Abstract
Deformable image registration between Computed Tomography (CT) images and Magnetic Resonance (MR) imaging is essential for many image-guided therapies. In this paper, we propose a novel translation-based unsupervised deformable image registration method. Distinct from other translation-based methods that attempt to convert the multimodal problem (e.g., CT-to-MR) into a unimodal problem (e.g., MR-to-MR) via image-to-image translation, our method leverages the deformation fields estimated from both: (i) the translated MR image and (ii) the original CT image in a dual-stream fashion, and automatically learns how to fuse them to achieve better registration performance. The multimodal registration network can be effectively trained by computationally efficient similarity metrics without any ground-truth deformation. Our method has been evaluated on two clinical datasets and demonstrates promising results compared to state-of-the-art traditional and learning-based methods.
Keywords: Multimodal Registration, Generative Adversarial Network, Unsupervised Learning
1. Introduction
Deformable multimodal image registration has become essential for many procedures in image-guided therapies, e.g., preoperative planning, intervention, and diagnosis. Due to substantial improvement in computational efficiency over traditional iterative registration approaches, learning-based registration approaches are becoming more prominent in time-intensive applications.
Related work.
Many learning-based registration approaches adopt fully supervised or semi-supervised strategies. Their networks are trained with ground-truth deformation fields or segmentation masks [5, 19, 13, 12, 16], and may struggle with limited or imperfect data labeling. A number of unsupervised registration approaches have been proposed to overcome this problem by training unlabeled data to minimize traditional similarity metrics, e.g., mean squared intensity differences [4, 26, 15, 11, 21, 17]. However, the performances of these methods are inherently limited by the choice of similarity metrics. Given the limited selection of multimodal similarity metrics, unsupervised registration approaches may have difficulties outperforming traditional multimodal registration methods as they both essentially optimize the same cost functions. A recent trend for multimodal image registration takes advantage of the latent feature disentanglement [18] and image-to-image translation [6, 20, 23]. Specifically, translation-based approaches use Generative Adversarial Network (GAN) to translate images from one modality into the other modality, thus are able to convert the difficult multimodal registration into a simpler unimodal task. However, being a challenging topic by itself, image translation may inevitably produce artificial anatomical features that can further interfere with the registration process.
In this work, we propose a novel translation-based fully unsupervised multimodal image registration approach. In the context of Computed Tomography (CT) image to Magnetic Resonance (MR) image registration, previous translation-based approaches would translate a CT image into an MR-like image (tMR), and use tMR-to-MR registration to estimate the final deformation field ϕ. In our approach, the network estimates two deformation fields, namely ϕs of tMR-to-MR and ϕo of CT-to-MR, in a dual-stream fashion. The addition of the original ϕo enables the network to implicitly regularize ϕs to mitigate certain image translation problems, e.g., artificial features. The network further automatically learns how to fuse ϕs and ϕo towards achieving the best registration accuracy.
Contributions and advantages of our work can be summarized as follows:
Our method leverages the deformation fields estimated from the original multimodal stream and synthetic unimodal stream to overcome the shortcomings of translation-based registration;
We improve the fidelity of organ boundaries in the translated MR by adding two extra constraints in the image-to-image translation model Cycle-GAN.
We evaluate our method on two clinically acquired datasets. It outperforms state-of-the-art traditional, unsupervised and translation-based registration approaches.
2. Methods
In this work, we propose a general learning framework for robustly registering CT images to MR images in a fully unsupervised manner.
First, given a moving CT image and a fixed MR image, our improved Cycle-GAN module translates the CT image into an MR-like image. Then, our dual-stream subnetworks, UNet_o and UNet_s, estimate two deformation fields ϕo and ϕs respectively, and the final deformation field is fused via a proposed fusion module. Finally, the moving CT image is warped via Spatial Transformation Network (STN) [14], while the entire registration network aims to maximize the similarity between the moved and the fixed images. The pipeline of our method is shown in Fig. 1.
Fig. 1.
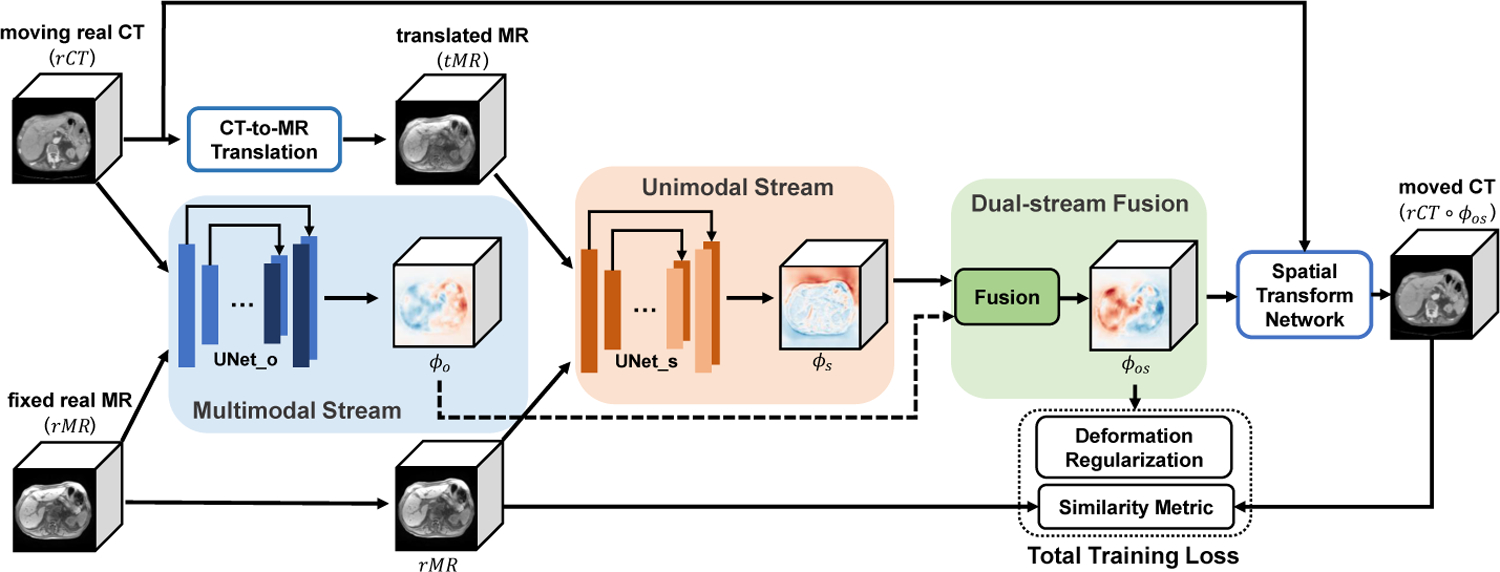
Illustration of the proposed method. The entire unsupervised network is mainly guided by the image similarity between rCT ○ ϕos and rMR.
2.1. Image-to-Image Translation with Unpaired Data
The CT-to-MR translation step consists of an improved Cycle-GAN with additional structural and identical constraints. As a state-of-the-art image-to-image translation model, Cycle-GAN [28] can be trained without pairwise aligned CT and MR datasets of the same patient. Thus, Cycle-GAN is widely used in medical image translation [25, 1, 9].
Our Cycle-GAN model is illustrated in Fig. 2. The model consists of two generators GMR and GCT, which can provide CT-to-MR and MR-to-CT translation respectively. Besides, it has two discriminators DCT and DMR. DCT is used to distinguish between translated CT(tCT) and real CT(rCT), and DMR is for translated MR(tMR) and real MR(rMR). The training loss of original Cycle-GAN only adopts two types of items: adversarial loss given by two discriminators ( and ) and cycle-consistency loss to prevent generators from generating images that are not related to the inputs (refer to [28] for details).
Fig. 2.
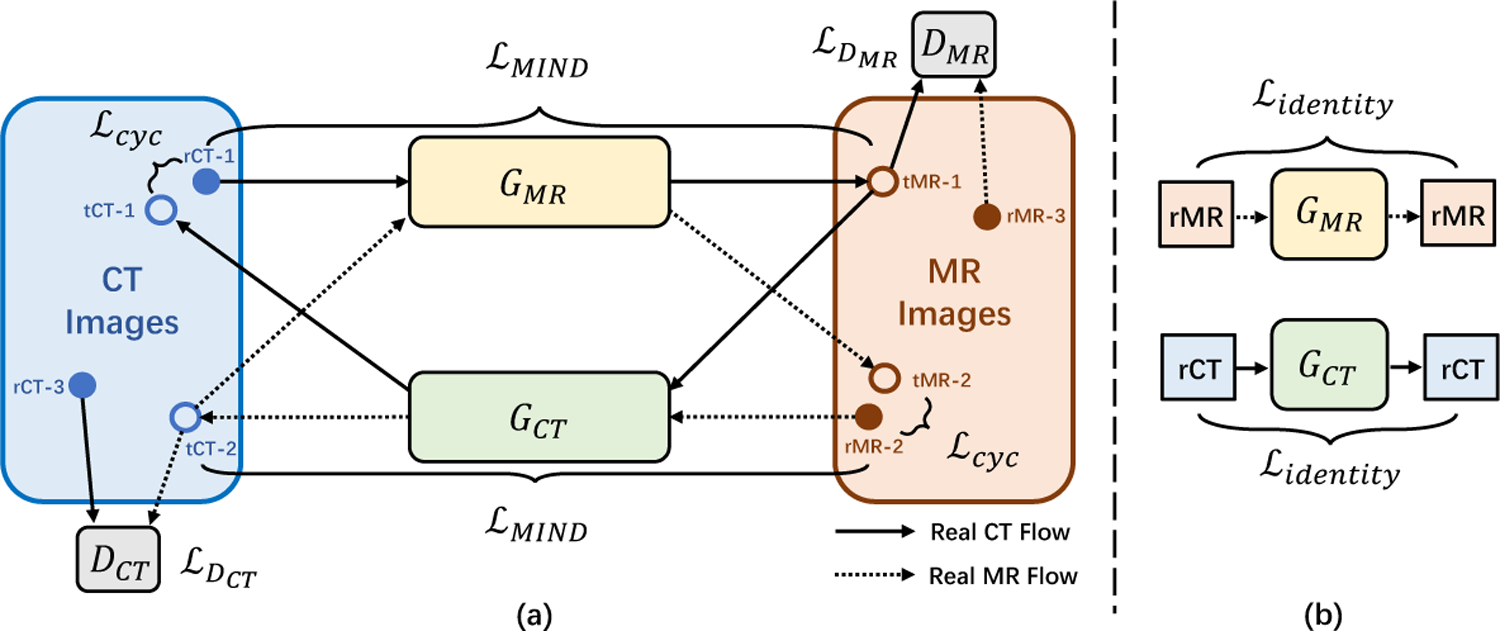
Schematic illustration of Cycle-GAN with strict constraints. (a) The workflow of the forward and backward translation; (b) The workflow of identity loss.
However, training a Cycle-GAN on medical images is difficult since the cycle-consistency loss is not enough to enforce structural similarity between translated images and real images (as shown in the red box in Fig. 3(b)). Therefore, we introduce two additional losses, structure-consistency loss and identity loss , to constrain the training of Cycle-GAN.
Fig. 3.
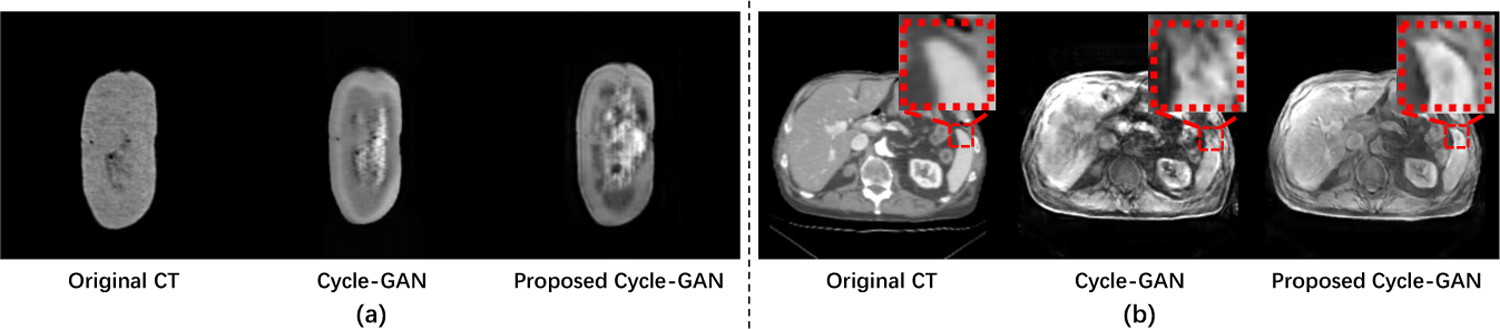
CT-to-MR translation examples of original Cycle-GAN and proposed Cycle-GAN tested for (a) pig ex-vivo kidney dataset and (b) abdomen dataset.
MIND (Modality Independent Neighbourhood Descriptor) [8] is a feature that describes the local structure around each voxel. Thus, we minimize the difference in MIND between translated images GCT (IrMR) or GMR(IrCT ) and real images IrMR or IrCT to enforce the structural similarity. We define as follows:
| (1) |
where M represents MIND features, NMR and NCT denote the number of voxels in IrMR and IrCT, and R is a non-local region around voxel x.
The identity loss (as shown in Fig. 2(b)) is included to prevent images already in the expected domain from being incorrectly translated to the other domain. We define it as:
| (2) |
Finally, the total loss of our proposed Cycle-GAN is defined as:
| (3) |
where λcyc, λidentity and λMIND denotes the relative importance of each term.
2.2. Dual-stream Multimodal Image Registration Network
As shown in Fig. 3, although our improved Cycle-GAN can better translate CT images into MR-like images, the CT-to-MR translation is still challenging for translating “simple” CT images to “complex” MR images. Most image-to-image translation methods will inevitably generate unrealistic soft-tissue details, resulting in some mismatch problems. Therefore, the registration methods that simply convert multimodal to unimodal registration via image translation algorithm are not reliable.
In order to address this problem, we propose a dual-stream network to fully use the information of the moving, fixed and translated images as shown in Fig. 1. In particular, we can use effective similarity metrics to train our multimodal registration model without any ground-truth deformation.
Network Details.
As shown in Fig. 1, our dual-stream network is comprised of four parts: multimodal stream subnetwork, unimodal stream subnetwork, deformation field fusion, and Spatial Transformation Network.
In Multimodal Stream subnetwork, original CT(rCT) and MR(rMR) are represented as the moving and fixed images, which allows the model to propagate original information to counteract mismatch problems in translated MR(tMR).
Through image translation, we obtain the translated MR(tMR) with similar appearance to the fixed MR(rMR). Then, in Unimodal Stream, tMR and rMR are used as moving and fixed images respectively. This stream can effectively propagate more texture information, and constrain the final deformation field to suppress unrealistic voxel drifts from the multimodal stream.
During the network training, the two streams constrain each other, while they are also cooperating to optimize the entire network. Thus, our novel dual-stream design allows us to benefit from both original image information and homogeneous structural information in the translated images.
Specifically, UNet_o and UNet_s adopt the same UNet architecture used in VoxelMorph [4] (shown in Fig. 4). The only difference is that UNet_o is with multimodal inputs but UNet_s is with unimodal inputs. Each UNet takes a single 2-channel 3D image formed by concatenating Im and If as input, and outputs a volume of deformation field with 3 channels.
Fig. 4.

Detailed architecture of UNet-based subnetwork. The encoder uses convolution with stride of 2 to reduce spatial resolution, while the decoder uses 3D upsampling layers to restore the spatial resolution.
After Uni- and Multi-model Stream networks, we obtain two deformation fields, ϕo (for rCT and rMR) and ϕs (for tMR and rMR). We stack ϕo and ϕs, and apply a 3D convolution with size of 3×3×3 to estimate the final deformation field ϕos, which is a 3D volume with the same shape of ϕo and ϕs.
To evaluate the dissimilarity between moved and fixed images, we integrate spatial transformation network (STN) [14] to warp the moving image using ϕos. The loss function consists of two components as shown in Eq. (4).
| (4) |
where λ is a regularization weight. The first loss is similarity loss, which is to penalize the differences in appearance between fixed and moved images. Here we adopt SSIM [22] for experiments. Suggested by [4], deformation regularization adopts a L2-norm of the gradients of the final deformation field ϕos.
3. Experiments and Results
Dataset and Preprocessing.
We focus on the application of abdominal CT-to-MR registration. We evaluated our method on two proprietary datasets since there is no designated public repository.
Pig Ex-vivo Kidney CT-MR Dataset. This dataset contains 18 pairs of CT and MRI kidney scans from pigs. All kidneys are manually segmented by experts. After preprocessing the data, e.g., resampling and affine spatial normalization, we cropped the data to 144 × 80 × 256 with 1mm isotropic voxels and arbitrarily divided it into two groups for training (15 cases) and testing (3 cases).
Abdomen (ABD) CT-MR Dataset. This 50-patient dataset of CT-MR scans was collected from a local hospital and annotated with anatomical landmarks. All data were preprocessed into 176 × 176 × 128 with the same resolution (1mm3) and were randomly divided into two groups for training (45 cases) and testing (5 cases).
Implementation.
We trained our model using the following settings: (1) The Cycle-GAN for CT-MR translation network is based on the existing implementation [27] with changes as discussed in Section 2.1. (2) The Uni- and Multi-modal stream registration networks were implemented using Keras with the Tensorflow backend and trained on an NVIDIA Titan X (Pascal) GPU.
3.1. Results for CT-to-MR Translation
We extracted 1792 and 5248 slices from the transverse planes of the Pig kidney and ABD dataset respectively to train the image translation network. Parameters λcyc, λidentity and λMIND were set to 10, 5, and 5 for training.
Since our registration method is for 3D volumes, we apply the pre-trained CT-to-MR generator to translate moving CT images into MR-like images slice-by-slice and concatenate 2D slices into 3D volumes. The qualitative results are visualized in Fig. 3. In addition, to quantitatively evaluate the translation performance, we apply our registration method to obtain aligned CT-MR pairs and utilize SSIM [22] and PSNR [10] to judge the quality of translated MR (shown in Table 1). In our experiment, our method predicts better MR-like images on both datasets.
Table 1.
Quantitative results for image translation.
| Pig Kidney | Method | PSNR | SSIM | ABD | Method | PSNR | SSIM |
| Cycle-GAN | 32.07 | 0.9025 | Cycle-GAN | 22.95 | 0.7367 | ||
| Ours | 32.74 | 0.9532 | Ours | 23.55 | 0.7455 |
3.2. Registration Results
Affine registration is used as the baseline method. For traditional method, only mutual information (MI) based SyN [2] is compared since it is the only metric (available in ANTs [3]) for multimodal registration. In addition to SyN, we implemented the following learning-based methods: 1) VM_MIND and VM_SSIM which extends VoxelMorph with similarity metrics MIND [8] and SSIM [22]. 2) M2U which is a typical translation-based registration method. It generates tMR from CT and converts the multimodal problem to tMR-to-MR registration. It’s noteworthy that the parameters of all methods are optimized to the best results on both datasets.
Two examples of the registration results are visualized in Fig. 5, where the red and yellow contours represent the ground truth and registered organ boundaries respectively. As shown in Fig. 5, the organ boundaries aligned by the traditional SyN method have a considerable amount of disagreement. Among all learning-based methods, our method has the most visually appealing boundary alignment for both cases. VM_SSIM performed significantly worse for the kidney. VM_MIND achieved accurate registration for the kidney, but its result for the ABD case is significantly worse. Meanwhile, M2U suffers from artificial features in the image translation, which leads to an inaccurate registration result.
Fig. 5.

Visualization results of our model compared to other methods. Upper: Pig Kidney. Bottom: Abdomen(ABD). The red contours represent the ground truth organ boundary while the yellow contours are the warped contours of segmentation masks.
The quantitative results are presented in Table 2. We compare different methods by the Dice score [7] and target registration error (TRE) [24]. We also provide the average run-time for each method. As shown in Table 2, our method consistently outperformed other methods and was able to register a pair of images in less than 2 seconds (when using GPU).
Table 2.
Quantitative results for Pig Kidney Dataset and Abdomen (ABD) Dataset.
| Metric | Organ | Affine | SyN | M2U | VM_SSIM | VM_MIND | Ours | |
|---|---|---|---|---|---|---|---|---|
| Dice (%) | Pig | kidney | 89.53 | 89.87 | 90.21 | 93.75 | 96.48 | 98.57 |
| ABD | kidney | 80.03 | 82.36 | 78.96 | 82.21 | 84.58 | 85.66 | |
| spleen | 79.58 | 80.38 | 77.76 | 81.79 | 83.11 | 87.01 | ||
| liver | 78.74 | 79.13 | 78.83 | 82.05 | 81.98 | 83.34 | ||
| TRE (mm) | ABD | spleen | 4.16 | 4.20 | 3.76 | 3.58 | 3.65 | 2.47 |
| liver | 6.55 | 5.61 | 5.91 | 4.72 | 4.87 | 3.64 | ||
| Time(s) GPU/CPU | Pig | -/103 | -/121 | 1.08/20 | 1.06/20 | 1.07/20 | 1.12/21 | |
| ABD | -/108 | -/137 | 1.23/24 | 1.22/22 | 1.21/23 | 1.27/24 | ||
3.3. The effect of each deformation field
In order to validate the effectiveness of the deformation field fusion, we compare ϕs, ϕo and ϕos together with warped images (shown in Fig. 6). The qualitative result shows that ϕs from the unimodal stream alleviates the voxel drift effect from the multimodal stream. While ϕo from the multimodal stream uses the original image textures to maintain the fidelity and reduce artificial features for the generated tMR image. The fused deformation field ϕos produces better alignment than both streams alone, which demonstrates the effectiveness of the joint learning step.
Fig. 6.
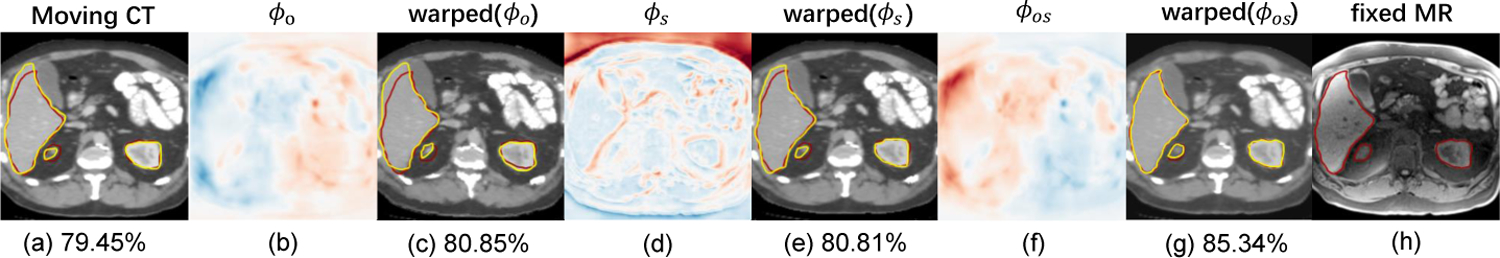
Visualizations of the deformation field fusion. (a) moving image; (h) fixed image; (b/d/f) deformation fields; (c/e/g) images warped by (b/d/f), corresponding average Dice scores (%) of all organs are calculated. The contours in red represent ground truth, while yellow shows the warped segmentation mask.
4. Conclusion
We proposed a fully unsupervised uni- and multi-modal stream network for CT-to-MR registration. Our method leverages both CT-translated-MR and original CT images towards achieving the best registration result. Besides, the registration network can be effectively trained by computationally efficient similarity metrics without any ground-truth deformation. We evaluated the method on two clinical datasets, and it outperformed state-of-the-art methods in terms of accuracy and efficiency.
Acknowledgement
This project was supported by the National Institutes of Health (Grant No. R01EB025964, R01DK119269, and P41EB015898) and the Overseas Cooperation Research Fund of Tsinghua Shenzhen International Graduate School (Grant No. HW201808).
References
- 1.Armanious K, Jiang C, Abdulatif S, Küstner T, Gatidis S, Yang B: Unsupervised medical image translation using cycle-medgan. In: 2019 27th European Signal Processing Conference (EUSIPCO). pp. 1–5. IEEE; (2019) [Google Scholar]
- 2.Avants BB, Epstein CL, Grossman M, Gee JC: Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis 12 1, 26–41 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC: A reproducible evaluation of ants similarity metric performance in brain image registration. NeuroImage 54, 2033–2044 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV: An unsupervised learning model for deformable medical image registration. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 9252–9260 (2018) [Google Scholar]
- 5.Cao X, Yang J, Zhang J, Nie D, Kim M, Wang Q, Shen D: Deformable image registration based on similarity-steered cnn regression. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 300–308. Springer; (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao X, Yang J, Wang L, Xue Z, Wang Q, Shen D: Deep learning based inter-modality image registration supervised by intra-modality similarity. In: International Workshop on Machine Learning in Medical Imaging. pp. 55–63. Springer; (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dice LR: Measures of the amount of ecologic association between species. Ecology 26(3), 297–302 (1945) [Google Scholar]
- 8.Heinrich MP, Jenkinson M, Bhushan M, Matin T, Gleeson FV, Brady M, Schnabel JA: Mind: Modality independent neighbourhood descriptor for multimodal deformable registration. Medical image analysis 16(7), 1423–1435 (2012) [DOI] [PubMed] [Google Scholar]
- 9.Hiasa Y, Otake Y, Takao M, Matsuoka T, Takashima K, Carass A, Prince JL, Sugano N, Sato Y: Cross-modality image synthesis from unpaired data using cyclegan. In: International workshop on simulation and synthesis in medical imaging. pp. 31–41. Springer; (2018) [Google Scholar]
- 10.Hore A, Ziou D: Image quality metrics: Psnr vs. ssim. In: 2010 20th International Conference on Pattern Recognition. pp. 2366–2369. IEEE; (2010) [Google Scholar]
- 11.Hu X, Kang M, Huang W, Scott MR, Wiest R, Reyes M: Dual-stream pyramid registration network. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 382–390. Springer; (2019) [Google Scholar]
- 12.Hu Y, Modat M, Gibson E, Ghavami N, Bonmati E, Moore CM, Emberton M, Noble JA, Barratt DC, Vercauteren T: Label-driven weakly-supervised learning for multimodal deformable image registration. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). pp. 1070–1074. IEEE; (2018) [Google Scholar]
- 13.Hu Y, Modat M, Gibson E, Li W, Ghavami N, Bonmati E, Wang G, Bandula S, Moore CM, Emberton M, et al. : Weakly-supervised convolutional neural networks for multimodal image registration. Medical image analysis 49, 1–13 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaderberg M, Simonyan K, Zisserman A, et al. : Spatial transformer networks. In: Advances in neural information processing systems. pp. 2017–2025 (2015) [Google Scholar]
- 15.Kuang D, Schmah T: Faim–a convnet method for unsupervised 3d medical image registration. In: International Workshop on Machine Learning in Medical Imaging. pp. 646–654. Springer; (2019) [Google Scholar]
- 16.Liu C, Ma L, Lu Z, Jin X, Xu J: Multimodal medical image registration via common representations learning and differentiable geometric constraints. Electronics Letters 55(6), 316–318 (2019) [Google Scholar]
- 17.Mahapatra D, Ge Z, Sedai S, Chakravorty R: Joint registration and segmentation of xray images using generative adversarial networks. In: International Workshop on Machine Learning in Medical Imaging. pp. 73–80. Springer; (2018) [Google Scholar]
- 18.Qin C, Shi B, Liao R, Mansi T, Rueckert D, Kamen A: Unsupervised deformable registration for multi-modal images via disentangled representations. In: International Conference on Information Processing in Medical Imaging. pp. 249–261. Springer; (2019) [Google Scholar]
- 19.Sedghi A, Luo J, Mehrtash A, Pieper S, Tempany CM, Kapur T, Mousavi P, Wells III WM: Semi-supervised image registration using deep learning. In: Medical Imaging 2019: Image-Guided Procedures, Robotic Interventions, and Modeling. vol. 10951, p. 109511G International Society for Optics and Photonics; (2019) [Google Scholar]
- 20.Tanner C, Ozdemir F, Profanter R, Vishnevsky V, Konukoglu E, Goksel O: Generative adversarial networks for mr-ct deformable image registration. arXiv preprint arXiv:1807.07349 (2018) [Google Scholar]
- 21.de Vos BD, Berendsen FF, Viergever MA, Sokooti H, Staring M, Išgum I : A deep learning framework for unsupervised affine and deformable image registration. Medical image analysis 52, 128–143 (2019) [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP: Image quality assessment: from error visibility to structural similarity. IEEE transactions on image processing 13(4), 600–612 (2004) [DOI] [PubMed] [Google Scholar]
- 23.Wei D, Ahmad S, Huo J, Peng W, Ge Y, Xue Z, Yap PT, Li W, Shen D, Wang Q: Synthesis and inpainting-based mr-ct registration for image-guided thermal ablation of liver tumors. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 512–520. Springer; (2019) [Google Scholar]
- 24.West JB, Fitzpatrick JM, Wang MY, Dawant BM, Maurer CR, Kessler RM, Maciunas RJ, Barillot C, Lemoine D, Collignon AM, et al. : Comparison and evaluation of retrospective intermodality image registration techniques. In: Medical Imaging 1996: Image Processing. vol. 2710, pp. 332–347. International Society for Optics and Photonics; (1996) [Google Scholar]
- 25.Yang H, Sun J, Carass A, Zhao C, Lee J, Xu Z, Prince J: Unpaired brain mr-to-ct synthesis using a structure-constrained cyclegan. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, pp. 174–182. Springer; (2018) [Google Scholar]
- 26.Zhao S, Lau T, Luo J, Eric I, Chang C, Xu Y: Unsupervised 3d end-to-end medical image registration with volume tweening network. IEEE journal of biomedical and health informatics (2019) [DOI] [PubMed] [Google Scholar]
- 27.Zhu JY, Park T, Isola P, Efros AA: Cyclegan (2017), https://github.com/xhujoy/CycleGAN-tensorflow
- 28.Zhu JY, Park T, Isola P, Efros AA: Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE international conference on computer vision. pp. 2223–2232 (2017) [Google Scholar]


