci 44–53 μm × 9–10 μm ( = 48 × 9, n = 30), 8-spored, unitunicate, sessile, elongate to clavate. Ascospores 10–12 × 2–3 μm ( = 11 × 2.5, n = 50), hyaline, two-celled, often 4-guttulate, with larger guttules at centre and smaller one at ends, elongated to elliptical. Asexual morph: Not observed.
Culture characteristics: Colonies covering entire PDA Petri dishes after 10 d at 25 °C producing abundant white aerial mycelium, reverse early yellow and turned to fuscous black.
Material examined: China, Guizhou Province, Guizhou Province, Suiyang broad water nature reserve, on decaying branch, April 2018. Y.Y. Chen (HKAS 107538, holotype), ex-type living culture CGMCC 3.20101 = GZCC 19-0145; ibid., Xingyi Wanfenglin, saprobic on decaying woody host, June 2019, Y. Y. Chen (GZAAS 19-2066, paratype), living culture GZCC 19-0343; ibid., saprobic on decaying branch, June 2018. Y.Y. Chen, GZAAS 19-2075, living culture GZCC19-0351.
Notes: In the combined phylogenetic tree, Diaporthe lenispora groups in a distinct clade with maximum support (ML/MP/BI = 100/100/1.0) and it appears to be most closely related to D. vawdreyi (Figure 1). Diaporthe lenispora can be distinguished from D. vawdreyi based on ITS, tef and tub loci (19/539 in ITS, 56/467 in tef and 23/453 in tub), cal and his gene regions are unavailable for D. vawdreyi. We are not able to compare the morphology of D. lenispora and D. vawdreyi as the latter has no reported sexual morph [74].
Diaporthe minima Y.Y. Chen, A.J. Dissanayake and Jian K. Liu sp. nov. (Figure 7)
Index Fungorum number: IF557393; Facesoffungi Number: FoF07858; MycoBank Number: MB836215
Etymology: Named for the small conidia.
Holotype: HKAS 107539
Saprobic on decaying woody branch. Sexual morph: Not observed. Asexual morph: Conidiomata up to 230 μm in diam., immersed, scattered on PDA, dark brown to black, globose, solitary or clustered in groups of 3–5 conidiomata. Conidiophores 9–13 × 1–2 μm ( = 11 × 1.5 μm) aseptate, cylindrical, straight or sinuous, densely aggregated, terminal, slightly tapered towards the apex. Alpha conidia 6.5–8.5 × 2–3 μm ( = 7 × 2 μm), biguttulate, hyaline, fusiform or oval, both ends obtuse. Beta conidia not observed.
Culture characteristics: Cultures incubated on PDA at 25 °C in darkness. Colony at first flat with white felty mycelium, becoming black in the center and black at the marginal area with 8 d, pycnidia not observed.
Material examined: China, Guizhou Province, Guiyang District, Huaxi Wetland Park, Saprobic on decaying branch, April 2017, Y.Y. Chen (HKAS 107539, holotype), ex-type living culture CGMCC 3.20097 = GZCC 19-0066; ibid., (GZAAS 19-1786, paratype), living culture GZCC19-0070; ibid., GZAAS 19-1787, living culture GZCC19-0061; ibid., GZAAS 19-1788, living culture GZCC19-0207.
Notes: The phylogenetic result showed that isolates of Diaporthe minima clustered closer to D. bohemiae, D. juglandicola and D. rostrata, and formed a distinct lineage (Figure 1) with maximum support (ML/MP/BI = 100/100/1.0). Diaporthe minima can be distinguished from the above closely related species based on ITS, tef, tub, cal and his loci for D. bohemiae (11/539 in ITS, 45/467 in tef, 14/453 in tub, 37/606 in cal, 34/513 in his), D. juglandicola (24/539 in ITS, 19/467 in tef, 12/453 in tub, 27/606 in cal and 47/513 in his) and D. rostrata (19/539 in ITS, 48/467 in tef, 13/453 in tub 17/606 in cal and 49/513 in his). Morphologically, Diaporthe minima differs from D. bohemiae, D. juglandicola and D. rostrata in having smaller alpha conidia (7 × 2 vs. 9 × 3 μm) (7 × 2 vs. 11 × 13 μm) [17,18].
Diaporthe minusculata Y.Y. Chen, A.J. Dissanayake and Jian K. Liu sp. nov. (Figure 8)
Index Fungorum number: IF557394; Facesoffungi Number: FoF07859; MycoBank Number: MB836216
Etymology: Name based on a Latin adjective minusculus, meaning rather small conidiomata.
Holotype: HKAS 107540
Saprobic on decaying branch. Sexual morph: Not observed. Asexual morph: Conidiomata up to 430 μm in diam., superficial, erumpent, scattered on PDA, dark brown to black, globose, solitary or clustered in groups of 3–5 pycnidia, with prominent necks 130–320 μm long. Conidiophores 11–18 × 1.5–2.5 μm ( = 14 × 2 μm), aseptate, cylindrical, straight or sinuous, densely aggregated, terminal, slightly tapered towards the apex. Alpha conidia 7–10 × 2–3 μm ( = 9 × 2 μm), biguttulate, hyaline, fusiform or oval, both ends obtuse. Beta conidia not observed.
Culture characteristics: Cultures incubated on PDA at 25 °C in darkness showed colony at first white, becoming pale brown with yellowish dots within 10 d, with dense and felted mycelium, visible solitary or aggregated pycnidia at maturity.
Material examined: China, Guizhou Province, Xingyi Wanfenglin, Saprobic on decaying branch, June 2019, Y.Y. Chen (HKAS 107540, holotype), ex-type living culture CGMCC 3.20098 = GZCC 19-0215; ibid., GZAAS 19-2072, living culture GZCC 19-0372; ibid., Suiyang broad water nature reserve, saprobic on decaying woody host, April 2018, Y. Y. Chen (GZAAS19-2062, paratype), living culture GZAAS19-2062; ibid., GZAAS19-2070, living culture GZCC 19-0366.
Notes: The phylogenetic results showe that Diaporthe minusculata clustered close to D. malorum and D. passiflorae, and formed a distinct lineage (Figure 1) with maximum support (ML/MP/BI = 100/100/1.0). Diaporthe minusculata can be distinguished from D. malorum (23/539 in ITS, 41/467 in tef, 29/453 in tub, 52/606 in cal and 35/513 in his) and from D. passiflorae (25/539 in ITS, 13/467 in tef, 11/453 in tub, 22/606 in cal and 17/513 in his). Morphologically, Diaporthe minusculata differs from D. malorum in having conidiomata with a long neck and differs from D. passiflorae in shorter conidiophores (14 × 2 vs. 26 × 4 μm) [16,17,18,66,74].
4. Discussion
Based on the phenotypic characters and the multi-locus phylogeny, the 31 isolates obtained in this study can be recognized as twelve species. Among the five species are previously known and seven species are new to science. These newly discovered species are Diaporthe constrictospora, D. ellipsospora, D. guttulata, D. irregularis, D. lenispora, D. minima and D. minusculata. The other taxa are identified as Diaporthe cercidis [42], D. cinnamomi [42], D. conica [42], D. nobilis [4] and D. sackstonii [70]. Morphological characters of the known species isolated in this study were compared with their original descriptions. Phylogenetically, there were no significant base pair differences between these and their type based combined gene alignments.
A phylogenetic tree derived from an alignment of ITS sequences is beneficial as a guide for the identification of isolates of Diaporthe species [65,75]. ITS sequences offer convincing proof for species demarcation where a limited number of taxa are analyzed, such as species associated with the same host [62,64,76]. However, confusion arises when a large number of species from an extensive range of host species are examined. Santos et al. [77] proposed that tef is a superior phylogenetic marker in Diaporthe than ITS, and has been commonly used as the secondary locus for phylogenetic studies [8,10,64,75]. Gomes et al. [4] studied five loci from 95 species and stated that tef poorly distinguished species, and recommended that his and tub were suitable possibilities as subordinate phylogenetic markers to accompany the authorized fungi barcode: the internal transcribed spacer region (ITS). Dissanayake et al. [10] reviewed the genus Diaporthe and provided a checklist for 171 species with available molecular data (from culture and fruiting body) and a phylogenetic tree using four gene regions (ITS, tef, tub and cal). According to Santos et al. [16], incorporation of a five-loci dataset (ITS, cal, his, tef, tub) was recommended as the best combination for species identification within the genus and recent studies seems to favor the selection of four or five genes [13,14,15,33,38,39,40,41,42,43,44]. Hence, the present study is conducted combining the five gene regions analyses of ITS, tef, tub, cal and his to reveal five known Diaporthe species and to assist in the introduction of seven new Diaporthe species.
Several studies have been conducted to reveal the association of Diaporthe species with various hosts in China. Huang et al. [60] revealed seven apparently undescribed endophytic Diaporthe species (Diaporthe biconispora, D. biguttulata, D. discoidispora, D. multigutullata, D. ovalispora, D. subclavata and D. unshiuensis) on Citrus. Gao et al. [63] identified four novel species (D. apiculata, D. compacta, D. oraccinii, D. penetriteum) and three known species (D. discoidispora, D. hongkongensis, D. ueckerae) associated with Camellia (tea). Gao et al. [78] showed eight new species of Diaporthe (Diaporthe acutispora, D. elaeagni-glabrae, D. incompleta, D. podocarpi-macrophylli, D. undulata, D. velutina, D. xishuangbanica and D. yunnanensis) from leaves of several hosts while Yang et al. [42] introduced twelve new Diaporthe species (Diaporthe acerigena, D. alangii, D. betulina, D. caryae, D. cercidis, D. chensiensis, D. cinnamomi, D. conica, D. fraxinicola, D. kadsurae, D. padina and D. ukurunduensis) from infected forest trees in Beijing, Heilongjiang, Jiangsu, Jiangxi, Shaanxi and Zhejiang Provinces. Three new Diaporthe species: Diaporthe anhuiensis, D. huangshanensis, D. shennongjiaensis and two other known species: D. citrichinensis and D. eres were described as endophytes by Zhou et al. [44]. Yang et al. [14] established three new species: D. albosinensis, D. coryli and D. shaanxiensis isolated from symptomatic twigs and branches at the Huoditang Forest Farm in Shaanxi Province, China. High diversity of Diaporthe species associated with pear shoot canker in China was observed by Guo et al. [15] representing thirteen known species (Diaporthe caryae, D. cercidis, D. citrichinensis, D. eres, D. fusicola, D. ganjae, D. hongkongensis, D. padina, D. pescicola, D. sojae, D. taoicola, D. unshiuensis and D. velutina) and six new species (Diaporthe acuta, D. chongqingensis, D. fulvicolor, D. parvae, D. spinosa and D. zaobaisu). However, the identification of Diaporthe species associated with hosts in nature reserves in China has rarely been studied. Thus, an investigation of Diaporthe species was conducted and this provides the first molecular phylogenetic frame of Diaporthe diversity in six nature reserves in the Karst region of Guizhou province, combined with morphological descriptions.
Among the twelve species identified in this study, four species have been previously isolated from China. Yang et al. [42] introduced Diaporthe cercidis from twigs and branches of Cercis chinensis in Jiangsu Province, D. cinnamomi from symptomatic twigs of Cinnamomum sp. in Zhejiang Province and D. conica from symptomatic branches of Alangium chinense in Zhejiang Province. Diaporthe nobilis has been isolated from Camellia sinensis in Guizhou Province [25]. The other known species: D. sackstonii [70] has been isolated from petioles of sunflower plants (Helianthus annuus) inAustralia. Based on the percentage of occurrence, Diaporthe irregularis sp. nov (13%), D. minima sp. nov (13%), and D. minusculata sp. nov (13%) were categorized as being frequent. Diaporthe cinnamomi, D. conica and D. sackstonii were ranked as infrequent, since only one isolate has been isolated for each species. Interestingly, the type species of the genus, D. eres Nitschke [79] was not observed in our survey. This species is one of the frequent species in most of the studies and appears with 365 Fungus–Host combinations [80].
The discovery of these species of Diaporthe from different nature reserves in Guizhou province as well as worldwide occurrence shows the polyphagous and cosmopolitan behavior of species in this genus. Certainly, it is obvious that performing complementary studies based on sequencing five gene regions of Diaporthe species is essential in order to support reliable species identification. The descriptions and molecular data of Diaporthe species provided in this study would serve as a resource for plant pathologists, plant quarantine officials and taxonomists for better identification of Diaporthe and its species boundaries. Such studies are necessary to investigate this group of fungi in different unexploited biomes, to reveal the degree of diversity and to support more suitable control measures to prevent their dissemination. Importantly, based on the Diaporthe taxa identification in this study coupled with previous studies, it could be concluded that almost all the known species isolated (Diaporthe cercidis, D. cinnamomi, D. conica, D. nobilis and D. sackstonii) as saprobes in this study were pathogenic on various host plants [25,42,70]. This could indicate that the seven newly introducing species could potentially be pathogens even though they were isolated from decaying woody hosts, and their pathogenicity should be evaluated in further studies with more samples (from other kinds of habitats and hosts, as well as the different distributions and substrates). In the meantime, we provided the culture details and deposited them in publicly accessible culture collections for further evaluation or comparison of the life modes of these taxa.
5. Conclusions
We carried out fungal diversity investigations with large-scale sampling in the Karst region of southwestern China and this is the first report of Diaporthe species isolated from nature reserves in Karst region of Guizhou province, China. The identification of twelve Diaporthe species (five known species and seven new species) associated with saprobic woody hosts is documented.
Acknowledgments
We are grateful to E.B. Gareth Jones for his valuable suggestions and comments on the manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/2309-608X/6/4/251/s1, Figure S1: Phylogram generated from maximum likelihood analysis of all available type species of Diaporthe, based on ITS sequence data.
Author Contributions
Conceptualization, J.-K.L. and Y.-Y.C.; methodology, Y.-Y.C. and A.J.D.; formal analysis, Y.-Y.C. and A.J.D.; resources, Y.-Y.C. and J.-K.L.; data curation, A.J.D. and J.-K.L.; writing—original draft preparation, A.J.D.; writing—review and editing, A.J.D. and J.-K.L.; supervision, J.-K.L.; project administration, J.-K.L.; funding acquisition, J.-K.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Joint Fund of the National Natural Science Foundation of China and the Karst Science Research Center of Guizhou province (Grant No. U1812401).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hyde K.D., Nilsson R.H., Alias S.A., Ariyawansa H.A., Blair J.E. One stop shop, backbone trees for important phytopathogenic genera, I. Fungal Divers. 2014;67:21–125. doi: 10.1007/s13225-014-0298-1. [DOI] [Google Scholar]
- 2.Maharachchikumbura S.S.N., Hyde K.D., Jones E.B.G., McKenzie E.H.C., Huang S.K. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 2015;72:199–301. doi: 10.1007/s13225-015-0331-z. [DOI] [Google Scholar]
- 3.Maharachchikumbura S.S.N., Hyde K.D., Jones E.B.G., McKenzie E.H.C., Bhat D.J. Families of Sordariomycetes. Fungal Divers. 2016;79:1–317. doi: 10.1007/s13225-016-0369-6. [DOI] [Google Scholar]
- 4.Gomes R.R., Glienke C., Videira S.I.R., Lombard L., Groenewald J.Z. Diaporthe; a genus of endophytic; saprobic and plant pathogenic fungi. Persoonia. 2013;31:1–41. doi: 10.3767/003158513X666844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossman A.Y., Adams G.C., Cannon P.F., Castlebury L.A., Crous P.W., Gryzenhout M., Jaklitsch W.M., Mejia L.C., Stoykov D., Udayanga D., et al. Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus. 2015;6:145–154. doi: 10.5598/imafungus.2015.06.01.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udayanga D., Castlebury L.A., Rossman A.Y., Hyde K.D. Species limits in Diaporthe; molecular re-assessment of D. citri; D. cytosporella; D. foeniculina and D. rudis. Persoonia. 2014;32:83–101. doi: 10.3767/003158514X679984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Udayanga D., Castlebury L.A., Rossman A.Y., Chukeatirote E., Hyde K.D. Insights into the genus Diaporthe; phylogenetic species delimitation in the D. eres species complex. Fungal Divers. 2014;67:203–229. doi: 10.1007/s13225-014-0297-2. [DOI] [Google Scholar]
- 8.Dissanayake A.J., Liu M., Zhang W., Chen Z., Udayanga D. Morphological and molecular characterization of Diaporthe species associated with grapevine trunk disease in China. Fungal Biol. 2015;11:283–294. doi: 10.1016/j.funbio.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Dissanayake A.J., Zhang W., Liu M., Hyde K.D., Zhao W.S. Diaporthe species associated with peach tree dieback in Hubei, China. Mycosphere. 2017;8:533–549. doi: 10.5943/mycosphere/8/5/2. [DOI] [Google Scholar]
- 10.Dissanayake A.J., Phillips A.J.L., Yan J.Y., Li X.H., Hyde K.D. The current status of species in Diaporthe. Mycosphere. 2017;8:1106–1156. doi: 10.5943/mycosphere/8/5/5. [DOI] [Google Scholar]
- 11.van Rensburg J.C.J., Lamprecht S.C., Groenewald J.Z., Castlebury L.A., Crous P.W. Characterization of Phomopsis spp. associated with dieback of rooibos (Aspalathus linearis) in South Africa. Stud. Mycol. 2006;55:65–74. doi: 10.3114/sim.55.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Udayanga D., Liu X.Z., McKenzie E.H.C., Chukeatirote E., Bahkali A.H. The genus Phomopsis: Biology, applications, species concepts and names of common pathogens. Fungal Divers. 2011;50:189–225. doi: 10.1007/s13225-011-0126-9. [DOI] [Google Scholar]
- 13.Yang Q., Du Z., Tian C.M. Phylogeny and morphology reveal two new species of Diaporthe from Traditional Chinese Medicine in Northeast China. Phytotaxa. 2018;336:159–170. doi: 10.11646/phytotaxa.336.2.3. [DOI] [Google Scholar]
- 14.Yang Q., Jiang N., Tian C.M. Three new Diaporthe species from Shaanxi Province; China. MycoKeys. 2020;67:1–18. doi: 10.3897/mycokeys.67.49483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y.S., Crous P.W., Bai Q., Fu M., Yang M.M., Wang X.H., Du Y.M., Hong N., Xu W.X., Wang G.P. High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia. 2020;45:132–162. doi: 10.3767/persoonia.2020.45.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos L., Phillips A.J.L., Crous P.W., Alves A. Diaporthe species on Rosaceae with descriptions of D. pyracanthae sp. nov. and D. malorum sp. nov. Mycosphere. 2017;8:485–511. doi: 10.5943/mycosphere/8/5/1. [DOI] [Google Scholar]
- 17.Fan X.L., Hyde K.D., Udayanga D., Wu X.Y. Diaporthe rostrata; a novel ascomycete from Juglans mandshurica associated with walnut dieback. Mycol. Prog. 2016;14:82. doi: 10.1007/s11557-015-1104-5. [DOI] [Google Scholar]
- 18.Yang Q., Fan X.L., Du Z., Tian C.M. Diaporthe juglandicola sp. nov. (Diaporthales; Ascomycetes) evidenced by morphological characters and phylogenetic analysis. Mycosphere. 2017;8:817–826. doi: 10.5943/mycosphere/8/5/3. [DOI] [Google Scholar]
- 19.Guarnaccia V., Groenewald J.Z., Woodha J. Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia. 2018;40:135–153. doi: 10.3767/persoonia.2018.40.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C., Wen Y. Study on Nature Reserve Management Strategy Based on Externality of Public Goods. Reform Strategy. 2011;3 [Google Scholar]
- 21.Xu Z.L. Dashahe Nature Reserve Assessment and Strategies for Protection and Management. For. Inventory Plan. 2003;1 [Google Scholar]
- 22.Yao Z.M., Yu D.L., Ran J.C. Study on the Ecotourism and Community Economic Development of Maolan National Natural Reserve in Guizhou Province. J. Anhui Agric. Sci. 2011;3:87–91. [Google Scholar]
- 23.Teng S.C. Fungi of China. Science Press; Beijing, China: 1963. pp. 1–808. (In Chinese) [Google Scholar]
- 24.Tai F.L. Sylloge Fungorum Sinicorum. Science Press; Beijing, China: 1979. [Google Scholar]
- 25.Li Y., Tan P., Zhao D.G. Diaporthe nobilis; a new record on Camellia sinensis in Guizhou Province; China. Mycosphere. 2017;8:1–8. doi: 10.5943/mycosphere/8/1/1. [DOI] [Google Scholar]
- 26.Long H., Zhang Q., Hao Y.Y., Shao X.Q., Wei X.X., Hyde K.D., Wang Y., Zhao D.G. Diaporthe species in south-western China. Mycokeys. 2019;57:113–127. doi: 10.3897/mycokeys.57.35448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J.K., Chomnunti P., Cai L., Phookamsak R., Chukeatirote E., Jones E.B.G., Moslem M., Hyde K.D. Phylogeny and morphology of Neodeightonia palmicola sp. nov. from palms. Sydowia. 2010;62:261–276. [Google Scholar]
- 28.Chomnunti P., Hongsanan S., Aguirre-Hudson B., Tian Q., Peršoh D., Dhami M.K., Alias A.S., Xu J.C., Hyde K.D. The sooty moulds. Fungal Divers. 2014;66:1–36. doi: 10.1007/s13225-014-0278-5. [DOI] [Google Scholar]
- 29.Rayner R.W. A Mycological Colour Chart. Commonwealth Mycological Institute; Kew, UK: 1970. [Google Scholar]
- 30.White T.J., Bruns T., Lee S., Taylor J. Amplifcation and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990;18:315–322. [Google Scholar]
- 31.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 32.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/AEM.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crous P.W., Gams W., Stalpers J.A. MycoBank: An online initiative to launch mycology into the 21st century. Stud. Mycol. 2004;50:19–22. [Google Scholar]
- 34.Hall T. Department of Microbiology. North Carolina State University; [(accessed on 29 March 2020)]. Bioedit. Available online: http://wwwmbioncsuedu/BioEdit/Bioedithtml. [Google Scholar]
- 35.Crous P.W., Wingfield M.J., Burgess T.I., Hardy G.E.J., Gené J. Fungal Planet description sheets: 716–784. Persoonia. 2018;40:240–393. doi: 10.3767/persoonia.2018.40.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crous P.W., Luangsa-ard J.J., Wingfield M.J., Carnegie A.J., Hernández-Restrepo M. Fungal Planet description sheets: 785–867. Persoonia. 2018;41:238–417. doi: 10.3767/persoonia.2018.41.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crous P.W., Carnegie A.J., Wingfield M.J., Sharma R., Mughini G. Fungal Planet description sheets: 868–950. Persoonia. 2019;42:291–473. doi: 10.3767/persoonia.2019.42.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guarnaccia V., Crous P.W. Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus. 2018;8:317–334. doi: 10.5598/imafungus.2017.08.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milagres C.A., Belisário R., Silva M.A., Lisboa D.O., Pinho D.B., Furtado G.Q. A novel species of Diaporthe causing leaf spot in Pachira glabra. Trop. Plant. Pathol. 2019;43:460–467. doi: 10.1007/s40858-018-0242-0. [DOI] [Google Scholar]
- 40.Ozawa K., Mochizuki K., Takagi D., Ishida K., Sunada A., Ohkusu K., Kamei K., Hashimoto A., Tanaka K. Identification and antifungal sensitivity of two new species of Diaporthe isolated. J. Infect. Chemother. 2018;25:96–103. doi: 10.1016/j.jiac.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Wanasinghe D.N., Phukhamsakda C., Hyde K.D., Jeewon R., Lee H.B. Fungal diversity notes 709–839, taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018;89:1–236. doi: 10.1007/s13225-018-0395-7. [DOI] [Google Scholar]
- 42.Yang Q., Fan X.L., Guarnaccia V., Tian C.M. High diversity of Diaporthe species associated with dieback diseases in China; with twelve new species described. Mycokeys. 2018;39:97–149. doi: 10.3897/mycokeys.39.26914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyde K.D., Tennakoon D.S., Jeewon R., Bhat D.J., Maharachchikumbura S.S.N. Fungal diversity notes 1036–1150, taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019;96:1–242. doi: 10.1007/s13225-019-00429-2. [DOI] [Google Scholar]
- 44.Zhou H., Hou C.L. Three new species of Diaporthe from China based on morphological characters and DNA sequence data analyses. Phytotaxa. 2019;422:157–174. doi: 10.11646/phytotaxa.422.2.3. [DOI] [Google Scholar]
- 45.Zapata M., Palma M.A., Aninat M.J., Piontelli E. Polyphasic studies of new species of Diaporthe from native forest in Chile; with descriptions of Diaporthe araucanorum sp. nov.; Diaporthe foikelawen sp. nov. and Diaporthe patagonica sp. nov. Int. J. Syst. Evol. Microbiol. 2020;70:3379–3390. doi: 10.1099/ijsem.0.004183. [DOI] [PubMed] [Google Scholar]
- 46.Katoh K., Toh H. Recent evelopments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2010;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 47.Swofford D.L. PAUP*, Phylogenetic Analysis Using Parsimony, * and Other Methods. Sinauer Associates; Sunderland, UK: 2003. Version 4.0b10. [Google Scholar]
- 48.Stamatakis A. RAxML-VI-HPC, maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 49.Ronquist F., Huelsenbeck J.P. MrBayes 3, Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 50.Nylander J.A.A. MrModeltest v2. Program Distributed by the Author Evolutionary Biology Centre. Uppsala University; Uppsala, Sweden: 2004. [Google Scholar]
- 51.Silvestro D., Michalak I. RaxmlGUI, a Graphical Front-End for RAxML. [(accessed on 29 March 2020)]; Available online: http://sourceforgenet/projects/raxmlgui/
- 52.Kishino H., Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data; and the branching order in Hominoidea. J. Mol. Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 53.Jayasiri S.C., Hyde K.D., Ariyawansa H.A., Bhat J., Buyck B., Cai L., Dai Y.C., Abd-Elsalam K.A., Ertz D., Hidayat I., et al. The Faces of Fungi database, fungal names linked with morphology; phylogeny and human impacts. Fungal Divers. 2015;74:3–18. doi: 10.1007/s13225-015-0351-8. [DOI] [Google Scholar]
- 54.Vasilyeva L.N., Rossman A.Y., Farr D.F. New species of the Diaporthales from eastern Asia and eastern North America. Mycologia. 2007;99:916–923. doi: 10.1080/15572536.2007.11832523. [DOI] [PubMed] [Google Scholar]
- 55.Crous P.W., Wingfield M.J., Schumacher R.K. Fungal Planet description sheets: 281–319. Persoonia. 2014;33:212–292. doi: 10.3767/003158514X685680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mostert L., Crous P.W., Kang J.C. Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa; morphological; cultural; molecular and pathological characterization. Mycologia. 2001;93:146–167. doi: 10.1080/00275514.2001.12061286. [DOI] [Google Scholar]
- 57.Castlebury L.A., Farr D.F., Rossman A.Y., Jaklitsch W.J. Diaporthe angelicae comb. nov.; a modern description and placement of Diaporthopsis in Diaporthe. Mycoscience. 2003;44:203–208. doi: 10.1007/S10267-003-0107-2. [DOI] [Google Scholar]
- 58.Hu D.M., Cai L., Hyde K.D. Three new ascomycetes from freshwater in China. Mycologia. 2012;104:1478–1489. doi: 10.3852/11-430. [DOI] [PubMed] [Google Scholar]
- 59.Lombard L., van Leeuwen G.C.M., Guarnaccia V. Diaporthe species associated with Vaccinium; with specific reference to Europe. Phytopathol. Mediterr. 2014;53:287–299. [Google Scholar]
- 60.Huang F., Udayanga D., Wang X., Hou X., Mei X. Endophytic Diaporthe associated with Citrus, A phylogenetic reassessment with seven new species from China. Fungal Biol. 2015;119:331–347. doi: 10.1016/j.funbio.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 61.Crous P.W., Wingfield M.J., Richardson D.M. Fungal Planet description sheets: 400–468. Persoonia. 2016;36:316–458. doi: 10.3767/003158516X692185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson S.M., Tan Y.P., Young A.J., Neate S.M., Aitken E.A. Stem cankers on sunflower (Helianthus annuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia. 2011;27:80–89. doi: 10.3767/003158511X617110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao Y.H., Liu F., Cai L. Unravelling Diaporthe species associated with Camellia. Syst. Biodivers. 2016;14:102–117. doi: 10.1080/14772000.2015.1101027. [DOI] [Google Scholar]
- 64.Santos J.M., Vrandecic K., Cosic J., Duvnjak T., Phillips A.J.L. Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia. 2011;27:9–19. doi: 10.3767/003158511X603719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan Y.P., Edwards J., Grice K.R.E., Shivas R.G. Molecular phylogenetic analysis reveals six new Diaporthe species from Australia. Fungal Divers. 2013;61:251–260. doi: 10.1007/s13225-013-0242-9. [DOI] [Google Scholar]
- 66.Crous P.W., Summerell B.W., Shivas R.G., Burgess T.I. Fungal Planet description sheets: 107–127. Persoonia. 2012;28:138–182. doi: 10.3767/003158512X652633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Niekerk J.M., Groenewald J.Z., Farr D.F., Fourie P.H., Halleen F., Crous P.W. Reassessment of Phomopsis species on grapevines. Australas. Plant. Pathol. 2005;34:27–39. doi: 10.1071/AP04072. [DOI] [Google Scholar]
- 68.Crous P.W., Wingfield M.J., Guarro J. Fungal Planet description sheets: 154–213. Persoonia. 2013;31:188–296. doi: 10.3767/003158513X675925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Udayanga D., Liu X.Z., MCkenzie E.H.C., Chukeatirote E., Hyde K.D. Multi-locus phylogeny reveals three new species of Diaporthe from Thailand. Cryptogam. Mycol. 2012;33:295–309. doi: 10.7872/crym.v33.iss3.2012.295. [DOI] [Google Scholar]
- 70.Thompson S.M., Tan Y.P., Shivas R.G., Neate S.M. Green and brown bridges between weeds and crops reveal novel Diaporthe species in Australia. Persoonia. 2015;35:39–49. doi: 10.3767/003158515X687506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Udayanga D., Castlebury L.A., Rossman A.Y. The Diaporthe sojae species complex; phylogenetic re-assessment of pathogens associated with soybean; cucurbits and other field crops. Fungal Biol. 2015;119:383–407. doi: 10.1016/j.funbio.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 72.Williamson P.M., Higher A.S., Gams W. Diaporthe toxica sp. nov. the cause of lupinosis in sheep. Mycol. Res. 1994;98:1364–1368. doi: 10.1016/S0953-7562(09)81064-2. [DOI] [Google Scholar]
- 73.Crous P.W., Wingfield M.J., Roux J.J., Richardson D.M. Fungal Planet description sheets; 371–399. Persoonia. 2015;35:264–327. doi: 10.3767/003158515X690269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crous P.W., Schumacher R.K., Wingfield M.J., Lombard L. Fungal Systematics and Evolution; FUSE 1. Sydowia. 2015;67:81–118. [Google Scholar]
- 75.Udayanga D., Liu X.Z., Crous P.W., McKenzie E.H.C. A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis) Fungal Divers. 2012;56:157–171. doi: 10.1007/s13225-012-0190-9. [DOI] [Google Scholar]
- 76.Santos J.M., Phillips A.J.L. Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Divers. 2009;34:111–125. [Google Scholar]
- 77.Santos J.M., Correia V.G., Phillips A.J.L. Primers for mating-type diagnosis in Diaporthe and Phomopsis, their use in teleomorph induction in vitro and biological species definition. Fungal Biol. 2010;114:255–270. doi: 10.1016/j.funbio.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 78.Gao Y., Liu F., Duan W., Crous P.W., Cai L. Diaporthe is paraphyletic. IMA Fungus. 2017;8:153–187. doi: 10.5598/imafungus.2017.08.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nitschke T. Pyrenomycetes Germanici 2. Eduard Trewendt; Breslau, Germany: 1870. pp. 161–320. [Google Scholar]
- 80.Farr D.F., Rossman A.Y. Fungal Databases; Systematic Mycology and Microbiology Laboratory; ARS; USDA. (SMML Database) [(accessed on 25 March 2020)]; Available online: https://nt.ars-grin.gov/fungaldatabases/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.