Abstract
Background
The potential benefit of risk stratification using a 4-miRNA signature in combination with MGMT promoter methylation in IDH1/2 wild-type glioblastoma patients was assessed.
Methods
Primary tumors from 102 patients with comparable treatment from the LMU Munich (n = 37), the University Hospital Düsseldorf (n = 33), and The Cancer Genome Atlas (n = 32) were included. Risk groups were built using expressions of hsa-let-7a-5p, hsa-let-7b-5p, hsa-miR-615-5p, and hsa-miR-125a-5p to assess prognostic performance in overall survival (OS). MGMT promoter methylation and age were considered as cofactors. Integrated miRNA, DNA methylome, and transcriptome analysis were used to explore the functional impact of signature miRNAs.
Results
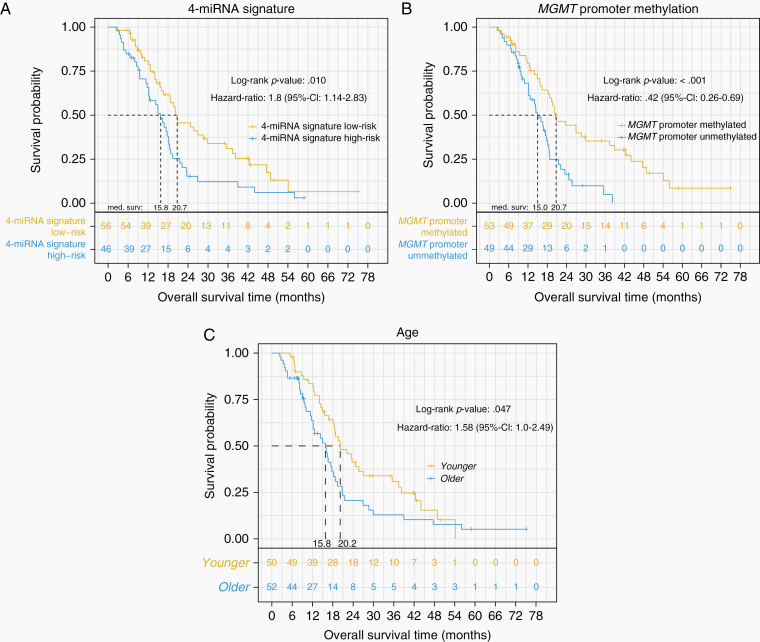
The 4-miRNA signature defined high-risk (n = 46, median OS: 15.8 months) and low-risk patients (n = 56, median OS: 20.7 months; univariable Cox proportional hazard analysis: hazard ratio [HR]: 1.8, 95% confidence interval [CI]: 1.14–2.83, P = .01). The multivariable Cox proportional hazard model including the 4-miRNA signature (P = .161), MGMT promoter methylation (P < .001), and age (P = .034) significantly predicted OS (Log-rank P < .0001). Likewise to clinical routine, analysis was performed for younger (≤60 years, n = 50, median OS: 20.2 months) and older patients (>60 years, n = 52, median OS: 15.8) separately. In younger patients, the 4-miRNA signature had prognostic value (HR: 1.92, 95% CI: 0.93–3.93, P = .076). Particularly, younger, MGMT methylated, 4-miRNA signature low-risk patients (n = 18, median OS: 37.4 months) showed significantly improved survival, compared to other younger patients (n = 32, OS 18.5 months; HR: 0.33, 95% CI: 0.15–0.71, P = .003). Integrated data analysis revealed 4-miRNA signature-associated genes and pathways.
Conclusion
The prognostic 4-miRNA signature in combination with MGMT promoter methylation improved risk stratification with the potential for therapeutic substratification, especially of younger patients.
Keywords: glioblastoma, MGMT promoter methylation, miRNA, prognostic signature, risk stratification
Key Points.
A prognostic 4-miRNA signature was validated in a multicenter cohort of IDH wild-type glioblastoma patients.
Combination of the 4-miRNA signature with MGMT promoter methylation status markedly improved prediction of survival, particularly in patients 60 years of age or younger at diagnosis.
Importance of the Study.
We confirm the prognostic significance of a 4-miRNA signature in IDH wild-type glioblastoma patients. The combination of the 4-miRNA signature with MGMT promoter methylation status improves stratification of younger glioblastoma patients and may enable personalized treatment decisions.
Glioblastomas lacking hotspot mutations in the isocitrate dehydrogenase (IDH) genes 1 or 2 (IDH wild-type glioblastomas) are the most common and most malignant glial tumors of the brain.1,2 Standard of care for IDH wild-type glioblastoma patients comprises neurosurgical tumor resection or stereotactical biopsy followed by chemoradiotherapy (CRT) with temozolomide (TMZ) and 6 cycles of maintenance TMZ chemotherapy.2 Despite this aggressive treatment, prognosis of patients with IDH wild-type glioblastoma remains poor as indicated by median overall survival (OS) times of only 15–18 months in contemporary clinical trials.2
Transcriptional silencing of the O6-DNA-methylguanine-DNA methyltransferase (MGMT) gene by aberrant promoter methylation is linked to better response to TMZ and longer survival of glioblastoma patients.3 However, even MGMT promoter-methylated glioblastomas invariably relapse after initial therapy, with recurrent tumors typically occurring within the high-dose region of initial radiotherapy.4 Patients with recurrent glioblastoma require second-line therapy, which is not as well standardized and may comprise repeated surgery, systemic chemotherapy, and/or reirradiation.2
Even though the need for glioblastoma treatment intensification is evident, trials on the dose escalation of primary radiotherapy did not result in a relevant survival benefit5,6 but caused significantly increased toxicity.7 Unfortunately, also treatments based on molecularly targeted pharmacological approaches have generally failed in glioblastoma to date.8 In this respect, a better discrimination of prognostically distinct subgroups of patients based on molecular biomarkers beyond MGMT promoter methylation might facilitate the development and clinical validation of novel personalized therapies. Especially in younger patients, in which MGMT promoter methylation is not used for treatment stratification,9 further biomarkers are needed.
We have established a 4-miRNA signature with prognostic significance in glioblastoma patients.10 Here, we confirm that IDH wild-type glioblastoma patients can be stratified into high-risk and low-risk groups with a significant difference in OS with the use of a risk score calculation using expression levels of 4 miRNAs, namely hsa-let-7a-5p, hsa-let-7b-5p, hsa-miR-125a-5p, and hsa-miR-615-5p, the Cox coefficients of the prognostic model and a threshold as defined in the discovery study.10 Moreover, we show that a combination of the 4-miRNA signature with MGMT promoter methylation further improves prognostic stratification of glioblastoma patients who were 60 years or younger at the time of diagnosis. For the purpose of the biological and mechanistic interpretation of the 4-miRNA signature, we assessed DNA methylation profiling data through gene set enrichment analysis (GSEA) in addition to integrated 4-miRNA signature, DNA methylation, and transcriptome analysis.
Materials and Methods
Clinical Characteristics of the Glioblastoma Patient Cohorts
All patients were treated by a standard of care CRT with concomitant TMZ.11,12 Data on relevant prognostic factors including the MGMT promoter methylation status, age at diagnosis, and the resection status were available for all patients. Histological tumor classification was performed at each center according to the revised World Health Organization (WHO) classification of central nervous system tumors of 2016.1 Fisher’s exact test and Kruskal–Wallis test were used to evaluate differences between the patients of the University Hospital of the Ludwig Maximilians University Munich (LMU), the University Hospital Düsseldorf (UKD), and The Cancer Genome Atlas (TCGA) subgroups. OS was calculated from the beginning of CRT until death or last follow-up. Median follow-up was calculated using the reverse Kaplan–Meier method.13 The ethics committees approved the project regarding the patients in Munich (LMU ethics approval number: 239-16) and Düsseldorf (approval number: 2017074384).
TCGA Validation Set
The institutional cohorts (LMU and UKD) were complemented by clinical and molecular data of 32 patients retrieved from the publicly available database of TCGA project (https://cancergenome.nih.gov/). The TCGA cohort included patients with available clinical follow-up data, MGMT promoter methylation status, postoperative treatment, as well as information on miRNA expression data. All data on primary tumors were retrieved from the TCGA legacy archive stored at GDC data portal in February 2018. The 32 patients are the subset of the TCGA validation set initially used in the work of Niyazi et al.,10 for which MGMT promoter methylation status was known and which were IDH wild-type.
Processing of Tissue Samples and Nucleic Acid Extraction
Formalin-fixed paraffin-embedded (FFPE) tissue sections (10–20 micron, 3–5 sections per case) from resected tissue samples or stereotactic biopsies of primary tumors were subjected to deparaffinization in xylene followed by washing with ethanol. Representative tumor areas marked by a neuropathologist were scraped off the deparaffinated sections using a scalpel. DNA and total RNA (including the small RNA fraction) were simultaneously extracted from the dissected specimens using the AllPrep DNA/RNA FFPE Kit (Qiagen) according to the manufacturer’s protocol.
Determination of MGMT Promoter Methylation and IDH Mutation
LMU cohort
MGMT promoter methylation in the LMU Munich cohort was assessed by methylation-specific PCR (MSP) and sequencing as reported.14 Briefly, after sodium bisulfite treatment of the DNA, MSP was performed using primers specific for methylated or unmethylated DNA,15 and PCR products were visualized using the FlashGelTM System (Lonza). For bisulfite sequencing, a 316 bp fragment with 25 CpG sites of the MGMT promoter region was amplified after bisulfite treatment.16 Sequencing of purified PCR products was subsequently performed on an ABI3130 sequencer (Applied Biosystems). The MGMT promoter sequence was considered “methylated” if ≥ 13 of the 25 CpG sites showed methylation-specific peaks, that is, at least 50% signal intensity of the corresponding thymine peak. IDH mutation status was determined as reported.14 In brief, the DNA segments containing the R132 (IDH1) and the R172 (IDH2) coding regions were amplified by PCR. Subsequent pyrosequencing was performed on a PyroMark System Q24 MDX (Qiagen) with the appropriate reagents according to the manufacturer’s instructions. Pyrograms were analyzed by the PyroMark Q24 software (Qiagen).
UKD cohort.
MGMT promoter methylation status was determined by MSP as reported.17IDH mutation status was determined by immunohistochemistry for IDH R132H and DNA pyrosequencing as reported before.17
TCGA cohort.
The MGMT promoter methylation status was determined by Illumina Human Methylation 450k Bead Array analysis according to Bady et al.18 The IDH mutation status was retrieved from the clinical data annotation provided by TCGA.
Quantitative Real-Time PCR Analysis of Signature miRNAs in the LMU and UKD Cohorts
Determination of expression levels of miRNAs hsa-let-7a-5p, hsa-let-7b-5p, hsa-miR-125a-5p, and hsa-miR-615-5p was carried out by quantitative real-time PCR (qRT-PCR) using the miScript II System (Qiagen) on a ViiA 7 Real-Time PCR System (Applied Biosystems). The small RNA SNORD68 was used as an endogenous control. For reverse transcription, 10 ng total RNA was used in a total reaction volume of 20 µL. For the qRT-PCR 1 µL of the cDNA was added to 9 µL of master mix and the reaction PCR cycled using the following program: 95°C for 15 min (initiation and denaturation), 94°C for 15 s, 55°C for 30 s, 66°C for 30 s for 40 cycles. The cycling conditions and primer concentration were optimized with regard to reaction efficiency. Reactions were carried out in duplicates and exponentiated delta Ct values reflecting miRNA expression relative to the SNORD68 control were calculated. All primers were part of the proprietary miScript Primer Assays as provided by Qiagen.
Determination of Signature miRNA Expression Levels in TCGA miRNA Microarray Data
The log2-transformed expression values for the 4 signature miRNAs were extracted from quantile normalized Agilent human miRNA array data as reported before.10
Calculation of Signature Risk Score and Risk Factor
The expression values (log2) of each subcohort were z-scaled per miRNA, and risk scores were calculated by building the scalar product of the z-scaled expression values and the Cox proportional hazard coefficients from the original 4-miRNA signature prognostic model.10 Patients were assigned to the high-risk or the low-risk group, using the cutoff value as described before.10
Uni- and Multivariable Survival Analysis
Univariable and multivariable Cox proportional hazard models were built for testing the prognostic significance of the 4-miRNA signature, MGMT promoter methylation status, age, sex, and resection status. According to the median age of the patients, which was 60 years, the cohort was dichotomized in patients older than 60 years and patients aged 60 years or younger. All independent variables were tested pairwise for possible associations. Moreover, possible statistical interactions of age or the MGMT promoter methylation status with the 4-miRNA signature were tested. In order to integrate the prognostic values of the 4-miRNA signature with MGMT promoter methylation status, 4 subgroups were built and analyzed by univariable and multivariable testing. Predictor variables that were significantly associated with OS in the univariable analysis were included in the multivariable models. The prognostic performance of multivariable models to be compared was assessed using C-index over time.19
Recursive Partitioning Analysis
Recursive partitioning analysis (RPA) was performed using the R package rpart for the endpoint OS and including the dichotomous variables 4-miRNA signature, MGMT promoter methylation status, age, and extent of resection status.20 The terminal nodes (leaves) had to contain at least 5 observations and cross-validation was repeated 1000 times.
DNA Methylome Profiling and GSEA for Differentially Methylated Genes
For patients included in the TCGA validation subcohort, the Illumina human methylation 450k DNA methylation profiles were downloaded from the GDC Data Portal (legacy archive) using the R package TCGAbiolinks. In case that 450k data were not available, the 27k profiles were downloaded. The beta values for probes that were present in both the 450k and 27k data were combined into one data set. For differential analysis of CpG sites, the DMP function of the R package ChAMP was used. Benjamini–Hochberg false discovery rate (FDR) multiple testing correction was applied to P values while FDRs smaller than 0.05 were considered as statistically significant.21 The top 10 CpG sites for each of the 4 signature miRNAs were plotted in heatmaps. For methylation GSEA, the Empirical Bayes GSEA method implemented in the R package ChAMP was applied to the methylation data, while the t-test statistics of signature miRNAs were used as a ranking parameter. Enrichment was performed with gene sets from the KEGG and Reactome pathway databases.22,23
Integrated Analysis of the Impact of Signature miRNAs and DNA Methylation on Gene Expression
We used a subset of the TCGA data set for which data from the miRNA, DNA methylation, and transcriptome levels were available. All data were downloaded from the GDC data portal using the R package TCGAbiolinks.24 For the identification of genes that are cooperatively regulated by the 4-miRNA signature and DNA methylation, we generated 2 linear regression models for each combination of miRNA and gene.25 One model only considered the additive effects of miRNA expression and DNA methylation and the second model also contained the interaction term. Using ANOVA F-test, the fit of the 2 models was compared and in case of statistical significance of the difference, we assumed that interaction of the miRNA with DNA methylation influences the expression of the gene of interest. With regard to the selection of genes, we followed 2 approaches. In one approach, we identified the predicted target genes for each signature miRNA by querying multiple miRNA-target prediction databases using the get_multimir function as implemented in the multiMiR R package.26 The top 60% predicted targets were used in the analysis. In a second approach, the genes as provided by the COSMIC Cancer Gene Census (https://cancer.sanger.ac.uk/census, downloaded on June 27, 2020) were tested for the cooperative effects of the 4-miRNA signature and DNA methylation.27 In order to check significantly interacting genes if published in the context of glioblastoma, a batch PMID query “glioblastoma[Title/Abstract] AND [candidate gene]” was conducted using the R package easyPubMed.
R Analysis and Supplementary Data File
The statistical analysis was conducted in R using the RStudio Server developer environment, which was run as a virtual machine on a high-performance computing server from a docker image (kristianunger/rstudio:neurooncologyadvances). All R analyses were conducted and documented using R Markdown (https://rmarkdown.rstudio.com/), which generates an html report file that accompanies this paper as Supplementary File 1.
Results
Patient Characteristics
Our study includes 102 patients with IDH wild-type glioblastoma. Clinical data and tissue samples of 70 patients were retrospectively collected at 2 different centers in Germany, namely the LMU Klinikum, LMU Munich, Munich (n = 37 patients) and the UKD, Heinrich Heine University, Düsseldorf (n = 33 patients).The median age at diagnosis was 61 years (range: 33–78 years). Sixty patients (58.8%) were male and 42 patients (41.2%) were female. Eighty-four patients (82.4%) underwent tumor resection (gross total or partial), while 18 patients (17.6%) underwent biopsy only. The median follow-up of the whole cohort was 42.7 months (interquartile range [IQR]: 29–58.8 months). Median follow-up times of the LMU cohort were 49.8 months (IQR: 41.7–58.8 months) and that of the UKD cohort was 42.7 months (IQR: 29–NA months). Median follow-up could not be estimated by reverse KM due to the small number of losses in the TCGA subcohort (n = 32) combined with the high number of events after the last loss in this subcohort. Details on clinical data are summarized in Table 1 and Supplementary File 1, Sections 4–6).
Table 1.
Patient Characteristics
| LMU (N = 37) | UKD (N = 33) | TCGA (N = 32) | Total (N = 102) | Difference Across Subcohorts (P value) | |
|---|---|---|---|---|---|
| 4-miRNA signature group | .810CS | ||||
| Low-risk | 20 (54.1%) | 17 (51.5%) | 19 (59.4%) | 56 (54.9%) | |
| High-risk | 17 (45.9%) | 16 (48.5%) | 13 (40.6%) | 46 (45.1%) | |
| MGMT promoter methylation status | .003CS | ||||
| Unmethylated | 10 (27.0%) | 22 (66.7%) | 17 (53.1%) | 49 (48.0%) | |
| Methylated | 27 (73.0%) | 11 (33.3%) | 15 (46.9%) | 53 (52.0%) | |
| Age categories | .094CS | ||||
| ≤60 years | 14 (37.8%) | 21 (63.6%) | 15 (46.9%) | 50 (49.0%) | |
| >60 years | 23 (62.2%) | 12 (36.4%) | 17 (53.1%) | 52 (51.0%) | |
| Age | .071KW | ||||
| Median | 65 | 59 | 61 | 61 | |
| Range | 47 - 76 | 39 - 78 | 33 - 76 | 33 - 78 | |
| Sex | .202CS | ||||
| Female | 19 (51.4%) | 10 (30.3%) | 13 (40.6%) | 42 (41.2%) | |
| Male | 18 (48.6%) | 23 (69.7%) | 19 (59.4%) | 60 (58.8%) | |
| Resection status | <.001CS | ||||
| Resection | 24 (64.9%) | 33 (100.0%) | 27 (84.4%) | 84 (82.4%) | |
| Biopsy | 13 (35.1%) | 0 (0.0%) | 5 (15.6%) | 18 (17.6%) |
CS, chi-square test; KW, Kruskal–Wallis test.
4-MiRNA Signature and MGMT Promoter Methylation Status-Defined Patient Groups and Association With Clinical Parameters
After determining the risk status according to Niyazi et al.10 45.1% of patients in the entire cohort of 102 patients were assigned to the high-risk group and 54.9% of the patients to the low-risk group (Figure 1A). Possible associations of the 4-miRNA signature groups with patient age (older than the cohort median age of 60 years vs 60 years or younger), MGMT promoter methylation status, sex, and resection status were tested. The signature risk groups were significantly associated with age (P = .031) and borderline significantly associated with resection status (P = .066). Proportionally, more patients older than 60 years belonged to the 4-miRNA signature high-risk group. 4-miRNA signature risk and MGMT promoter methylation status-defined groups were not significantly associated (odds ratio 1.85, 95% confidence interval [CI]: 0.79–4.43, P = .16) and did not show interaction in the multivariable Cox proportional hazard analysis (hazard ratio [HR]: 0.80, 95% CI: 0.32–2.02, P = .640). MGMT promoter methylation status showed association with sex (odds ratio 2.77, 95% CI: 1.14–6.95, P = .016) with proportionally more male patients in the MGMT promoter-unmethylated group. Details are presented in Supplementary File 1, Sections 7–9.
Figure 1.
Kaplan–Meier analysis for the endpoint overall survival. (A) 4-miRNA signature defined risk groups, (B) MGMT promoter methylation defined risk groups, (C) age defined risk groups. P values were calculated using the log-rank test.
Univariable Analyses and Stratified Subgroups
The 5 risk factors 4-miRNA signature risk group, MGMT promoter methylation status, sex, resection status, and age were univariably tested for association with OS. The 4-miRNA signature (P = .01, HR: 1.8, 95% CI: 1.14–2.83, Figure 1A), MGMT promoter methylation status (P < .001, HR: 0.42, 95% CI: 0.26–0.69, Figure 1B), and age (P = .048, HR: 1.58, 95% CI: 1.0–2.49, Figure 1C) were significantly associated with OS. Median OS was 20.7 versus 15.8 months for patients with 4-miRNA signature low-risk versus high-risk, 20.7 versus 15.0 months for methylated versus unmethylated MGMT promoter, and 20.2 versus 15.8 months for younger (≤60 years) and older (>60 years) patients, respectively.
In patients aged 60 years or younger, the combination of a methylated MGMT promotor and a low-risk 4-miRNA signature was associated with more than 2 times longer median OS (37.4 months), when compared to the corresponding 4-miRNA signature high-risk group (14.9 months) and the MGMT promoter-unmethylated groups (MGMT promoter-unmethylated/4-miRNA signature low-risk 18.6 months, MGMT promoter-unmethylated/4-miRNA signature high-risk 18.3 months; log-rank P = .019). Of note, in younger patients, the 4-miRNA signature high- and low-risk groups did not significantly differ in age (P = .078).
In patients older than 60 years of age, substratification of patients by the 4-miRNA signature and MGMT promoter methylation status resulted in median OS times of 20.7 for the MGMT promoter-methylated/4-miRNA signature low-risk-group, 18.2 months for the MGMT promoter-methylated/4-miRNA signature high-risk group, 14.1 months for the MGMT promoter-unmethylated/4-miRNA signature low-risk group, and 9.5 months for the MGMT promoter-unmethylated/4-miRNA signature high-risk group (log-rank P = .012). In older patients, the 4-miRNA signature high- and low-risk groups did not significantly differ in age (P = .577).
Multivariable Analyses
For the multivariable Cox analysis, the parameters 4-miRNA signature, MGMT promoter methylation status, and age were considered, which showed prognostic significance in univariable analysis on OS. Age was statistically associated with the 4-miRNA signature (see above) but did not show statistical interaction with the 4-miRNA signature in the Cox model (Chi-square test, P = .636). The multivariable Cox model including MGMT promoter methylation status, the 4-miRNA signature, and age significantly predicted OS (log-rank P value < .0001). In line with clinical practice and in order to rule out age-dependent effects, multivariable analysis including MGMT promoter methylation status and the 4-miRNA signature was carried out separately for younger (≤60 years) and older (>60 years) patients. A further prognostic impact of age was neither detectable in the younger (age as continuous variable: log-rank P > .3, age as median-split variable: log-rank P > .07) nor in the older patients (age as continuous variable: log-rank P > .5, age as median-split variable: log-rank P > .1). Details are presented in Table 2 and Supplementary File 1, Section 8.
Table 2.
Univariable and Multivariable Survival Analysis
| HR | 95% CI | P | ||
|---|---|---|---|---|
| Lower | Upper | |||
| Univariable models | ||||
| 4-miRNA signature | 1.8 | 1.14 | 2.83 | .01 |
| MGMT promoter methylation | 0.42 | 0.26 | 0.69 | <.001 |
| Age (categorized) | 1.58 | 1.0 | 2.49 | .047 |
| Sex | 1.31 | 0.82 | 2.08 | .261 |
| Resection status | 1.39 | 0.80 | 2.42 | .238 |
| Multivariable model younger patients | ||||
| 4-miRNA signature | 1.92 | 0.94 | 3.93 | .076 |
| MGMT promoter methylation | 0.45 | 0.21 | 0.95 | .036 |
| Multivariable model older patients | ||||
| 4-miRNA signature | 1.24 | 0.65 | 2.38 | .516 |
| MGMT promoter methylation | 0.34 | 0.16 | 0.71 | .004 |
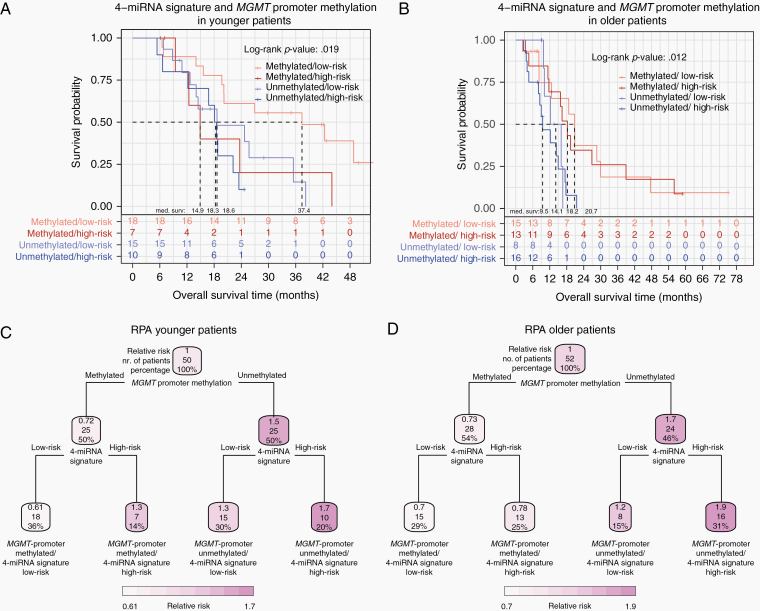
In younger patients of less than or equal to 60 years of age, the model including the 4-miRNA signature and MGMT promoter methylation status was significantly prognostic (log-rank P = .007, 4-miRNA signature: P = .076, HR 1.92, 95% CI: 0.94–3.93; MGMT promoter methylation: P = .036, HR 0.45, 95% CI: 0.21–0.95; Table 2 and Figure 2A). MGMT promoter-methylated/4-miRNA signature low-risk patients significantly performed better with regard to OS (median OS: 37.4 months) compared with the 3 remaining groups who had a median OS of 18.5 months (log-rank P = .0032, HR: 0.33, 95% CI: 0.15–0.71, Supplementary File 1, Section 17). The concordance index (C-Index) of the multivariable model including MGMT promoter methylation and the 4-miRNA signature was higher (0.62) compared to that of the univariable Cox model including the 4-miRNA signature (C-index: 0.59) or MGMT promoter methylation (C-index: 0.59) alone (Supplementary File 1, Section 18).
Figure 2.
Kaplan–Meier and recursive partitioning analysis (RPA) for younger (aged 60 years or younger) and older (aged >60 years) glioblastoma patients. (A) Kaplan–Meier analysis of combined 4-miRNA signature and MGMT promoter methylation defined risk groups in younger patients. (B) Kaplan–Meier analysis of combined 4-miRNA signature and MGMT promoter methylation defined risk groups in older patients. (C) RPA in younger patients, based on 4-miRNA signature and MGMT promoter methylation defined risk groups for the endpoint overall survival. (D) RPA in older patients, based on 4-miRNA signature and MGMT promoter methylation defined risk groups for the endpoint overall survival.
In patients older than 60 years of age, the model including the 4-miRNA signature and MGMT promoter methylation status was also significantly prognostic (log-rank P = .006, 4-miRNA signature: P = .516, HR 1.24, 95% CI: 0.65–2.38; MGMT promoter methylation: P = .004, HR 0.34, 95% CI: 0.16–0.71; Table 2 and Figure 2B). However, the contribution of the 4-miRNA signature in older patients was less prominent compared to younger patients. The multivariable Cox model including the 4-miRNA signature and MGMT promoter methylation had a C-index of 0.64 and the univariable models including MGMT promoter methylation C-indices of 0.62 and 0.56, respectively (Supplementary File 1, Section 18).
In both age groups, combining MGMT promoter methylation status with the 4-miRNA signature improved prognostic prediction accuracy.
Recursive Partitioning Analysis
For the generation of an RPA decision tree model, the parameters 4-miRNA signature, and MGMT promoter methylation status in combination with OS were given as input and the analysis was conducted separately for younger (≤60 years of age) and older patients (>60 years of age). For both age groups, the first hierarchical level of the decision tree was given by MGMT promoter methylation status, and the second level was given by the 4-miRNA signature.
In younger patients, the relative risk (predicted probability) of death was lower in MGMT promoter-methylated/4-miRNA signature low-risk patients (0.61) compared to the other patient groups (MGMT promoter-methylated/4-miRNA signature high-risk: 1.3, MGMT promoter-unmethylated/4-miRNA signature low-risk: 1.3 and MGMT promoter-unmethylated/4-miRNA signature high-risk: 1.7). In older patients, relative risks were markedly different in patients with MGMT promoter-methylated (0.73) versus -unmethylated (1.7) tumors, while further stratification by the 4-miRNA signature only led to moderate differences of relative risks between the groups (MGMT promoter-methylated/4-miRNA signature low-risk: 0.7, MGMT promoter-methylated/4-miRNA signature high-risk: 0.78, MGMT promoter-unmethylated/4-miRNA signature low-risk: 1.2, MGMT promoter-unmethylated/4-miRNA signature high-risk: 1.9). However, the 4-miRNA signature consistently identified subgroups with corresponding increased and decreased risks for all combinations of age group and methylation status. Details on the RPA results are shown in Figure 2C and D.
DNA Methylome Profiles and GSEA for Differentially Methylated Genes
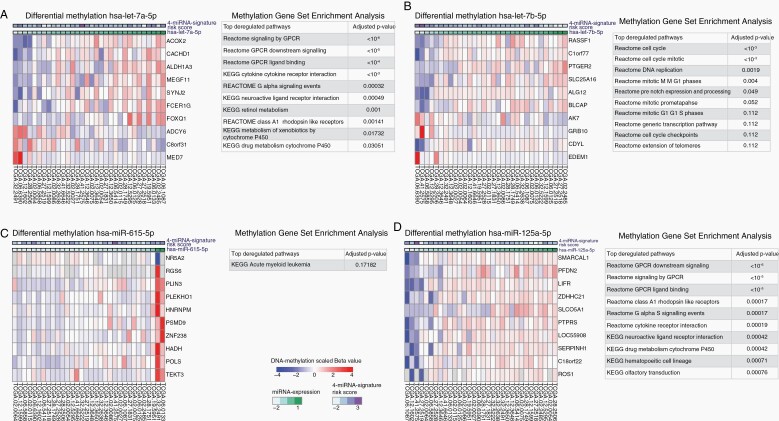
In order to gain insights into potential biological associations underlying the prognostic effect of the signature miRNAs, we utilized large-scale DNA methylation data from the TCGA database for analyses of differential gene methylation followed by gene set analysis for the identification of differentially methylated genes and corresponding pathways associated with the expression of each of the 4 signature miRNAs. The genes of the top 10 most differentially methylated CpG sites for each signature miRNA are shown as heatmaps in Figure 3A–D.
Figure 3.
Differential methylation and methylation gene set enrichment analysis (GSEA) for the signature miRNAs in the TCGA validation subcohort. (A–D) Heatmaps of the top 10 differentially methylated CpG sites (left in each panel) and up to 10 of the most significant (multiple testing corrected) enriched gene sets from the KEGG and Reactome pathway databases (right in each panel) for the miRNAs hsa-let-7a-5p, hsa-let-7b-5p, hsa-miR-615-5p, and hsa-miR-125a-5p.
Moreover, for each of the 4 signature miRNAs, enriched gene sets were identified. For hsa-miR-615-5p, only one enriched pathway (KEGG acute myeloid leukemia) was found. For hsa-let-7a-5p, hsa-let-7b-5p, and hsa-miR-125a-5p, several enriched pathways were identified, while all miRNAs showed deregulation of G-protein-coupled receptor signaling associated pathways, neuroactive ligand-receptor interaction, and cytokine–cytokine receptor interaction. Results of gene set enrichment for differentially methylated genes are summarized for each miRNA in Figure 3A–D.
Integrated Analysis of the Impact of Signature miRNAs and DNA Methylation on Gene Expression
For 23 patients, miRNA-, DNA methylation, and transcriptome data were available. A list of TCGA patient/sample IDs included in the analysis is presented in Supplementary File 2. In total, 36 genes were identified to be cooperatively regulated by DNA methylation and signature miRNAs. In detail for hsa-let-7a-5p 8 genes, for hsa-miR-125b-5p 16 genes, hsa-miR-615-5p 3 genes, and for hsa-let-7b-5p 9 genes were identified. The analysis of cooperative effects of signature miRNAs and DNA methylation on the expression of cancer genes as defined by the COSMIC Cancer Gene Consensus revealed for hsa-let-7a-5p genes SF3B1 and PTEN, for hsa-miR-125a-5p genes MDM4, PTEN, SND1, WT1, BRCA1, LMO2, ESR1, DNM2, MLLT10, and for hsa-miR-615-5p the CSF1R gene. The list of genes along with a functional description and their roles as cancer hallmarks defined by Hanahan and Weinberg (2011) can be found in Supplementary File 2. With regard to the relevance of identified genes, reflected by the number of PubMed-listed articles containing the keyword “glioblastoma” and the gene name of interest, we conducted a batch query of the PubMed database. Among genes that are predicted targets of the 4-miRNA signature, PAX6 regulated by hsa-let-7b-5p (29 articles), TCF4 regulated by hsa-let-7a-5p (15 articles), RRM2 regulated by hsa-miR-125a-5p (12 articles), EFEMP1 regulated by hsa-let-7b-5p (8 articles), and BCAN regulated by hsa-let-7a-5p (8 articles) were the most important. Among the Cancer Gene Consensus genes, PTEN regulated by hsa-miR-125a-5p and hsa-let-7a-5p (769 articles), BRCA1 regulated by hsa-miR-125a-5p (38 articles), WT1 regulated by hsa-miR-125a-5p (29 articles), MDM4 regulated by hsa-miR-125a-5p (17 articles), CSF1R regulated by hsa-miR-615-5p (9 articles), and ESR1 regulated by hsa-miR-125a-5p (6 articles) were the most important. The results of the integrated analysis are summarized in Supplementary File 2.
Discussion
MGMT promoter methylation is still the only clinically relevant biomarker in patients with IDH wild-type glioblastomas.2 It is commonly used for stratification of patients in clinical trials, may guide individual treatment decisions, in particular in older patients,2,28 and is linked to survival in patients treated with TMZ.2 However, additional predictive and prognostic biomarkers especially for younger glioblastoma patients are urgently needed for personalized treatment stratification and improved prediction of outcome.
The prognostic value of the 4-miRNA signature was confirmed in our cohort. Moreover, our data point to an additional prognostic value of the 4-miRNA signature in younger patients (aged 60 years or younger), when combined with information on MGMT promoter methylation. Here, the combination of the 4-miRNA signature with MGMT promoter methylation showed particularly beneficial results for patients with a methylated MGMT promoter status and low-risk 4-miRNA signature profile compared to all other groups, with 37.5 months median OS versus 14.9–18.6 months.
In contrast to previously published prognostic miRNA signatures for glioblastoma patients,29–31 the 4-miRNA signature was applied to an independent cohort without further adaptation of the underlying prognostic model, therefore, providing a high level of external validity. Even though the UKD subcohort comprised a selected group of glioblastoma patients, who all underwent surgery at recurrence, the split into 4-miRNA signature high-risk and low-risk patients and the combination with the MGMT promoter methylation status could be achieved in the same way as in the LMU and TCGA subcohorts.
In order to gain first insights into biological mechanisms underlying the prognostic association of the 4-miRNA signature, we performed a GSEA of the TCGA subcohort based on differentially methylated genes identified by array-based epigenetic profiling. In an analogous approach, carried out in the frame of the study that discovered the 4-miRNA signature, expression of the 4 signature miRNAs was correlated to global gene expression and revealed involvement of the immune response, extracellular matrix organization, neural growth factor, Wnt and G-protein-coupled receptors (GPCR) signaling.10
In the herein presented DNA methylation-based GSEA analysis, the most prominent molecular mechanism was the deregulation of GPCR-mediated signaling, which was identified for 3 of the 4 miRNAs, except hsa-miR-615-5p. GPCR signaling was thus found by both mRNA expression and DNA methylation profiling. It has been described as being of major importance for cell cycle regulations, cell death, and infiltration consisting of migration and invasion of glioma cells.32 Another pathway that was present in all gene set enrichment data except hsa-miR-615-5p was the neuroactive ligand-receptor interaction pathway. This pathway shows a high overlap with the GPCR pathway. Xi et al.33 reported that the neuroactive ligand-receptor interaction pathway is regulated by the upregulation of the miRNA hsa-let-7b-5p in human glioma, which leads to its transcriptional repression.
Moreover, our DNA methylation-based GSEA showed deregulation of the KEGG acute myeloid leukemia pathway, which integrates a number of cancer pathways including Pi3K-Akt, apoptosis, mTOR signaling, and cell cycle pathways. Sun et al.34 have identified the Gab2 gene as a target of hsa-miR-125a-5p and a functional association with cytoskeleton rearrangement and matrix metalloproteinases expression, which inhibited invasiveness of human glioma cells. In addition, we performed an analysis in which cooperative effects of the expression of the 4-miRNA signature and DNA methylation on predicted miRNA-target genes and on known cancer genes from the Cancer Gene Census were investigated. A number of known and new genes associated with glioblastoma were discovered. Among the 4-miRNA signature target genes, SF3B1 (splicing factor 3b subunit 1), ONECUT2 (one cut homeobox 2), and PAWR (pro-apoptotic WT1 regulator) showed the strongest interaction effects. SF3B1 is involved in alternative splicing, frequently mutated in myeloid neoplasms and appeared also as most statistically significant in the analysis of Cancer Gene Census gene and has not been published in the context of glioblastoma so far.35 ONECUT2, a transcription factor of the onecut family, is involved in cell differentiation was shown to be down-regulated in glioblastoma. PAWR is a known tumor-suppressor gene involved in the regulation of apoptosis and has been suggested as a prognostic risk factor in glioblastoma.36 The genes PAX6, TCF4, RRM2, EFEMP1, and BCAN also appeared to be cooperatively affected by the 4-miRNA signature and DNA methylation and are well known to be involved in glioblastoma tumorigenesis, reflected by their numbers of PubMed records. Among analyzed cancer genes and in addition to the aforementioned SF3B1 gene, the suppressor gene PTEN, which is involved in various cancers including glioblastoma, showed the second strongest statistical significance. These are followed by MDM4, CSF1R, SND1, WT1, BRCA1, LMO2, ESR1, DNM2, MLLT10, all of which, except MLLT10, known glioblastoma genes. Due to the small sample size (n = 23) of the integrated data set, the results can only be indicative, but they provide potentially meaningful hypotheses for further investigation.
In univariable and multivariable analyses, we detected the MGMT promoter methylation status, the 4-miRNA signature, and age as statistically significant prognostic parameters and an association between the 4-miRNA signature and age. In order to account for the association of the 4-miRNA signature with age, separate RPAs for younger and older patients were performed, as in the setting of clinical decision-making. When looking separately at the stratified age cohorts of younger and older patients, no further statistically significant stratification by age was observed, while the combination of MGMT promoter methylation with 4-mRNA signature was able to sub-stratify both age groups.
As expected, in the first hierarchy level of the RPA through stratification by the MGMT promoter methylation status, a comparable risk difference was present for both age groups. In the second hierarchy level, in the case of an unmethylated MGMT promoter, further risk stratification by the 4-miRNA signature was possible for younger and older patients. In case of a methylated MGMT promoter, an especially large risk difference between the 4-miRNA signature defined high-risk and low-risk group was found for younger patients, whereas for older patients only a marginal difference was present. This finding might be of special interest for further risk stratification, as the group of younger patients with MGMT promoter-methylated glioblastoma and a 4-miRNA signature high-risk constellation might benefit most from therapy intensification, such as radiation dose escalation or additional targeted medication given concomitantly to CRT or as maintenance therapy.
As recently reported, combined TMZ and lomustine (CCNU) chemotherapy prolongs survival of glioblastoma patients with a methylated MGMT promoter status, but also increases toxicity.37 Therefore, it is of particular interest whether the 4-miRNA signature could be applied to identify those glioblastoma patients with an MGMT promoter-methylated status who benefit most from treatment intensification through additional alkylating chemotherapy.
In MGMT promoter-unmethylated glioblastoma patients who show weaker response to alkylating chemotherapy, it has been investigated in small-scale studies, whether these patients might benefit from dose escalation of radiotherapy exceeding the standard dose of 60 Gy in 2 Gy fractions.38,39 Following the results of our study, in the case of an unmethylated MGMT promoter, patients with a 4-miRNA signature low-risk constellation could possibly have a greater benefit from dose escalation. Prospective clinical trials are warranted to assess the survival benefit of dose escalation for this specific subgroup of glioblastoma patients.
Yet, and comparable to the detection of the high relevance of IDH mutation, it remains to be determined further which molecular alterations are driving the prognostic value of the 4-miRNA signature. To address this question, comprehensive integrated in vitro and in vivo studies of the signature miRNAs are needed.40,41 In addition, validation in prospectively collected glioblastoma cohorts treated according to the standard of care will be the next step toward clinical application of the 4-miRNA signature. The influence of sample modality (snap-frozen vs FFPE) and intratumoral heterogeneity on the stability of the 4-miRNA signature are also important issues to be investigated in future studies.
In summary, the 4-miRNA signature in combination with the MGMT promoter methylation status improves individual prognosis prediction of IDH wild-type glioblastoma patients treated with standard CRT, particularly in patients aged 60 years or younger at diagnosis.
Supplementary Material
Acknowledgments
The authors would like to thank Aaron Selmeier, Claire Innerlohinger, and Laura Dajka for their excellent technical support. M.M. would like to thank the Luxembourg National Research Fund (FNR) for support (FNR PEARL P16/BM/11192868 grant). Parts of the reported results are based on data generated by the TCGA Research Network (http://cancergenome.nih.gov/).
Funding
No funding source to be declared.
Conflict of interest statement. A patent application for the 4-miRNA signature as a prognostic marker in glioblastoma has been filed and is pending in the United States and the European Union (WO2017/109089, K.U., H.Z., C.B., M.N., M.M.). G.R. has received honoraria for advisory boards from Abbvie. M.N. has received research grants from Roche, speaker honoraria from Novocure, as well as honoraria for advisory boards from Brainlab. The other authors have no conflicts of interest to declare.
Authorship statement. K.U., D.F.F., C.B., and M.N. developed the study concept and supervised the study; K.U., D.F.F., V.R., J.F., D.P., D.S., Ju.H., M.M., K.L., W.B., M.P.S., C.R., G.R., Jo.H., J.-C.T., H.Z., C.B., and M.N. contributed to its implementation; K.U., D.F.F., D.S., Ju.H., and M.N. performed the statistical evaluations; and K.U., D.F.F., K.L., J.-C.T., H.Z., C.B., and M.N. contributed to the interpretation of the data. K.U. and D.F.F. wrote the manuscript with the help of the other authors. All authors read and approved the final manuscript.
References
- 1. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2. Weller M, van den Bent M, Tonn JC, et al. ; European Association for Neuro-Oncology (EANO) Task Force on Gliomas European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18(6):e315–e329. [DOI] [PubMed] [Google Scholar]
- 3. Hegi ME, Diserens AC, Gorlia T, et al. . MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 4. Brandes AA, Tosoni A, Franceschi E, et al. . Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: correlation With MGMT promoter methylation status. J Clin Oncol. 2009;27(8):1275–1279. [DOI] [PubMed] [Google Scholar]
- 5. Tsien C, Moughan J, Michalski JM, et al. ; Radiation Therapy Oncology Group Trial 98-03 Phase I three-dimensional conformal radiation dose escalation study in newly diagnosed glioblastoma: Radiation Therapy Oncology Group Trial 98-03. Int J Radiat Oncol Biol Phys. 2009;73(3):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Piroth MD, Pinkawa M, Holy R, et al. . Prognostic value of early [18F]fluoroethyltyrosine positron emission tomography after radiochemotherapy in glioblastoma multiforme. Int J Radiat Oncol Biol Phys. 2011;80(1):176–184. [DOI] [PubMed] [Google Scholar]
- 7. Fitzek MM, Thornton AF, Rabinov JD, et al. . Accelerated fractionated proton/photon irradiation to 90 cobalt gray equivalent for glioblastoma multiforme: results of a phase II prospective trial. J Neurosurg. 1999;91(2):251–260. [DOI] [PubMed] [Google Scholar]
- 8. Le Rhun E, Preusser M, Roth P, et al. . Molecular targeted therapy of glioblastoma. Cancer Treat Rev. 2019;80:101896. [DOI] [PubMed] [Google Scholar]
- 9. Hegi ME, Stupp R. Withholding temozolomide in glioblastoma patients with unmethylated MGMT promoter—still a dilemma? Neuro Oncol. 2015;17(11):1425–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niyazi M, Pitea A, Mittelbronn M, et al. . A 4-miRNA signature predicts the therapeutic outcome of glioblastoma. Oncotarget. 2016;7(29):45764–45775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stupp R, Hegi ME, Mason WP, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 12. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 13. Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials. 1996;17(4):343–346. [DOI] [PubMed] [Google Scholar]
- 14. Eigenbrod S, Trabold R, Brucker D, et al. . Molecular stereotactic biopsy technique improves diagnostic accuracy and enables personalized treatment strategies in glioma patients. Acta Neurochir (Wien). 2014;156(8):1427–1440. [DOI] [PubMed] [Google Scholar]
- 15. Esteller M, Garcia-Foncillas J, Andion E, et al. . Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. [DOI] [PubMed] [Google Scholar]
- 16. Möllemann M, Wolter M, Felsberg J, Collins VP, Reifenberger G. Frequent promoter hypermethylation and low expression of the MGMT gene in oligodendroglial tumors. Int J Cancer. 2005;113(3):379–385. [DOI] [PubMed] [Google Scholar]
- 17. Felsberg J, Thon N, Eigenbrod S, et al. ; German Glioma Network Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer. 2011;129(3):659–670. [DOI] [PubMed] [Google Scholar]
- 18. Bady P, Sciuscio D, Diserens AC, et al. . MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerds TA, Kattan MW, Schumacher M, Yu C. Estimating a time-dependent concordance index for survival prediction models with covariate dependent censoring. Stat Med. 2013;32(13):2173–2184. [DOI] [PubMed] [Google Scholar]
- 20. Breiman L, Friedman J, Stone CJ, Olshen RA.. Classification and Regression Trees. Abingdon, UK: Taylor & Francis; 1984. [Google Scholar]
- 21. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 22. Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jassal B, Matthews L, Viteri G, et al. . The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48(D1):D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mounir M, Lucchetta M, Silva TC, et al. . New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput Biol. 2019;15(3):e1006701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shivakumar M, Lee Y, Bang L, Garg T, Sohn KA, Kim D. Identification of epigenetic interactions between miRNA and DNA methylation associated with gene expression as potential prognostic markers in bladder cancer. BMC Med Genomics. 2017;10(Suppl 1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ru Y, Kechris KJ, Tabakoff B, et al. . The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014;42(17):e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer. 2018;18(11):696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malmström A, Grønberg BH, Marosi C, et al. ; Nordic Clinical Brain Tumour Study Group (NCBTSG) Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13(9):916–926. [DOI] [PubMed] [Google Scholar]
- 29. Sana J, Radova L, Lakomy R, et al. . Risk score based on microRNA expression signature is independent prognostic classifier of glioblastoma patients. Carcinogenesis. 2014;35(12):2756–2762. [DOI] [PubMed] [Google Scholar]
- 30. Hayes J, Thygesen H, Tumilson C, et al. . Prediction of clinical outcome in glioblastoma using a biologically relevant nine-microRNA signature. Mol Oncol. 2015;9(3):704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hermansen SK, Sørensen MD, Hansen A, et al. . A 4-miRNA signature to predict survival in glioblastomas. PLoS One. 2017;12(11):e0188090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cherry AE, Stella N. G protein-coupled receptors as oncogenic signals in glioma: emerging therapeutic avenues. Neuroscience. 2014;278:222–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xi X, Chu Y, Liu N, et al. . Joint bioinformatics analysis of underlying potential functions of hsa-let-7b-5p and core genes in human glioma. J Transl Med. 2019;17(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun L, Zhang B, Liu Y, Shi L, Li H, Lu S. MiR125a-5p acting as a novel Gab2 suppressor inhibits invasion of glioma. Mol Carcinog. 2016;55(1):40–51. [DOI] [PubMed] [Google Scholar]
- 35. Hershberger CE, Moyer DC, Adema V, et al. . Complex landscape of alternative splicing in myeloid neoplasms. Leukemia. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deshmukh H, Yu J, Shaik J, et al. . Identification of transcriptional regulatory networks specific to pilocytic astrocytoma. BMC Med Genomics. 2011;4:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Herrlinger U, Tzaridis T, Mack F, et al. ; Neurooncology Working Group of the German Cancer Society Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): a randomised, open-label, phase 3 trial. Lancet. 2019;393(10172):678–688. [DOI] [PubMed] [Google Scholar]
- 38. Tini P, Nardone V, Pastina P, et al. . Patients affected by unmethylated O(6)-methylguanine-DNA methyltransferase glioblastoma undergoing radiochemotherapy may benefit from moderately dose-escalated radiotherapy. Biomed Res Int. 2017;2017:9461402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zschaeck S, Wust P, Graf R, et al. . Locally dose-escalated radiotherapy may improve intracranial local control and overall survival among patients with glioblastoma. Radiat Oncol. 2018;13(1):251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. . Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parsons DW, Jones S, Zhang X, et al. . An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.