Abstract
The scope and trajectory of today’s escalating antimicrobial resistance (AMR) crisis is inadequately captured by existing surveillance systems, particularly those of lower income settings. AMR surveillance systems typically collate data from routine culture and susceptibility testing performed in diagnostic bacteriology laboratories to support healthcare. Limited access to high quality culture and susceptibility testing results in the dearth of AMR surveillance data, typical of many parts of the world where the infectious disease burden and antimicrobial need are high. Culture and susceptibility testing by traditional techniques is also slow, which limits its value in infection management. Here, we outline hurdles to effective resistance surveillance in many low-income settings and encourage an open attitude towards new and evolving technologies that, if adopted, could close resistance surveillance gaps. Emerging advancements in point-of-care testing, laboratory detection of resistance through or without culture, and in data handling, have the potential to generate resistance data from previously unrepresented locales while simultaneously supporting healthcare. Among them are microfluidic, nucleic acid amplification technology and next-generation sequencing approaches. Other low tech or as yet unidentified innovations could also rapidly accelerate AMR surveillance. Parallel advances in data handling further promise to significantly improve AMR surveillance, and new frameworks that can capture, collate and use alternate data formats may need to be developed. We outline the promise and limitations of such technologies, their potential to leapfrog surveillance over currently available, conventional technologies in use today and early steps that health systems could take towards preparing to adopt them.
Keywords: other diagnostic or tool, epidemiology, health systems, medical microbiology, other infection, disease, disorder, or injury
Summary box.
Surveillance is essential for qualifying and quantifying antimicrobial resistance but is inadequately conducted in many low-income and middle-income settings due to technical hurdles.
Our review of the current implementation and development landscape reveals that leapfrog technologies that could overcome some of these hurdles are in existence or could soon become available.
Low- and middle-income countries settings with few surveillance facilities should be encouraged and supported to embrace out-of-the-Petri dish innovations rather than model their surveillance systems solely on difficult-to-implement-and-quality-assure traditional methods.
Lower income health systems can and should position themselves to adopt promising leapfrog technologies by upgrading financial and procurement systems, moving to digital health records and integrating molecular biology into medical laboratory science education.
Introduction
Leapfrog technologies, innovations that overcome seemingly intractable barriers to progress, can have extraordinary impact in low- and middle-income countries (LMIC). For example, the African mobile phone explosion removed the need for networks of telephone cabling and led to huge advances in data sharing and the availability of banking. Similarly, localised solar and wind technologies are emerging as practical alternatives for electrification of distant locations.
There is a global consensus that antimicrobial resistance (AMR) is a critical global health problem. The systematic identification of bacterial pathogens and their associated AMR patterns allows clinicians to use antimicrobials judiciously in individuals and to design locally relevant antimicrobial stewardship guidelines, reducing selective pressure on the emergence of multidrug-resistant strains of the major species infecting humans and livestock. This information is also essential for estimating the burden that AMR adds to poor patient outcomes and healthcare costs. AMR surveillance is therefore a critical, often neglected, component of AMR control that is needed, but least available, in LMIC settings. What innovations, now and in the future, could leapfrog these problems?
Hurdles to antimicrobial resistance surveillance in resource-limited settings
The gold standard approach to bacterial resistance surveillance remains culturing organisms from clinical specimens in quality assured microbiology laboratories, testing their susceptibility against a limited panel of antimicrobial drugs, and disseminating the resulting data to surveillance networks and healthcare professionals. Even where available, this slow process may not provide information fast enough to influence immediate care or guide antimicrobial stewardship. Despite the challenges of culture-based methods, they remain the main tool supporting patient care and informing empirical antimicrobial selection and surveillance and much-needed surveillance data is therefore lacking.1–3 There are model LMIC settings where surveillance is strong but establishing quality assured laboratory networks has proven too complex or costly to be implemented in many resource-limited parts of the world, particularly within Africa.4 Where this approach has been successful, it has often been in the context of extramurally-supported academic or industrial ventures. There is currently new commitment, support and activity towards building new culture-based LMIC surveillance systems. However, scale-up is slow,5 6 sustainability remains in question and an outside-the-Petri-dish review of surveillance possibilities is overdue.
In many resource-limited settings, formidable hurdles hamper implementation of AMR surveillance based on traditional culture and susceptibility testing. These fall into three broad categories (table 1): (1) pre-laboratory, which prevent taking a sample and adequate metadata and delivering these to a quality assured testing in a usable condition, (2) laboratory, which prevent optimal specimen processing, and (3) data dissemination, which reduce the utility of susceptibility information that is generated. As indicated in table 1, there are advances to eliminate or at least lower many of these hurdles in existence or in development but a truly leapfrog technology would eliminate multiple steps in the traditional pathway to surveillance (figure 1), eliminate a roadblock for which no solution exists (eg, invasive sampling) or create an entirely new pathway to surveillance.
Table 1.
Common antimicrobial surveillance hurdles in low-resource and remote settings
| The hurdle | Structure of the hurdle | Potential solutions |
| Access to testing |
|
|
| Invasive sampling and associated biohazards |
|
|
| Data-entry and handling | Paper records that cannot be queried or viewed from more than one location |
|
| Extensive, non-integrated quality assurance requirements |
|
Integrated quality assurance such as read quality analyses for WGS |
| In-lab processing time | Long incubation time makes direct benefits for testing patients requiring urgent care, or who cannot return to the laboratory for test results doubtful |
|
| Subtyping |
|
Rapid WGS with microfluidic DNA preparation, which can provide fine subtyping information along with identification and susceptibility information at no extra cost. To reduce costs, other methods are often batched at the species level, leading to longer turn-around times. WGS is as effective without species batching |
| Quality assurance | Access to external quality assurance is difficult in many resource settings |
|
| The cold-chain | Current testing requires a large number of different reagents requiring cold storage to be shipped to testing laboratories |
|
| Human resources | There is a shortage of qualified laboratory technicians and a wide range of skills is needed for broad surveillance |
|
| Infrastructure | Electricity, running water and internet access are often unavailable or only intermittently available at remote testing sites |
|
| Data sharing and connectivity | Personal data needs to be protected and be compliant with General Data Protection Regulation and pseudonymised before sharing. Robust protocols are not always in place |
|
CSF, cerebrospinal fluid; LIMS, laboratory information management systems; WGS, whole-genome sequencing.
Figure 1.
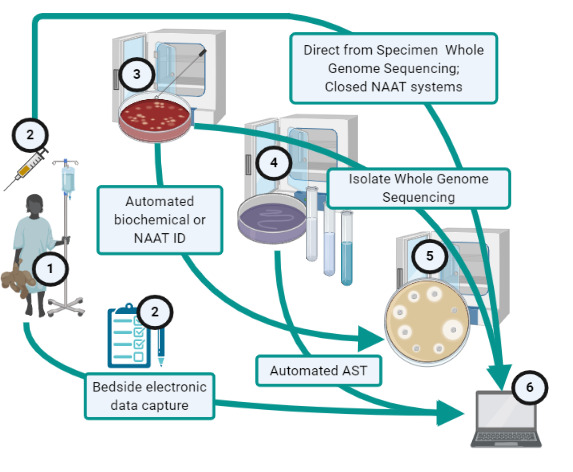
Existing options for leapfrogging across the path from patient (1) to surveillance data (6) that traditionally includes manual health-worker collection of specimen and patient data (2), isolate recovery (3), identification (4) and antimicrobial susceptibility testing (AST) (5), with incubation during steps 2 to 5, followed by data entry and processing (6). NAAT, nucleic acid amplification technology.
Closing the distance between patients and testing
Patients and their healthcare providers are often distant from labs that typically test using expensive equipment, labile consumables and highly trained staff, all of which are in short supply.7 In high-income countries (HICs), fewer resource constraints allow better decentralisation of laboratory services. Remote access to specialised testing is a global problem solved in HICs by tiered systems linked by specimen transport couriers to centralised facilities. Courier services are inadequately networked, dependent on poorly developed road infrastructure and/or unreliable in many LMIC settings and therefore not optimal for routine or urgent sample transport. Specimen collection riders that can meet (and where necessary sample) individuals at remote locations represent a significant and tailored advance but still require some road infrastructure and significant expense for the most remote settings.8 Drones, already a successful alternative to road-based transportation in medical supply chains (Rwanda) or blood products (Malawi), could potentially represent a truly leapfrog alternative that can reach the most remote settings if cost and patient sampling barriers can be overcome.3
Point-of-care tests could make sample transport unnecessary if they could identify bacterial infections in a manner that is affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free and deliverable: ‘ASSURED’.9 In reality, tests that meet these criteria and identify resistant infections remain to be deployed. A target product profile (TPP) for such a point-of-care test would specify that it should be equipment-free, require no electricity to ship, store or perform, and produce reliable results in under 30 min. It would discriminate multiple pathogens associated with a specific syndrome and would be able to read out, or be paired with a second rapid test that identifies, AMR. TPPs along these lines have been issued for gonococcal clinical diagnostics and surveillance10 and similar profiles would be applicable for febrile illnesses, meningitis and invasive diarrhoea. A decade ago, it would have been challenging or impossible to respond to such a TPP. Today, technological innovations with the power to promote point-of-care testing at the hardest-to-reach locations along these lines exist, as has been reviewed extensively elsewhere.11–13 The products themselves remain to be finalised in part because they represent relatively new innovations still in development, but also because their markets, in a world where gold-standard AMR data are considered to be an output of quality assured diagnostic microbiology laboratories, is presently uncertain.
To effectively link bedside caregiving to surveillance systems, testing innovations need to be paired with data handling innovations.14 Ideally, health facilities would record patient, clinical microbiology and treatment data in real time, and warning systems would alert healthcare workers to collect clinical specimens for bacteriology investigations before antibiotic prescription orders are filled. Further data would only be entered once. As smartphones with camera and video functions are increasingly available, technologies to assist clinical practice and decisions should be but a simple app away. However, data handling is a foremost surveillance challenge and one that might be easier to address with point-of-care testing that provides results immediately to clinicians. However, it may be challenging to link aetiologic information from point-of-care testing to AMR surveillance because current bacterial resistance surveillance systems are isolate-based.
Leapfrogging within laboratories, or the hare and the tortoise revisited
AMR surveillance information is typically generated from sequential culture-based methods of bacterial isolation, identification and susceptibility testing (figure 1). Most bacterial culture media are comprised of cheap components and other requirements are simple; modest upgrades from the time of Robert Koch.3 Superficially, this might appear to be an appropriate technology when resources are constrained given its cost, apparent simplicity and long history, but in practice this is not the case.7 Outcomes from LMIC Quality Assurance programmes indicate that, when performed, identification and susceptibility testing is associated with high error rates outside of highly controlled environments such as provided by quality assured microbiology laboratories.7 15 16 To date, establishing quality-assured laboratory networks has proven complex and costly for many resource-limited parts of the world, including many sites in Africa.4 5 This, in turn, limits diagnostic stewardship in which clinicians and healthcare workers understand recommended guidelines, perform appropriate diagnostic tests (particularly bacterial culture if indicated), and use local AMR surveillance data to provide the most optimal treatment.17
1. Shortening turn-around times
The first step in traditional bacteriology is pathogen recovery. Paradoxically, waiting for ‘fast-growing’ pathogens to multiply is the major reason why routine bacterial culture is so slow. For some pathogen-specimen combinations, ‘Gold-Standard’ culture methods only recover the pathogen in a fraction of instances, so leapfrog solutions with greater sensitivity will have global value. Cases in point include cerebrospinal fluid processing in meningitis, particularly in pretreated patients, and low-abundance blood-borne bacteria like Salmonella enterica serovariety Typhi, a key AMR priority pathogen. To address these limitations, which are not large in well-resourced settings, technology has largely used ‘tortoise’ advancements to culture-based protocols which marginally extend the sensitivity, specificity and speed attainable for identifying a huge diversity of bacteria and AMR phenotypes. Even the most innovative ‘tortoise’ advancements speed up or eliminate one or more incubation steps but typically still require some growth.18–20 More sensitive or alternate growth-detection technologies can drastically reduce turn-around times and represent longer stride tortoise strategies, often with unfortunately hefty price tags. Automated culture, identification and sensitivity testing systems such as BACTEC, BacT/Alert or VersaTREK (blood culture) and VITEK and VITEK-MS, Phoenix, Bruker MALDI Biotyper or Sensititre (identification and/or susceptibility testing), as well as DNA-leakage or early growth-detection tests18–20 accomplish this and, in some cases, are easier to quality assure than traditional protocols. Even shorter incubation times could be possible from ultrasensitive detection platforms in development.21 22 Because they still require basic specimen and isolate management and are expensive to institute and run, these generally do not constitute leapfrog technologies for LMICs although some have been deployed.
Challenging or dangerous-to-culture pathogens have provided intrinsic motivation for bypassing culture and provide leapfrog models. Analogous technologies for detecting antibacterial resistance largely do not exist but these new methods could serve as important templates. A recently described Lab-on-a-Chip, built from complementary metal-oxide-semiconductor technology coupled with loop-mediated isothermal amplification addresses common Plasmodium resistance single nucleotide polymorphisms (SNPs) in malaria23 and could feasibly do the same for bacterial resistance conferred by SNPs. While culture-free technologies offer significant promise, many alternatives to culture narrow the range of organisms that can be identified and most have the added disadvantage of not capturing the infecting organism for archiving and future research.
Antimicrobial panels for susceptibility testing are selected based on the identity of infectious pathogens and naming and subtyping are the most complicated and least generalisable protocols by traditional testing. It is sometimes possible to base identification on bacterial cell membrane composition, or its impact on the host immune system. These options open the possibility of antigen-based or antibody-based diagnostics, which, in turn, present the possibility of field-ready platforms such as microfluidic systems, simpler mobile-readable lateral-flow devices or at least protocols to remove one or more of the sequential steps needed for routine identification by culture.24 25 In a few instances, a surface-expressed resistance-conferring protein can serve as a marker for AMR but those represent the exception.26 All of these approaches could be difficult to multiplex and until that challenge is addressed, their use for multiaetiologic syndromes where bacteria predominate as pathogens will be limited.
2. Promises and pitfalls of nucleic acid amplification tests
One oft-cited leapfrog technology for AMR surveillance that offers the opportunity to bundle pathogen identification, subtyping, and in many cases resistance profiling too, is nucleic acid amplification technology (NAAT), for example by PCR, in which oligonucleotide primers bind to a complementary DNA target, permitting specific amplification and thence detection of one or more signature fragments. All nucleic acid-based tests, including NAAT and whole-genome sequencing (WGS; discussed in the next section), have the disadvantage of identifying resistance genotypes rather than phenotypes. As some genes, including many encoding resistance, are not expressed and new resistance genes can only be identified after they are known, nucleic acid-based AMR surveillance cannot completely replace phenotypic methods, which must continue to be performed at some level in each health system.
In spite of this caveat, NAAT is widely used in research, has long since superseded culture in diagnostic virology laboratories but as with culture, PCR is an easy thing to do badly. NAATs often depend on samples being appropriately taken from patients and delivered in a timely fashion to a quality assured facility, although NAAT is certainly faster than culture. The limitations of NAAT are exquisitely illustrated by the challenges seen in both LMIC and HIC settings during the first few months of the COVID-19 pandemic when real-timePCR was the only diagnostic option for the disease and could only be performed in very few diagnostic laboratories. However, the subsequent scale-up in testing in virtually every country in the world demonstrates the potential of these technologies for select applications.
When performed from first principles, NAAT tests are exceptionally prone to contamination, which is then difficult to detect, but some of this shortfall can be addressed by test format innovation, and microfluidics advances have made it possible to automate temperamental preparatory steps.27 Lab-on-chip, discs and cards can effectively bypass inadequate human resources, infrastructure and sometimes cold-chains.28 The GeneXpert MTB/RIF platform is an example of one such leapfrog technology and the BioFire FilmArray represents another. The former has been more widely deployed in LMIC settings and consists of automated PCR system initially and most commonly used for tuberculosis diagnostics and rifampicin sensitivity testing. As with BioFire, it requires less training than conventional PCR as all the specimen processing steps are completed within the cartridge and has therefore circumvented many of these problems. GeneXpert MTB/RIF has been widely deployed in a range of LMIC settings and appears to be cost-effective in at least some of them.29–31 However, even this system shares the challenge presented by low copy number bacteria, which may certainly be the case in paucibacillary mycobacterial disease. Potential solutions to insensitivity include new technologies and applications of nucleic acid-based methods such as single-cell whole genome sequence Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-based detection32 as well as free DNA capture as applied in fetal abnormality and cancer screening tests.33
For relatively difficult-to-culture bacteria, including mycobacteria, and sexually-transmitted bacteria, NAAT-based tests have overcome diagnostic and surveillance roadblocks in LMIC laboratories. Xpert and BioFire tests that detect priority resistant pathogens that are easier to culture, such as methicillin-resistant Staphylococcus aureus and carbapenemase-producing Enterobacteriaceae are commercially available and approved for use at the point of care.34–37 However, these are not marketed or prioritised in low-income settings even when the Xpert hardware already exists in admittedly overstretched tuberculosis programmes, where it has been applied to other diagnostics, most notably for Ebola and now COVID-19.38 Even in the best resourced settings, NAAT cannot compete with culture in terms of the diversity of bacteria and AMR types it can identify as there is a limit to the degree to which assays are multiplexed. In AMR surveillance, NAAT is comparable to the hare that raced ahead of culture, but which has yet to cross the line into routine surveillance on a large scale and may well be overtaken by other technologies.11
3. Whole-genome sequencing-based surveillance in LMICs
Beyond those bacterial pathogens that are challenging to culture, today’s exemplar leapfrog technology remains WGS, which has been revolutionised by the introduction of inexpensive, rapid and high throughput next-generation sequencing technology. Such equipment still requires significant laboratory infrastructure and advanced equipment maintenance and information technology support.39 40 Nonetheless, second-generation sequencing technologies have been used in local outbreak situations, such as the Ebola, Zika, COVID-19 and Lassa outbreaks of the current decade where portable MinION sequencers generated actionable information,41 41–43 and this technology is eminently suitable for surveillance of AMR. The diversity of sequencing options may well grow but for genomic surveillance, the present challenge is increasing their applicability both by increasing the number and types of patient specimens that reach this type of analysis as well as the scope of stakeholder epidemiologists that can access and use the information.
Sequencing requires upstream DNA preparation and manipulation steps, which currently limit its use in some settings, but progress has been made in miniaturising and automating these using microfluidic devices.27 44 While we can expect these to continue to improve and converge into combined, portable devices, field-ready WGS platforms still have the same challenges as culture and NAAT systems; a given patient must have an adequate sample taken to confer high sensitivity. Furthermore, WGS creates large data files which need analysis at source by highly skilled bioinformaticians and adequate internet connectivity. Despite these challenges, WGS combines advantages of culture and NAAT, operating in the absence of bacteriology-specific laboratory infrastructure while retaining the relative speed of NAAT, and the diverse diagnostic capability of culture WGS provides finetyping information for any species along with identification and susceptibility data at no additional cost.45
Current routine application of WGS in AMR surveillance uses sequence derived from isolates. A bigger leap can be made by sequencing direct-from-specimen (figure 1). This has been accomplished for LMIC specimens, generating in the case of meningitis, valuable information that would not otherwise have been obtained.46 It is, however, costlier because specimens contain a lot of reaction inhibitors, including host and commensal cells, all of which contain non-target DNA. Therefore, direct-from-specimen sequencing requires enriching for pathogen DNA in silico after expensively generating total metagenomic DNA or enriching for pathogen DNA in clinical specimens before sequencing, an approach that is still in development.
Data collection and sharing
Acquiring and managing metadata is often a more difficult challenge than obtaining rich laboratory data, even in resource-rich settings.47 Many LMIC health facilities use handwritten patient records in hard copy files, making it challenging to retrieve and use meta-data associated with laboratory bacterial isolates. A first prerequisite for the leap into effective surveillance is electronic record-keeping in clinics and labs. Given the range of tests and possible results, the technical requirements for microbiology laboratory information management systems (LIMS) are somewhat broader than those required for the blood sciences.48 Purpose-built electronic microbiology LIMS systems are uncommon5 12 and there are few comprehensive open source or low cost options available. Thus, generating pathogen-susceptibility data sets or reports for sharing locally, nationally and internationally remains challenging.49 In the absence of a LIMS, WHONET provides a robust solution for standardised AMR data capture, deduplication, analysis and sharing. Where automated analysis tools are in use, some instruments can be connected directly to the internet or converted using BacLink to WHONET50 and fed into surveillance.
When paper records are still used, the supplementary task of data entry by overstretched health workers can be automated by technologies like DataFax (Clinical DataFax Systems, Canada). Encryption can be added into the communication between mobile phones and a local server (which could be just a single computer) in the hospital to ensure that personal information of patients remain secure. Collection of patient metadata in the context of global antimicrobial resistance surveillance system-based AMR surveillance was successfully implemented recently at a tertiary centre in Thailand.51 App-based collection of clinical data by surveillance staff took an average of 10 min per patient and led the authors to conclude that such surveillance might be best conducted periodically, rather than continuously, to monitor trends. Further work is ongoing to define the optimal clinical variable data set to efficiently determine key patient-level impacts of AMR.52
Data generated from instruments (eg, GeneXpert BioFire, Phoenix, Sensititre or VITEK) or from bioinformatic sequence analysis can be connected to surveillance systems. This reduces the burden of surveillance on diagnostic services, and can also serve as early-warning systems for outbreaks by reducing the time lag between data collection and reporting.14 53 However, integrating data from different platforms can sometimes be challenging and is also an area where concerted innovation is necessary. Verification and linkage of clinical and laboratory data using open source middleware/ automated scripts, coupled with visualisation via user-friendly reports and interactive dashboards, permits standardised preparation of data for sharing and improved understanding of the local AMR situation and its impacts. Several such tools are under development, for example the AutoMated tool for Antimicrobial resistance Surveillance System.54 With the right system, resistance surveillance data can be shared with clinicians directly via data visualisations/reports and indirectly via development of local antimicrobial treatment apps, which provide an opportunity for further buy-in.49 55
Community surveillance: leapfrogging patients
We began this article by outlining pre-laboratory, laboratory and dissemination hurdles that need to be overcome for AMR surveillance. Surveillance of the environment has the advantage of bypassing many hurdles entirely but does not take advantage of, or contribute to clinical patient management. An international sewage study recently mapped geographical distribution of AMR genes garnered from sewage system metagenomes and another metagenomic study is surveilling resistance via public transportation systems.56 57 Community surveillance disconnects surveillance from clinical practice (with the possible exception of those that surveil health facilities58) and, in low-income countries, privileges better resourced communities, which have sewerage, public transport networks and concentrate resistance genes at tertiary care referral health centres. It also does not deconvolute human and animal resistance reservoirs, which may not be the same.59 However, community surveillance can use specimens other than sewage, and stations and may be an inexpensive and non-obtrusive way to measure and map resistance, disease or both.58 60–62 Rationale and justification needs careful consideration: building community surveillance along these lines does not simultaneously enhance patient care, which makes it less justifiable in parts of the world where patient testing is highly desirable but barely available.
Elephants in the room: cost and logistical hurdles
Traditional culture and susceptibility testing carries many indirect and hidden costs; but the cost of sequence-based and disposable point-of-care tests are still presently higher and need to fall before these methods can be broadly applied. If the rate of cost decline for sequencing continues, it will become cheaper than conventional bacteriology within a decade. However, culture and susceptibility resources are almost certainly likely to be needed to support leapfrog technologies that could broaden access to bacteriology diagnostics. For this reason, we are not proposing that efforts to develop and strengthen laboratories along the traditional paradigm should be halted or slowed. Preparing to add on leapfrog technologies could lead to overall system strengthening that will benefit current and future practices for AMR surveillance.
As with financial systems, lab-clinic interfaces and data handling in many low-income settings have changed marginally, if at all, since colonial times when patient and surveillance needs in no way constituted an actual health system.63 Health systems and the laboratories within them need to move to efficient, patient-centred electronic data systems. Cheaper end-user hardware like netbooks and tablets makes it possible to do so without heavy infrastructure investment. Internet connectivity, an overarching hurdle that must be scaled for many other purposes, may be the greatest barrier to data access and use in some settings.
It can take relatively well-prepared and resourced LMIC tertiary care systems years from goal conception to generating local sequence-based surveillance data. Therefore, with the anticipated drop in prices, less endowed systems would be wise to begin to make essential but inexpensive preparations to shift to or, in the event that they have no surveillance, adopt newer technologies. Some of the most useful ways to poise to leap include:
Update financial practices and supply chains: Outdated systems do not serve present-day needs and would not be agile enough to procure library preparation and sequencing reagents in bulk, the best way to secure competitive pricing. Financial system development that has begun in academia to address these gaps for research (https://www.aasciences.africa/ggc/standard) needs to extend to health system procurement, human resource and financial management.
Applying and exploiting molecular testing requires a foundation in molecular sciences that is often absent or inadequate in LMIC curricula and continuing education programmes. Developing this expertise now would make it easier for LMIC systems to make rapid and informed transitions to or adoptions of new technologies.
Health systems need to move to electronic record keeping for patient care and in labs, exploiting free approaches and tools such as WHONET and ACORN.50 52
Many of the cost and access issues surrounding materials required for leapfrog science arise because of the present dependence on external resources. Development of local systems and implementation science to better adapt existing systems is not only possible, but an urgent health system need. In performing clinical diagnostics and surveillance of dolutegravir-resistant HIV, Seatla et al developed a ‘home brew’ that allowed them to identify mutations with slightly elevated sensitivity and only a fraction of the cost of commercial kits. In doing so, they turned a technically appropriate LMIC technology to one that was economically feasible, analogous to the development of low cost smart phones that are targeted directly at LMIC markets and have leapfrogged access to the internet in many of these settings. Similar research and innovation may have to precede more widespread adoption of leapfrog technologies.
Conclusion
Traditional methods for AMR surveillance can be challenging to establish and truly representative, high quality surveillance may be easier to achieve by combining those approaches with new innovations or exploring entirely novel paths to usable resistance information. Surveillance systems that are starting from scratch have the best opportunity to adopt next generation of AMR surveillance technologies and should be encouraged and supported to embrace newer approaches rather than to automatically model surveillance systems on age-old frameworks. Where immediate adoption may not yet be feasible, active preparation to integrate cost-effective technologies should be an immediate priority.
Acknowledgments
We are grateful to Francesca Chiara, Jamie Nunn and El-Shama Monu-Nwoko for administrative support.
Footnotes
Handling editor: Seye Abimbola
Twitter: @iruka_okeke
Collaborators: Surveillance and Epidemiology of Drug-resistant Infections Consortium (SEDRIC)
Contributors: INO, SP, DL, JP, AH and NF co-conceived the article at a Surveillance and Epidemiology of Drug-resistant Infections Consortium (SEDRIC) meeting. INO, NF, DL, JP, PT and GP wrote significant portions of the text. The manuscript’s content was honed by co-author discussions at further meetings, including the SEDRIC annual conference by INO, NF, SP, DL, JP, AH, and PT. INO made the initial synthetic draft, with significant input from INO, NF, DL, JP and SP. INO, NF, SP, DL, JP, AH, PT and GP edited drafts and approved the final version.
Funding: The Wellcome Drug Resistant Infection Priority Programme supports the Surveillance and Epidemiology of Drug-resistant Infections Consortium (SEDRIC). I. N. O. is an African Research Leader supported by the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement that is also part of the EDCTP2 programme supported by the EU. The funders had no involvement in the content, writing or submission of this paper.
Competing interests: JP reports personal fees from NextGen Diagnostic, outside the submitted work; other authors have no competing interests to declare.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer-reviewed.
Data availability statement: There are no data in this work.
References
- 1.WHO Antimicrobial resistance: global report on surveillance 2014 2014:257.
- 2.Hay SI, Rao PC, Dolecek C, et al. . Measuring and mapping the global burden of antimicrobial resistance. BMC Med 2018;16:78. 10.1186/s12916-018-1073-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okeke IN. Divining without seeds : the case for strengthening laboratory medicine in Africa. Ithaca: ILR/ Cornell University Press, 2011. [Google Scholar]
- 4.Leski TA, Ansumana R, Malanoski AP, et al. . Leapfrog diagnostics: demonstration of a broad spectrum pathogen identification platform in a resource-limited setting. Health Res Policy Syst 2012;10:22. 10.1186/1478-4505-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ibrahim RA, Teshal AM, Dinku SF, et al. . Antimicrobial resistance surveillance in Ethiopia: implementation experiences and lessons learned. Afr J Lab Med 2018;7:770. 10.4102/ajlm.v7i2.770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Opintan JA. Leveraging donor support to develop a national antimicrobial resistance policy and action plan: Ghana's success story. Afr J Lab Med 2018;7:825. 10.4102/ajlm.v7i2.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ombelet S, Ronat J-B, Walsh T, et al. . Clinical bacteriology in low-resource settings: today's solutions. Lancet Infect Dis 2018;18:e248–58. 10.1016/S1473-3099(18)30093-8 [DOI] [PubMed] [Google Scholar]
- 8.Moran ZR, Frimpong AB, Castañeda-Casado P, et al. . Tropical laboratory initiative: an innovative model for laboratory medicine in rural areas. Afr J Lab Med 2019;8:922. 10.4102/ajlm.v8i1.922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peeling RW, Mabey D, Herring A, et al. . Why do we need quality-assured diagnostic tests for sexually transmitted infections? Nat Rev Microbiol 2006;4:S7–19. 10.1038/nrmicro1569 [DOI] [PubMed] [Google Scholar]
- 10.Ferreyra C, Osborn J, Moussy F, et al. . Developing target product profiles for Neisseria gonorrhoeae diagnostics in the context of antimicrobial resistance: an expert consensus. PLoS One 2020;15:e0237424. 10.1371/journal.pone.0237424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guy RJ, Causer LM, Klausner JD, et al. . Performance and operational characteristics of point-of-care tests for the diagnosis of urogenital gonococcal infections. Sex Transm Infect 2017;93:S16–21. 10.1136/sextrans-2017-053192 [DOI] [PubMed] [Google Scholar]
- 12.Sayed S, Cherniak W, Lawler M, et al. . Improving pathology and laboratory medicine in low-income and middle-income countries: roadmap to solutions. Lancet 2018;391:1939–52. 10.1016/S0140-6736(18)30459-8 [DOI] [PubMed] [Google Scholar]
- 13.Schito M, Peter TF, Cavanaugh S, et al. . Opportunities and challenges for cost-efficient implementation of new point-of-care diagnostics for HIV and tuberculosis. J Infect Dis 2012;205 Suppl 2:S169–80. 10.1093/infdis/jis044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gous NM, Onyebujoh PC, Abimiku Alash'le, et al. . The role of connected diagnostics in strengthening regional, National and continental African disease surveillance. Afr J Lab Med 2018;7:775. 10.4102/ajlm.v7i2.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayeni FA, Odumosu BT. False identification of other microorganisms as Staphylococcus aureus in Southern Nigeria. Tropical Journal of Pharmaceutical Research 2016;15:1941–5. 10.4314/tjpr.v15i9.19 [DOI] [Google Scholar]
- 16.Jesumirhewe C, Ogunlowo PO, Olley M, et al. . Accuracy of conventional identification methods used for Enterobacteriaceae isolates in three Nigerian hospitals. PeerJ 2016;4:e2511. 10.7717/peerj.2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO Diagnostic stewardship: a guide to implementation in antimicrobial resistance surveillance sites; 2018.
- 18.Savela ES, Schoepp NG, Cooper MM, et al. . Surfactant-enhanced DNA accessibility to nuclease accelerates phenotypic β-lactam antibiotic susceptibility testing of Neisseria gonorrhoeae. PLoS Biol 2020;18:e3000651. 10.1371/journal.pbio.3000651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoepp NG, Liaw EJ, Winnett A, et al. . Differential DNA accessibility to polymerase enables 30-minute phenotypic β-lactam antibiotic susceptibility testing of carbapenem-resistant Enterobacteriaceae. PLoS Biol 2020;18:e3000652. 10.1371/journal.pbio.3000652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Shin DJ, Zheng S, et al. . Direct-qPCR assay for coupled identification and antimicrobial susceptibility testing of Neisseria gonorrhoeae. ACS Infect Dis 2018;4:1377–84. 10.1021/acsinfecdis.8b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cansizoglu MF, Tamer YT, Farid M, et al. . Rapid ultrasensitive detection platform for antimicrobial susceptibility testing. PLoS Biol 2019;17:e3000291. 10.1371/journal.pbio.3000291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Besant JD, Sargent EH, Kelley SO. Rapid electrochemical phenotypic profiling of antibiotic-resistant bacteria. Lab Chip 2015;15:2799–807. 10.1039/C5LC00375J [DOI] [PubMed] [Google Scholar]
- 23.Malpartida-Cardenas K, Miscourides N, Rodriguez-Manzano J, et al. . Quantitative and rapid Plasmodium falciparum malaria diagnosis and artemisinin-resistance detection using a CMOS Lab-on-Chip platform. Biosens Bioelectron 2019;145:111678. 10.1016/j.bios.2019.111678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peeters M, Chung P, Lin H, et al. . Diagnostic accuracy of the InBiOS AMD rapid diagnostic test for the detection of Burkholderia pseudomallei antigen in grown blood culture broth. Eur J Clin Microbiol Infect Dis 2018;37:1169–77. 10.1007/s10096-018-3237-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuijpers LMF, Chung P, Peeters M, et al. . Diagnostic accuracy of antigen-based immunochromatographic rapid diagnostic tests for the detection of Salmonella in blood culture broth. PLoS One 2018;13:e0194024. 10.1371/journal.pone.0194024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu D, Stach L, Newland JG, et al. . Integrating a rapid diagnostic test and antimicrobial stewardship: optimizing discharge antibiotics in skin and soft tissue infections. Pediatr Infect Dis J 2016;35:1362–4. 10.1097/INF.0000000000001332 [DOI] [PubMed] [Google Scholar]
- 27.Boykin LM, Sseruwagi P, Alicai T, et al. . Tree lab: portable genomics for early detection of plant viruses and pests in sub-Saharan Africa. Genes 2019;10:632. 10.3390/genes10090632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Manzano J, Moser N, Malpartida-Cardenas K, et al. . Rapid detection of mobilized colistin resistance using a nucleic acid based Lab-on-a-Chip diagnostic system. Sci Rep 2020;10:8448. 10.1038/s41598-020-64612-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vassall A, Siapka M, Foster N, et al. . Cost-Effectiveness of Xpert MTB/RIF for tuberculosis diagnosis in South Africa: a real-world cost analysis and economic evaluation. Lancet Glob Health 2017;5:e710–9. 10.1016/S2214-109X(17)30205-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feasey NA, Banada PP, Howson W, et al. . Evaluation of Xpert MTB/RIF for detection of tuberculosis from blood samples of HIV-infected adults confirms Mycobacterium tuberculosis bacteremia as an indicator of poor prognosis. J Clin Microbiol 2013;51:2311–6. 10.1128/JCM.00330-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sagili KD, Muniyandi M, Nilgiriwala KS, et al. . Cost-Effectiveness of GeneXpert and LED-FM for diagnosis of pulmonary tuberculosis: a systematic review. PLoS One 2018;13:e0205233. 10.1371/journal.pone.0205233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Y, Zhang F, Zhang X, et al. . High-Throughput single-cell whole-genome amplification through centrifugal emulsification and eMDA. Commun Biol 2019;2:147. 10.1038/s42003-019-0401-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thakral D, Das N, Basnal A, et al. . Cell-Free DNA for genomic profiling and minimal residual disease monitoring in Myeloma- are we there yet? Am J Blood Res 2020;10:26–45. [PMC free article] [PubMed] [Google Scholar]
- 34.Coppens J, Van Heirstraeten L, Ruzin A, et al. . Comparison of GeneXpert MRSA/SA ETA assay with semi-quantitative and quantitative cultures and nuc gene-based qPCR for detection of Staphylococcus aureus in endotracheal aspirate samples. Antimicrob Resist Infect Control 2019;8:4. 10.1186/s13756-018-0460-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pannala R, Baldwin B, Aluru V, et al. . Prospective study of the feasibility of point-of-care testing strategy for carbapenem-resistant organism detection. Endosc Int Open 2018;6:E58–63. 10.1055/s-0043-122141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cwengros LN, Mynatt RP, Timbrook TT, et al. . Minimizing time to optimal antimicrobial therapy for Enterobacteriaceae bloodstream infections: a retrospective, hypothetical application of predictive scoring tools vs rapid diagnostics tests. Open Forum Infect Dis 2020;7:ofaa278. 10.1093/ofid/ofaa278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang C-M, Chen X-J, Chih C-C, et al. . Rapid identification of bloodstream bacterial and fungal pathogens and their antibiotic resistance determinants from positively flagged blood cultures using the BioFire FilmArray blood culture identification panel. J Microbiol Immunol Infect 2020. 10.1016/j.jmii.2020.03.018. [Epub ahead of print: 02 Apr 2020]. [DOI] [PubMed] [Google Scholar]
- 38.Butler D. Speedy Ebola tests help contain Africa's latest outbreak. Nature 2018;558:172–3. 10.1038/d41586-018-05389-2 [DOI] [PubMed] [Google Scholar]
- 39.Gwinn M, MacCannell D, Armstrong GL. Next-Generation sequencing of infectious PathogensNext-Generation sequencing of infectious PathogensNext-Generation sequencing of infectious pathogens. JAMA 2019;321:893–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Köser CU, Ellington MJ, Cartwright EJP, et al. . Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog 2012;8:e1002824. 10.1371/journal.ppat.1002824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arias A, Watson SJ, Asogun D, et al. . Rapid outbreak sequencing of Ebola virus in Sierra Leone identifies transmission chains linked to sporadic cases. Virus Evol 2016;2:vew016. 10.1093/ve/vew016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siddle KJ, Eromon P, Barnes KG, et al. . Genomic analysis of Lassa virus during an increase in cases in Nigeria in 2018. N Engl J Med 2018;379:1745–53. 10.1056/NEJMoa1804498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoenen T, Groseth A, Rosenke K, et al. . Nanopore sequencing as a rapidly Deployable Ebola outbreak tool. Emerg Infect Dis 2016;22:331–4. 10.3201/eid2202.151796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burklund A, Petryk JD, Hoopes PJ, et al. . Microfluidic enrichment of bacteria coupled to contact-free lysis on a magnetic polymer surface for downstream molecular detection. Biomicrofluidics 2020;14:034115. 10.1063/5.0011908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reuter S, Ellington MJ, Cartwright EJP, et al. . Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med 2013;173:1397–404. 10.1001/jamainternmed.2013.7734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saha S, Ramesh A, Kalantar K, et al. . Unbiased metagenomic sequencing for pediatric meningitis in Bangladesh reveals Neuroinvasive Chikungunya virus outbreak and other Unrealized pathogens. mBio 2019;10. 10.1128/mBio.02877-19. [Epub ahead of print: 17 12 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green ED, Rubin EM, Olson MV. The future of DNA sequencing. Nature 2017;550:179–81. 10.1038/550179a [DOI] [PubMed] [Google Scholar]
- 48.Rhoads DD, Sintchenko V, Rauch CA, et al. . Clinical microbiology informatics. Clin Microbiol Rev 2014;27:1025–47. 10.1128/CMR.00049-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vong S, Anciaux A, Hulth A, et al. . Using information technology to improve surveillance of antimicrobial resistance in South East Asia. BMJ 2017;358:j3781. 10.1136/bmj.j3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Brien TF, Clark A, Peters R, et al. . Why surveillance of antimicrobial resistance needs to be automated and comprehensive. J Glob Antimicrob Resist 2019;17:8–15. 10.1016/j.jgar.2018.10.011 [DOI] [PubMed] [Google Scholar]
- 51.Sirijatuphat R, Sripanidkulchai K, Boonyasiri A, et al. . Implementation of global antimicrobial resistance surveillance system (GLASS) in patients with bacteremia. PLoS One 2018;13:e0190132. 10.1371/journal.pone.0190132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner P, Ashley EA, Celhay OJ, et al. . ACORN (A Clinically-Oriented antimicrobial resistance surveillance network): a pilot protocol for case based antimicrobial resistance surveillance. Wellcome Open Res 2020;5:13. 10.12688/wellcomeopenres.15681.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rawson TM, Hernandez B, Moore LSP, et al. . A real-world evaluation of a case-based Reasoning algorithm to support antimicrobial prescribing decisions in acute care. Clin Infect Dis 2020. 10.1093/cid/ciaa383. [Epub ahead of print: 04 Apr 2020]. [DOI] [PubMed] [Google Scholar]
- 54.Lim C, Miliya T, Chansamouth V, et al . Automating the generation of antimicrobial resistance surveillance reports: proof-of-concept study involving seven hospitals in seven countries. J Med Internet Res 2020;22:e19762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fox-Lewis A, Takata J, Miliya T, et al. . Antimicrobial resistance in invasive bacterial infections in hospitalized children, Cambodia, 2007-2016. Emerg Infect Dis 2018;24:841–51. 10.3201/eid2405.171830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hendriksen RS, Munk P, Njage P, et al. . Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat Commun 2019;10:1124. 10.1038/s41467-019-08853-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Afshinnekoo E, Meydan C, Chowdhury S, et al. . Geospatial resolution of human and bacterial diversity with City-Scale Metagenomics. Cell Syst 2015;1:97–97.e3. 10.1016/j.cels.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 58.Chng KR, Li C, Bertrand D, et al. . Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment. Nat Med 2020;26:941–51. 10.1038/s41591-020-0894-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ludden C, Raven KE, Jamrozy D, et al. . One health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. mBio 2019;10. 10.1128/mBio.02693-18. [Epub ahead of print: 22 01 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raven KE, Ludden C, Gouliouris T, et al. . Genomic surveillance of Escherichia coli in municipal wastewater treatment plants as an indicator of clinically relevant pathogens and their resistance genes. Microb Genom 2019;5. 10.1099/mgen.0.000267. [Epub ahead of print: 20 05 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saha S, Tanmoy AM, Andrews JR, et al. . Evaluating PCR-Based Detection of Salmonella Typhi and Paratyphi A in the Environment as an Enteric Fever Surveillance Tool. Am J Trop Med Hyg 2019;100:43–6. 10.4269/ajtmh.18-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Erlich Y. A vision for ubiquitous sequencing. Genome Res 2015;25:1411–6. 10.1101/gr.191692.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaughan M. Curing their ills: colonial power and African illness. Cambridge, UK: Polity Press, 1991. [Google Scholar]


