Abstract
It is now well recognized that the information processing machineries of archaea are far more closely related to those of eukaryotes than to those of their prokaryotic cousins, the bacteria. Extensive studies have been performed on the structure and function of the archaeal DNA replication origins, the proteins that define them, and the macromolecular assemblies that drive DNA unwinding and nascent strand synthesis. The results from various archaeal organisms across the archaeal domain of life show surprising levels of diversity at many levels—ranging from cell cycle organization to chromosome ploidy to replication mode and nature of the replicative polymerases. In the following, we describe recent advances in the field, highlighting conserved features and lineage-specific innovations.
Keywords: archaea, DNA polymerase, DNA primase, CMG, Sulfolobus, helicase
ARCHAEAL REPLICON ARCHITECTURE
Although, like many bacteria, archaea possess simple, circular chromosomes, distinct archaea can exploit strikingly diverse modes of genome replication. Thus far, all bacteria characterized have a single replication origin per chromosome. Initial radiolabeling studies in the euryarchaeon Pyrococcus abyssi provided the first characterization of replication initiation in archaea and supported the existence of a single origin in the Pyrococcus chromosome (38). Subsequent fine-mapping experiments confirmed the activity of an origin of replication, termed oriC, in the vicinity of the gene for the candidate initiator protein, a relative of the eukaryotic Orc1 and Cdc6 proteins (33). In eukaryotes, Orc1 is a highly conserved component of the origin replication complex (ORC), which defines replication origins. ORC leads to the recruitment of the replicative helicase [MCM(2–7) in eukaryotes] via the actions of two coloader proteins, Cdc6 and Cdt1 (6). Interestingly, eukaryotic Cdc6 is related in sequence to Orc1, suggesting they evolved from a common ancestor via gene duplication and diversification. Such a progenitor-like molecule is found in archaea and even some early-branching eukaryotes such as trypanosomes (58). The relationship between the archaeal protein and both Orc1 and Cdc6 has led to a confusing nomenclature, with individual archaeal genome sequencing projects arbitrarily assigning names of Cdc6 or Orc1 to the candidate initiator proteins. We refer to the archaeal proteins as Orc1/Cdc6 generically but use the individual genes’ names when referring to a given species in the discussion below. The situation is further complicated by the existence of genes for multiple Orc1/Cdc6 paralogs in various archaeal species. For example, while Pyrococcus encodes a single Orc1/Cdc6 protein, Sulfolobus encodes three and members of the Haloarchaea encode ten or more.
In Pyrococcus, the single Orc1/Cdc6 protein is encoded by a gene located immediately adjacent to oriC, a situation reminiscent of that seen in many bacteria where the analogous initiator protein, DnaA, is encoded adjacent to the bacterial oriC (33). This cis relationship between initiator protein gene and origin of replication is found in other archaea, too. However, although the single replication origin of Pyrococcus was the first to be mapped and characterized, it transpires to be a rather anomalous replication mode for archaea. Indeed, the majority of archaea subsequently characterized have multiple replication origins per chromosome. Members of the Sulfolobus genus have three replication origins, each of which fires once per cell per cell cycle (29, 49, 50). Another crenarchaeon, Pyrobaculum calidifontis, has four replication origins (45), while the main chromosomes of members of the genus Haloferax have three origins. Interestingly, however, a laboratory-adapted strain possesses a fourth main chromosome replication origin courtesy of an integrated extrachromosomal element, the origin of which remains active in its new ectopic location (18).
Genetic studies in Sulfolobus demonstrated that while wild-type cells utilize all three replication origins, none is individually required for viability (54). Indeed, strains with only a single replication origin are fully viable, albeit with extended doubling time and a longer S phase than their wild-type counterparts. Notably, replication termination in the origin-deficient strains repositioned to midway between the remaining active replication origin(s), indicating that, unlike bacteria, Sulfolobus does not possess a dedicated site-specific replication termination system. An even more remarkable situation was observed in Haloferax volcanii, where cells remained fully viable, even having accelerated growth compared to wild type, when all of the main chromosome’s origins of replication were deleted (18). Genome-wide replication profiling by marker frequency analysis unambiguously demonstrated that the origin-free population did not utilize unique start sites. Further, viability of this strain was dependent on the presence of the RadA recombinase (the archaeal ortholog of bacterial RecA and eukaryotic Rad51). Thus, the origin-free strain was likely using a recombination-based mechanism to ensure genome replication. Intriguingly, a study in a closely related species, Haloferax mediterranei, also demonstrated that it is possible to delete all main-chromosome replication origins in that species. However, the resultant population utilized a previously inactive dormant replication origin on the main chromosome (62). The remarkable plasticity of the Haloferax genome has been underscored by a recent study in which it was shown that deletion of a single orc1/cdc6 paralog, termed orc5, leads to fission of the main chromosome into two distinct replicons via homologous recombination between two near-identical copies of genes for superoxide dismutase (1).
The ability of Haloferax to survive (indeed thrive) under laboratory conditions in the absence of replication origins is likely linked to the extremely high ploidy of this organism—exponentially growing cells contain approximately 15 copies of the genome (56). This high copy number may help promote survival by recombination-mediated mechanisms. While wild-type Haloferax cells make use of the replication origins, a more recent study in Thermococcus kodakarensis has suggested that the strain TS559 does not exclusively use the predicted single oriC in the genome during logarithmic growth (15). While this strain possesses a number of mutations throughout the genome relative to the wild-type isolate, it is presumed to have growth parameters reflective of the natural isolate. The lack of origin usage, as determined by marker frequency, was underscored by the lack of essentiality of the single orc1/cdc6 gene in this strain. In contrast, the radA gene was required for viability, again pointing to a recombination-based mechanism for genome propagation in this species. Notably, as with Haloferax, Thermococcus is a highly-oligoploid member of the euryarchaea, possessing 7–19 copies of the genome per cell. In contrast to the case of Haloferax, however, deletion of oriC resulted in a reduction in long-term viability, particularly under conditions of nutrient deprivation.
Thus, the emerging rules, albeit from a very small sampling of the broad archaeal taxonomic diversity, suggest that the highly oligoploid euryarchaea can effectively propagate their genomes via recombination-based mechanisms under laboratory-based conditions. It is worth noting that both Thermococcus kodakarensis and H. volcanii have been selected as model organisms in part because of the relative ease with which they can be genetically modified, a testament to their robustly active homologous recombination systems. However, the near universality of orc1/cdc6 genes in archaeal genomes points to a selective advantage for their retention across the diversity of archaeal lineages.
In contrast to the euryarchaeal species described above, members of the crenarchaea, including Sulfolobus, have organized cell cycles with defined gap phases and cycle between one and two copies of their chromosome (7, 47). This cell cycle organization, with a G1-phase bottle-neck during which cells have a single copy of the chromosome, effectively precludes homologous recombination–based initiation from occurring. While it is formally possible that genomes could be replicated via R-loop-mediated initiation, the apparent requirement for at least one replication origin in Sulfolobus suggests that this mechanism is not readily employed (54).
REPLICATION ORIGIN FUNCTION
With the characterization of the Sulfolobus replication origins, it was recognized that many archaea share a common type of origin (50), typified by Sulfolobus oriC1 and the single Pyrococcus oriC (for a recent review, see 3). Biochemical, chromatin immunoprecipitation, and/or structural studies revealed that the Pyrococcus Orc1/Cdc6 and its sequence orthologs in other archaeal species (Orc1–1 in Sulfolobus) bind specifically to the oriC1-type replication origins and, moreover, bind specifically to sequence elements within the origin termed origin recognition box (ORB) elements (14, 33, 50) (Figure 1a). ORB elements possess dyad symmetry with an asymmetric G-rich sequence on one side of the dyad element. Each ORB element is bound by a single monomer of Orc1–1 with a defined polarity. The Orc1/Cdc6 proteins possess an N-terminal AAA+ fold and C-terminal winged-helix (wH) domain (13, 14, 28). The wH domain recognizes the dyad element of the ORB element, and an α-helical-rich signature motif within the AAA+ domain, termed the initiator-specific motif (ISM), recognizes the asymmetric G-rich motif. While the number of ORB elements present in oriC1-type origins varies across species, a common feature is that two ORBs are present in inverted orientation relative to one another and separated by minimally 75 base pairs of typically AT-rich DNA—a candidate duplex unwinding element (DUE) (3). In Sulfolobus, it has been demonstrated that this pair of inverted ORB elements is necessary and sufficient for maximal levels of replicative helicase recruitment by Orc1–1 in vitro and for oriC1 function in vivo (53, 57). The consequence of this unique polarity of binding of Orc1–1 to the two inverted ORB elements is that the wH domains of two Orc1/Cdc6s will face each other, separated by an extensive AT-rich region that serves as a candidate DUE. In vitro reconstitution experiments revealed that Orc1–1 recruits the replicative helicase to oriC1 dependent on a surface-exposed α-helical region (MCM recruitment motif, MRM) on the wH-proximal face of the Orc1–1 AAA+ domain. Importantly, this interaction was dependent on the nucleotide cofactor bound by the Orc1–1 AAA+ domain (53). More specifically, when ATP was stabilized in the active site, for example by mutation of the protein’s Walker B glutamate (E147), a residue involved in ATP hydrolysis, then Orc1–1 E147A could support MCM recruitment in vitro and support oriC1 replication in vivo. In contrast, when the active site was occupied by ADP, the ability to interact with MCM was abrogated. These observations, coupled with the facts that Orc1–1 is a rather unusual single-turnover ATPase and transcription of the orc1–1 gene is restricted to the immediately prereplicative phase of the cell cycle, suggest that one level of the control of origin firing is at the level of availability of the ATP-bound form of Orc1–1 (53, 54).
Figure 1.
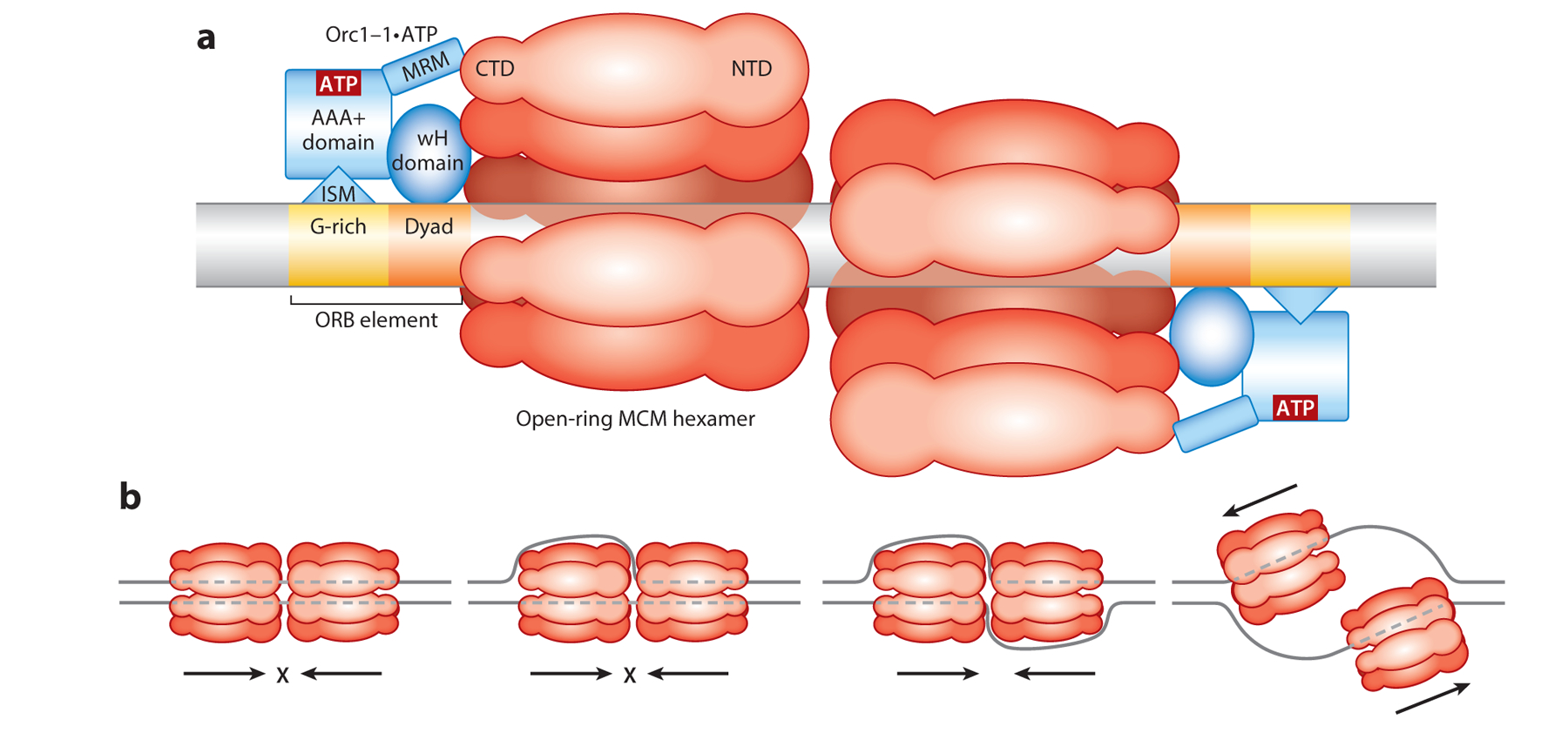
(a) Schematic of the loading of two open-ring homohexamers of MCM onto a replication origin by Orc1–1•ATP. The origin contains two cis-acting elements—ORB elements—that are presented in inverted orientation. Orc1–1 binds as a monomer, with the G-rich motif (yellow) in the ORB being recognized by the ISM motif present in the AAA+ ATPase domain of Orc1–1 and a dyad-containing sequence (orange) in the ORB bound by the wH domain of Orc1–1. When bound to ATP, Orc1–1 contacts the C-terminal domain of an MCM subunit via the MRM within Orc1–1’s AAA+ ATPase domain. (b) Model for origin melting by MCM. MCM proceeds with NTDs leading, and thus when one or two hexamers are double-stranded-DNA-bound, they will act as impediments to encroachment by the other hexamer (two left panels); dashed lines indicate DNA obscured from view in the central cavities of the MCM hexamers. However, once both hexamers have transitioned to binding single-stranded DNA following full melting, they will be able to pass one another, thus establishing bidirectional replication fork progression. Not shown in the illustration are the CG complexes that stimulate MCM’s helicase activity, incorporated at a currently unknown point in the origin activation process. Abbreviations: CTD, C-terminal domain; ISM, initiator-specific motif; MRM, MCM-recruitment motif; NTD, N-terminal domain; ORB, origin recognition box; wH, winged helix.
The archaeal replicative helicase MCM is a ring-shaped homohexamer MCM (for more details of the architecture of MCM, see the following section). How is the ring opened and MCM loaded onto DNA? In the eukaryotic replication apparatus, the association of the accessory protein Cdt1 helps crack the MCM ring [a heterohexameric assembly of MCM(2–7) in eukaryotes] at the interface between the MCM2 and MCM5 subunits. Interestingly, early electron microscopy studies revealed that archaeal MCM (from Methanothermobacter thermautotrophicus) intrinsically adopts an open-ring conformation at the organism’s physiological growth temperature (17). Analogous treatment of the Sulfolobus MCM significantly elevated MCM loading in vitro, suggesting the recruitment of MCM to origin DNA did not require active ring-opening by Orc1–1 (53). The ultimate consequence of MCM recruitment by the two inversely juxtaposed Orc1–1•ATP will be the formation of a double hexamer of MCM encircling the origin DNA. Thus far, evidence in the Sulfolobus system is compatible with MCM being loaded onto double-stranded DNA with origin melting occurring at a later stage (Figure 1a,b). It is notable that recent work in the eukaryotic budding yeast system has also invoked symmetrically opposed ORC proteins directing the formation of an MCM(2–7) double hexamer, suggesting a conserved mechanism for replicative helicase assembly in archaea and eukaryotes. Recent studies have demonstrated that Sulfolobus MCM in isolation is rather promiscuous with respect to DNA binding (46)—with the homohexamer being able to bind to a single-stranded tail of a short oligonucleotide model substrate in both orientations (i.e., with either the N-terminal or C-terminal domain facing the duplex DNA). Indeed, for binding, the C-terminal-domain-facing-duplex orientation is favored, in agreement with earlier studies (35, 51). However, productive translocation and DNA melting appear to be effected by the minority conformation, with N-terminal domains leading during helicase movement. Such an orientation is in agreement with the prevalent model for translocation by the eukaryotic Cdc45•MCM(2–7)•GINS (CMG) complex and provides an elegant mechanism to ensure bidirectional replication from replication origins (12, 16, 41) (Figure 1b).
THE ARCHAEAL MCM HOMOHEXAMER AND THE CMG COMPLEX
The MCM hexamer is a 3′-to-5′ helicase that melts duplex DNA as it translocates 3′ to 5′ (5). As a member of the AAA+ ATPase superfamily, MCM has activities that are driven by ATP hydrolysis, release, and rebinding (5, 52). In archaea, the MCM homohexamer is assembled of identical protomer subunits, and in eukaryotes, the MCM(2–7) hexamer is assembled of MCM paralogs reflecting duplications of the ancestral MCM (30). The fundamental MCM architecture consists of an N-terminal domain, the AAA+ domain, and a wH domain (36). Additionally, eukaryotic Mcm protomers have terminal extension posttranslation-modification targets involved with regulation and recruitment (5). The homohexameric MCM ring complex is tiered into N-terminal and AAA+ levels, possesses a DNA-accommodating central pore lined with β hairpins coordinated with ATP hydrolysis, and possesses interprotomer allosteric communication (36). Notably, ATP binding occurs at the subunit interface, and the protomers act with localized cooperativity (37).
In eukaryotes, Mcm2–7 associates with GINS (Sld5, Psf1–3) and Cdc45 to form the replicative helicase assembly CMG (6). Archaeal GINS homologs and Cdc45 orthologs have been identified (30). The majority of archaea possess one GINS subunit, termed Gins23, that is related to Psf2 and Psf3 and a second subunit, termed Gins15 (or Gins51 in some species) that is related to Psf1 and Sld5. The archaeal GINS assembly is thus a dimer of dimers containing two copies each of Gins23 and Gins15 (31). Studies within the last three years have characterized biochemical properties of archaeal CMG and its subassemblies (39, 42, 59), as well as genetic or physical interactions of archaeal Cdc45 or GINS (10, 26, 39, 41, 43, 44, 59).
ARCHAEAL Cdc45 AND GINS ASSOCIATE TO FORM THE STABLE (Cdc45)2•(GINS) COMPLEX
In eukaryotes, the regulated and sequential association of Cdc45 and then that of GINS with origin-loaded MCM are key regulatory steps in the initiation of DNA replication. In the Sulfolobus acidocaldarius system, Gins23 and Gins15 copurify with Cdc45 (59). Further, using Sulfolobus islandicus, it was found that his-tagged Cdc45 forms a stable complex with recombinant Gins15 and Gins23 (59). As determined by both gel filtration analysis and native electrospray ionization mass spectrometry, the reconstituted complex Cdc45:Gins23:Gins15 has a stoichiometric ratio of 2:2:2. The Sulfolobales GINS is a dimer of dimers [(Gins23)2 •(Gins15)2], and thus two Cdc45 proteins with one GINS comprise the archaeal CG complex (59). Similarly and more recently using the Thermoplasma acidophilum system, which has an unusual homotetrameric GINS, recombinant Cdc45 ortholog TaRecJ2 and TaGins51-tetramer were shown to form a complex at a 2:1 ratio (42). The Cdc45-GINS interface has been characterized for both the Sulfolobus solfataricus and Thermococcus kodakarensis complexes. In both cases, the Cdc45 homolog interacts with the C-terminal B-domain of Gins15 (44, 59). The crystal structure of C-terminal Gins15•Cdc45 from T. kodakarensis (Figure 2) highlights the features of the interface: Gins15-C and the N terminus of the DHH domain of Cdc45 bind through hydrophobic and hydrophilic interactions, including the formation of an interprotein β sheet (44). The conserved Gins51-C•Cdc45/RecJ binding interface has also been confirmed in T. acidophilum and in Pyrococcus furiosus (26, 42). Notably, the archaeal CG complex is remarkably stable (59) and characterized by high Cdc45-Gins15 binding affinity (39, 42, 44). Reconstituted S. islandicus CG withstands 8-M urea washes (59). In agreement, surface plasmon resonance studies with recombinant T. kodakarensis Cdc45 (GAN) with GINS or Gins51-C exhibit rapid binding saturation and minimal to no disassociation (40, 44). Similarly, the generated binding curve of recombinant T. acidophilum TaRecJ2 and TaGINS is characteristic of high-affinity binding (42). The conserved nature of a constitutive CG complex in archaea contrasts with the highly regulated and sequential assembly of Cdc45 then GINS with MCM(2–7) in eukaryotes and likely reflects the increased requirement for regulation of DNA replication initiation in eukaryotes.
Figure 2.
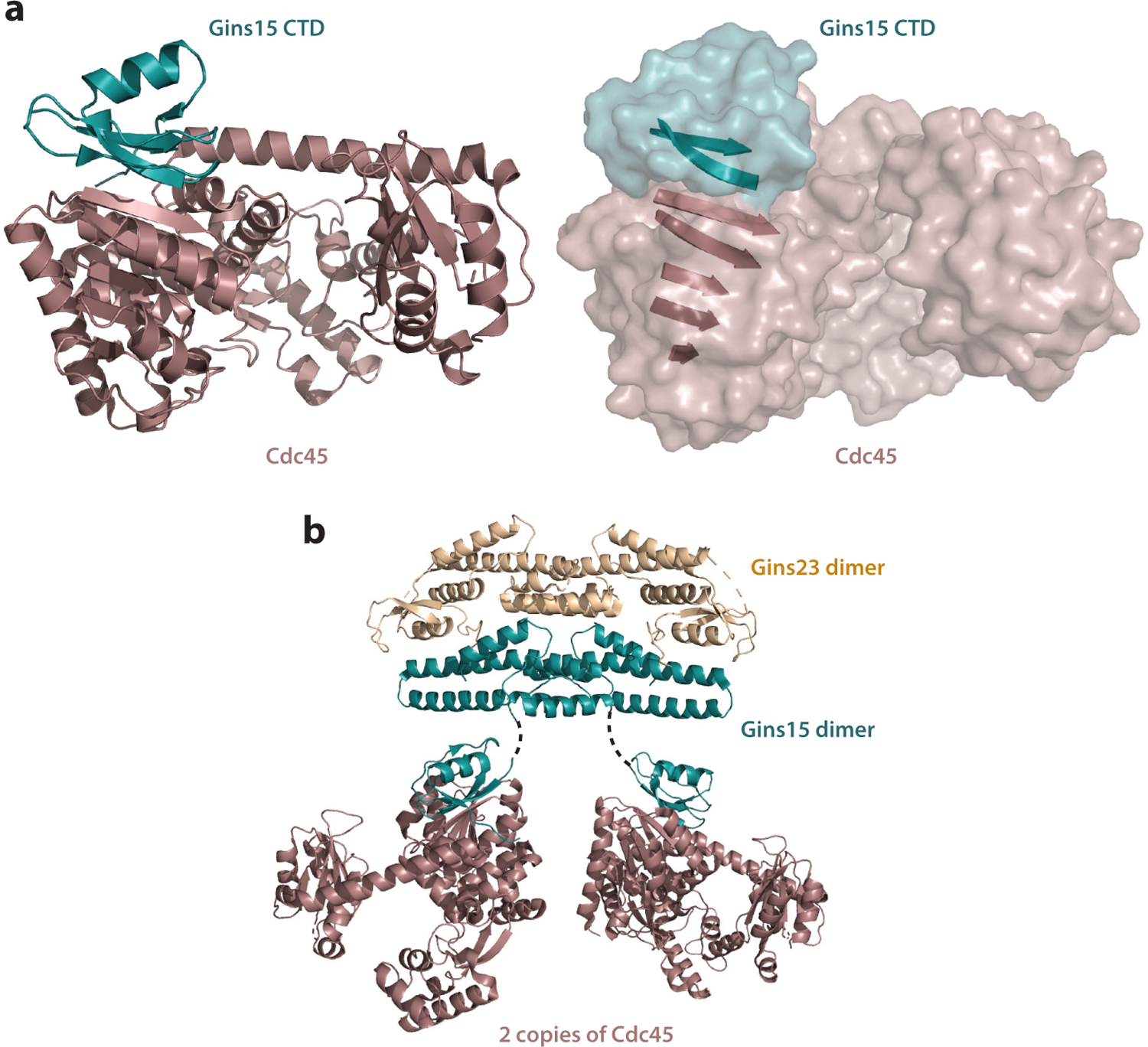
(a) The interaction interface between Gins15 and Cdc45 (aka GAN). The right-hand panel emphasizes the extended β sheet that lies at the heart of the interface. Data from PDB file 5GHS (44). Rendered with PyMOL. (b) Illustrative, but likely inaccurate, model for the interaction of two Cdc45s (GANs) with a GINS complex. The coordinates for the GINS complex were obtained from PDB 3ANW (43). Gins23 is shown in wheat, Gins15 in teal, and Cdc45 (GAN) in brown. The position of Cdc45 (GAN) relative to GINS in solution is currently unknown beyond the primary interaction interface with Gins15’s CTD. Rendered with PyMOL. Abbreviations: CTD, C-terminal domain; PDB, Protein Data Bank.
DIVERGENT ARCHAEA HAVE Cdc45 (GAN) WITH EXONUCLEASE ACTIVITY STIMULATED BY GINS IN VITRO
The structure of Cdc45 (GAN) of T. kodakarensis reveals a conserved DHH architecture suitable for divalent cation coordination and DNA binding, thus suggesting a catalytic DHH domain conferring nuclease activity following the RecJ family orthologs (44). Indeed, the recombinant Cdc45 (GAN) possesses nuclease activity that is stimulated by GINS or Gins51 addition in vitro (44). Specifically, the basal 5′-to-3′ processive exonuclease activity is stimulated 1.5-fold by GINS addition and prefers divalent manganese over magnesium (39). Subsequent structural and biochemical work in P. furiosus established that the GAN ortholog PfuRecJ also has an intact catalytic architecture: In vitro, it also is a 5′-to-3′ exonuclease that prefers manganese and is stimulated by GINS (26). TaRecJ2 of Thermoplasma acidophilum exhibits 3′−5′ exonuclease activity towards both DNA or RNA substrates, requires manganese, and is stimulated by TaGINS addition as determined by recombinant in vitro assays (42). Interestingly, P. furiosus PfuRecJ can also act as a 3′-to-5′ RNA exonuclease in vitro (26). In contrast, the DHH domain of S. solfataricus Cdc45 is degenerate, similar to the situation of eukaryotic Cdc45 (30), thus hinting that archaeal Cdc45 ultimately has a structural role similar to that of the eukaryotic counterpart as opposed to a catalytic RecJ role. Notably, T. kodakarensis gan deletion mutants exhibit a growth defect that is accentuated at higher temperatures, but insertion of catalytically inactive gan into the strain rescues normal cell growth (39). [Note, however, that in other work, a T. kodakarensis gan deletion strain was observed to have wild-type growth (10).] Furthermore, the T. kodakarensis intracellular divalent magnesium and manganese concentration levels are incompatible with GAN exonuclease activity in vitro (39). Thus, the in vivo significance, if any, of the Cdc45/GAN nuclease activity observed under non-physiological conditions in vitro is unclear.
ARCHAEAL CG STIMULATES MCM HELICASE ACTIVITY IN VITRO
Recently, biochemical assays confirmed that archaeal MCM has basal helicase activity (39, 42, 59). CG addition stimulates MCM helicase activity, whereas Cdc45 alone does not in recombinant in vitro assays derived from S. islandicus (59), T. kodakarensis (39), and T. acidophilum (42). CG addition also stimulates the ATPase rate of single-stranded-DNA-bound MCM (39, 59) and increases MCM binding to a Y-shaped oligonucleotide substrate (mimicking the DNA template at the replication fork) (59). The physical CG-MCM interaction is necessary for the stimulation: Truncated MCMΔA maintaining the helicase domain while lacking the A-domain mediating CG interaction preserves basal helicase activity but is not stimulated by CG addition (59). There is some discrepancy in the influence of GINS versus CG on MCM activity among the archaea in vitro. Stimulation of MCM helicase activity by CG addition does not exceed the stimulation by GINS addition alone in biochemical assays of the T. kodakarensis (39) and T. acidophilum (42) systems. In contrast, there is no helicase stimulation at all by GINS addition alone for the S. islandicus system (59). Interestingly, it was found earlier that GINS alone does stimulate MCM for the closely related species S. solfataricus (25). Ultimately as discussed above, the archaeal Cdc45•GINS is an extremely stable assembly in S. islandicus (59), T. kodakarensis (39, 44), and T. acidophilum (42), and thus assembled CG appears to be constitutive and the physiologically relevant species.
ARCHAEAL CG ASSOCIATES WITH MCM TO FORM THE CMG REPLISOME CORE
In the archaeal system, the components Cdc45, MCM, and GINS associate to form the CMG assembly. T. kodakarensis recombinant surface plasmon resonance experiments with Cdc45 immobilized to the chip and sequential addition of GINS and MCM reveal that the three constituents can form a single assembly (39). Further, work in various species confirms that GINS is required to bridge between MCM and Cdc45 (39, 42, 59). There is evidence that Cdc45, MCM, and GINS are bound together in vivo. In whole-cell extracts of exponentially growing T. kodakarensis and T. acidophilum cells, Cdc45 (GAN), MCM, and GINS all coimmunoprecipitate (39, 42). Importantly, there is also evidence supporting the replisome core function of archaeal CMG. Intracellular concentrations of Cdc45, MCM, and GINS in exponentially growing T. kodakarensis are suggestive of them primarily functioning in concert (39). Additionally, chromatin immunoprecipitation in Sulfolobus indicates that both Cdc45 and MCM are enriched at the origins of replication in G1/early S phase, are relatively enriched at interorigin regions during mid–S phase, and are relatively enriched at what has been established as the last replicated region during G2 phase (59). Thus, archaeal Cdc45 was consistently found to chromosomally colocalize with MCM, a hallmark of the replication fork (59).
GENETIC INTERACTIONS IMPLICATE THERMOCOCCUS Cdc45 (GAN) IN DNA PROCESSING PATHWAYS
Genetic studies with archaeal Cdc45 (GAN) reveal DNA-repair-related interactions. In T. kodakarensis, deletion strains reveal synthetic lethality for gan and fen1 deletions (10). Fen1 could be involved in several DNA repair pathways as well as in Okazaki fragment maturation, and thus synthetic lethality suggests redundant roles for GAN and Fen1 in one or more of those pathways (10). Because catalytically inactive gan is also synthetically lethal with the fen1 deletion, the catalytic activity of GAN would be required for these proposed roles. Double deletion of gan and RNase HII also produces synthetic lethality. Oddly enough, the fen1 and RNase HII double-deletion strain is viable (10). However, as mentioned above, the nuclease activity of GAN is not supported at the physiologically relevant divalent cation concentrations found intracellularly (39). Also using T. kodakarensis, Nagata et al. (40) found that the double deletion of gan and han, coding for a nuclease associated with the archaeal ortholog of FANCM identified as Hef, did not grow at temperatures at which the wild type normally would grow. Given this observation and that Hef-HAN function in the repair of stalled replication forks, it is therefore postulated that there is increased replication fork stalling at higher temperatures and that GAN contributes to replication fork stability in T. kodakarensis (40).
THE EVOLUTIONARY CONTEXT AND FUNCTION OF ARCHAEAL CMG
In eukaryotes, it is well established that the CMG assembly contains a single copy of the Cdc45 protein. In contrast, as detailed above, archaeal CG and, by inference, CMG have two copies of the archaeal Cdc45 homolog. This stoichiometry arises from the observation that archaeal GINS is typically a dimer of dimers and thus has two Cdc45-binding domains (59). Significantly, it has been proposed that eukaryotic Cdc45 resulted from gene duplication and subsequent fusion of the ancestral archaeal Cdc45 resulting in a pseudodimer (59).
While the identity and existence of archaeal CMG as the active form of the replicative helicase have now been established, open questions remain about how the assembly of the complex is effected in vivo. In particular, whether the assembly and thus activation of the CMG helicase is a key regulatory step in archaea as in eukaryotes remains to be determined (6).
INITIATING DNA SYNTHESIS—THE ROLE OF THE ARCHAEAL PRIMASE
The steps that mediate and regulate the melting of DNA at archaeal DNA replication origins are not yet determined at the biochemical level. However, it is clear that the initiation of synthesis ultimately depends on the recruitment of the replication-dedicated DNA-dependent RNA polymerase, DNA primase. The archaeal replicative primase is a dedicated, template-dependent, low-processivity RNA polymerase that synthesizes an oligoribonucleotide primer that is then transferred to a DNA polymerase for extension. In principle, primase activity is required once for leading strand synthesis but once per ∼100–200 nucleotides on the lagging strand for initiation of every Okazaki fragment (34). In Sulfolobus, primase has been shown to interact with the GINS complex, thus providing a mechanism to coordinate priming activity with processive replication fork progression (31). The archaeal primase has two subunits that are conserved across the archaeal domain of life—PriS and PriL. PriS contains the active site of the enzyme. Further, PriS and PriL have orthologs in eukaryotes that form the primase module of the DNA polymerase α−primase complex. Thus, PriSL is sometimes referred to as the archaeal eukaryotic primase (AEP). Interestingly, AEP relatives are also found in a range of extrachromosomal elements in both bacteria and archaea as well as in the dedicated eukaryotic enzyme termed PrimPol that is thought to play a role in DNA damage tolerance (for recent reviews see 4, 22). The PriSL heterodimeric assembly has been biochemically characterized in a number of archaeal species and shown to possess both RNA and DNA de novo synthetic capability. With regard to the latter activity, studies of the euryarchaeal Archaeoglobus and Pyrococcus primases have demonstrated that they possess the ability to synthesize past a range of lesions in DNA, suggesting that they may play moonlighting roles in DNA repair, in addition to having the canonical primase function (21, 24).
STRUCTURAL STUDIES OF PRIMASE
Initial studies of the Sulfolobus PriSL heterodimer demonstrated weak RNA and DNA synthesis capability. A key study by Li Huang and colleagues revealed that the native Sulfolobus primase possessed a hitherto unidentified third subunit that they termed PriX (27). Importantly, addition of PriX to PriSL formed a stable heterotrimer with massively elevated activity compared to PriSL. Initial structural studies demonstrated that PriX formed a helical bundle and also established a structural relationship between PriX and a domain of eukaryotic PriL that was missing from the Sulfolobus PriL homolog (27). Thus, the combination of PriX and Sulfolobus PriL corresponds to the full-length eukaryotic PriL protein. Significantly, this helical bundle region of human PriL had been demonstrated to interact with the 5′ end of the RNA primer in a primer-template complex (2). Additionally, mutation of a conserved arginine residue in this domain abrogated the ability of eukaryotic primase to initiate synthesis but did not impact its ability to elongate a preformed primer (23). Additional insight into the role of this conserved helical domain came with the determination of the structure of the Sulfolobus PriSLX assembly in the presence of the ATP analog AMPCPP (19). One AMPCPP molecule was bound to the elongation site within PriS, but a second AMPCPP was found to be coordinated by residues within the PriX helical bundle (Figure 3a). Importantly, nucleotide binding by PriX was effected by interactions between the 5′ triphosphate of the nucleotide and residues in PriX that include an arginine that occupies the position orthologous to that of the eukaryotic helical bundle arginine, mutation of which abrogated initiation but not elongation. Analogous effects on initiation specifically, and not elongation, were observed upon mutation of the PriX residues involved in triphosphate coordination (19). Thus, PriX and, by inference, the helical bundle domain of eukaryotic PriL serve as the binding site for the initiating nucleotide in primer synthesis. More recently, a similar mechanism has been proposed for an extrachromosomal element-encoded AEP, ORF904 of the archaeal plasmid pRN1 (8). In both pRN1 primase and the cellular Sulfolobus PriSLX, the connection between the main body of the enzyme and the helical bundle initiation site appears quite flexible (8, 19). This flexibility may be important for both initiation, during which the initiation site and the catalytic elongation site are presumably juxtaposed, and elongation. The exclusive interactions of the helical bundle with the 5′ triphosphate have important implications for how primer length is determined (61). The 5′ triphosphate serves as a unique signifier of the 5′ end of the RNA primer, and if the helical bundle remains associated with the 5′ end as synthesis is ongoing, then the primase could serve as a caliper to measure the distance, and thus chain length, between the initiation and elongation sites (Figure 3b). A series of experiments with model substrates and mutant forms of the primase provided evidence that supports this caliper model (61). The caliper could function simply by measuring length, but it also could be influenced by the helical geometry of the heteroduplex as the RNA chain is elongated. Structural studies on elongation complexes with defined primer lengths will be of profound importance in illuminating this model.
Figure 3.
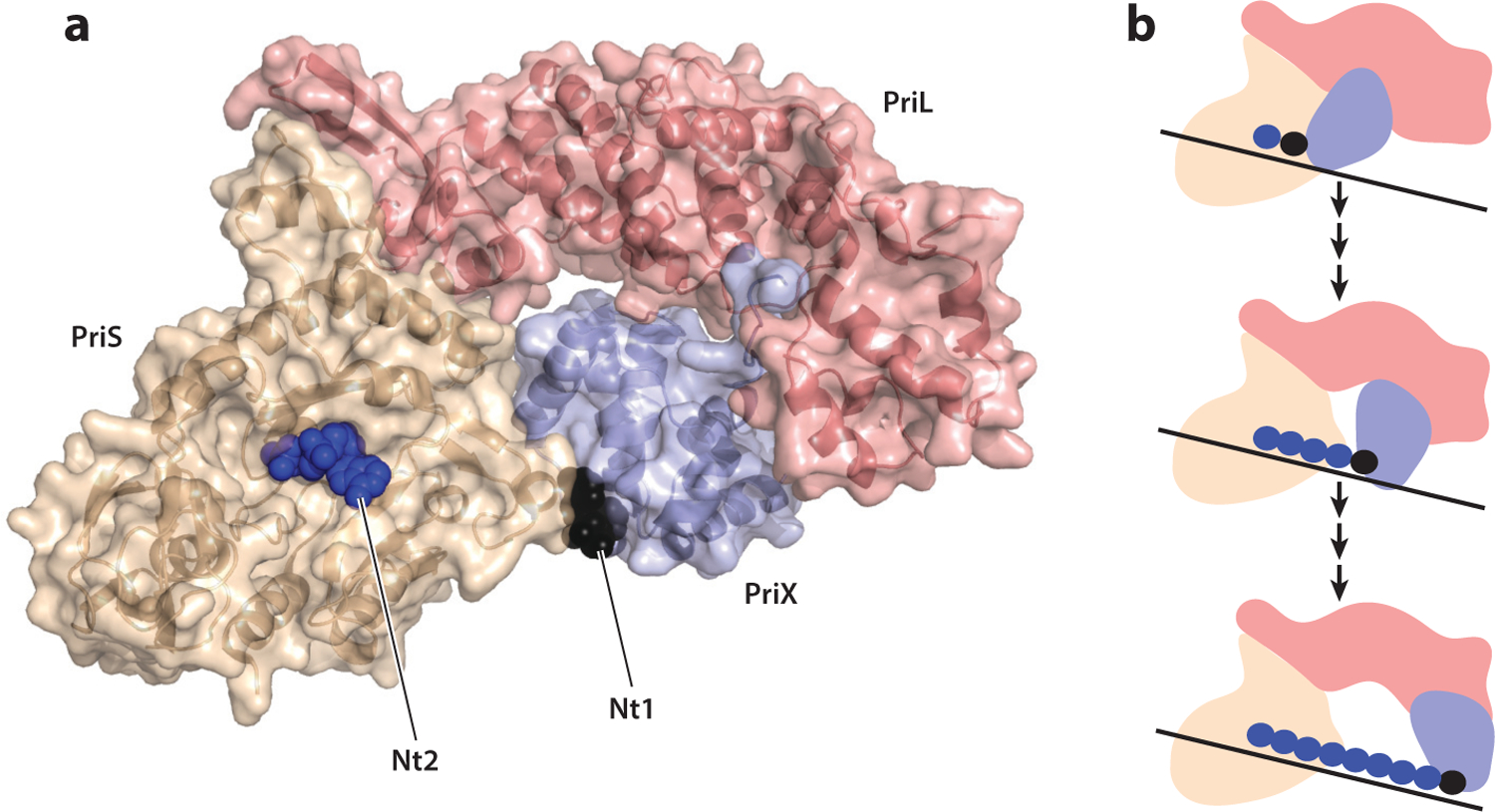
(a) Structure of Sulfolobus PriSLX with the initiating nucleotide (Nt1) shown in black and the nucleotide in the catalytic elongation site (Nt2) in blue. Prepared from Protein Data Bank file 5OF3. Rendered with PyMOL. (b) Schematic of the caliper model for primer length determination. As in panel a, PriS is shown in wheat, PriL in salmon, and PriX in blue. Juxtaposition of initiation and elongation sites allows dinucleotide formation. Subsequent nucleotide monophosphates are incorporated at the elongation site while the initiation site retains a grip on the 5′ end of the primer. Eventually, a maximal length of primer is reached, dictated by the maximal distance tolerated between initiation and elongation sites. This constraint could be imposed by distance and/or helical rotation of the heteroduplex.
ARCHAEAL DNA POLYMERASES
In the late 1990s, Cann and Ishino made the startling discovery that Euryarchaea possess a novel DNA polymerase, termed PolD (11, 20). PolD was shown to be phylogenetically distinct from all known cellular DNA polymerases and to be the founding member of a new family of DNA polymerases, the D-family. Subsequently, D-family polymerases have been shown to be present in all archaeal phyla with the exception of the crenarchaea. PolD is a two-subunit enzyme, and genetic studies have shown the genes to be essential for viability in a number of archaeal species. Notably, many PolD-containing archaea also possess a distinct DNA polymerase belonging to the B-family. The cellular replicases of eukaryotic genomes are centered around three distinct B-family enzymes: Polα, Polδ, and Polε. However, in the PolD-containing archaea, PolB enzymes have generally been found to be dispensable for viability, suggesting that PolD is the true replicative DNA polymerase and the B-family enzyme plays an ancillary role, perhaps in DNA repair (for a more detailed discussion see 48).
As is found in other replicative polymerases, PolD has both polymerase and proofreading exonuclease activities. These properties are attributable to distinct subunits—DP1 confers the proofreading ability, while polymerization is effected by DP2. Structural studies have revealed that DP1 belongs to the calcineurin-like phosphodiesterase family, and its catalytically active fold is unique to the PolD family of replicative polymerases (48, 55). However, the overall structure of DP1 is closely related to the accessory B-subunit of the eukaryotic B-family DNA polymerases. Intriguingly, however, the eukaryotic B-subunit proteins have lost the catalytic residues associated with exonuclease activity, and the eukaryotic B-subunit proteins lack any catalytic activity. Instead, proofreading by eukaryotic Polδ and Polε is conferred by a DnaQ-like domain in the same large subunit that contains the DNA polymerase catalytic subunit. An even more striking finding was that the structure of the DP2 DNA polymerase catalytic subunit is unique to the PolD family of enzymes and in fact resembles the catalytic center of RNA polymerases (RNAPs) (55). Other cellular DNA polymerases possess a classical right-hand morphology with the catalytic center residing in the so-called palm domain. The palm domain can be split into two subtypes based on the topology of the active-site fold. They can be either Klenow-type, as found in the A-, B-, and Y-family DNA polymerases, or PolB-like, as in the X- and C-family polymerases (9). Uniquely, PolD possesses a double-psi beta-barrel fold that is clearly related to the catalytic center of multisubunit cellular RNAPs, RNAPs involved in RNA silencing, and viral RNAPs (55). Thus, it appears that, in the evolutionary gulf between archaea and eukaryotes, the eukaryotic B-subunit may have evolved from an archaeal-like DP1-subunit and, in doing so, lost the catalytic activity of the archaeal proofreading site while retaining the overall fold of the subunit (Figure 4). The situation with the catalytic subunit is more complex. PolD’s DP2 differs from the eukaryotic Polδ and Polε in both the fold of the catalytic center and the presence of a catalytic-subunit-associated proofreading nuclease activity in the eukaryotic enzymes (48). It is open to speculation whether the D-family double-psi barrel fold or the B-family Klenow-type fold was present in the last common ancestor between these two domains. Regardless of the evolutionary pathway leading to its generation, the available evidence, with regard to both genetic essentiality and the documented interactions with a number of components of the euryarchaeal replication machinery, strongly implicates PolD as a key component of the archaeal replisome.
Figure 4.
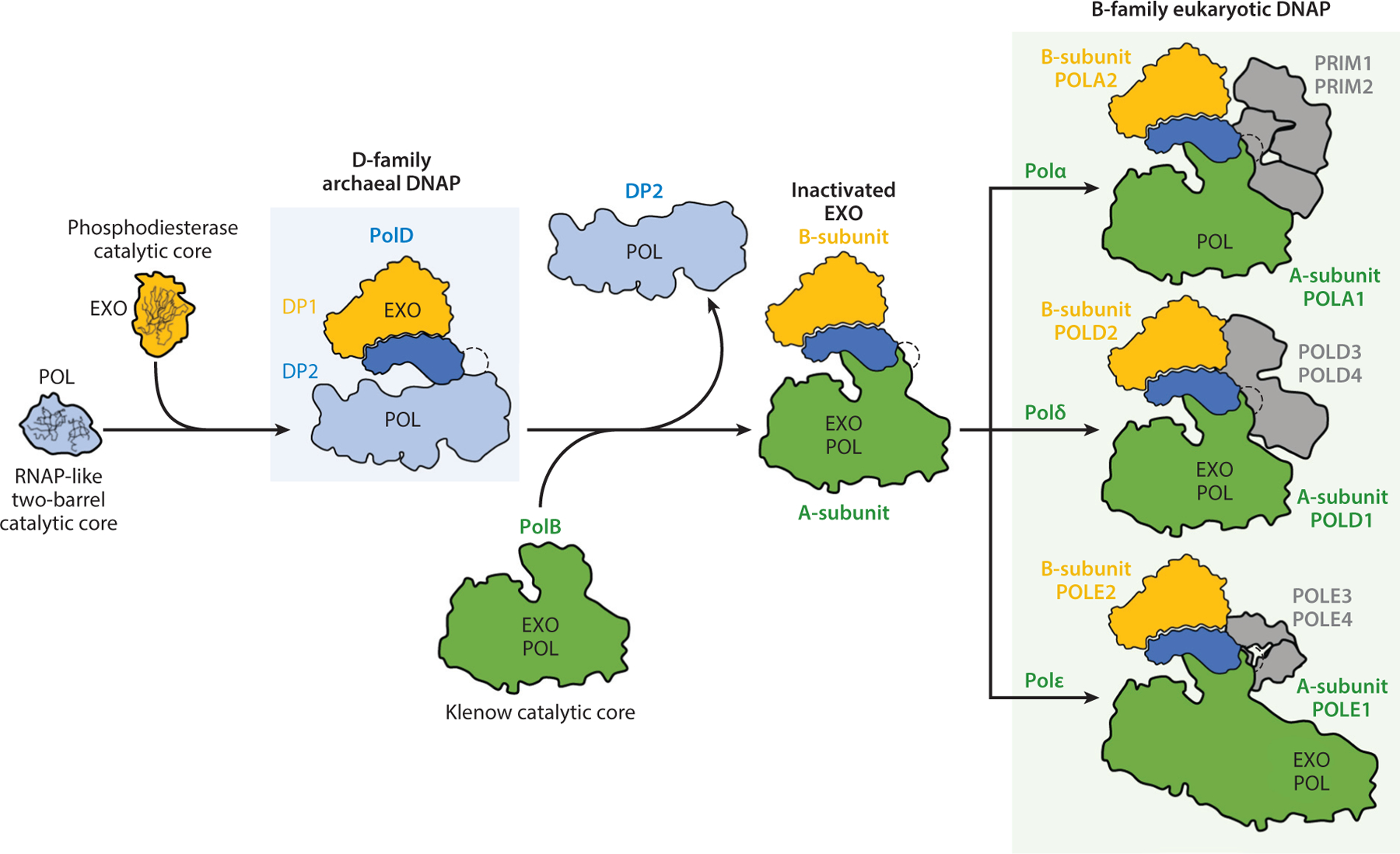
Schema for the evolution of PolD and eukaryotic DNA polymerases α, δ, and ε. Abbreviations: DNAP, DNA polymerase; EXO, exonuclease domain; POL, DNA polymerase domain; RNAP, RNA polymerase. Adapted from Reference 48 (CC-BY-4.0).
However, as mentioned above, the D-family DNA polymerases are absent from the crenarchaeal phylum. Typically, crenarchaea encode multiple B-family polymerases. For example, Sulfolobus species encode three, termed PolB1, PolB2, and PolB3. A recent study revealed that neither PolB2 nor PolB3 was required for viability in Sulfolobus—this agrees with their rather modest sequence conservation, their weak in vitro activities, and the presence of amino acid substitutions in otherwise highly conserved regions of the polymerase (32). It seems likely that PolB2 and PolB3 play roles in DNA damage tolerance pathways. In contrast, the gene for PolB1 is essential. Although PolB1 was initially identified in the 1980s and studied by many laboratories as a recombinant single-subunit enzyme, recent work has revealed that two small subunits, now termed PBP1 and PBP2, were overlooked in the initial characterizations (60). PolB1 was demonstrated to exist in cells as a PolB1•PBP1•PBP2 heterotrimeric holoenzyme assembly, PolB1-HE. Reconstitution experiments revealed that PolB1-HE was more thermostable than the catalytic subunit alone. Further, PBP1 played a key role in mitigating PolB1’s strand-displacement activity by enhancing polymerase recycling. A consequence of this property of PBP1 was to improve the efficiency of Okazaki fragment maturation by minimizing wasteful resynthesis of DNA during the Okazaki-fragment-processing pathway. Whether the small subunits play roles in facilitating the integration of PolB1-HE into higher-order replisome assemblies is currently under investigation.
CONCLUSIONS
Over the last two decades, numerous laboratories have contributed to our understanding of the structures and mechanisms of action of components of the archaeal DNA replication machinery. Importantly, the field has moved beyond simple characterizations of homologs of known eukaryotic replication factors and, through application of gene-tagging and affinity purification methodologies, has revealed the identity of novel archaea-specific components of the replication apparatus. With the recent development of in vitro assays for MCM loading at replication origins and reconstitution of partial replisome assemblies, it is anticipated that a fully reconstituted archaeal DNA replication system will be defined in the near future. Clear goals are to understand the higher-order architectures of replisome assemblies and to determine the mechanisms that ensure appropriate regulation of archaeal chromosome replication in the context of the cell cycle.
ACKNOWLEDGMENTS
S.D.B.’s laboratory is supported by grants R01GM135178 and R01GM125579 from the National Institutes of Health and by the College of Arts and Sciences, Indiana University.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Ausiannikava D, Mitchell L, Marriott H, Smith V, Hawkins M, et al. 2018. Evolution of genome architecture in archaea: spontaneous generation of a new chromosome in Haloferax volcanii. Mol. Biol. Evol 35:1855–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranovskiy AG, Zhang Y, Suwa Y, Gu J, Babayeva ND, et al. 2016. Insight into the human DNA primase interaction with template-primer. J. Biol. Chem 291:4793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell SD. 2017. Initiation of DNA replication in the archaea. Adv. Exp. Med. Biol 1042:99–115 [DOI] [PubMed] [Google Scholar]
- 4.Bell SD. 2019. Initiating DNA replication: a matter of prime importance. Biochem. Soc. Trans 47:351–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell SD, Botchan MR. 2013. The minichromosome maintenance replicative helicase. Cold Spring Harb. Perspect. Biol 5:a012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell SP, Labib K. 2016. Chromosome duplication in Saccharomyces cerevisiae. Genetics 203:1027–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernander R, Poplawski A. 1997. Cell cycle characteristics of thermophilic archaea. J. Bacteriol 179:4963–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boudet J, Devillier JC, Wiegand T, Salmon L, Meier BH, et al. 2019. A small helical bundle prepares primer synthesis by binding two nucleotides that enhance sequence-specific recognition of the DNA template. Cell 176:154–66.e13 [DOI] [PubMed] [Google Scholar]
- 9.Braithwaite DK, Ito J. 1993. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res 21:787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burkhart BW, Cubonova L, Heider MR, Kelman Z, Reeve JN, Santangelo TJ. 2017. The GAN exonuclease or the flap endonuclease Fen1 and RNase HII are necessary for viability of Thermococcus kodakarensis. J. Bacteriol 199(13):e00141–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cann IK, Komori K, Toh H, Kanai S, Ishino Y. 1998. A heterodimeric DNA polymerase: evidence that members of Euryarchaeota possess a distinct DNA polymerase. PNAS 95:14250–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas ME, Ali FA, Costa A, Diffley JFX. 2018. The mechanism of eukaryotic CMG helicase activation. Nature 555:265–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dueber EL, Corn JE, Bell SD, Berger JM. 2007. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317:1210–13 [DOI] [PubMed] [Google Scholar]
- 14.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. 2007. Structural basis of DNA replication origin recognition by an ORC protein. Science 317:1213–16 [DOI] [PubMed] [Google Scholar]
- 15.Gehring AM, Astling DP, Matsumi R, Burkhart BW, Kelman Z, et al. 2017. Genome replication in Thermococcus kodakarensis independent of Cdc6 and an origin of replication. Front. Microbiol 8:2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgescu R, Yuan Z, Bai L, Santos RDLA, Sun J, et al. 2017. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. PNAS 114:E697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez-Llorente Y, Fletcher RJ, Chen XS, Carazo JM, San Martin C. 2005. Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J. Biol. Chem 280:40909–15 [DOI] [PubMed] [Google Scholar]
- 18.Hawkins M, Malla S, Blythe MJ, Nieduszynski CA, Allers T. 2013. Accelerated growth in the absence of DNA replication origins. Nature 503:544–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holzer S, Yan J, Kilkenny ML, Bell SD, Pellegrini L. 2017. Primer synthesis by a eukaryotic-like archaeal primase is independent of its Fe-S cluster. Nat. Commun 8:1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishino Y, Komori K, Cann IK, Koga Y. 1998. A novel DNA polymerase family found in Archaea. J. Bacteriol 180:2232–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jozwiakowski SK, Borazjani Gholami F, Doherty AJ. 2015. Archaeal replicative primases can perform translesion DNA synthesis. PNAS 112:E633–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazlauskas D, Sezonov G, Charpin N, Venclovas C, Forterre P, Krupovic M. 2018. Novel families of archaeo-eukaryotic primases associated with mobile genetic elements of bacteria and archaea. J. Mol. Biol 430:737–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilkenny ML, Longo MA, Perera RL, Pellegrini L. 2013. Structures of human primase reveal design of nucleotide elongation site and mode of Pol α tethering. PNAS 110:15961–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Killelea T, Palud A, Akcha F, Lemor M, L’haridon S, et al. 2019. The interplay at the replisome mitigates the impact of oxidative damage on the genetic integrity of hyperthermophilic Archaea. eLife 8:e45320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang S, Huang L. 2015. The Sulfolobus solfataricus GINS complex stimulates DNA binding and processive DNA unwinding by minichromosome maintenance helicase. J. Bacteriol 197:3409–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li MJ, Yi GS, Yu F, Zhou H, Chen JN, et al. 2017. The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45. Nucleic Acids Res 45:12551–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B, Ouyang S, Makarova KS, Xia Q, Zhu Y, et al. 2015. A primase subunit essential for efficient primer synthesis by an archaeal eukaryotic-type primase. Nat. Commun 6:7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Smith CL, DeRyckere D, DeAngelis K, Martin GS, Berger JM. 2000. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell 6:637–48 [DOI] [PubMed] [Google Scholar]
- 29.Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. 2004. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. PNAS 101:7046–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makarova KS, Koonin EV. 2013. Archaeology of eukaryotic DNA replication. Cold Spring Harb. Perspect. Biol 5:a012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, Bell SD. 2006. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep 7:539–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Alvarez L, Deng L, Peng X. 2017. Formation of a viral replication focus in Sulfolobus cells infected by the rudivirus Sulfolobus islandicus rod-shaped virus 2. J. Virol 91:e00486–17. Erratum. 2018. J. Virol. 92:e01991–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsunaga F, Forterre P, Ishino Y, Myllykallio H. 2001. In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. PNAS 98:11152–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsunaga F, Norais C, Forterre P, Myllykallio H. 2003. Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep 4:154–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. 2005. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol 12:756–62 [DOI] [PubMed] [Google Scholar]
- 36.Miller JM, Enemark EJ. 2015. Archaeal MCM proteins as an analog for the eukaryotic Mcm2–7 helicase to reveal essential features of structure and function. Archaea 2015:305497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. 2007. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol. Cell 28:304–14 [DOI] [PubMed] [Google Scholar]
- 38.Myllykallio H, Lopez P, Lopez-Garcia P, Heilig R, Saurin W, et al. 2000. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science 288:2212–15 [DOI] [PubMed] [Google Scholar]
- 39.Nagata M, Ishino S, Yamagami T, Ogino H, Simons JR, et al. 2017. The Cdc45/RecJ-like protein forms a complex with GINS and MCM, and is important for DNA replication in Thermococcus kodakarensis. Nucleic Acids Res 45:10693–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagata M, Ishino S, Yamagami T, Simons JR, Kanai T, et al. 2017. Possible function of the second RecJ-like protein in stalled replication fork repair by interacting with Hef. Sci. Rep 7:16949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Donnell ME, Li H. 2018. The ring-shaped hexameric helicases that function at DNA replication forks. Nat. Struct. Mol. Biol 25:122–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogino H, Ishino S, Kohda D, Ishino Y. 2017. The RecJ2 protein in the thermophilic archaeon Thermoplasma acidophilum is a 3′–5′ exonuclease that associates with a DNA replication complex. J. Biol. Chem 292:7921–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oyama T, Ishino S, Fujino S, Ogino H, Shirai T, et al. 2011. Architectures of archaeal GINS complexes, essential DNA replication initiation factors. BMC Biol 9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oyama T, Ishino S, Shirai T, Yamagami T, Nagata M, et al. 2016. Atomic structure of an archaeal GAN suggests its dual roles as an exonuclease in DNA repair and a CMG component in DNA replication. Nucleic Acids Res 44:9505–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelve EA, Lindas AC, Knoppel A, Mira A, Bernander R. 2012. Four chromosome replication origins in the archaeon Pyrobaculum calidifontis. Mol. Microbiol 85:986–95 [DOI] [PubMed] [Google Scholar]
- 46.Perera HM, Trakselis MA. 2019. Amidst multiple binding orientations on fork DNA, Saccharolobus MCM helicase proceeds N-first for unwinding. eLife 8:e46096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poplawski A, Bernander R. 1997. Nucleoid structure and distribution in thermophilic Archaea. J. Bacteriol 179:7625–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raia P, Carroni M, Henry E, Pehau-Arnaudet G, Brule S, et al. 2019. Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases. PLOS Biol 17:e3000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson NP, Blood KA, McCallum SA, Edwards PA, Bell SD. 2007. Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO J 26:816–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD. 2004. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell 116:25–38 [DOI] [PubMed] [Google Scholar]
- 51.Rothenberg E, Trakselis MA, Bell SD, Ha T. 2007. MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J. Biol. Chem 282:34229–34 [DOI] [PubMed] [Google Scholar]
- 52.Sakakibara N, Kelman LM, Kelman Z. 2009. Unwinding the structure and function of the archaeal MCM helicase. Mol. Microbiol 72:286–96 [DOI] [PubMed] [Google Scholar]
- 53.Samson RY, Abeyrathne PD, Bell SD. 2016. Mechanism of archaeal MCM helicase recruitment to DNA replication origins. Mol. Cell 61:287–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samson RY, Xu Y, Gadelha C, Stone TA, Faqiri JN, et al. 2013. Specificity and function of archaeal DNA replication initiator proteins. Cell Rep 3:485–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauguet L, Raia P, Henneke G, Delarue M. 2016. Shared active site architecture between archaeal PolD and multi-subunit RNA polymerases revealed by X-ray crystallography. Nat. Commun 7:12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soppa J 2011. Ploidy and gene conversion in Archaea. Biochem. Soc. Trans 39:150–54 [DOI] [PubMed] [Google Scholar]
- 57.Takemata N, Samson RY, Bell SD. 2019. Physical and functional compartmentalization of archaeal chromosomes. Cell 179:165–79.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tiengwe C, Marques CA, McCulloch R. 2014. Nuclear DNA replication initiation in kinetoplastid parasites: new insights into an ancient process. Trends Parasitol 30:27–36 [DOI] [PubMed] [Google Scholar]
- 59.Xu Y, Gristwood T, Hodgson B, Trinidad JC, Albers SV, Bell SD. 2016. Archaeal orthologs of Cdc45 and GINS form a stable complex that stimulates the helicase activity of MCM. PNAS 113:13390–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan J, Beattie TR, Rojas AL, Schermerhorn K, Gristwood T, et al. 2017. Identification and characterization of a heterotrimeric archaeal DNA polymerase holoenzyme. Nat. Commun 8:15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan J, Holzer S, Pellegrini L, Bell SD. 2018. An archaeal primase functions as a nanoscale caliper to define primer length. PNAS 115:6697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang H, Wu Z, Liu J, Liu X, Wang L, et al. 2015. Activation of a dormant replication origin is essential for Haloferax mediterranei lacking the primary origins. Nat. Commun 6:8321. [DOI] [PMC free article] [PubMed] [Google Scholar]


