Abstract
Porphyromonas gingivalis, a periodontal pathogen, translocates many virulence factors including the cysteine proteases referred to as gingipains to the cell surface via the type IX secretion system (T9SS). Expression of the T9SS component proteins is regulated by the tandem signaling of the PorXY two-component system and the ECF sigma factor SigP. However, the details of this regulatory pathway are still unknown. We found that one of the T9SS conserved C-terminal domain-containing proteins, PGN_0123, which we have designated PorA, is involved in regulating expression of genes encoding T9SS structural proteins and that PorA can be translocated onto the cell surface without the T9SS translocation machinery. X-ray crystallography revealed that PorA has a domain similar to the mannose-binding domain of Escherichia coli FimH, the tip protein of Type 1 pilus. Mutations in the cytoplasmic domain of the sensor kinase PorY conferred phenotypic recovery on the ΔporA mutant. The SigP sigma factor, which is activated by the PorXY two-component system, markedly decreased in the ΔporA mutant. These results strongly support a potential role for PorA in relaying a signal from the cell surface to the PorXY-SigP signaling pathway.
Subject terms: Microbiology, Molecular biology
Introduction
Chronic periodontitis is caused by bacterial infection and the subsequent immune response. A gram-negative anaerobe, Porphyromonas gingivalis, is a keystone pathogen of chronic periodontitis1. Gingipains are cysteine proteases located on the cell surface, which are central to virulence and hemagglutinating activity2,3. P. gingivalis has two types of gingipains; arginine-specific proteases, RgpA and RgpB, and lysine-specific protease, Kgp. Gingipains are translocated to the cell surface or in milieu by using the Type IX secretion system (T9SS)4,5. Approximately 30 proteins, including gingipains, Hbp35, TapA, PepK, PPAD, Mfa5, PorU, and PorZ, are secreted via the T9SS6–8 and are called T9SS cargo proteins. However, most of the T9SS cargo proteins have not been characterized.
The T9SS cargo proteins share a conserved C-terminal domain (CTD) composed of about 80 amino acid residues as a secretion signal for the T9SS. Sequence analysis of CTDs have revealed that T9SS CTDs belong to either protein domain family TIGR04183 (type A CTDs) or TIGR04131 (type B CTDs)9. Most of the T9SS cargo proteins have type A CTDs, but Flavobacterium johnsoniae has many T9SS cargo proteins with type B CTDs10. P. gingivalis has only one T9SS cargo protein with type B CTD (PGN_1317) and the others with type A CTDs. P. gingivalis T9SS cargo proteins can be classified into three groups according to their anchoring modes on the cell surface2,6,8,11–21. Group I proteins, which include RgpB, Hbp35, TapA, PepK, and PPAD, are covalently attached to anionic polysaccharide-containing lipopolysaccharides (A-LPS) after removal of their CTDs by the PorU sortase22. The attachment to A-LPS yields diffuse protein bands of the group I proteins in SDS-PAGE. Group II proteins are associated with other proteins, not A-LPS on the cell surface after removal of their CTDs by PorU, which include Mfa5. Mature/processed RgpA and Kgp may be categorized in this group; however, the proteins are generated by the proteolytic processing of RgpA and Kgp precursors. Group III proteins possess their CTDs on cell surface and are not associated with A-LPS. PorU and PorZ, which are T9SS component proteins, belong to group III11,16. PorU cleaves CTD of groups I and II cargo proteins, but no common sequence has been found at the cleavage sites23. It is still unknown how PorU recognizes the cleavage site.
Porphyromonas gingivalis colonies grown on blood agar plates are black-pigmented, and many genes responsible for this unique characteristic of the bacterium have been identified. The pigmentation is caused by release of heme from hemoglobin by Kgp24,25 and subsequent accumulation of a heme derivative, μ-oxo heme dimer, on the cell surface26. Transposon mutagenesis revealed that porR and porT genes are involved in the pigmentation4,27. PorR is prerequisite to form A-LPS, and PorT is one of the T9SS component proteins. Our exhaustive random transposon mutagenesis analysis on pigmentation has revealed that 19 genes are involved in the A-LPS synthesis, 15 genes are T9SS component proteins, and 3 genes, porX, porY and sigP are involved in gene regulation of the T9SS component proteins28–31. The porX and porY genes encode a response regulator and a sensor kinase of two-component system, and the sigP gene encodes an ECF sigma factor5,29. However, more genes may relate to the pigmentation because we analyzed only non-pigmented mutants and not the slightly-pigmented mutants, which may have defects in pigmentation-related genes31.
A T9SS CTD-containing protein, PorA (PGN_0123), was reported to form diffuse protein bands with molecular masses of more than 35 kDa7 and its CTD cleavage site was determined11. Here we show that PorA is located in the upstream of the PorXY-SigP signaling cascade, which impacts the T9SS expression. PorA is translocated to the cell surface without the T9SS translocation machinery and localized on the cell surface in both A-LPS bound diffuse form and 23-kDa CTD-containing form, and thus PorA is a novel type of T9SS CTD-containing protein. PorA is 246 amino acids in length and is composed of an N-terminal domain and CTD with an N-terminal signal sequence consisting of 27 residues. The structure of the N-terminal domain resembles the ligand binding domain of FimH. On the basis of the structure and the results of the functional analyses, we propose a plausible mechanism for how PorA influences the T9SS expression.
Results
Pigmentation, hemagglutination, and gingipain activities of the ΔporA mutant
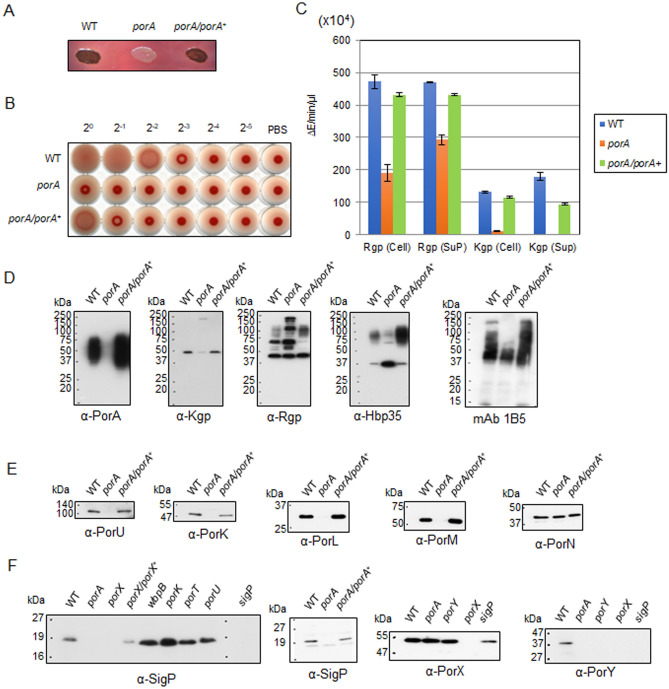
PGN_0123, PGN_0654, and PGN_1770 are small CTD-containing proteins, but their functions are not known. We constructed deletion mutants of each protein and examined their phenotypes. The PGN_0123 mutant showed less pigmentation on blood agar plates (Fig. 1A) as expected by Klein et al.12, whereas PGN_0654 and PGN_1770 mutants were pigmented (Fig. S1A). Thus, we designated the gene of PGN_0123 as porA. The ΔporA mutation decreased the hemagglutinating activity and the gingipain activities of the cells (Fig. 1B,C). These functional defects were recovered by expression of porA+ from plasmid in the ΔporA mutant cells, indicating that PorA is required for pigmentation and virulence. In this complementation experiment, we used a shuttle vector, pTCB, and expressed the porA gene with the promoter of the catalase gene (cat) in Porphyromonas gulae (see in Supplemental text).
Figure 1.
Characterization of P. gingivalis ΔporA mutant. (A) Pigmentation of the ΔporA mutant on blood agar plates for 3 days. (B) Hemagglutinating activity of the ΔporA mutant. (C) Kgp and Rgp activities of the ΔporA mutant. The cell lysates (cell) and vesicle-containing culture supernatants (sup) were subjected to the assay. Lane 1: wild type, Lane 2: ΔporA, Lane 3: ΔporA/porA+. (D) Immunoblot analyses of T9SS CTD-containing proteins and A-LPS in the ΔporA mutant. Cell lysates of the wild type (lane 1), ΔporA (lane 2), and ΔporA/porA+ (lane3) were analyzed by SDS-PAGE, followed by immunoblot analyses of T9SS CTD-containing proteins and A-LPS using antibodies against PorA (α-PorA), Kgp (α-Kgp), Rgp (α-Rgp) and Hbp35 (α-Hbp35), and monoclonal antibody against A-LPS (mAb 1B5). (E) Cell lysates of the wild type (lane 1), ΔporA (lane 2), and ΔporA/porA+ (lane3) were analyzed by SDS-PAGE, followed by immunoblot analyses of T9SS component proteins using antibodies against PorU (α-PorU), PorK (α-PorK), PorL (α-PorL), PorM (α-PorM), and PorN (α-PorN). (F) Immunoblot analyses of the T9SS regulatory proteins SigP, PorY, and PorX in various T9SS-related mutants. Cells of P. gingivalis strains were lysed with 1% N-Dodecyl-β-d-maltopyranoside (DDM) (vol/vol) and then subjected to SDS-PAGE, followed by immunoblot analysis using antibodies against SigP (α-SigP), PorX (α-PorX), and PorY (α-PorY). Full-length blots/gels of (E) and (F) are presented in Supplementary Figure S11.
PorA-mediated expression of T9SS component proteins
Previous studies revealed that the cause of no or less colony pigmentation is divided into two categories: one cause is defined by defects in T9SS and the other is defined by defects in A-LPS synthesis4,27. In the T9SS-deficient mutants, such as the porK and porT mutants, T9SS cargo proteins are not associated with A-LPS; they are detected in the periplasm only as unprocessed and unmodified forms lacking N-terminal signal peptides, and mAb 1B5, a monoclonal antibody against A-LPS, reacts to produce diffuse bands with apparent molecular masses lower than those of the wild type in SDS-PAGE32. On the other hand, in the A-LPS-deficient mutants such as the porR and wbpB mutants, most of the T9SS cargo proteins are secreted into culture supernatants and mAb 1B5 shows no reaction to their cell lysates33. To elucidate whether the PorA deficit affects the T9SS or the A-LPS synthesis, we conducted immunoblot analysis of the ΔporA mutant. In the ΔporA mutant, unprocessed and unmodified form bands of Kgp and Rgp were detected at significant amounts in addition to their mature/processed form bands, and the diffuse bands were markedly reduced for Hbp35 compared to those of the wild type and the complemented strains (Fig. 1D). The immunoblot of SDS-PAGE gels of the ΔporA mutant with mAb 1B5 showed diffuse bands with their apparent molecular masses lower than those of the wild type and the complemented strain (Fig. 1D). These properties of the ΔporA mutant were more similar to those of the T9SS-deficient mutants than the A-LPS-deficient mutants, suggesting that the ΔporA mutation does not affect the A-LPS synthesis but induces a partial defect in the T9SS.
We next investigated the levels of several T9SS component proteins in the ΔporA mutant by immunoblot analysis. PorU, PorK, PorL, and PorM were almost absent in the ΔporA mutant (Fig. 1E); however, PorK, PorL, and PorM were still produced at a certain level compared to corresponding null mutants (Fig. S2). In contrast, ΔporA mutation did not affect the amount of PorN (Fig. 1E). Since production of these Por proteins is regulated by the PorXY-SigP signaling pathway, we analyzed the amounts of PorX, PorY, and SigP in the ΔporA mutant (Fig. 1F). No SigP was detected in the ΔporA mutant, which was also the case for the ΔporX mutant29. Amounts of PorX in the ΔporA and ΔporY mutants were comparable to that of the wild type, whereas that in the ΔsigP mutant exhibited nearly 70% and 72% reduction in mRNA and protein levels, respectively (Fig. 1F, Fig. S3). Intriguingly, we found that no PorY protein was observed in the ΔporA, ΔporX, or ΔsigP mutant (Fig. 1F, Fig. S4A). qRT-PCR analysis revealed that the porY mRNA levels of the ΔporA, ΔporX, and ΔsigP mutants were reduced by 50%, 30%, and 80%, respectively, compared to that of the wild type (Fig. S4B). Therefore, marked decrease of the PorY protein in the ΔporA, ΔporX, and ΔsigP mutants may be caused not only at the mRNA level but also at the protein level.
Analysis of gain-of-function mutants from the ΔporA mutant
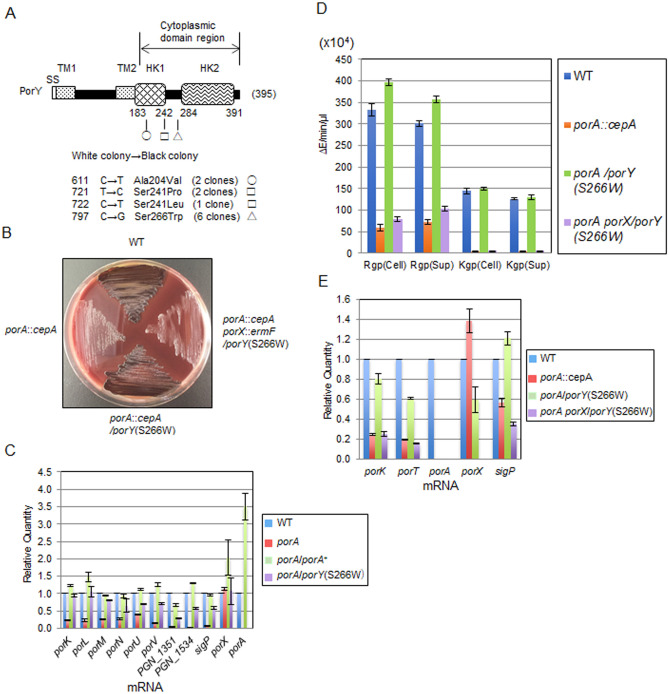
We isolated gain-of-function mutants from the ΔporA mutant by repeating inoculation of the ΔporA mutant on blood agar plates. Black-pigmented colonies appeared after three times successive inoculation. Two pseudo-revertants were subjected to genome analysis. Both of the two strains had the same two mutations that caused the conversion of the 241st Ser of PorY and the 295th Val of PGN_1053 (putative phospho-2-dehydro-3 deoxyheptonate aldolase/chorismate mutase) to Leu and Glu, respectively. These pseudo-revertants appeared to be siblings, so that we accounted the two strains as one clone (Fig. 2A). Next, we sequenced the porY gene regions from ten pseudo-revertants that were obtained independently, and found that all the strains possessed amino acid-substituted mutations in the putative cytoplasmic domain region of the porY gene (Fig. 2A). To confirm whether the porY mutations compensate the defects of the ΔporA mutant, the porY gene carrying one of the mutations, porY(S266W), was introduced into the ΔporA mutant by a shuttle plasmid. The resulting strain ΔporA/porY(S266W) showed a black-pigmented phenotype, as did the wild type (Fig. 2B). PorY is a component of the two-component signal transduction system involved in gene regulation of the T9SS component proteins28–31. PorY is a sensor kinase and PorX is a response regulator of the two-component system. The PorXY system positively regulates the T9SS expression by influencing SigP, an extra-cytoplasmic function (ECF) sigma factor5,29. Taken together with marked decrease of PorY and SigP in the ΔporA mutant, we speculated that PorA influences the expression of the T9SS related genes at mRNA level through the PorXY-SigP signaling pathway. To confirm this hypothesis, qRT-PCR analysis of the ΔporA mutant was performed. The ΔporA mutant showed markedly reduced mRNA levels of PGN_1351, PGN_1534, and T9SS component genes such as porK, porL, porM, porN, porU, and porV (Fig. 2C), all of which are reported to be down-regulated in the porX and sigP mutants29. Notably, the mRNA level of sigP was also markedly decreased in the ΔporA mutant compared to that of the wild type. Complemented strain ΔporA/porA+ and strain ΔporA/porY(S266W) recovered the expression of all these genes to the wild type level. We also confirmed that the ΔporA/porA+ strain recovered the SigP protein expression to the wild type level (Fig. 1F). To examine whether the ΔporA/porY(S266W) strain still utilizes the PorXY-SigP signaling pathway for T9SS regulation, a porX mutation was introduced into the chromosome of the ΔporA/porY(S266W) strain. The resulting strain ΔporA ΔporX/porY(S266W) showed a non-pigmented phenotype, indicating that PorY(S266W) also requires PorX for the phenotypic recovery (Fig. 2B,D,E). Furthermore, we constructed a ΔporA ΔporY mutant and introduced the porY(S266W) gene-carrying shuttle plasmid to the ΔporA ΔporY mutant and found that the resulting ΔporA ΔporY/porY(S266W) strain shows pigmented, produces PorY(S266W), and has Rgp and Kgp activities (Figs. S1B, S5, and S6). In addition, the ΔporA ΔporY mutant was stably non-pigmented after several passages and exhibited significantly reduced Kgp activity (Figs. S1B, S5, and S6). These results suggest that PorA is located upstream of the PorXY-SigP signaling pathway.
Figure 2.
Pseudo-revertants from the porA deletion mutant. (A) Mutations in porY of the pseudo-revertants. Mutations causing amino acid substitutions were found in the cytoplasmic domain-encoding region of porY. HK, histidine kinase domain; SS, signal sequence; TM, transmembrane region. Circle, square, and triangle indicate amino acid substitutions at 204, 241, and 266, respectively. (B) Colony pigmentation of the wild type, ΔporA, ΔporA/porY(S266W), and ΔporA ΔporX/porY(S266W) strains on the blood agar plate for 6 days. (C) qRT-PCR expression analysis of various T9SS-related genes in the wild type, ΔporA, ΔporA/porA+ and ΔporA/porY(S266W) strains. The mean of expression of each wild type gene was regarded as 1. (D) Gingipain activities of the wild type, ΔporA, ΔporA/porY(S266W), and ΔporA ΔporX/porY(S266W) strains. (E) Expression of the porK, porT, porA, porX, and sigP genes in the wild type, ΔporA, ΔporA/porY(S266W), and ΔporA ΔporX/porY(S266W) strains. RNA samples of the strains were subjected to qRT-PCR analysis. The mean expression of each wild type gene was regarded as 1.
Translocation of PorA onto the cell surface in a T9SS translocation machinery-independent manner
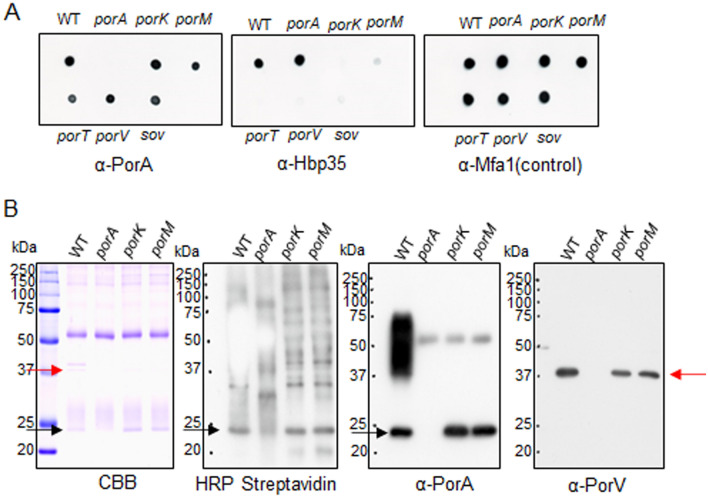
Since the C-terminal amino acid sequence of PorA (D163-K246) shows significant homology to the T9SS CTD sequence6, PorA is expected to be secreted to the cell surface through the T9SS. We therefore examined the PorA exposure on the cell surface by a dot blot analysis of intact cells with anti-PorA antibody (α-PorA) and with anti-Hbp35 antibody (α-Hbp35) reacting to Hbp35, a typical group I T9SS cargo protein. The wild-type cells reacted to both α-PorA and α-Hbp35, suggesting that PorA is present on the cell surface (Fig. 3A). α-Hbp35 did not react to the cell surface of T9SS-deficient mutant (porK, porM, porT, porV, or sov), whereas α-PorA reacted to those of the T9SS-deficient mutants (Fig. 3A)15. The result suggests that PorA can be translocated on the cell surface through a pathway without the T9SS translocation machinery. Hbp35 was detected on the cell surface in the ΔporX and the ΔporY mutants as well as the ΔporA mutant, but not in the porK mutant, indicating that the phenotype of the ΔporA mutant is similar to those of the ΔporX and ΔporY mutants (Fig. S7).
Figure 3.
Cell surface localization of PorA in P. gingivalis cells. (A) Cells of P. gingivalis wild type, ΔporA, porK, porM, porT, porV, and sov strains were blotted on nylon membranes and immunodetected by antibodies against PorA (α-PorA), Hbp35 (α-Hbp35), and Mfa1 (α-Mfa1). (B) Biotin-labeled cells of P. gingivalis wild type, ΔporA, porK, and porM strains were lysed and then immunoprecipitated by α-PorA. The immunoprecipitated samples were subjected to SDS-PAGE, followed by immunoblot analyses using HRP conjugated streptavidin, α-PorA, and antibody against PorV (α-PorV). CBB: Coomassie Brilliant Blue staining. Red and black arrows indicate PorV and PorA proteins, respectively.
Cell surface exposure of PorA was further examined by biotin labelling assay. Proteins exposed on the cell surfaces of porK, porM, ΔporA mutant and the wild-type cells were labeled with biotin. The cells were lysed, and the lysates were immunoprecipitated with α-PorA. The immunoprecipitated samples were loaded in SDS-PAGE and immunodetected with HRP conjugated streptavidin (HRP-S) and α-PorA (Fig. 3B). The sample from the wild-type cells showed diffuse bands and a thick band of about 23 kDa in both HRP-S and α-PorA staining gels. The diffuse bands are typical for the group I cargo proteins covalently bound to A-LPS on the cell surface. The theoretical molecular mass of PorA without the N-terminal signal sequence is 23,426. Therefore, these results suggest that PorA is located on the cell surface in two forms, an A-LPS bound form and a 23-kDa CTD-containing form. On the other hand, the porK and porM mutant samples showed the 23-kDa band but no diffuse bands in the HRP-S and α-PorA staining gels, indicating that PorA is present only in the 23-kDa CTD-containing form on the cell surfaces of the T9SS-deficient mutants.
The PorV protein can bind several T9SS CTD-containing proteins34,35. Glew et al.35 reported interaction between PorA and PorV. To confirm the interaction, the samples immuno-precipitated with α-PorA were subjected to immunoblot analysis with α-PorV. A protein band with 37 kDa reacted to α-PorV, indicating interaction of PorA and PorV (Fig. 3B).
Interaction of CTD of PorA with PorV
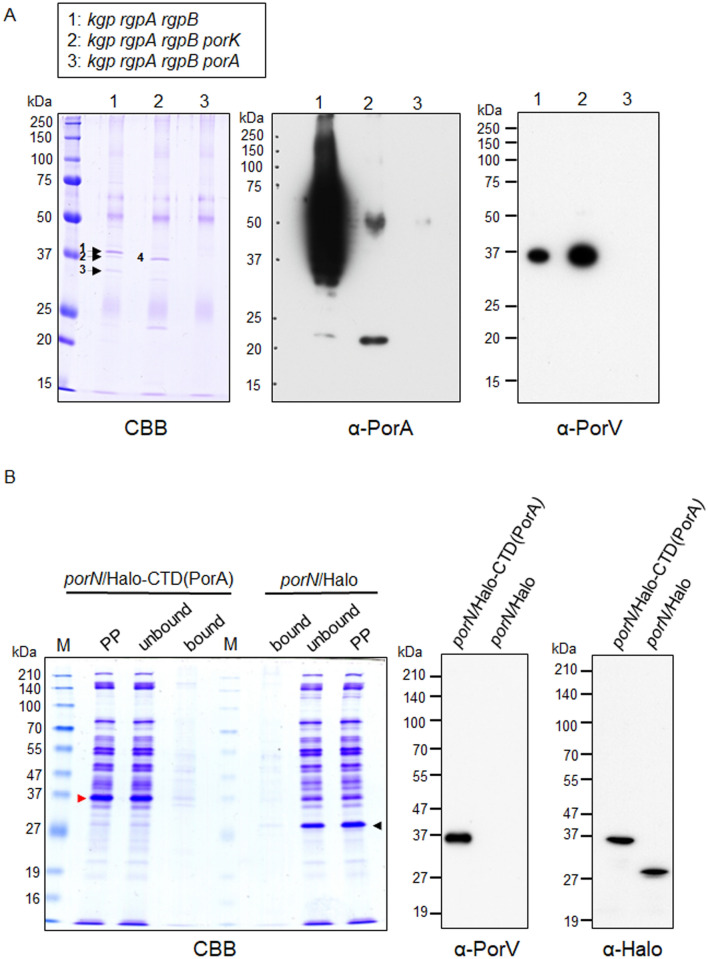
We performed peptide mass fingerprinting to identify proteins immuno-precipitaed with α-PorA. In the experiment, we used derivatives of a gingipain-null mutant (kgp rgpA rgpB) to avoid excessive proteolysis during sample preparation. Membrane fractions of kgp rgpA rgpB, kgp rgpA rgpB porK, and kgp rgpA rgpB ΔporA cells treated with a chemical crosslinking reagent, dimethyl 3,3′-dithiobispropionimidate (DTBP) were prepared, immuno-precipitated with α-PorA and subjected to SDS-PAGE. Protein bands on the gel indicated by numerals were subjected to peptide mass fingerprinting using MALDI TOF–MS (Fig. 4A and Table 1). The gel was also subjected to immunoblot analyses with α-PorA and α-PorV (Fig. 4A). The sample of kgp rgpA rgpB showed diffuse bands and a weak band with 23 kDa that reacted to α-PorA, and a band with 37 kDa that reacted to α-PorV, while the sample of kgp rgpA rgpB porK showed relatively strong bands with 23 kDa and 50 kDa that reacted to α-PorA and a band with 37 kDa that reacted to α-PorV. The 50-kDa band reacting to α-PorA appeared to be non-specific because the ΔporA and kgp rgpA rgpB ΔporA mutants also showed the 50-kDa band (Figs. 3B and 4A). However, we did not find any PorA-PorV heterodimer covalently cross-linked by treatment with DTBP even at non-reduced conditions, suggesting that the DTBP treatment did not work between PorA and PorV. Therefore, we performed the immuno-precipitation analysis with α-PorA in the absence of DTBP to examine whether there was stable interaction between PorA and PorV. Membrane fractions of the wild-type, porK, and ΔporA cells were prepared, immuno-precipitated with α-PorA, and subjected to SDS-PAGE and native PAGE, and the following immunoblot analyses with α-PorA and α-PorV (Fig. S8). The SDS-PAGE analysis without the DTBP treatment showed essentially the same results as that with the DTBP treatment (Fig. 4A, Fig. S8A). The native PAGE analysis showed diffuse protein bands with high molecular masses that reacted to both α-PorA and α-PorV in the samples of wild-type and porK cells (Fig. S8B). These results suggest that the 23-kDa CTD-containing PorA interacts with PorV.
Figure 4.
Interaction between PorA and PorV. (A) Immunoblot analysis with α-PorA and α-PorV. The α-PorA-immunoprecipitated proteins from the membrane fractions of P. gingivalis cells treated with a chemical crosslinker, dimethyl 3,3′-dithiobispropionimidate (DTBP) were separated by SDS-PAGE, followed by immunoblot analyses using α-PorA and α-PorV. Protein bands in the CBB-stained gel indicated by numerals were subjected to peptide mass fingerprinting analysis (Table 1). CBB: Coomassie Brilliant Blue staining. (B) Immunoblot analysis of Halo-CTD(PorA) chimera protein fraction with α-PorV. Cells of the porN mutants expressing Halo-CTD(PorA) chimera protein (porN/Halo-CTD(PorA)) and Halo protein (porN/Halo), respectively, were treated with DTBP. Partially purified samples (PP) were obtained from the cells and added to HaloLink Resin. After centrifugation, the supernatants (unbound) were removed and the resin was washed three times with HaloTag purification buffer. After centrifugation, the resin was dissolved with SDS sample buffer containing dithiothreitol (bound). The resulting ‘bound’ samples from porN/Halo-CTD(PorA) and porN/Halo were heat denatured and subjected to SDS-PAGE and immunoblot analyses with α-PorV and antibody against Halo tag (α-Halo). CBB: Coomassie Brilliant Blue staining. Red and black arrowheads indicate Halo-CTD(PorA) and Halo proteins, respectively.
Table 1.
Peptide mass fingerprinting (PMF) analysis.
| Strain | Band | Gene | Annotation | Mascot score |
|---|---|---|---|---|
| kgp rgpA rgpB | 1 | PGN_0728 | OmpA | 43 |
| kgp rgpA rgpB | 2 | PGN_0023 | PorV | 87 |
| kgp rgpA rgpB | 3 | PGN_1484 | Methylated-DNA-protein-cysteine methyltransferase | 59 |
| kgp rgpA rgpB porK | 4 | PGN_0023 | PorV | 61 |
Proteins in the bands with numerals in the CBB stained gel of Fig. 4A were identified by PMF analysis using MALDI TOF–MS.
To determine whether the PorV protein can bind the CTD of PorA, we constructed a gene encoding a chimera protein consisting of the N-terminal signal peptide of Kgp, the Halo tag, and the CTD of PorA and expressed it in the porN mutant. The chimera protein was purified using HaloLink Resin after protein crosslinking with DTBP and subjected to SDS-PAGE and immunodetection (Fig. 4B). The result suggested that PorV can bind the PorA CTD. Furthermore, we examined whether the CTDs of other T9SS cargo proteins interact with PorV using the same method. The results showed that the CTDs of RgpB, Mfa5, PPAD, and Kgp interacted with PorV although the strength of interaction was different one another, and the CTD of Hbp35 did not (Fig. S9).
Formation of the PorA A-LPS-bound form in co-culture of the porK and porR mutants
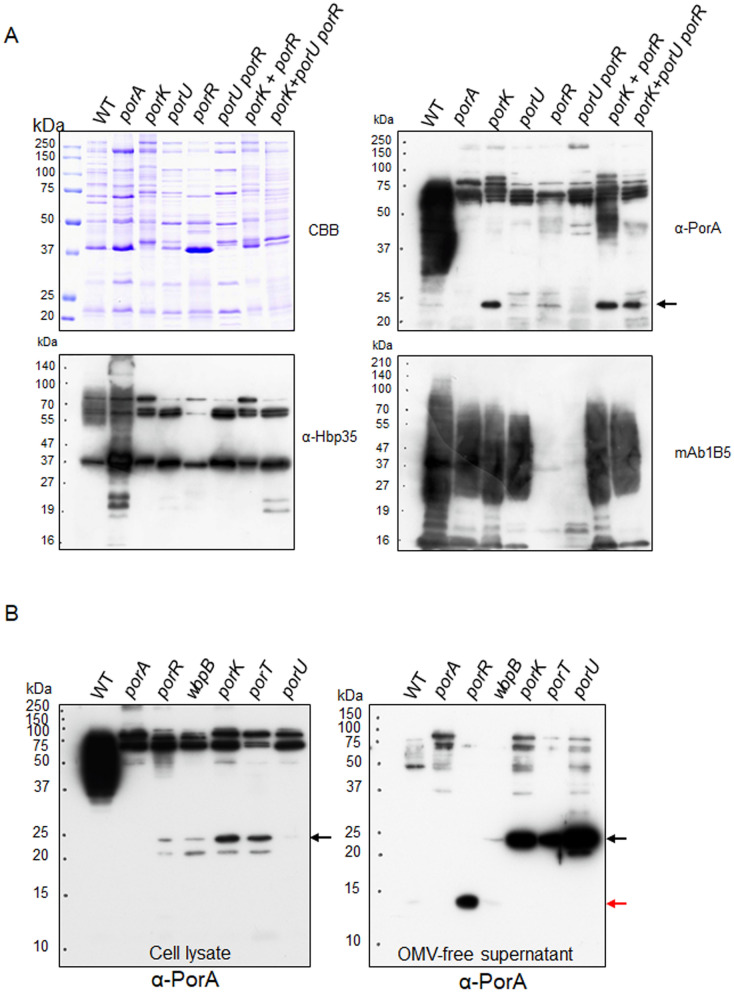
Group I CTD cargo proteins are covalently bound to A-LPS following the cleavage of their CTDs by the PorU sortase secreted to the cell surface through the T9SS. We analyzed whether the 23-kDa CTD-containing PorA that is translocated to the cell surface without passing through the T9SS translocation machinery in the porK mutant can be bound to A-LPS after removal of CTD by PorU. The porK mutant cells were mixed with cells of a ΔporR mutant and the mixture was co-cultured for two days. PorK is an essential component of the T9SS translocation machinery, so the porK mutant has no T9SS, but produces A-LPS. PorR is an enzyme involved in A-LPS synthesis, so the ΔporR mutant does not synthesize A-LPS, but has normal T9SS including PorU on the cell surface. If the 23-kDa CTD-containing PorA on the cell surface can be a substrate of PorU, the CTD of PorA in the porK mutant would be cleaved and the remaining part of PorA would be bound to A-LPS by PorU in the ΔporR mutant. Such reaction may take place between the outer membrane vesicles (OMV) of the porK and ΔporR mutants. OMV fractions of the culture were subjected to SDS-PAGE followed by immunoblot analysis with α-PorA (Fig. 5A). Diffuse bands appeared in the OMV fraction of the co-culture of the porK and the ΔporR mutants, but no such pattern was detected in that of the individual culture of the mutants and in that of the co-culture of the porK and the porU ΔporR mutants. In contrast, the OMV fraction of the co-culture of the porK and the ΔporR mutants showed no diffuse bands with α-Hbp35 because Hbp35 is not secreted on the cell surface without the T9SS. These results also suggest that the 23-kDa CTD-containing PorA is present on the cell surfaces of the T9SS-deficient mutants and can be converted to the A-LPS-bound diffuse form in the presence of PorU. In this analysis, we found that there was a markedly reduced amount of 23-kDa CTD-containing PorA in the OMV fraction of the porU mutant compared to that of the porK mutant (Fig. 5A). The amount of the 23-kDa CTD-containing PorA was almost equivalent to that in the OMV fraction of the wild type. The wild type had a large amount of A-LPS-bound diffuse PorA. Next, we examined the presence and location of PorA in the porU mutant (Fig. 5B). The cell lysate samples showed the results similar to those of the OMV samples, whereas the porU mutant showed a large amount of the 23-kDa CTD-containing PorA in the OMV-free culture supernatant samples.
Figure 5.
Appearance of A-LPS-bound PorA in co-culture of the porK and porR mutants. (A) OMV fractions of cultures of the wild type, ΔporA, porK, porU, ΔporR, and porU ΔporR strains and co-cultures of the porK and ΔporR strains, and the porK and porU ΔporR strains were subjected to SDS-PAGE, followed by immunoblot analyses using α-PorA, α-Hbp35, and mAb 1B5. CBB: Coomassie Brilliant Blue staining. The black arrow indicates the 23-kDa CTD-containing PorA. (B) Cell lysates (left) and OMV-free culture supernatants (right) of the wild type, ΔporA, ΔporR, wbpB, porK, porT, and porU strains were subjected to SDS-PAGE, followed by immunoblot analysis with α-PorA, respectively. The black and red arrows indicate the 23-kDa CTD-containing PorA and the 14-kDa CTD-lacking PorA, respectively.
The porK mutant, which created the 23-kDa CTD-containing PorA but not the A-LPS-bound diffuse PorA on the cell surface (Fig. 3B), produced SigP equal or higher levels than the wild type (Fig. 1F). SigP produced in the porK mutant appears to be functional because the mutant showed expression of SigP-regulated genes at both mRNA and protein levels (Fig. S10). These results suggest that at least the 23-kDa CTD-containing PorA on the cell surface has the ability to activate the PorXY-SigP signaling pathway of the T9SS expression to yield the functional SigP.
Crystal structure of the N-terminal domain of PorA
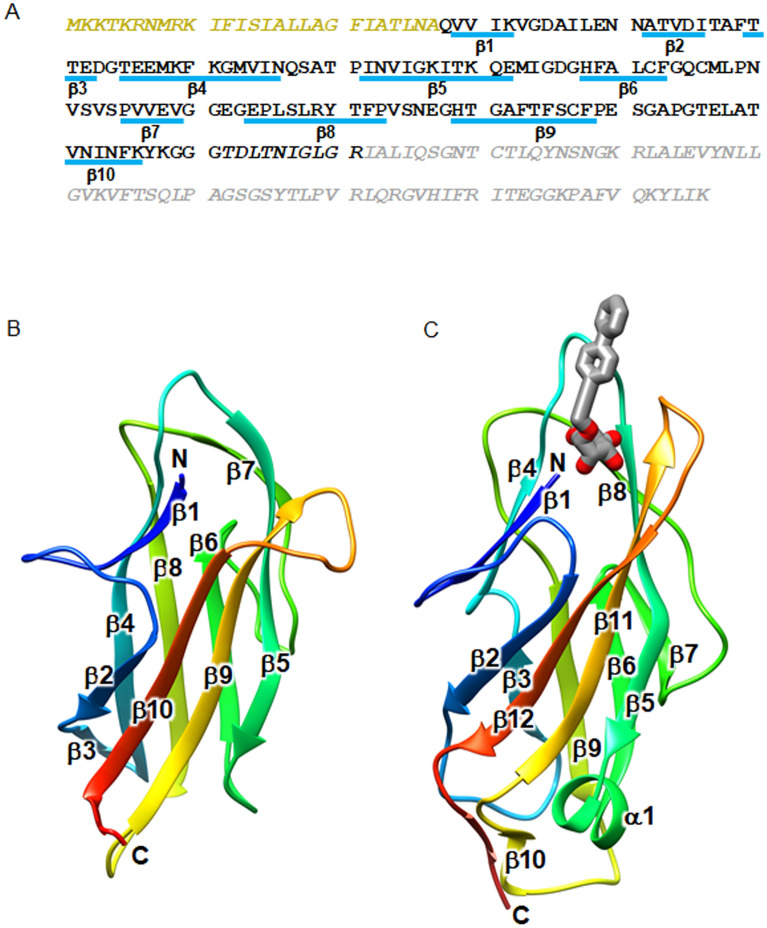
We next conducted the X-ray crystal structure analysis of PorA. Initially, we tried to determine the structure of the 23-kDa CTD-containing PorA, but no crystal was obtained. Therefore, we determined the core region of PorA by limited proteolysis of purified PorA with trypsin. The 23-kDa CTD-containing PorA was digested into a fragment composed of Q28 to R171 (Fig. 6A), and the C-terminal 75 residues, which corresponds to CTD, were degraded into small peptides. Thus, we purified and crystallized the trypsin-resistant fragment, termed PorA-N. The structure of PorA-N was determined at 1.3 Å resolution (Fig. 6B). The C-terminal 10 residues were invisible in the electron density map, and therefore, residues Q28-G161 were modeled. The crystal belongs to the space group C2 and contains a single PorA-N molecule in an asymmetric unit. The structure of PorA-N adopts an immunoglobulin-like fold composed of 10 β-strands (Fig. 6B). The Dali database search revealed that FimH from Escherichia coli in complex with propynyl biphenyl α-d-mannoside, a ligand analogue, gave the best structural similarity (PDB ID: 4av5, Z-score: 8.4) to PorA-N (Fig. 6C). FimH is a tip protein of Type 1 pilus and binds to mannose. This structural similarity suggests that PorA-N may bind an unknown molecule. The mannose binding site of FimH is located at the top of the barrel, and the N-terminus, the loop connecting β4 and β5 and the loop connecting β11 and β12 (the 11–12 loop) surround the ligand and form hydrogen bonds with the ligand. In contrast, the barrel top of PorA-N shows a wide-open structure. The loop connecting β9 and β10 corresponding to the 11–12 loop of FimH extends outward. Thus, the barrel top of PorA-N is exposed to the outside, implying that interaction with the unknown binding partner may induce a structural change of PorA-N.
Figure 6.
Crystal structure analysis of PorA. (A) Amino acid sequence of PorA. The core fragment used for crystallization is shown in black and the region removed by protease is in gray. The signal peptide region is colored in yellow. β-strands are indicated by blue bars with labels. The residues not included in the structure model are shown in italics. (B) Structure of the core fragment of PorA. The model is color coded from blue to red from the N- to the C-terminus. (C) Structure of the mannose-binding lectin domain of FimH in complex with propynyl biphenyl α-d-mannoside (PDB ID: 4av5). The model is color coded from blue to red from the N- to the C-terminus. The ligand molecule is drawn in stick model with oxygen atoms colored red and carbon atoms colored gray.
Discussion
We found in this study that PorA impacts the PorXY-SigP signaling pathway that regulates expression of the T9SS in P. gingivalis. PorA is the first T9SS CTD-containing protein involved in gene regulation of the T9SS component proteins. Like other T9SS CTD-containing proteins, PorA is secreted on the cell surface. Previously characterized T9SS CTD-containing proteins such as Hbp35 are not secreted on the cell surface in the absence of the T9SS secretion machinery. In contrast, PorA is located on the cell surface of the T9SS-deficient mutants (porK, porM, porT, porV and sov) as its CTD-containing form (Fig. 3A). Lauber et al.36 reported that SprA, a Sov homolog of Flavobacterium johnsoniae, forms a water-filled conduit of sufficient size to allow the passage of folded proteins across the outer membrane. Although most T9SS CTD-containing proteins are secreted via the T9SS in F. johnsoniae, a small CTD-containing protein with 112 amino acid residues, Fjoh_0547, is secreted in the culture supernatants of sprA and porV mutants36, suggesting that Fjoh_0547 can be secreted without passing through the SprA channel. In this context, PorA, the second smallest CTD-containing protein (246 amino acid residues) in P. gingivalis, may be translocated to the cell surface through a similar pathway as Fjoh_0547 without passing through the Sov channel.
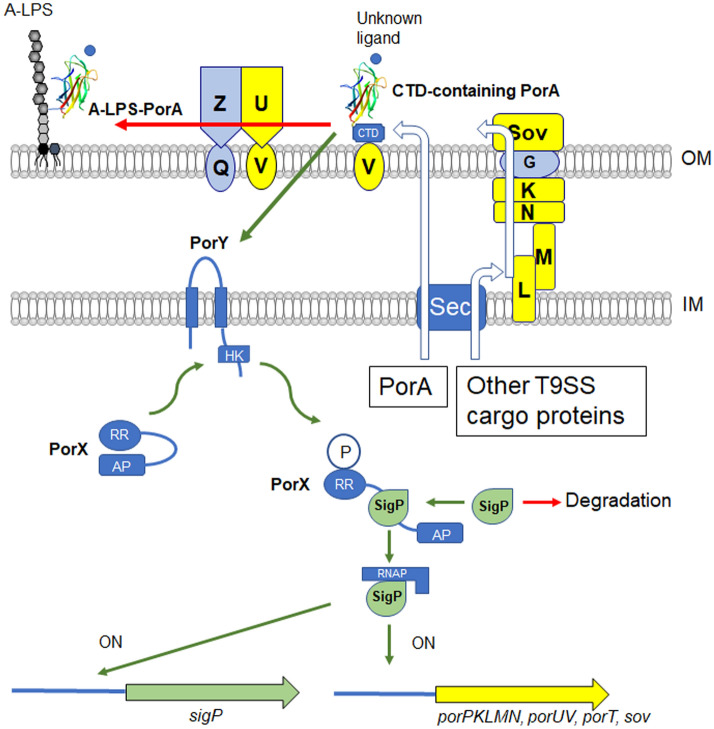
Previously characterized T9SS cargo proteins belong to one of the three groups; I, II and III (Table 2). CTDs of groups I and II proteins are removed by PorU. A-LPS is then covalently bound to the group I proteins, but not to the group II proteins. Group III proteins retain their CTDs on the cell surface and do not bind A-LPS. PorA has both characteristics of groups I and III. PorA is present on the cell surface in both the A-LPS bound diffuse form and the 23-kDa CTD-containing form. Thus, PorA is a novel type of T9SS CTD-containing protein. P. gingivalis T9SS consists of the translocation machinery (PorG, PorK, PorL, PorM, PorN, and Sov), the attachment complex (PorQ, PorU, PorV, and PorZ), the PorV shuttle protein, and other components with unknown functions (PorE, PorF, PorP, PorT, and PorW) (Fig. 7)37. PorA appears to use the PorV shuttle protein and the attachment complex for covalently binding to A-LPS, but not to use the T9SS translocation machinery for secretion to the cell surface.
Table 2.
T9SS cargo proteins in P. gingivalis.
| PGN_number | PG_number | Annotation | Amino acidsa | Groupb | References |
|---|---|---|---|---|---|
| 0014c | 0018c | Hypothetical protein | 748/748 | ND | |
| 0022 | 0026 | PorU | 1158/1158 | III | 11 |
| 0123 | 2172 | PorA | 246/248 | I and III | This study, 12 |
| 0152 | 2102 | TapA, immunoreactive 61 kDa antigen PG91 | 540/540 | I | 13 |
| 0291 | 0182 | Mfa5, von Willebrand factor type A domain protein | 1288/1226 | II | 14 |
| 0295c | Lack | C-terminal domain of Arg- and Lys-gingipain proteinase | 293/– | ND | |
| 0335 | 0232 | Cpg70, zinc carboxypeptidase | 821/821 | I | 15 |
| 0458c | Lack | Hypothetical protein | 508/– | ND | |
| 0509 | 1604 | PorZ, immunoreactive 84 kDa PG93 | 776/776 | III | 16 |
| 0561 | 1548 | PrtT, trypsin like proteinase | 840/frameshift | ND | |
| 0654 | 0611 | Putative lipoprotein | 313/316 | ND | This study |
| 0657 | 0614 | Hypothetical protein | 308/326 | ND | |
| 0659 | 0616 | Hbp35 | 344/344 | I | 15,17 |
| 0693 | 0654 | Hypothetical protein | 390/390 | ND | |
| 0795 | 0769 | Fibronectin type III domain protein | 713/540 | ND | |
| 0852 | 1374 | Immunoreactive 47 kDa antigen PG97 LRR protein | 428/428 | ND | |
| 0898 | 1424 | PPAD, peptidylarginine deiminase | 556/556 | I | 18 |
| 0900 | 1427 | Periodontain, thiol protease/hemagglutinin PrtT precursor | 843/843 | ND | |
| 1115 | 1326 | Hemagglutinin, putative | 369/377 | ND | |
| 1317c | 1035c | Hypothetical protein | 496/496 | ND | |
| 1321 | 1030 | Hypothetical protein | 446/446 | ND | |
| 1416 | 0553 | PepK, extracellular protease | 940/940 | I | 19 |
| 1466 | 0506 | RgpB, arginine-specific cysteine proteinase | 736/736 | I | 6 |
| 1476 | 0495 | Hypothetical protein | 464/469 | ND | |
| 1556 | 0411 | Hemagglutinin, putative | 925/925 | ND | |
| 1611 | 0350 | Internalin-related protein LRR protein | 485/484 | ND | |
| 1728 | 2024 | Kgp, lysine-specific cysteine proteinase | 1723/1706 | II | 2 |
| 1733 | 1837 | HagA, hemagglutinin protein | 2628/2164 | II | 2 |
| 1767 | 1798 | Immunoreactive 46 kDa antigen PG99 RHS repeat protein | 423/405 | ND | |
| 1770 | 1795 | Hypothetical protein | 262/273 | ND | This study, 12 |
| 1817c | Lack | Hypothetical protein | 145/– | ND | |
| 1970 | 2024 | RgpA, hemagglutinin protein HagE | 1703/1706 | II | 20,21 |
| 2065 | 2198 | Immunoreactive 32 kD antigen PG25 | 293/293 | ND | |
| 2080 | 2216 | Hypothetical protein | 576/576 | ND | 12 |
| Lack | 0183 | Lipoprotein, putative | –/2204 | ND | |
| Lack | 0410 | Hypothetical protein gingipain-like peptidase C25 | –/1294 | ND | |
| Lack | 0626 | Hypothetical protein | –/288 | ND | |
| Lack | 1102c | Hypothetical protein | –/925 | ND | |
| Lack | 1969 | Hypothetical protein | –/300 | ND | |
| Lack | 2100 | TapC, immunoreactive 63 kDa antigen PG102 | –/554 | ND |
aLeft and right numbers indicate amino acids from ATCC 33277 and W83, respectively.
bGroup I indicates that C-terminal domain is cleaved and then processed protein forms diffuse bands in SDS-PAGE. Group II indicates that C-terminal domain is cleaved but processed protein does not form diffuse bands in SDS-PAGE. Group III indicates that C-terminal domain is not cleaved. ND, not determined.
cNot reported in Lasica et al.8.
Figure 7.
Proposed model of regulation of the T9SS expression in P. gingivalis. The PorA protein is located in the outer membrane. In the proposed model, the 23-kDa CTD-containing PorA activated by an unknown ligand activates the PorY sensor kinase of the PorXY two-component system directly or indirectly. Alternatively, the 23-kDa CTD-containing PorA may activate PorY without any environmental substances. The PorA-activated PorY activates the PorX response regulator. The PorY-activated PorX activates the SigP sigma protein and the resulting active SigP upregulates its own gene and several T9SS component genes.
The porA deletion mutant formed non-pigmented colonies on the blood agar plate, decreased hemagglutination and gingipain activities, and reduced expression of T9SS component genes such as porK, porL, porM, and porU, indicating that the ΔporA mutant is deficient in the T9SS. However, the ΔporA mutant is not a T9SS-null mutant, but still expresses a small amount of T9SS component proteins. We detected Hbp35 on the cell surface of the ΔporA mutant. This phenotype is very similar to the ΔporX and ΔporY mutants, supporting that PorA influences the PorXY-SigP signaling pathway.
PorY and PorX proteins are the sensor histidine kinase and the response regulator of the two-component system, respectively, which regulates the T9SS29. What activates the PorY sensor kinase remains to be determined. Since the amino acid substitution in the cytoplasmic domain of PorY compensated the defective phenotype of the ΔporA mutant, it is plausible that PorA activates the PorY sensor kinase. In our preliminary experiments, however, direct interaction between PorA and PorY was not detected. PorA is located on the cell surface whereas PorY is in the inner membrane, so other factor(s) should mediate between PorA and PorY.
ECF sigma factor was first recognized as a distinct subgroup of σ70-like factor in 199438 and after that it has been found in many bacterial species39. In most cases, ECF sigma factor is regulated by its cognate anti-sigma factor protein, which is encoded by a downstream gene on the same transcriptional unit39. In P. gingivalis, there are six ECF sigma factors such as PGN_0274 (SigP)29,40, PGN_0319 (SigCH)41, PGN_0450, PGN_0970, PGN_110842, and PGN_1740 (SigH)43. Their downstream CDSs such as PGN_0320, PGN_0451, PGN_0969, and PGN_1107 may encode anti-sigma factors; however, it has not been elucidated whether they are anti-sigma factors. SigP regulates gene expression of the T9SS components29. PorX binds SigP but not DNA29. PorX has the PglZ domain, which may function as a phosphatase, phosphomutase, or phosphodiesterase44, in its C-terminal region. Thus, PorX activated by PorY binds and activates SigP. The sigP gene is auto-regulated by its own gene product, SigP40. We previously found that the sigP gene was down-regulated in the ΔporX mutant29. This may be because the PorX-activated SigP, not the intact SigP, induces the expression of the sigP gene. In this study, down-regulation of the sigP gene was also found in the ΔporA mutant, that is consistent with the idea that PorA is located upstream of the PorXY-SigP signaling pathway (Fig. 7).
Most of T9SS cargo proteins share a common structural architecture composed of a signal peptide, a functional domain(s), an Ig-like domain and CTD45,46. The role of the Ig-like domain is believed to stabilize the functional domain46. However, PorA is composed of a signal peptide, the Ig-like domain and CTD. The Ig-like domain structure of PorA differs from those of other T9SS cargo proteins, such as Hbp35, Kgp and Rgp, but resembles the mannose-binding lectin domain of FimH. The structural similarity to FimH suggests that on the cell surface PorA binds an unknown environmental substance that stimulates PorA to impact the PorXY-SigP signaling pathway.
There are two forms of PorA on the cell surface: the A-LPS-bound diffuse form and the 23-kDa CTD-containing form. Production of functional SigP in mutants deficient in T9SS components such as the porK and porT mutants suggests that the 23-kDa CTD-containing PorA on the cell surface has the ability to activate the PorXY-SigP pathway. The 23-kDa CTD-containing PorA is cleaved by the PorU protease in the attachment complex and the resulting mature/processed PorA is covalently bound to A-LPS. If the A-LPS-bound diffuse form has no ability to activate the PorXY-SigP pathway, the attachment complex would determine amounts of PorA with the activating ability. Because PorU and PorZ in the attachment complex are secreted by the T9SS translocation machinery, the attachment complex on the cell surface is a reflection of the functionality of the T9SS translocation machinery37. This may imply the following feedback system in the regulation of the T9SS: When operation of the T9SS translocation machinery decreases, amounts of PorU and PorZ are reduced on the cell surface, resulting in increase of the 23-kDa CTD-containing PorA on the cell surface. The 23-kDa CTD-containing PorA activates the PorXY-SigP pathway and increases expression of T9SS component genes to yield increase of the T9SS operation. Further investigation is needed to reveal the role of PorA in activation of the PorXY-SigP signaling pathway.
In conclusion, we found that PorA, a T9SS CTD-containing protein, impacts the PorXY-SigP signaling pathway that is responsible for expression of the T9SS. Putative protein-encoding genes homologous to porA with BLAST E-value of < 0.0001 have been found only in the genus Porphyromonas, indicating that the presence of PorA is restricted to a very narrow range of taxa. Therefore, the cell surface protein PorA may be a good target for pharmacotherapeutically controlling the periodontal pathogen P. gingivalis. This study provides a new insight into the signaling pathway that regulates the T9SS.
Methods
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Supplemental Table S14,5,15,29,31,33,47–49.
Media and conditions for bacterial growth
Media and conditions for bacterial growth was described previously2. Briefly, P. gingivalis strains were grown anaerobically (80% N2, 10% CO2, 10% H2) in enriched brain–heart infusion (BHI) broth (Becton Dickinson, Franklin Lakes, NJ) or on enriched tryptic soy agar plates (Nissui, Tokyo, Japan) supplemented with 5 μg/ml hemin (Sigma, St. Louis, MO) and 0.5 μg/ml menadione (Sigma). For blood agar plates, defibrinated laked sheep blood was added to enriched tryptic soy agar at 5%. Luria–Bertani (LB) broth and LB agar plates were used for growth of Escherichia coli strains. Antibiotics were used at the following concentrations: ampicillin (Ap; 100 μg/ml for E. coli, 10 μg/ml for P. gingivalis), erythromycin (Em; 10 μg/ml for P. gingivalis), gentamicin (Gm; 50 μg/ml for P. gingivalis), and tetracycline (Tc; 0.7 μg/ml for P. gingivalis).
Chemicals
The proteinase inhibitors Nα-p-tosyl-l-lysine chloromethyl ketone hydrochloride (TLCK) and leupeptin were purchased from Wako (Osaka, Japan) and Peptide Institute (Osaka, Japan), respectively. Iodoacetamide was purchased from Wako (Osaka, Japan).
Construction of bacterial mutant strains
Construction of P. gingivalis mutants was described in detail in Supplemental text. Primers used in this study are listed in Supplemental Table S2.
Construction of a complemented strain of the ΔporA mutant
Construction of a complemented strain of the ΔporA mutant was described in detail in Supplemental Text5,47,49.
Electrotransformation of pTIO-tetQ-porY(S266W) in the ΔporA mutant
Complementation of pTIO-tetQ-porY(S266W) was described in Supplemental Text.
Enzymatic assay
Kgp and Rgp activities were determined as previously described30. The determination is described in detail in Supplemental Text.
Hemagglutinating activity
Hemagglutinating activities were determined as previously described33. The determination is described in detail in Supplemental Text.
Preparation of vesicle fraction
Vesicle fraction was obtained as described previously50. The determination is described in detail in Supplemental Text.
Antibodies
His6-tagged recombinant proteins of PorA, PorV, SigP, PorL, PorM, and PorN were overexpressed in E. coli BL21(DE3) carrying an expression plasmid. The expression plasmid was constructed as follows. DNA fragments of porA, porV, sigP, porL, porM, and porN gene were PCR amplified with primer sets PorA-15bF/PorA-15bR, PorV-32bF/PorV-32bR, SigP-15bF/SigP-15bR, PorL-15bF/PorL-15bR, PorM-22bF/PorM-22bR, and PorN-22bF/PorN-22bR, respectively, using P. gingivalis ATCC 33277 chromosomal DNA as a template. The amplified DNA of porA was digested with NdeI plus BglII, the amplified DNA of sigP and porL was digested with NdeI plus BamHI, and the resulting DNA fragments were inserted into the NdeI-BamHI site of pET-15b (Novagen). The amplified DNA of porM was digested with BamHI plus XhoI and then inserted into the BamHI-XhoI site of pET-22b(+) (Novagen). The amplified DNA of porN was digested with SalI plus XhoI and then inserted into the SalI-XhoI site of pET-22b(+). The amplified DNA of porV was digested with EcoRV plus XhoI and then inserted into the EcoRV-XhoI site of pET-32b(+) (Novagen). E coli BL21 (DE3) harboring the resulting plasmids were grown on LB broth at 30 °C, and PorA, PorV, SigP, PorL, PorM, and PorN were induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside. His6-tagged recombinant proteins were purified by using a HiTrap Chelating HP column (GE Healthcare Life Sciences). To raise antiserum against PorA, SigP, PorV, PorL, PorM, and PorN, rabbits were immunized with PorA-His6 and guinea pigs were immunized with the SigP-His6 by EveBioscience Co., Ltd. (Wakayama, Japan). PorV, PorL, PorM, and PorN-His6 protein were mixed with TiterMax Gold (TiterMax), and the mixtures were injected into mice (BALB/c) subcutaneously, resulting in α-PorV, α-PorL, α-PorM, α-PorN antiserum. An α-Kgp rabbit polyclonal antibody51, an α-PorK rabbit polyclonal antibody5, an α-Rgp mouse polyclonal antibody51, an α-PorU mouse polyclonal antibody52, an α-Hbp35 rabbit polyclonal antibody53, an α-Mfa1 rabbit polyclonal antibody54, an α-PorX rabbit polyclonal antibody29, and an α-PorY rabbit polyclonal antibody29 were used to detect Kgp, PorK, Rgp, PorU, Hbp35, Mfa1, PorX, and PorY, respectively. A monoclonal antibody 1B5 (mAb 1B5), which recognizes the anionic polysaccharide of A-LPS, was kindly provided by Prof. M. A. Curtis55. An α-Halo Tag rabbit polyclonal antibody was purchased from Promega.
Gel electrophoresis and immunoblot analysis
SDS-PAGE and immunoblot analysis were performed as previously described15,17.
Dot blot analysis
Dot blot analysis was performed as previously described15.
Biotinylation of surface proteins of P. gingivalis
Biotinylation of surface proteins in P. gingivalis cells was performed as previously described56. The procedure is described in detail in Supplemental Text.
Immunoprecipitation with α-PorA antibody
Immunoprecipitation assay were described in detail in Supplemental Text.
MS analysis and database search for protein identification
Proteins were identified by peptide mass fingerprinting (PMF) after in-gel tryptic digestion as previously described5.
Whole genome sequencing of the ΔporA pseudo-revertants
Porphyromonas gingivalis ΔporA pseudo-revertants were grown on the enriched tryptic soy plates. Chromosomal DNA was extracted from the ΔporA pseudo-revertants using the MasterPure DNA purification kit for Blood Version II (Epicentre) and concentrated using ethanol precipitation then suspended in Tris–EDTA buffer pH8.0. Whole genome sequencing was performed at the Nagasaki University Genome Research Facility using the Ilumina HiSeq 2500 Sequencer (Illumina, San Diego, Calif). The sequence data were analyzed by using a standard pipeline. To identify small indels and single-nucleotide variants, sequence reads were aligned against P. gingivalis ATCC 33277 genome (AP009380)57 modified with the insertion of ermF to porA. We also used information of the P. gingivalis W83 genome (GenBank under accession number AE015924)58.
Quantification of gene expression by quantitative RT (qRT)-PCR
qRT-PCR analysis was performed as previously described5. Primers used in this study are listed in Supplemental Table S2.
Purification of Halo-CTD(PorA) chimera protein with DTBP-mediated crosslinking
Purification of Halo-CTD(PorA) chimera protein with DTBP-mediated crosslinking was described in Supplemental Text59.
Purification of His-PorA(Q28-K246)
Purification od His-PorA(Q28-K246) was described in Supplemental Text.
Limited proteolysis
Limited proteolysis was performed as previously described60. Purified His-PorA(Q28-K246) solution was mixed with trypsin at the protease/protein ratio of 1/100 (w/w) for 90 min at 27 °C. The products were analyzed by SDS-PAGE and MALDI-TOF Mass Spectrometry using sinapinic acid or α-Cyano-4-hydroxycinnamic acid as matrix reagents.
Purification of PorA-N and Se-Met PorA-N
Purification procedure of PorA-N and Se-Met PorA-N was described in Supplemental Text.
Crystallization, data collection and structure determination
Procedures of crystallization, data collection and structure determination were described in detail in Supplemental Text61–64. Structural refinement statistics are summarized in Supplemental Table S3.
Data deposition
The atomic coordinates have been deposited in Protein Data Bank, http://www.pdb.org (PDB ID code 6KJK).
Supplementary information
Acknowledgements
This work was supported by grants from the Japan Society for the Promotion of Science (Grant-in-Aid for Challenging Exploratory Research, No. 16K15776 and Grant-in-Aid for Scientific Research (C), No. 20K09921 to K.N.). We thank T. Ogi and M. Shimada (Nagoya University and Nagasaki University) for the whole genome sequencing analysis and the beamline staff of SPring-8 for technical help with data collection. We thank Editage (http://www.editage.com) for English language editing.
Author contributions
H.Y., M.S., K.I., and K.N. wrote the main manuscript text. H.Y., M.S., K.S., and M.N. prepared Figs. 1, 2, 3, 4, 5, supplemental Figures S1–S13, and Table 1. Y.H., and K.I. prepared Fig. 6 and supplemental Table S3. H.Y., M.S., and K.N. prepared Fig. 7 and Table 2 and supplemental Tables S1, S2. All authors reviewed the manuscript.
Data availability
The atomic coordinate of the crystal structure has been deposited into the Protein Data Bank (PorA: PDB ID 6KJK). Additional raw data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-77987-y.
References
- 1.Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 2012;91:816–820. doi: 10.1177/0022034512453589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Y, et al. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 3.Sakai E, et al. Construction of recombinant hemagglutinin derived from the gingipain-encoding gene of Porphyromonas gingivalis, identification of its target protein on erythrocytes, and inhibition of hemagglutination by an interdomain regional peptide. J. Bacteriol. 2007;189:3977–3986. doi: 10.1128/JB.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato K, et al. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J. Biol. Chem. 2005;280:8668–8677. doi: 10.1074/jbc.M413544200. [DOI] [PubMed] [Google Scholar]
- 5.Sato K, et al. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seers CA, et al. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 2006;188:6376–6386. doi: 10.1128/JB.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veith PD, et al. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J. Proteome. Res. 2013;12:4449–4461. doi: 10.1021/pr400487b. [DOI] [PubMed] [Google Scholar]
- 8.Lasica AM, Ksiazek M, Madej M, Potempa J. The type IX secretion system (T9SS): Highlights and recent insights into its structure and function. Front. Cell. Infect. Microbiol. 2017;7:215. doi: 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulkarni SS, Zhu Y, Brendel CJ, McBride MJ. Diverse C-terminal sequences involved in Flavobacterium johnsoniae protein secretion. J. Bacteriol. 2017;199:e00884-16. doi: 10.1128/JB.00884-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulkarni SS, Johnston JJ, Zhu Y, Hying ZT, McBride MJ. The carboxy-terminal region of Flavobacterium johnsoniae SprB facilitates its secretion by the type IX secretion system and propulsion by the gliding motility machinery. J. Bacteriol. 2019;201:e00218-19. doi: 10.1128/JB.00218-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glew MD, et al. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J. Biol. Chem. 2012;287:24605–24617. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein BA, et al. Using Tn-seq to identify pigmentation-related genes of Porphyromonas gingivalis: Characterization of the role of a putative glycosyltransferase. J. Bacteriol. 2017;199:e00832-16. doi: 10.1128/JB.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo Y, et al. Tetratricopeptide repeat protein-associated proteins contribute to the virulence of Porphyromonas gingivalis. Infect. Immun. 2010;78:2846–2856. doi: 10.1128/IAI.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasegawa Y, et al. Role of Mfa5 in expression of Mfa1 fimbriae in Porphyromonas gingivalis. J. Dent. Res. 2016;95:1291–1297. doi: 10.1177/0022034516655083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoji M, et al. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS ONE. 2011;6:e21372. doi: 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasica AM, et al. Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. Sci. Rep. 2016;6:37708. doi: 10.1038/srep37708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoji M, et al. Characterization of hemin-binding protein 35 (HBP35) in Porphyromonas gingivalis: Its cellular distribution, thioredoxin activity and role in heme utilization. BMC Microbiol. 2010;10:152. doi: 10.1186/1471-2180-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabarrini G, et al. Dropping anchor: Attachment of peptidylarginine deiminase via A-LPS to secreted outer membrane vesicles of Porphyromonas gingivalis. Sci. Rep. 2018;8:8949. doi: 10.1038/s41598-018-27223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nonaka M, et al. Analysis of a Lys-specific serine endopeptidase secreted via the type IX secretion system in Porphyromonas gingivalis. FEMS Microbiol. Lett. 2014;354:60–68. doi: 10.1111/1574-6968.12426. [DOI] [PubMed] [Google Scholar]
- 20.Pike R, McGraw W, Potempa J, Travis J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J. Biol. Chem. 1994;269:406–411. [PubMed] [Google Scholar]
- 21.Kadowaki T, et al. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 1998;273:29072–29076. doi: 10.1074/jbc.273.44.29072. [DOI] [PubMed] [Google Scholar]
- 22.Gorasia DG, et al. Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog. 2015;11:e1005152. doi: 10.1371/journal.ppat.1005152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou XY, Gao JL, Hunter N, Potempa J, Nguyen KA. Sequence-independent processing site of the C-terminal domain (CTD) influences maturation of the RgpB protease from Porphyromonas gingivalis. Mol. Microbiol. 2013;89:903–917. doi: 10.1111/mmi.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto K, et al. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 25.Lewis JP, Dawson JA, Hannis JC, Muddiman D, Macrina FL. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J. Bacteriol. 1999;181:4905–4913. doi: 10.1128/JB.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smalley JW, Silver J, Marsh PJ, Birss AJ. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the mu-oxo dimeric form: An oxidative buffer and possible pathogenic mechanism. Biochem. J. 1998;331:681–685. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoji M, et al. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: Cell surface polysaccharide as an anchorage for gingipains. Microbiology. 2002;148:1183–1191. doi: 10.1099/00221287-148-4-1183. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama K. Porphyromonas gingivalis and related bacteria: From colonial pigmentation to the type IX secretion system and gliding motility. J. Periodontal. Res. 2015;50:1–8. doi: 10.1111/jre.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadowaki T, et al. A two-component system regulates gene expression of the type IX secretion component proteins via an ECF sigma factor. Sci. Rep. 2016;6:23288. doi: 10.1038/srep23288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoji M, Nakayama K. Glycobiology of the oral pathogen Porphyromonas gingivalis and related species. Microb. Pathog. 2016;94:35–41. doi: 10.1016/j.micpath.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Naito M, Tominaga T, Shoji M, Nakayama K. PGN_0297 is an essential component of the type IX secretion system (T9SS) in Porphyromonas gingivalis: Tn-seq analysis for exhaustive identification of T9SS-related genes. Microbiol. Immunol. 2019;63:11–20. doi: 10.1111/1348-0421.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoji M, et al. Identification of an O-antigen chain length regulator, WzzP, Porphyromonas gingivalis. Microbiologyopen. 2013;2:383–401. doi: 10.1002/mbo3.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shoji M, Sato K, Yukitake H, Naito M, Nakayama K. Involvement of the Wbp pathway in the biosynthesis of Porphyromonas gingivalis lipopolysaccharide with anionic polysaccharide. Sci. Rep. 2014;4:5056. doi: 10.1038/srep05056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saiki K, Konishi K. Porphyromonas gingivalis C-terminal signal peptidase PG0026 and HagA interact with outer membrane protein PG27/LptO. Mol. Oral Microbiol. 2014;29:32–44. doi: 10.1111/omi.12043. [DOI] [PubMed] [Google Scholar]
- 35.Glew MD, et al. PorV is an outer membrane shuttle protein for the type IX secretion system. Sci. Rep. 2017;7:8790. doi: 10.1038/s41598-017-09412-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lauber F, Deme JC, Lea SM, Berks BC. Type 9 secretion system structures reveal a new protein transport mechanism. Nature. 2018;564:77–82. doi: 10.1038/s41586-018-0693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veith PD, Glew MD, Gorasia DG, Reynolds EC. Type IX secretion: The generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol. Microbiol. 2017;106:35–53. doi: 10.1111/mmi.13752. [DOI] [PubMed] [Google Scholar]
- 38.Lonetto MA, Brown KL, Rudd KE, Buttner MJ. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sineva E, Savkina M, Ades SE. Themes and variations in gene regulation by extracytoplasmic function (ECF) sigma factors. Curr. Opin. Microbiol. 2017;36:128–137. doi: 10.1016/j.mib.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dou Y, Aruni W, Muthiah A, Roy F, Wang C, Fletcher HM. Studies of the extracytoplasmic function sigma factor PG0162 in Porphyromonas gingivalis. Mol. Oral Microbiol. 2016;31:270–283. doi: 10.1111/omi.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ota K, et al. SigCH, an extracytoplasmic function sigma factor of Porphyromonas gingivalis regulates the expression of cdhR and hmuYR. Anaerobe. 2017;43:82–90. doi: 10.1016/j.anaerobe.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Kikuchi Y, et al. Porphyromonas gingivalis mutant defective in a putative extracytoplasmic function sigma factor shows a mutator phenotype. Oral Microbiol. Immunol. 2009;24:377–383. doi: 10.1111/j.1399-302X.2009.00526.x. [DOI] [PubMed] [Google Scholar]
- 43.Yanamandra SS, Sarrafee SS, Anaya-Bergman C, Jones K, Lewis JP. Role of the Porphyromonas gingivalis extracytoplasmic function sigma factor, SigH. Mol. Oral Microbiol. 2012;27:202–219. doi: 10.1111/j.2041-1014.2012.00643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galperin MY. Structural classification of bacterial response regulators: Diversity of output domains and domain combinations. J. Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Diego I, et al. The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a C-terminal β-sandwich domain. Sci. Rep. 2016;6:23123. doi: 10.1038/srep23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato K, et al. Immunoglobulin-like domains of the cargo proteins are essential for protein stability during secretion by the type IX secretion system. Mol. Microbiol. 2018;110:64–81. doi: 10.1111/mmi.14083. [DOI] [PubMed] [Google Scholar]
- 47.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram negative bacteria. Biotechnology. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 48.Sato K, et al. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol. Lett. 2013;338:68–76. doi: 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- 49.Nagano K, et al. Characterization of RagA and RagB in Porphyromonas gingivalis: Study using gene-deletion mutants. J. Med. Microbiol. 2007;56:1536–1548. doi: 10.1099/jmm.0.47289-0. [DOI] [PubMed] [Google Scholar]
- 50.Potempa J, Pike R, Travis J. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect. Immun. 1995;63:1176–1182. doi: 10.1128/IAI.63.4.1176-1182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takii R, Kadowaki T, Baba A, Tsukuba T, Yamamoto K. A functional virulence complex composed of gingipains, adhesins, and lipopolysaccharide shows high affinity to host cells and matrix proteins and escapes recognition by host immune systems. Infect. Immun. 2005;73:883–893. doi: 10.1128/IAI.73.2.883-893.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taguchi Y, et al. Involvement of an Skp-like protein, PGN_0300, in the type IX secretion system of Porphyromonas gingivalis. Infect. Immun. 2015;84:230–240. doi: 10.1128/IAI.01308-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abiko Y, et al. Cloning of a Bacteroides gingivalis outer membrane protein gene in Escherichia coli. Arch. Oral Biol. 1990;35:689–695. doi: 10.1016/0003-9969(90)90091-N. [DOI] [PubMed] [Google Scholar]
- 54.Xu Q, et al. A distinct type of pilus from the human microbiome. Cell. 2016;165:690–703. doi: 10.1016/j.cell.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Curtis MA, et al. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 1999;67:3816–3823. doi: 10.1128/IAI.67.8.3816-3823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shoji M, et al. The major structural components of two cell surface filaments of Porphyromonas gingivalis are matured through lipoprotein precursors. Mol. Microbiol. 2004;52:1513–1525. doi: 10.1111/j.1365-2958.2004.04105.x. [DOI] [PubMed] [Google Scholar]
- 57.Naito M, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 2008;15:215–225. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nelson KE, et al. Complete genome sequence of the oral pathogenic Bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen YY, et al. The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol. Microbiol. 2011;79:1380–1401. doi: 10.1111/j.1365-2958.2010.07530.x. [DOI] [PubMed] [Google Scholar]
- 60.Imada K. Design and preparation of the fragment proteins of the flagellar components suitable for X-ray crystal structure analysis. Methods Mol. Biol. 2017;1593:97–103. doi: 10.1007/978-1-4939-6927-2_7. [DOI] [PubMed] [Google Scholar]
- 61.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinate of the crystal structure has been deposited into the Protein Data Bank (PorA: PDB ID 6KJK). Additional raw data that support the findings of this study are available from the corresponding author upon reasonable request.