Abstract
Chronic recalcitrant dermatophytoses, due to Trichophyton (T.) mentagrophytes Type VIII are on the rise in India and are noteworthy for their predominance. It would not be wrong to assume that travel and migration would be responsible for the spread of T. mentagrophytes Type VIII from India, with many strains resistant to terbinafine, to other parts of the world. From September 2016 until March 2020, a total of 29 strains of T. mentagrophytes Type VIII (India) were isolated. All patients were residents of Germany: 12 females, 15 males and the gender of the remaining two was not assignable. Patients originated from India (11), Pakistan (two), Bangladesh (one), Iraq (two), Bahrain (one), Libya (one) and other unspecified countries (10). At least two patients were German-born residents. Most samples (21) were collected in 2019 and 2020. All 29 T. mentagrophytes isolates were sequenced (internal transcribed spacer (ITS) and translation elongation factor 1-α gene (TEF1-α)). All were identified as genotype VIII (India) of T. mentagrophytes. In vitro resistance testing revealed 13/29 strains (45%) to be terbinafine-resistant with minimum inhibitory concentration (MIC) breakpoints ≥0.2 µg/mL. The remaining 16 strains (55%) were terbinafine-sensitive. Point mutation analysis revealed that 10/13 resistant strains exhibited Phe397Leu amino acid substitution of squalene epoxidase (SQLE), indicative for in vitro resistance to terbinafine. Two resistant strains showed combined Phe397Leu and Ala448Thr amino acid substitutions, and one strain a single Leu393Phe amino acid substitution. Out of 16 terbinafine-sensitive strains, in eight Ala448Thr, and in one Ala448Thr +, new Val444 Ile amino acid substitutions were detected. Resistance to both itraconazole and voriconazole was observed in three out of 13 analyzed strains. Treatment included topical ciclopirox olamine plus topical miconazole or sertaconazole. Oral itraconazole 200 mg twice daily for four to eight weeks was found to be adequate. Terbinafine-resistant T. mentagrophytes Type VIII are being increasingly isolated. In Germany, transmission of T. mentagrophytes Type VIII from the Indian subcontinent to Europe should be viewed as a significant public health issue.
Keywords: dermatophytoses, terbinafine-resistant, squalene epoxidase, point mutation, transmission, Itraconazole, Ciclopirox, Miconazole
1. Introduction
There is a veritable epidemic of varieties of chronic recalcitrant dermatophytoses due to Trichophyton (T.) mentagrophytes Type VIII in India [1]. A wide variation in clinical features is seen. Tinea corporis, tinea cruris, tinea faciei and their combinations are the most common presentations. Lesions often show a minimal to a high degree of inflammation, and large lesions with a tendency to coalesce and spread are common. Severe itching is common [2]. There has been an undeniable association between the occurrence of extensive and hard to treat tinea and long-term abuse of potent and super-potent topical corticosteroids, predominantly clobetasol propionate [1,3].
Extensive travel and migration are considered vital in the spread of dermatophytoses. Especially the terbinafine-resistant strains of T. mentagrophytes Type VIII, detected as the causative genotype, are now increasingly isolated in Germany and other European countries. The main criterion to identify this particular infection is the very noticeable treatment failure with topical and oral terbinafine.
2. Patients and Methods
2.1. Patients
Patients discussed herein were predominantly German residents with chronic dermatophytoses who had been failing treatment with terbinafine and, therefore, were suspected to harbour T. mentagrophytes Type VIII. They were subjected to mycological diagnostics including Blancophor® preparation, fungal culture and molecular biological fungal DNA detection. Skin scrapings taken from suspicious skin sites were investigated. Additionally, a few fungal cultures isolated in other laboratories were sent to our center for precise identification of the fungal species and internal transcribed spacer (ITS) genotype has been included in the epidemiological investigation (Table 1).
Table 1.
Overview of the 29 patients with dermatomycoses due to T. mentagrophytes of (ITS) genotype VIII (India) diagnosed all over Germany from 2016–2020. Abbreviations: SQLE, squalene epoxidase; ITS, internal transcribed spacer; TEF1-α, translation elongation factor 1-α; MIC, minimum inhibitory concentration; NCBI, National Center for Biotechnology Information, Bethesda, Maryland; DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen (German Collection of Microorganisms and Cell Cultures), Braunschweig, Germany; f, female; m, male.
| Strain Number | Collection No. DSMZ | Gender | Age in Years/ Months |
Pathology | Additional Information/ Remarks |
MIC Terbinafine [µg/mL] | Drug Resistance to Terbinafine | Amino Acid Substitution within the Squalene Epoxidase | Codon Change in SQLE | MIC itraconazole [µg/mL] | Drug Resistance to Itraconazole | MIC Voriconazole [µg/mL] | Drug Resistance to Voriconazole | GenBank Accession Number ITS Gene | GenBank Accession Number TEF1-α Gene (NCBI) | GenBank Accession Number SQLE Gene (NCBI) |
Sample Date | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 214677/16 | DSM 108903 | f | 35 | Tinea pedis | Indian | 0.2 | resistant | Phe397Leu | TTC → CTC | 0.06 | sensitive | 0.03 | sensitive | MT330252 | MT340499 | MT700499 | 2016 |
| 2 | 216377/17 | DSM 108902 | f | 29 | Tinea | Foreigners | 0.2 | resistant | Phe397Leu | TTC → CTC | 0.06 | sensitive | 0.03 | sensitive | MT330249 | MT340500 | MT700500 | 2017 |
| 3 | 211542/18 | DSM 108901 | m | 19 | Tinea corporis | Asylum office Herford | 8 | resistant | Phe397Leu | TTC → CTC | 0.087 | sensitive | 0.088 | sensitive | MT330250 | MT340501 | MT700501 | 2018 |
| 4 | 211564/18 | DSM 108900 | f | 30 | Tinea unguium et corporis | Indian | <0.2 | sensitive | Ala448Thr | GCT → ACT | 0.067 | sensitive | 0.088 | sensitive | MT330248 | MT340502 | MT700502 | 2018 |
| 5 | 218160/18 | DSM 108899 | m | 35 | Tinea corporis et cruris | Indian | <0.2 | sensitive | wild type | - | 0.125 | sensitive | 0.03 | sensitive | MT330253 | MT340503 | MT700503 | 2018 |
| 6 | 218360/18 | DSM 108898 | f | 25 | Tinea corporis | married couple on a pilgrimage in Saudi Arabia | <0.2 | sensitive | Ala448Thr | GCT → ACT | 0.5 | resistant | 0.5 | resistant | MT330255 | MT340504 | MT700504 | 2018 |
| 7 | 218691/18 | DSM 108897 | m | 58 | Tinea faciei | Visit India | 8 | resistant | Phe397Leu | TTC → CTC | 0.125 | sensitive | 0.03 | sensitive | MT330254 | MT340505 | MT700505 | 2018 |
| 8 | 901538/18 | DSM 108896 | m | 4 | Tinea corporis | German name | <0.2 | sensitive | Ala448Thr | GCT → ACT | 0.25 | sensitiv | 0.25 | resistant | MT330251 | MT340506 | MT700506 | 2018 |
| 9 | 200618/19 | DSM 109024 | f | 6 months | Tinea corporis | Family came from Bahrain to visit Germany | 0.2 | resistant | Phe397Leu | TTC → TTA | not applied | not applied | MT330285 | MT345568 | MT700507 | 2019 | ||
| 10 | 202953/19 | DSM 109747 | m | 27 | Tinea | Libyan, for three years in Germany, German girlfriend also affected | 8 | resistant | Phe397Leu | TTC→CTC | 0.5 | resistant | 0.25 | resistant | MT330284 | MT340507 | MT700508 | 2019 |
| 11 | 203513/19 | DSM 109748 | f | 25 | Tinea cruris et inguinalis | Pakistani | <0.2 | sensitive | wild type | - | 0.0312 | sensitive | 0.0312 | sensitive | MT330280 | MT340508 | MT700509 | 2019 |
| 12 | 600174/19 | DSM 109749 | f | 34 | Tinea corporis | Indian, was in India when the skin symptoms started | <0.2 | sensitive | Ala448Thr | GCT → ACT | 0.125 | sensitive | 0.0312 | sensitive | MT330279 | MT340517 | MT700510 | 2019 |
| 13 | 205666/19 | DSM 109751 | m | 32 | Tinea cruris | Iraqi couple (see no. 14) | <0.2 | sensitive | Ala448Thr | GCT → ACT | 0.125 | sensitive | 0.125 | sensitive | MT330281 | MT340516 | MT700511 | 2019 |
| 14 | 205667/19 | DSM 109750 | f | 31 | Tinea corporis | Iraqi couple (see no. 13) | <0.2 | sensitive | Ala448Thr | GCT → ACT | 0.5 | resistant | 0.5 | resistant | MT330283 | MT340515 | MT700512 | 2019 |
| 15 | 208737/19 | DSM 110678 | f | 29 | Tinea corporis | Indian name | 8 | resistant | Phe397Leu | TTC → TTA | not applied | - | not applied | - | MT328783 | MT340514 | MT700513 | 2019 |
| 16 | 209934/19 | DSM 110677 | m | 28 | Tinea corporis gluteal | Indian name | <0.2 | sensitive | Ala448Thr | GCT → ACT | not applied | - | not applied | - | MT330278 | MT340513 | MT700514 | 2019 |
| 17 | 600231/19 | DSM 110676 | m | 40 | Tinea corporis | Family, large plaques | 16 | resistant | Leu393Phe | TTA → TTC | not applied | - | not applied | - | MT330282 | MT340512 | MT700515 | 2019 |
| 18 | 214174/19 | DSM 110675 | m | 37 | Tinea faciei and corporis: cheeks, neck, forehead, and ear helix | German | 8 | resistant | Phe397Leu Ala448Thr |
TTC → CTC GCT → ACT | not applied | - | not applied | - | MT330289 | MT340511 | MT700516 | 2019 |
| 19 | 216532/19 | DSM 110674 | m | 27 | Tinea corporis | Indian | <0.2 | sensitive | Ala448Thr | GCT → ACT | not applied | - | not applied | - | MT330288 | MT340510 | MT700517 | 2019 |
| 20 | 217201/19 | DSM 110673 | m | 24 | Tinea corporis | Indian | <0.2 | sensitive | Val444 Ile Ala448Thr |
GTA → ATAGCT → ACT | not applied | - | not applied | - | MT330286 | MT340509 | MT700518 | 2019 |
| 21 | 218676/19 | - | m | 20 | Tinea corporis | Foreigner | <0.2 | sensitive | wild type | - | not applied | - | not applied | - | MT330256 | MT340518 | MT700519 | 2019 |
| 22 | 219238/19 | - | f | 27 | Tinea corporis | Pakistani | <0.2 | sensitive | wild type | - | not applied | - | not applied | - | MT330290 | MT340519 | MT700520 | 2019 |
| 23 | 600380/19 | - | - | Tinea | Indian | <0.2 | sensitive | wild type | - | not applied | - | not applied | - | MT330291 | MT340520 | MT700521 | 2019 | |
| 24 | 220575/19 | - | f | 24 | Tinea corporis gluteal | Student from Bangladesh | <0.2 | sensitive | wild type | - | not applied | - | not applied | - | MT330287 | MT340521 | MT700522 | 2019 |
| 25 | 600002/20 | - | m | Tinea cruris | German patient with stay in Thailand and India | 16 | resistant | Phe397Leu | TTC → CTC | not applied | - | not applied | - | MT333227 | MT340522 | MT700523 | 2020 | |
| 26 | 101549/20 | - | m | 30 | Tinea | Foreigner | 16 | resistant | Phe397Leu Ala448Thr |
TTC → CTCGCT → ACT | not applied | - | not applied | - | MT333225 | MT340523 | MT700524 | 2020 |
| 27 | 200874/20 | - | m | 27 | Tinea corporis et manum | Foreigner | <0.2 | sensitive | wild type | not applied | - | not applied | - | MT333228 | MT345569 | MT700525 | 2020 | |
| 28 | 900138/20 | - | m | 51 | Tinea corporis | Foreigner | 16 | resistant | Phe397Leu | TTC → CTC | not applied | - | not applied | - | MT333226 | MT340524 | MT700526 | 2020 |
| 29 | 204532/20 | - | f | 23 | Tinea corporis | Originating and migrating for studying from Bangladesh, her father also affected. She was pretreated in her home country by oral voriconazole, without success. | 0.2 | resistant | Phe397Leu | TTC → CTC | not applied | - | not applied | - | MT333242 | MT340525 | MT700527 | 2020 |
2.2. Conventional Cultural Diagnostics
In mycological routine diagnostics, scrapings from the active edges of centrifugally spreading lesions of the free skin, as well as hair roots from lesions of the capillitium in some patients, were cultured on Sabouraud´s 4% dextrose agar (Sifin, Berlin, Germany) and, additionally, on cycloheximide (Actidione®-containing Sabouraud´s dextrose agar, Becton Dickinson, Heidelberg, Germany). Fungal isolates showing fast-growing, flat radiating fungal colonies with a white periphery, and sometimes bright yellowish centre typical for T. mentagrophytes, were further analyzed. Microscopic lactophenol cotton blue preparations were performed from such colonies.
2.3. Molecular Biological Characteristics of Trichophyton mentagrophytes ITS Type VIII
2.3.1. PCR-ELISA for Molecular Identification of Dermatophytes
DNA from either skin scrapings or fungal isolates (for identification of submitted fungal cultures) was extracted according to the manufacturer´s protocol using the QIAamp® DNA Mini Kit (Qiagen, Hilden, Germany). Samples were analyzed using a validated and standardized in-house developed enzyme linked immunoassay (PCR-ELISA) to detect dermatophyte DNA [4,5]. Specific probes detecting the following relevant dermatophytes were used: T. rubrum, T. interdigitale/T. mentagrophytes, Microsporum canis, and T. benhamiae (formerly referred to as T. anamorph or Arthroderma benhamiae).
All DNA samples extracted either from skin scrapings or from fungal cultures were positive in the PCR-ELISA for T. interdigitale/T. mentagrophytes. As the differentiation between T. interdigitale and the T. mentagrophytes complex was not possible by PCR-ELISA, the ITS regions of rDNA genes and the translation elongation factor (TEF)1-α gene were sequenced.
2.3.2. Sequencing of the ITS Regions of rDNA Genes for Species Identification of Trichophyton mentagrophytes Type VIII
For confirmation of the suspected dermatophyte species, Sanger sequencing of the ITS regions of rDNA genes (mainly the regions ITS 1, 5.8 S rRNA, ITS 2) and TEF1-α gene was performed for all isolates [6,7,8,9]. This required PCR amplification of a ∼900 bp DNA fragment using universal primers that bind to flanking pan-fungal sequence regions: V9G (5’-TTACGTCCCTGCCCTTTGTA-3’) and LS266 (5’-GCATTCCCAAACAACTCGACTC-3’).
The length of the analyezd region in the TEF1-α gene varied from 709 to 769 nucleotides among the various dermatophyte species. Primers EF1a-F (5’-CACATTAACTTGGTCGTTATCG-3’) and EF1a-R (5’-CATCCTTGGAGATACCAGC-3’) were used for sequencing [6].
2.3.3. Phylogenetic Analysis of Trichophyton mentagrophytes Type VIII
Both reference strains and clinically isolated wild type strains were used for comparative molecular analysis, and the generation of the phylogenetic tree based on the ITS region and the TEF 1α gene is listed in Table 2. In addition, GenBank numbers of all sequences used for generating phylogenetic trees are provided in Table 2.
Table 2.
Reference strains and clinical isolates used to generate the phylogenetic tree based on sequencing of the ITS region of rDNA and the TEF1-α gene. GenBank accession numbers of the nucleotide sequences used in this study are available at the NCBI. Abbreviations: NCBI, National Center for Biotechnology Information, Bethesda, Maryland; ITS, internal transcribed spacer; rDNA, ribosomal DNA; TEF1-α, translation elongation factor 1-α; CBS, Centraal Bureau voor Schimmelcultures (today, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands); DSM, T. mentagrophytes Deutsche Sammlung von Mikroorganismen und Zellkulturen (German Collection of Microorganisms and Cell Cultures), Braunschweig, Germany; ATCC, American Type Culture Collection (Manassas, VA, USA). * means a subtype of the genotype II.
| Species/Genotype | Strain | Collection | Accession No. NCBI ITS | Accession No. NCBI TEF1-α |
|---|---|---|---|---|
| T. interdigitale ITS genotype I | 208223/17 | DSM 108620 | MK447595 | MK460538 |
|
T. interdigitale ITS genotype I deposited in the NCBI as T. interdigitale |
Isolate 2 | KC595991 | ||
| T. interdigitale ITS genotype II | 200070/17 | DSM 108621 | MK447596 | MK460539 |
|
T. interdigitale ITS genotype II deposited in the NCBI as T. interdigitale |
RCPF-1301 | KP308373 | ||
|
T. interdigitale ITS genotype II deposited in the NCBI as T. interdigitale |
CBS 475.93 | MF926357 | ||
|
T. interdigitale ITS genotype II deposited in the NCBI as T. interdigitale |
CBS 428.63 | NR_144900 | ||
|
T. interdigitale ITS genotype II deposited in the NCBI as T. mentagrophytes |
CBS 647.73 | KT155955 | ||
|
T. mentagrophytes deposited in the NCBI as T. mentagrophytes |
CBS 102.68 | KM678062 | ||
| T. interdigitale ITS genotype II * | 212063/17 | DSM 108905 | MK630684 | MK751367 |
| T. interdigitale ITS genotype II * | 250016/18 | MN886818 | MN886231 | |
| T. mentagrophytes | CBS 116916 | KM678130 | ||
| T. interdigitale | CBS 130940 | KM678173 | ||
| T. mentagrophytes ITS genotype III | 217704/15 | DSM 108630 | MK450325 | MK460541 |
| T. mentagrophytes ITS genotype III | 200002/16 | DSM 103451 | KX866689 | MK460540 |
| ITS genotype III deposited in the NCBI as T. cf. mentagrophytes |
ATCC 60612 | KJ606099 | ||
|
T. mentagrophytes ITS genotype III deposited in the NCBI as T. mentagrophytes |
RCPF-1207 | KT253559 | ||
| T. mentagrophytes ITS genotype III * | 217907/15 | DSM 108628 | MK447605 | MK460542 |
| T. mentagrophytes ITS genotype III * | 218893/16 | DSM 108629 | MK447604 | MK460543 |
| T. mentagrophytes ITS genotype III * | 900120/17 | DSM 108632 | MK447606 | MK460544 |
|
T. mentagrophytes ITS genotype III * deposited in the NCBI as T. interdigitale |
CZE 4473 | LN736306 | ||
| T. mentagrophytes ITS genotype IV | 200602/17 | DSM 108631 | MK447607 | MK467447 |
| T. mentagrophytes ITS genotype IV | 200617/17 | DSM 108627 | MK447608 | MK467446 |
| T. mentagrophytes ITS genotype IV | 204543/17 | DSM 108626 | MK447609 | MK467445 |
|
T. mentagrophytes ITS genotype IV deposited in the NCBI as T. mentagrophytes |
CBS 304.38 | MF926360 | ||
|
T. mentagrophytes ITS genotype IV deposited in the NCBI as T. interdigitale |
SJEK 4836 | FM986773 | ||
|
T. mentagrophytes ITS genotype V deposited in the NCBI as T. cf. mentagrophytes |
ATCC 46950 | KJ606098 | ||
| T. mentagrophytes ITS genotype V | 600014/20 | MT374268 | MT375511 | |
| T. mentagrophytes ITS genotype V | 600024/20 | MT374269 | MT375512 | |
| T. mentagrophytes ITS genotype V | 600184/19 | MT374259 | MT375508 | |
| T. mentagrophytes ITS genotype V | 600197/19 | MT374258 | MT375509 | |
| T. mentagrophytes ITS genotype V | 600316/19 | MT374257 | MT375510 | |
|
T. mentagrophytes ITS genotype VI deposited in the NCBI as T. mentagrophytes |
D15P161/17 | MK722518 | - | |
| T. mentagrophytes ITS genotype VII | 210363/16 | DSM 108625 | MK450323 | MK467450 |
| T. mentagrophytes ITS genotype VII | 218904/16 | DSM 108622 | MK450322 | MK467448 |
| T. mentagrophytes ITS genotype VII | 200128/17 | DSM 108623 | MK447611 | MK460545 |
| T. mentagrophytes ITS genotype VII | 215003/16 | DSM 108624 | MK450324 | MK467449 |
|
T. mentagrophytes ITS genotype VII deposited in the NCBI as T. mentagrophytes |
NBRC 5809 | JN134101 | ||
| T. mentagrophytes ITS genotype VIII | 211509/17 | DSM 107597 | MH791420 | MH802491 |
| T. mentagrophytes ITS genotype VIII | 200095/18 | DSM 107602 | MH791425 | MH802496 |
| T. mentagrophytes ITS genotype VIII deposited in the NCBI as T. interdigitale |
VPCI 390/P/17 | - | MH990852 | |
| T. mentagrophytes ITS genotype IX | 214691/17 | DSM 108357 | MK447613 | MK467444 |
| T. mentagrophytes ITS genotype XXV | 218292/17 | - | MN886815 | MN886229 |
| T. mentagrophytes ITS genotype XXV | 201341/18 | - | MN886816 | MN886230 |
| T. quinckeanum | 218251/16 | |||
| T. quinckeanum | 210314/16 | |||
| T. quinckeanum | ATCC 32457 | KJ606088 | ||
| T. quinckeanum | 216686/15 | KY680503 | KY680502 |
2.3.4. Statistical Method for Generating Phylogenetic Trees
Phylogenetic relationships between dermatophyte species were generated using the software Mega X: Analysis Statistical Method Maximum Likelihood, Phylogeny Test Bootstrap Method Replications–1000, Substitution Model Maximum Composite Likelihood [10,11].
2.3.5. Deposition of the Isolates in Strain Collections and Gene Databases
Both ITS and TEF1 α gene sequences of all 29 strains/isolates are deposited at the database of the National Centre for Biotechnology Information (NCBI) in Bethesda, MD, USA (Table 1). The strains themselves were deposited at the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany).
2.4. Antifungal Resistance Testing
2.4.1. In Vitro Antifungal Susceptibility Testing
Isolated dermatophytes growing on culture media were tested for growth on Sabouraud’s dextrose agar containing 0.2 µg/mL terbinafine, as described by previous research [12]. The concentration of terbinafine was equivalent to twice that of the minimal inhibitory concentration (MIC) for T. mentagrophytes and T. rubrum under these conditions [13]. Fungal growth was examined after seven and 14 days. Growing strains were recorded as resistant. MICs of itraconazole and voriconazole were determined according to the broth microdilution method of the Clinical and Laboratory Standards Institute as previously described [14]. Based on epidemiological cut-off values (ECOFFs) from previous research, strains were classified as resistant or sensitive to itraconazole and voriconazole (ECOFF ≥ 0.5 µg/mL for itraconazole; ECOFF ≥ 0.25 µg/mL for voriconazole) [15].
2.4.2. Squalene Epoxidase Gene Analysis
Trichophyton total DNA was extracted from fresh fungal cultures on Sabouraud´s dextrose agar using a DNeasy Plant Minikit (Qiagen, Hilden, Germany). A square shaped area of approximately 1.0 mm2 of growing culture was used. The squalene epoxidase (SQLE) gene of the terbinafine-resistant clinical isolates was amplified by PCR with ReadyMix Taq PCR Reaction Mix (Sigma Aldrich, Merck). The primer pair TrSQLE-F2 (5’ ATGGTTGTAGAGGCTCCTCCC 3’) and TrSQLE-R1 (5’ CTAGCTTTGAAGTTCGGCAAA 3’) was used and chromosomal DNA served as the template. In some cases where fungal cultures were not obtained, SQLE gene fragments were analyzed from scale DNA as described [16] using primer pairs TmSQLEF4 (5’ AACGGCTTTGCGAATGGCTCC 3’) and TmSQLER4 (5’ GATGACCCTGCAGGCAGTAAG 3’). Sequences were aligned and screened for missense mutations using MEGA version 10.0.5 [10,11].
3. Results
3.1. Patients
Twenty-nine patients (all out-patients) with different clinical variants of dermatophytoses caused by T. mentagrophytes Type VIII (India) were diagnosed all over Germany (Table 1) between September 2016 and March 2020. The detection was based on both routine diagnostics performed in the laboratory Mölbis, Germany, and from cultures sent for fungal species identification of T. mentagrophytes Type VIII (India). The microbiological as well as molecular diagnosis of T. mentagrophytes Type VIII was possible in all 29 patients.
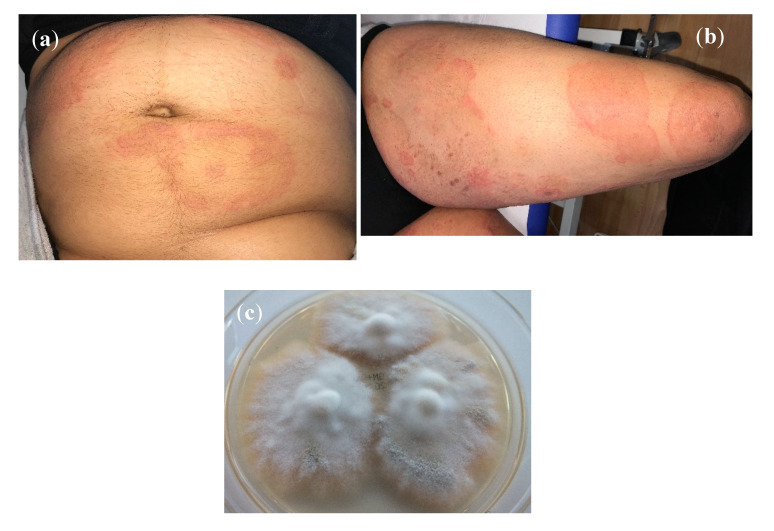
Males (n = 15) outnumbered females (n = 12). Gender was not specified for two patients. Patients’ age ranged from six months to 58 years. The mean age was 26 years (n= 26, for three patients the age was not known). Patients included two children aged six months and four years. Tinea corporis was the predominant variant of dermatophytosis (n = 14). Other variants of dermatophytoses were unspecified tinea (n = 3), tinea corporis and tinea cruris (n = 3), tinea cruris (n = 2), tinea corporis (including tinea glutealis) (n = 2), tinea faciei (n = 2), tinea corporis and tinea unguium (n = 1), tinea pedis (n = 1), and tinea corporis and tinea manuum (n = 1) (Table 1, Figure 1a,b). Both toddlers presented with tinea corporis (including tinea glutealis).
Figure 1.
(a) Tinea corporis due to T. mentagrophytes VIII in a 34-year old female from India residing in Germany. Patient No. 12. Laboratory number of the fungal isolate: 600174/2019. (b) Accompanying large area tinea of the thighs of the same patient. (c) Subculture of T. mentagrophytes VIII isolated from the patient´s skin scrapings on Sabouraud´s dextrose agar; developed fast growing white, flat, radiating and granular colonies.
3.2. Some Striking Clinical Presentations
(1) A six months-old-female infant from Bahrain visiting Germany with her family for a holiday was seen by us for extensive dermatophytosis of the back, buttocks, chest and groin [17]. Topical treatment by terbinafine for over two months was unsuccessful. Other family members, including adults and children, were treated in Bahrain with topical antifungals and oral voriconazole which was not helpful. The girl was successfully treated by topical miconazole and later by ciclopirox olamine.
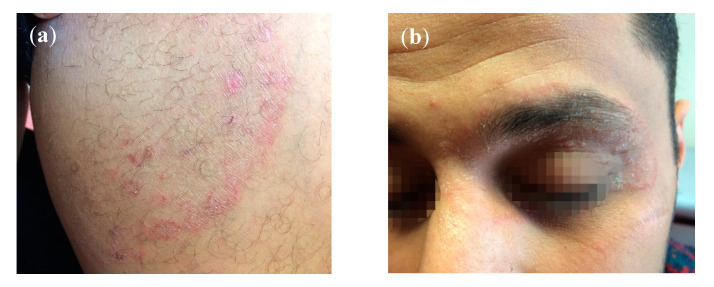
(2) A 28-year old male from Libya, living for three years in Germany, suffered from tinea cruris and tinea faciei involving the left upper and lower eyelids (Figure 2a,b). Treatment by oral fluconazole and terbinafine had failed. His German girlfriend was also affected by the dermatophytosis, though her child was spared. The patient had no contact with India, Indians or Arabs and had not visited Libya in the past few years. The man, however, regularly went to the gym. Treatment with itraconazole orally 400 mg daily for eight weeks cured him.
Figure 2.
(a) Tinea cruris due to T. mentagrophytes VIII in a 28-year old male from Libya residing in Germany for about three years. Patient No. 10. Laboratory number of the fungal isolate: 202953/2019. (b) Accompanying tinea faciei of the same patient involving the left upper and lower eyelids.
(3) A pregnant German woman presented with tinea cruris et corporis after a trip to Saudi Arabia. Her husband was also affected. Topical treatment was started by clotrimazole alone.
(4) An Iraqi couple living in Germany for a long time suffered from chronic recalcitrant dermatophytosis of the groin, thighs and buttocks for at least two years (Figure 3). Repeated topical treatments by fixed-dose combination creams (FDCs), also known as combination creams, (fluprednidene 21-acetate + miconazole nitrate, betamethasone dipropionate + gentamicin sulphate, and betamethasone dipropionate + clotrimazole) had failed. Topical antifungal therapy (ciclopirox olamine, sertaconazole) given for five to six weeks acted very slowly and they stopped treatment due to progress of the disease. Oral itraconazole 200 mg daily was started for four weeks leading to resolution.
Figure 3.
Tinea corporis und tinea glutealis due to T. mentagrophytes VIII in a woman originating from Iraq and living for a long time in Germany. Typical inflammatory and itching erythematosquamous plaques were observed involving the buttocks, the groin and the thighs. Patient No. 14. Laboratory number of the fungal isolate: 205667/2019.
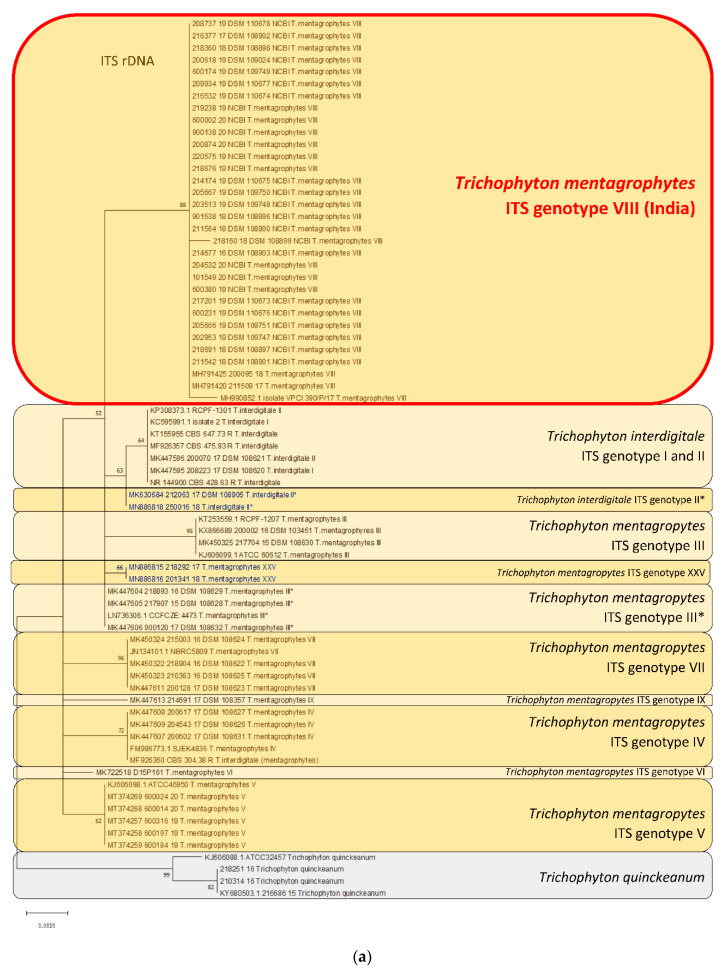
Phylogenetic analysis of Trichophyton mentagrophytes Type VIII in comparison to other genotypes.
Species identification was confirmed for all 29 isolates by sequencing of the ITS regions of rDNA genes. Molecular relationships of these 29 isolates with other genotypes within the species T. mentagrophytes, and with closely related dermatophytes, were depicted in a phylogenetic tree/dendrogram of the sequences (Figure 4a). All 29 isolates belonged to the same cluster, called ITS Type VIII, referred to as Indian variant. The isolates of T. mentagrophytes ITS Type VIII formed their own phylogenetic cluster. This genotype was clearly different from other already known genotypes of T. mentagrophytes, e.g., zoophilic strains isolated from human dermatophytoses and from animals, including a snow leopard at a zoo garden, and from T. mentagrophytes ITS genotype VII (Thai variant). The anthropophilic T. interdigitale could be distinguished clearly from zoophilic T. mentagrophytes clusters.
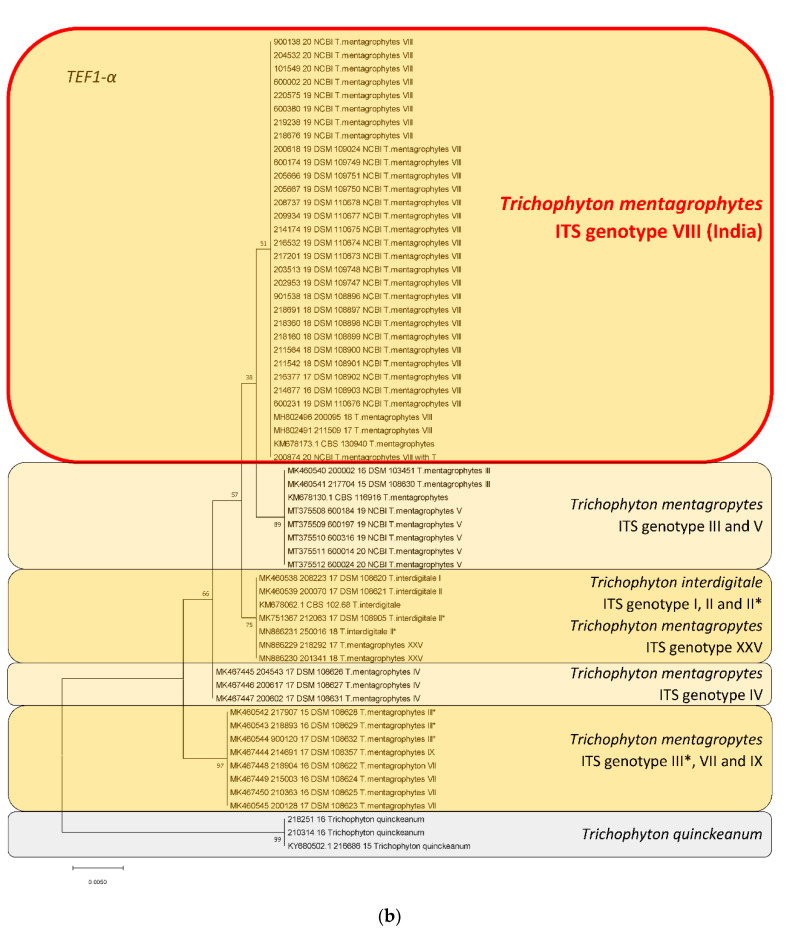
Figure 4.
Phylogenetic analysis of the dermatophyte isolates based on the ITS regions of rDNA and the TEF1-α gene (Table 2).The evolutionary history was inferred by using the maximum likelihood method and Tamura-Nei model [10]. The tree with the highest log likelihood (−907.70) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 28 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + noncoding. There were a total of 1086 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [11]. (a) Phylogenetic tree of T. mentagrophytes based on sequencing of the ITS regions of rDNA genes. By sequencing, a 100% concordance with NCBI reference strains (accession numbers MH791420, MH791425, MH990852) was found for all 29 isolates. All these isolates formed their own cluster, which is now called the ITS genotype VIII (India) of T. mentagrophytes. These isolates (T. mentagrophytes Type VIII or clade) were clearly discriminated from already known T. mentagrophytes genotypes, e.g., II, V, VII. Rooted with Trichophyton quinckeanum. (b) Phylogenetic tree of T. mentagrophytes based on sequencing of the TEF1-α gene. Used NCBI reference sequences (TEF1-α gene) were MH802491 and MH802496 (accession numbers). Within the phylogenetic tree, all T. mentagrophytes ITS Type VIII strains from Germany formed their own clade, which is clearly discriminated from the other, above mentioned, T. mentagrophytes genotypes. Rooted with T. quinckeanum.
The phylogenetic tree based on sequencing of the TEF1-α gene revealed a 100% concordance of all 29 isolates belonging to genotype VIII of T. mentagrophytes (Figure 4b). Within the phylogenetic tree, all T. mentagrophytes isolates from this study formed their own clade, which was clearly differentiated from other T. mentagrophytes genotypes, and from T. interdigitale.
The origin of all reference strains, and clinical or animal isolates and their sequences used here for comparison, is presented in Table 2.
3.3. Antifungal Resistance Testing and Point Mutation Analysis
In vitro resistance testing revealed that 13 (45%) out of 29 strains were terbinafine-resistant with breakpoints ≥ 0.2 µg/mL. The remaining 16 strains (55%) were terbinafine-sensitive (Table 1). Point mutation analysis revealed that among 13 resistant strains, 10 exhibited Phe397Leu amino acid substitution of the SQLE, indicative of in vitro resistance to terbinafine. Two resistant strains showed combined Phe397Leu and Ala448Thr amino acid substitutions, and one strain a single Leu393Phe amino acid substitution. Out of 16 terbinafine-sensitive strains, Ala448Thr was detected in nine strains, one of which also showed a new Val444Ile substitution. The remainder of the sensitive strains exhibited no substitution. Out of 13 strains tested for triazole sensitivity, nine proved to be sensitive to both itraconazole and voriconazole. Three strains revealed resistance to both triazoles, with one strain also showing resistance to terbinafine, while the remaining strain exhibited resistance to voriconazole but not itraconazole. The accession numbers of sequences of the SQLE gene of all 29 investigated T. mentagrophytes strains are deposited at the NCBI database (Table 1).
4. Discussion
4.1. Trichophyton mentagrophytes Type VIII (India)
ITS genotype VIII of T. mentagrophytes seems to have grown rapidly in recent years to become the predominant dermatophyte in India [18]. In contrast, the previously predominant T. rubrum for decades was isolated with much less frequency [19]. A currently published epidemiological study of a total of 402 Indian patients with extensive dermatophytoses revealed culture growth of T. mentagrophytes in 289 (71.9%) of samples [15]. T. rubrum was cultivated from only 19 (4.7%) samples. It was possible to identify T. interdigitale/T. mentagrophytes complex in 235/265 (88.7%) of samples by PCR-ELISA. DNA sequencing enabled identification of T. mentagrophytes ITS Type VIII in 311 (77%) samples, unspecified species of T. interdigitale/T. mentagrophytes complex in 21 (5%), and T. rubrum in 19 (5%) samples.
It is interesting to note that T. mentagrophytes Type VIII was not initially found in India, but was previously isolated in Oman, Iran and also in Australia under a different species name, T. interdigitale, in accordance with the old taxonomy of dermatophytes that was valid only until 2016 [20,21,22,23]. Currently, the species found in India and several other countries globally is referred to as T. mentagrophytes Type VIII in the dermatomycological community [24]. T. mentagrophytes Type VIII is only one variety within the cluster of a large number of genotypes of the T. mentagrophytes/T. interdigitale complex [16,21,25,26,27,28,29,30] (Figure 5a–d). Therefore, it does not appear justifiable to attempt assigning this single genotype VIII of T. mentagrophytes to a brand new species, so hurriedly, on the basis of just two isolated terbinafine-resistance isolates of T. mentagrophytes, disregarding the plethora of literature on a well-accepted taxonomy and nuances of this genotype, as has just happened in an isolated publication from Japan [31].
Figure 5.
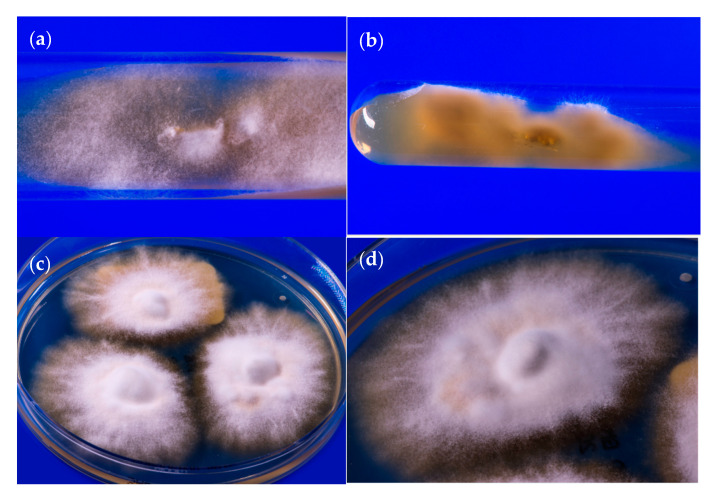
58-year old male originating from India suffering from tinea faciei due to T. mentagrophytes Type VIII. (a) Primary culture on slant agar tubes containing Sabouraud´s dextrose agar. Granular white, flat, fast-growing fungal colonies. (b) Yellow to brown stained reverse side of colonies on slant agar tubes. (c) Subculture of T. mentagrophytes Type VIII on Sabouraud´s dextrose agar petri dish agar. White, radiating and granular colonies with slightly yellowish stained centre of the colonies. (d) Single colony of T. mentagrophytes Type VIII on Sabouraud´s dextrose agar petri dish.
4.2. Spread of T. mentagrophytes Type VIII (India) to Other Parts of the World
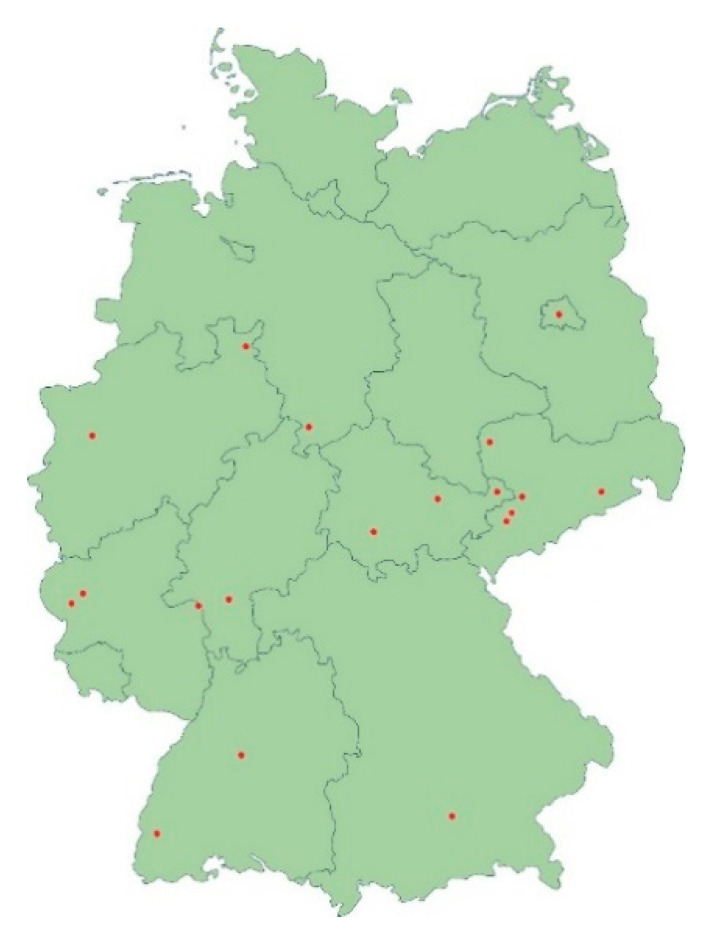
Numerous reports and findings from our own laboratory suggest that the frequently terbinafine-resistant dermatophyte T. mentagrophytes Type VIII is becoming increasingly prevalent in countries other than Germany and beyond. T. mentagrophytes Type VIII has been isolated from skin scrapings of patients in Iraq, Switzerland, Cambodia, Finland, Estonia and Poland [32,33]. We have isolated only individual strains of T. mentagrophytes Type VIII since 2016, albeit initially under a different name of the species, owing to the classification at that time. There has been a significant increase in the frequency of detecting T. mentagrophytes Type VIII in the past two years in Germany and we regularly see a strain of T. mentagrophytes type VIII in routine diagnostics about every two to three weeks, which appears significant. The isolates of T. mentagrophytes Type VIII originate from all over Germany. Patients with dermatophytoses, due to the Indian genotype of T. mentagrophytes, have been identified in large cities like Berlin, Munich and Leipzig, as well as small towns and rural areas of Germany (Figure 6).
Figure 6.
Geographical distribution of patients with dermatophytoses due to the Indian genotype of T. mentagrophytes identified in this study. Strains of T. mentagrophytes Type VIII were isolated in nearly all parts of Germany, in large cities like Berlin, Munich and Leipzig as well as in small towns and rural areas.
In retrospect we feel that patients of chronic dermatophytoses caused by T. mentagrophytes Type VIII, as seen here, have existed in Germany for years, though in smaller numbers. This genotype seems to be isolated preferentially in migrant patients. Many originally hail from the Indian subcontinent, including India, Bangladesh and Pakistan, but also from Arab countries such as Saudi Arabia, Iraq and Libya. The infection is found to be relatively easily transmitted within the family, especially to the spouse and partner.
4.3. Antifungal Resistance In Vitro and Point Mutation Analysis of the Squalene Epoxidase Gene
A significant percentage of the original Indian T. mentagrophytes strains was resistant to terbinafine both in vitro and due to genetic point mutations in the SQLE gene. Some strains were also found to be partially resistant against itraconazole and voriconazole. Several single point mutations in the fungal SQLE gene, which encodes the target for terbinafine, have also been recorded in T. rubrum and T. mentagrophytes/T. interdigitale. These mutations have led to substitutions at one of the four amino acid positions Leu393, Phe397, Phe415 and His440, and have been associated with terbinafine resistance [12,34]. 71% of isolates of T. mentagrophytes Type VIII from India were found to be resistant to terbinafine [15]. The amino acid substitution Phe397Leu in the squalene epoxidase of resistant T. mentagrophytes was found to be highly prevalent (91%) [15]. Two novel substitutions in resistant Trichophyton strains isolated in our currently published epidemiological study in India, Ser395Pro and Ser443Pro, were detected. In contrast, a missense substitution, Ala448Thr, was found in terbinafine-sensitive and resistant isolates. Among the 29 strains isolated in Germany, 13 strains (45%) were terbinafine-resistant with breakpoints > 0.2 µg/mL. The other 16 strains showed normal terbinafine susceptibility in vitro against terbinafine with breakpoints < 0.2 µg/mL. It is not clear if all cases of terbinafine resistance occurred after long-term treatment with the drug, or if they had primary resistances. Some patients with resistance were pretreated by oral or topical terbinafine; unfortunately, because the biggest part of our patient´s data on pretreatment were not available. Indeed, however, transmission from affected people to family members who were definitely not pretreated, occurred.
It has been observed that despite in vitro susceptibility, there is often a poor clinical response to terbinafine, as seen in a significant number of patients reported herein. There are some indications of a lack of in vivo correlation of in vitro resistance in dermatophytosis [35]. On the other hand, it was demonstrated that the odds of achieving a cure with terbinafine MIC < 1 µg/mL strains were 2.5 times the odds of achieving a cure with strains exhibiting MIC ≥ 1 µg/mL, suggesting a good in vitro and in vivo correlation [36]. Further studies are recommended to understand this complex problem of the in vitro/in vivo discordance.
We demonstrated that in vitro resistance to triazoles cannot only be observed in T. mentagrophytes Type VIII isolated in India, but also in strains from patients residing in Germany. Remarkably, three out of four strains showing resistance to voriconazole were also resistant to itraconazole, strengthening evidence that these strains share a common mechanism of resistance against triazoles [15]. Triazole resistance was recently associated with the substitution Ala448Thr in squalene epoxidase [15,30] and such a tendency could be observed in the current collection of strains. However, statistical significance could not be proven due to the relatively small sample size. Further investigations, especially on the role of SQLE double mutants on antifungal susceptibility [30], need to be carried out.
4.4. Treatment of Chronic Recalcitrant Dermatophytoses Due to T. mentagrophytes Type VIII
Patients described in this study represented the first reports on an infection due to a terbinafine-resistant T. mentagrophytes strain of the ITS genotype VIII from Germany and the Indian subcontinent. We aimed to highlight the recalcitrance to even long-term oral and topical treatment with terbinafine observed in our patients of tinea corporis and tinea cruris caused by T. mentagrophytes Type VIII. Unlike the scenario reported from India, the disease in German patients, seemed to respond to simple topical antifungal therapy other than terbinafine, as exemplified in the case of the baby originating from Bahrain with extensive tinea corporis [17]. It is interesting to note that the child showed significant improvement in the lesions after only one week of local treatment with miconazole and ciclopirox olamine, finally leading to resolution of all lesions with the same topical therapy for a total of four weeks. However, the lesions recurred after reaching Bahrain, where they stopped applying the topical antifungal agents. This case scenario has been experienced by several other patients with chronic, refractory dermatophytoses caused by T. mentagrophytes Type VIII. Treatment included topical ciclopirox olamine plus miconazole, sertaconazole or luliconazole with patients reporting more benefit with creams containing newer topical azoles. Many such creams are, unfortunately, not approved for use in Germany and other countries in Europe [37]. It seems appropriate to treat such cases with an oral antifungal agent like itraconazole in its adult dose of 100 mg twice daily after a meal for at least four to eight weeks. Some dermatologists recommend higher doses of the drug for a longer duration in widespread disease in patients who have abused topical steroid antifungal combinations for long periods.
Acknowledgments
We thank Esther Klonowski, biologist from Leipzig, for excellent support in preparing and formatting the manuscript. Uwe Schossig, photographer from Leipzig, Germany, provided beautiful pictures of fungal colonies. We thank Christine Scholz, Mycological laboratory, Department of Dermatology, University Freiburg, Germany, for continuous support for providing clinical and mycological data on patients described in this study.
Author Contributions
Conceptualization, P.N., M.M. and S.U.; Data curation, A.E., M.M., K.S. (Karine Salamin) and S.U.; Funding acquisition, P.N.; Investigation, P.N., S.B.V., A.E., A.S. (Anke Süß), E.F., E.A., S.D., W.H., S.S. (Simone Schmidt), K.N., R.R., S.S. (Sirius Sohl), U.H., U.K., H.-C.W., A.S. (Annegret Staginnus), J.S., V.M., C.T., M.G., K.S. (Katja Schubert), Z.A., R.S., A.F., C.S., C.R., C.O., T.N., A.K., S.K., M.S., B.W., T.W., L.K., M.A., U.W., M.M., K.S. (Karine Salamin), A.B., D.K., C.K. and S.U.; P.N., A.E., M.M. and S.U.; Project administration, P.N., S.B.V., M.M. and S.U.; Software, P.N., A.E. and S.U.; Supervision, P.N., S.B.V., A.E. and S.U.; Validation, P.N., A.E., M.M. and S.U.; Visualization, P.N., S.B.V., A.E., A.S. (Anke Süß), E.F., E.A., S.D., W.H., S.S. (Simone Schmidt), K.N., R.R., S.S. (Sirius Sohl), U.H., U.K., H.-C.W., A.S. (Annegret Staginnus), J.S., V.M., C.T., M.G., K.S. (Katja Schubert), Z.A., R.S., A.F., C.S., C.R., C.O., T.N., A.K., S.K., M.S., B.W., T.W., L.K., M.A., U.W., M.M., K.S. (Karine Salamin), A.B., D.K., C.K. and S.U.; Writing—original draft, P.N., S.B.V., A.E. and S.U.; Writing—review and editing, P.N., S.B.V., A.E. and S.U. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Verma S.B., Madhu R. The great Indian epidemic of superficial dermatophytosis: An appraisal. Indian J. Dermatol. 2017;62:227–236. doi: 10.4103/ijd.IJD_206_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma S., Vasani R., Reszke R., Matusiak Ł., Szepietowski J.C. Prevalence and clinical characteristics of itch in epidemic-like scenario of dermatophytoses in India: A cross-sectional study. J. Eur. Acad Dermatol. Venereol. 2020;34:180–183. doi: 10.1111/jdv.15877. [DOI] [PubMed] [Google Scholar]
- 3.Verma S.B. Comment on: Emergence of recalcitrant dermatophytosis in India. Lancet Infect. Dis. 2018;18:718–719. doi: 10.1016/S1473-3099(18)30338-4. [DOI] [PubMed] [Google Scholar]
- 4.Winter I., Uhrlaß S., Krüger C., Herrmann J., Bezold G., Winter A., Barth S., Simon J.C., Gräser Y., Nenoff P. Molecular biological detection of dermatophytes in clinical samples when onychomycosis or tinea pedis is suspected. A prospective study comparing conventional dermatomycological diagnostics and polymerase chain reaction. Hautarzt. 2013;64:283–289. doi: 10.1007/s00105-013-2562-9. [DOI] [PubMed] [Google Scholar]
- 5.Beifuss B., Bezold G., Gottlöber P., Borelli C., Wagener J., Schaller M., Korting H.C. Direct detection of five common dermatophyte species in clinical samples using a rapid and sensitive 24-h PCR-ELISA technique open to protocol transfer. Mycoses. 2011;54:137–145. doi: 10.1111/j.1439-0507.2009.01771.x. [DOI] [PubMed] [Google Scholar]
- 6.Mirhendi H., Makimura K., De Hoog G.S., Rezaei-Matehkolaei A., Najafzadeh M.J., Umeda Y., Ahmadi B. Translation elongation factor 1-α gene as a potential taxonomic and identification marker in dermatophytes. Med. Mycol. 2015;53:215–224. doi: 10.1093/mmy/myu088. [DOI] [PubMed] [Google Scholar]
- 7.Kargl A., Kosse B., Uhrlaß S., Koch D., Krüger C., Eckert K., Nenoff P. Hedgehog fungi in a dermatological office in Munich: Case reports and review. Hautarzt. 2018;69:576–585. doi: 10.1007/s00105-018-4134-5. [DOI] [PubMed] [Google Scholar]
- 8.Uhrlaß S., Schroedl W., Mehlhorn C., Krüger C., Hubka V., Maier T., Gräser Y., Paasch U., Nenoff P. Molecular epidemiology of Trichophyton quinckeanum-a zoophilic dermatophyte on the rise. J. Dtsch. Dermatol. Ges. 2018;16:21–32. doi: 10.1111/ddg.13408. [DOI] [PubMed] [Google Scholar]
- 9.Wiegand C., Mugisha P., Mulyowa G.K., Elsner P., Hipler U.-C., Gräser Y., Uhrlaß S., Nenoff P. Identification of the causative dermatophyte of tinea capitis in children attending Mbarara Regional Referral Hospital in Uganda by PCR-ELISA and comparison with conventional mycological diagnostic methods. Med. Mycol. 2017;55:660–668. doi: 10.1093/mmy/myw112. [DOI] [PubMed] [Google Scholar]
- 10.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada T., Maeda M., Alshahni M.M., Tanaka R., Yaguchi T., Bontems O., Salamin K., Fratti M., Monod M. Terbinafine resistance of Trichophyton clinical isolates caused by specific point mutations in the squalene epoxidase gene. Antimicrob. Agents Chemother. 2017;61:e00115-17. doi: 10.1128/AAC.00115-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mock M., Monod M., Baudraz-Rosselet F., Panizzon R.G. Tinea capitis dermatophytes: Susceptibility to antifungal drugs tested in vitro and in vivo. Dermatology (Basel) 1998;197:361–367. doi: 10.1159/000018032. [DOI] [PubMed] [Google Scholar]
- 14.Laurent A., Monod M. Production of Trichophyton rubrum microspores in large quantities and its application to evaluate amorolfine/azole compound interactions in vitro. Mycoses. 2017;60:581–586. doi: 10.1111/myc.12632. [DOI] [PubMed] [Google Scholar]
- 15.Ebert A., Monod M., Salamin K., Burmester A., Uhrlaß S., Wiegand C., Hipler U.-C., Krüger C., Koch D., Wittig F., et al. Alarming India-wide phenomenon of antifungal resistance in dermatophytes: A multicentre study. Mycoses. 2020;63:717–728. doi: 10.1111/myc.13091. [DOI] [PubMed] [Google Scholar]
- 16.Burmester A., Hipler U.-C., Hensche R., Elsner P., Wiegand C. Point mutations in the squalene epoxidase gene of Indian ITS genotype VIII T. mentagrophytes identified after DNA isolation from infected scales. Med. Mycol. Case Rep. 2019;26:23–24. doi: 10.1016/j.mmcr.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Süß A., Uhrlaß S., Verma S.B., Ludes A., Monod M., Krüger C., Nenoff P. Extensive tinea corporis due to a terbinafine resistant Trichophyton mentagrophytes isolate of the ‘Indian genotype’ in a young infant from Bahrain in Germany. Hautarzt. 2019;70:888–896. doi: 10.1007/s00105-019-4431-7. [DOI] [PubMed] [Google Scholar]
- 18.Nenoff P., Verma S.B., Vasani R., Burmester A., Hipler U.-C., Wittig F., Krüger C., Nenoff K., Wiegand C., Saraswat A., et al. The current Indian epidemic of superficial dermatophytosis due to Trichophyton mentagrophytes—A molecular study. Mycoses. 2019;62:336–356. doi: 10.1111/myc.12878. [DOI] [PubMed] [Google Scholar]
- 19.Pathania S., Rudramurthy S.M., Narang T., Saikia U.N., Dogra S. A prospective study of the epidemiological and clinical patterns of recurrent dermatophytosis at a tertiary care hospital in India. Indian J. Dermatol. Venereol. Leprol. 2018;84:678–684. doi: 10.4103/ijdvl.IJDVL_645_17. [DOI] [PubMed] [Google Scholar]
- 20.Rezaei-Matehkolaei A., Rafiei A., Makimura K., Gräser Y., Gharghani M., Sadeghi-Nejad B. Epidemiological aspects of dermatophytosis in Khuzestan, southwestern Iran, an Update. Mycopathologia. 2016;181:547–553. doi: 10.1007/s11046-016-9990-x. [DOI] [PubMed] [Google Scholar]
- 21.Taghipour S., Pchelin I.M., Zarei-Mahmoudabadi A., Ansari S., Katiraei F., Rafiei A., Shokohi T., Abastabar M., Taraskina A.E., Kermani F., et al. Trichophyton mentagrophytes and T. interdigitale genotypes are associated with particular geographic areas and clinical manifestations. Mycoses. 2019;69:1084–1091. doi: 10.1111/myc.12993. [DOI] [PubMed] [Google Scholar]
- 22.Nenoff P., Herrmann J., Gräser Y. Trichophyton mentagrophytes sive interdigitale? A dermatophyte in the course of time. J. Dtsch. Dermatol. Ges. 2007;5:198–202. doi: 10.1111/j.1610-0387.2007.06180.x. [DOI] [PubMed] [Google Scholar]
- 23.Nenoff P., Krüger C., Schaller J., Ginter-Hanselmayer G., Schulte-Beerbühl R., Tietz H.-J. Mycology-an update. Part 2: Dermatomycoses: Clinical picture and diagnostics. J. Dtsch. Dermatol. Ges. 2014;12:749–777. doi: 10.1111/ddg.12420. [DOI] [PubMed] [Google Scholar]
- 24.Nenoff P., Verma S.B., Uhrlaß S., Burmester A., Gräser Y. A clarion call for preventing taxonomical errors of dermatophytes using the example of the novel Trichophyton mentagrophytes genotype VIII uniformly isolated in the Indian epidemic of superficial dermatophytosis. Mycoses. 2019;62:6–10. doi: 10.1111/myc.12848. [DOI] [PubMed] [Google Scholar]
- 25.Pchelin I.M., Azarov D.V., Churina M.A., Scherbak S.G., Apalko S.V., Vasilyeva N.V., Taraskina A.E. Species boundaries in the Trichophyton mentagrophytes/T. interdigitale species complex. Med. Mycol. 2019;57:781–789. doi: 10.1093/mmy/myy115. [DOI] [PubMed] [Google Scholar]
- 26.Shamsizadeh F., Pchelin I.M., Makimura K., Alshahni M.M., Satoh K., Katiraee F., Ahmadi B., Rezaei-Matehhkolaei A. DNA Topoisomerase 2 Gene Polymorphism in Dermatophytes. Mycoses. 2020 doi: 10.1111/myc.13086. [DOI] [PubMed] [Google Scholar]
- 27.De Hoog G.S., Dukik K., Monod M., Packeu A., Stubbe D., Hendrickx M., Kupsch C., Stielow J.B., Freeke J., Göker M., et al. Toward a novel multilocus phylogenetic taxonomy for the dermatophytes. Mycopathologia. 2017;182:5–31. doi: 10.1007/s11046-016-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma S., Vasani R., Gupta S. Involvement of little discussed anatomical locations in superficial dermatophytosis sundry observations and musings. Indian Dermatol. Online J. 2020;11:419–424. doi: 10.4103/idoj.IDOJ_612_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taghipour S., Shamsizadeh F., Pchelin I.M., Rezaei-Matehhkolaei A., Zarei Mahmoudabadi A., Valadan R., Ansari S., Katiraee F., Pakshir K., Zomorodian K., et al. Emergence of terbinafine resistant Trichophyton mentagrophytes in Iran, harboring mutations in the squalene epoxidase (SQLE) gene. Infect. Drug Resist. 2020;13:845–850. doi: 10.2147/IDR.S246025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burmester A., Hipler U.-C., Uhrlaß S., Nenoff P., Singal A., Verma S.B., Elsner P., Wiegand C. Indian T. mentagrophytes squalene epoxidase erg1 double mutants show high proportion of combined fluconazole and terbinafine resistance. Mycoses. 2020 doi: 10.1111/myc.13150. [DOI] [PubMed] [Google Scholar]
- 31.Kano R., Kimura U., Kakurai M., Hiruma J., Kamata H., Suga Y., Harada K. Trichophyton indotineae sp. nov.: A new highly terbinafine-resistant anthropophilic dermatophyte species. Mycopathologia. 2020 doi: 10.1007/s11046-020-00455-8. [DOI] [PubMed] [Google Scholar]
- 32.Järv H., Uhrlaß S., Simkin T., Nenoff P., Alvarado Ramirez E., Chryssanthou E., Monod M. Terbinafine resistant Trichophyton mentagrophytes genotype VIII, Indian type, isolated in Finland. J. Fungi. 2019;5:117. [Google Scholar]
- 33.Uhrlass S., Sithach M., Koch D., Wittig F., Muetze H., Krueger C., Nenoff P. Trichophyton mentagrophytes—A new genotype in Cambodia. J. Fungi. 2019;5:460. [Google Scholar]
- 34.Singh A., Masih A., Khurana A., Singh P.K., Gupta M., Hagen F., Meis J.F., Chowdhary A. High terbinafine resistance in Trichophyton interdigitale isolates in Delhi, India harbouring mutations in the Squalene epoxidase (SQLE) gene. Mycoses. 2018;61:477–484. doi: 10.1111/myc.12772. [DOI] [PubMed] [Google Scholar]
- 35.Khurana A., Sardana K., Chowdhary A. Antifungal resistance in dermatophytes: Recent trends and therapeutic implications. Fungal. Genet. Biol. 2019;132:103255. doi: 10.1016/j.fgb.2019.103255. [DOI] [PubMed] [Google Scholar]
- 36.Khurana A., Masih A., Chowdhary A., Sardana K., Borker S., Gupta A., Gautam R.K., Sharma P.K., Jain D. Correlation of in vitro susceptibility based on MICs and SQLE mutations with clinical response to terbinafine in patients with tinea corporis/cruris. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.01038-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw D., Singh S., Dogra S., Jayaraman J., Bhat R., Panda S., Chakrabarti A., Anjum N., Chowdappa A., Nagamoti M., et al. MIC and upper limit of wild-type distribution for 13 antifungal agents against a Trichophyton mentagrophytes-Trichophyton interdigitale complex of indian origin. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.01964-19. [DOI] [PMC free article] [PubMed] [Google Scholar]