Abstract
Chromoblastomycosis is a chronic, cutaneous or subcutaneous mycosis characterized by the presence of muriform cells in host tissue. Implantation disease is caused by melanized fungi related to black yeasts, which, in humid tropical climates, are mainly members of the genus Fonsecaea. In endemic areas of Brazil, F. pedrosoi and F. monophora are the prevalent species. The current hypothesis of infection is traumatic introduction via plant materials, especially by plant thorns. However, isolation studies have demonstrated a low frequency of the agents in environmental substrates. The present study aimed to detect F. pedrosoi and F. monophora in shells of babassu coconuts, soil, plant debris, and thorns from endemic areas of chromoblastomycosis in Maranhão state, northern Brazil, using Rolling Circle Amplification (RCA) with padlock probes as a new environmental screening tool for agents of chromoblastomycosis. In addition to molecular screening, the environmental samples were analyzed by fungal isolation using mineral oil flotation. The limit of detection of the RCA method was 2.88 × 107 copies of DNA per sample for the used padlock probes, indicating that this represents an efficient and sensitive molecular tool for the environmental screening of Fonsecaea agents. In contrast, with isolation from the same samples using several selective methods, no agents of chromoblastomycosis were recovered.
Keywords: Black yeast, padlock probe, Fonsecaea pedrosoi, Fonsecaea monophora
1. Introduction
Melanized fungi in the family Herpotrichiellaceae (order Chaetothyriales) are involved in persistent (sub) cutaneous human infections [1]. These fungi have a complex ecological preference and life cycle, which are still poorly understood. They may be found in adverse and extreme conditions, such as on rocks, in arid and hot climates, and in toxic habitats, and also occur as opportunistic pathogens [2,3]. Virulence factors such as the presence of melanin and carotene, thick cell walls, differentiation in muriform cells, yeast phases, osmotolerance, adhesion, hydrophobicity, the assimilation of aromatic hydrocarbons, and the production of siderophores are shared by many members of the family and therefore contribute to the opportunistic nature of these infectious agents [4,5].
Chromoblastomycosis (CBM) is an uncommon and chronic, cutaneous and subcutaneous disease that mostly occurs in immunocompetent patients, resulting in nodular deformations and the presence of muriform cells in tissue, leading to a granulomatous immune response [6]. CBM is considered an occupational disease, especially among agricultural workers, and is caused by the transcutaneous implantation of plant material in tropical and subtropical climate zones around the world [6]. CBM lesions are usually recalcitrant and extremely difficult to eradicate. The disease mainly affects the poorest populations that often live in remote rural areas and have little political support to achieve priority in public health systems. Therefore, the disease has received the status of a neglected tropical disease by the World Health Organization (WHO) [7]. Species that have been described to cause cutaneous infection are classified as Exophiala, Cyphellophora, Phialophora, and Rhinocladiella, but the main agents of CBM belong to Cladophialophora and Fonsecaea. Members of the latter genera have a different epidemiology: Fonsecaea species are primarily found in humid areas, whereas Cladophialophora carrionii is prevalent in semiarid climates [6,8,9,10]. In Brazil, the Amazonian region is considered an endemic area, with the Maranhão state being hyperendemic, exhibiting the largest number of records of the disease [9,11,12]. According to the literature, Fonsecaea pedrosoi is the main etiological agent in these areas, followed by Fonsecaea monophora, which is also known from brain infections [12].
Epidemiological data on mycosis suggest an environmental origin [7,11]. The infection is frequently reported following the occurrence of skin trauma, mainly by plant thorns or wood fragments [13]. Knowledge of the environmental occurrence of pathogenic agents is important in understanding infection pathways, but methods of isolation remain an obstacle [14]. Studies have shown that species recovered from living plants or plant debris often belong to non-pathogenic relatives of the agents of chromoblastomycosis [11,15,16]. A comparative genomic analysis of Fonsecaea species demonstrated that environmental and opportunistic species share gene domains associated with the invasion of plant tissue, supporting the hypothesis of traumatic inoculation from plant material [17].
Molecular markers provide a reliable tool for characterizing the habitat of clinical species, presenting a high specificity, reproducibility, and sensitivity. Through the extraction of total DNA from environmental samples, it becomes possible to evaluate the fungal diversity in the sample, enhancing phylogenetic interference, the taxonomic delimitation of species, and identification [18,19,20,21]. In order to elucidate epidemiological aspects, efficient methods are required to recover and characterize pathogenic agents [6,18,22]. Padlock probes are oligonucleotides containing around 100 bp that recognize single point polymorphisms (SNPs) in target DNA in large populations of over 500 individuals [22,23,24]. Ligation products of the padlock probe can be amplified by isothermal amplification in Rolling Circle Amplification (RCA), which represents a sensitive and specific molecular method for the detection of Fonsecaea species [25].
The present study proposes the use of RCA padlock probes, previously described in the literature [25], as a new strategy for the environmental screening of F. pedrosoi and F. monophora, which are the main agents of chromoblastomycosis in endemic areas of Maranhão state, Brazil, in order to elucidate the routes of infection and ecological niches of species related to the disease.
2. Materials and Methods
2.1. Study Area and Samples
A total of 87 environmental samples were collected randomly in the living environment of symptomatic patients, i.e., five samples of each environmental source (soil, decomposing plant material, and living plants), of each one of the four regions in the north of Maranhão state (Figure 1). Plant materials of Solanum paniculatum (Jurubeba tree), Astrocaryum vulgare (Tucum tree), Platonia insignis (Bacuri tree), Scoparia dulcis (Vassourinha tree), Murraya paniculata (Murta tree), and Urtica spp. were divided according to leaves, stems, and thorns (when present). About 30 g of each sample was collected separately in sterilized paper bags [12,26]. In addition, 20 DNA samples of the babassu coconut shell, which is a known source of melanized fungi [2] and which has been suggested as a possible risk factor for CBM [26,27], were included.
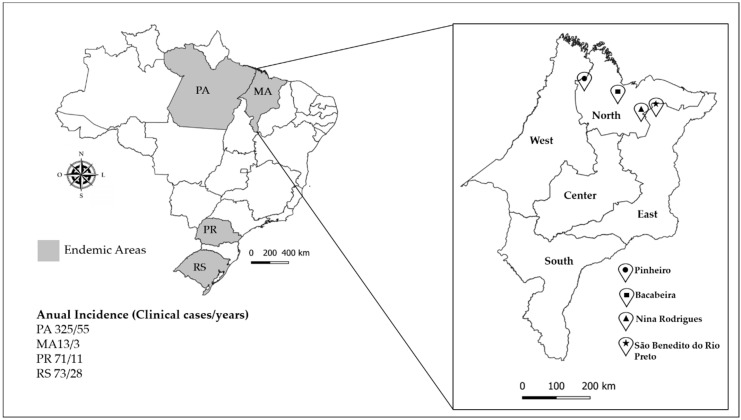
Figure 1.
Study area of Maranhão state. The annual incidence of chromoblastomycosis in Maranhão state and endemic areas is shown in gray on the map [6]. The area of study was located in the North and East of Maranhão state. Bacabeira, latitude: 02° 58′ 15″ S; longitude: 44° 18′ 56″ W; altitude: 44 m; area: 650 km2. São Benedito do Rio Preto, latitude: 3° 19′ 59″ S; longitude: 43° 31′ 40″ W; altitude: 22 m; area: 931.48 km2. Pinheiro, latitude: 02° 31′ 17″ S; longitude: 45° 04′ 57″ W; altitude: 15 m; area: 1559 km2. Nina Rodrigues, latitude: 3° 27′ 53″ S; longitude: 43° 54′ 19″ W; altitude: 33 m; area: 572.5 km2.
2.2. Padlock Probes
2.2.1. DNA Extraction of Environmental Samples
About 250 mg of each environmental sample was transferred to a 2 mL microtube containing 300 µL cetyltrimethylammonium bromide (CTAB) and about 80 mg of a silica mixture. Cells were grinded manually with a sterile pestle for approximately 5 min. Subsequently, 700 µL CTAB buffer was added. The mixture was vortexed for 5 min and incubated for 60 min at 65 °C. Then, 600 μL 24:1 chloroform: isoamylalcohol was added, mixed carefully, and centrifuged for 10 min at 12,000× g force. The supernatant was transferred to a new tube and 800 µL ice-cold 100% isopropyl alcohol was added. DNA was allowed to precipitate for 45 min at −20 °C and then centrifuged again for 15 min at 12,000× g. The pellet was washed twice with 500 µL cold 70% ethanol and once with 500 µL of cold 100% ethanol. After drying at room temperature, samples were resuspended in 100 µL of ultrapure water. The purity and integrity of the DNA were evaluated by spectrophotometry (NanoDrop®, Thermo Scientific, Waltham, MA, EUA) and on agarose gel 1% [16,28]. Total DNA extraction from the soil samples was performed using the EZNA Soil DNA kit (Omega Bio-Tek, Norcross, GA, USA).
2.2.2. DNA Amplification
Reaction mixtures had a total volume of 12.5 μL, comprising 1× PCR buffer, 2.0 mM MgCl2, 25 μM deoxynucleoside triphosphates (dNTPs), 0.5 μM of each forward and reverse primers ITS 1 and ITS 4 [29], 1 U of Taq DNA polymerase (Ludwing Biotec, Bela Vista, Brazil), and 20 ng of genomic DNA. Amplification was performed in an ABI Prism 2720 thermocycler (Applied Biosystems, Foster City, CA, USA), as follows: 95 °C for 4 min, followed by 35 cycles consisting of 95 °C for 45 s, 52 °C for 30 s, and 72 °C for 2 min, and a delay at 72 °C for 7 min. For some samples, annealing temperatures were changed from 50 to 55 °C.
2.2.3. Ligation of Padlock Probes
The padlock probes FOP (F. pedrosoi) and FOM (F. monophora) used in this study were previously designed by Najafzadeh et al. in 2011 [25]. One microliter of ITS amplicon was mixed with 2 U pfu DNA ligase (Agilent Technologies, Santa Clara, CA, USA) and 0.1 µmol l−1 padlock probe in 20 mmol l−1 Tris-HCl (pH 7.5), 20 mmol l−1 KCl, 10 mmol l−1 MgCl2, 0.1% Igepal, 0.01 mmol l−1 rATP, and 1 mmol l−1 DTT, with a total reaction volume of 10 µL. Padlock probe ligation was conducted with one cycle of denaturation for 5 min at 94 °C, followed by five cycles of 94 °C for 30 s and 4 min ligation at 50 °C. An exonucleolysis step was not required.
2.2.4. Rolling Circle Amplification Reaction
One microliter of ligation product was used as a template for RCA. The total volume was 12 µL−l, containing 8 U Bst DNA polymerase (New England Biolabs, Ipswich, MA, EUA), 400 µmol L−l deoxynucleoside triphosphate mix, 25 μmol deoxynucleoside triphosphates (dNTPs), and 10 pmol of each RCA primer (RCA1 5′-ATGGGCACCGAAGAAGCA-3′ and RCA2 5′-CGCGCAGACACGATA-3′) in distilled water. Probe signals were amplified by incubation at 65 °C for 60 min, and the accumulation of double stranded DNA products was visualized on a 2% agarose gel to verify the specificity of probe template binding. Positive reactions exhibited a ladder-like pattern, whereas negative reactions displayed a clean background.
2.2.5. Specificity and Detection Limit of RCA Padlock Probes In Vitro and In Vivo
The specificity of the padlock probes was tested for the detection of F. pedrosoi and F. monophora in environmental samples (Table 1). The DNA of F. pedrosoi (CBS 271.37) and F. monophora (CBS 269.37) was used as a positive control, and the DNA of Fonsecaea erecta (CBS 125760) was employed as a negative control. The effectivity of RCA padlock probes was demonstrated using artificial DNA sample mixtures containing plant debris or soil (20 ng/µL) with fungal DNA (0.5 ng/µL). In vivo, suspensions of a concentration of 105 cells/mL of the positive and negative controls mentioned above were inoculated by direct injection into the stem of the Bactris gasipaes (Peach palm). The plants were cultivated in a vase; after 60 days, stem fractions of the plant were collected for total DNA extraction and analysis by RCA.
Table 1.
In vitro specificity analyzes of the FOP and FOM probe species.
| DNA Test | Species | Collection Number | Source/Geography | Padlock Probe | |
|---|---|---|---|---|---|
| FOP | FOM | ||||
| Fungal DNA | F. pedrosoi | CBS271.37 T | Chromoblastomycosis, South America | (+) | (−) |
| F. monophora | CBS269.37 T | Chromoblastomycosis, South America | (−) | (+) | |
| F. erecta | CBS125763 T | Spine of Japecanga plant, Brazil, Bacabeira | (−) | (−) | |
| F. nubica | CBS125.198 T | Chromoblastomycosis, Cameroon | (−) | (−) | |
| F. pugnacious | CMRP1343 T | Chromoblastomycosis, South America | (−) | (−) | |
| C. albicans | CMRP816 | Human, Brazil | (−) | (−) | |
| P. citrinum | CMRP1538 | Metal, Tucuruí, Brazil | (−) | (−) | |
| A. nidulans | CMRP2338 | - | (−) | (−) | |
| Environmental test DNA samples | M. pudica | Plant in vitro | (−) | (−) | |
| M. pudica with F. pedrosoi | CBS271.37 T | Plant in vitro (20 ng/µL) with 0.5 ng/µL of F. pedrosoi | (+) | (−) | |
| M. pudica with F. monophora | CBS269.37 T | Plant in vitro (20 ng/µL) with 0.5 ng/µL of F. monophora | (−) | (+) | |
| M. pudica with F. erecta | CBS125763 T | Plant in vitro (20 ng/µL) with 0.5 ng/µL of F. erecta | (−) | (−) | |
| B. gasipaes with F. pedrosoi | CBS271.37 T | Plant inoculated with 105 spores of F. pedrosoi | (+) | (−) | |
| B. gasipaes with F. monophora | CBS269.37 T | Plant inoculated with 105 spores of F. monophora | (−) | (+) | |
| B. gasipaes with F. erecta | CBS125763 T | Plant inoculated with 105 spores of F. erecta | (−) | (−) | |
T, type strain; FOP, Fonsecaea pedrosoi padlock probe; FOM, Fonsecaea monophora padlock probe; (−), negative sample; (+), positive sample.
The sensitivity of padlock probes was tested using different dilutions of the internal transcribed region (ITS) amplicons. ITS concentrations of F. pedrosoi and F. monophora were determined with a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, EUA) and diluted to a final concentration of 2 ng/μL. Copy numbers were calculated with an online tool based on Avogadro’s number (http://cels.uri.edu/gsc/cndna.html). For calculation, an amplicon length of 645 bp was assumed for F. pedrosoi and 644 bp for F. monophora. We evaluated the sensitivity of the padlock probes to ensure reliable amplification at low levels of target DNA. We performed 10-fold serial dilutions of ITS DNA, starting with 2.88 × 109 copies per tube and ending with 28.8 copies per tube.
2.3. Isolation and Molecular Identification
2.3.1. Isolation
Selective isolation was performed [16,30] for positive samples in the RCA padlock probe essay. Approximately 20 g from each sample was processed for fungal isolation. Samples were incubated at room temperature for 30 min in 100 mL of a sterilized saline solution containing 200 U penicillin, 200 μg/L streptomycin, 200 μg/L chloramphenicol, and 500 μg/L cycloheximide. Twenty milliliters of sterilized mineral oil was added to the solution, vigorously shaken for 5 min, and left to settle for 40 min. The oil–water interphase was then collected, inoculated onto Mycosel Agar (Difco), and incubated for 4 weeks at 28 °C, with five replicates per sample. One hundred and seven samples were collected (20 of shell of the babassu coconut, 20 of soil, 20 of plant debris, 20 of leaves, 20 of stems, and 7 of thorns), with a total of 435 replicates.
2.3.2. Molecular Identification
The DNA extraction of fungi isolates was performed [11]. About 1 cm2 mycelium of 20 to 30-d-old cultures was transferred to a 2 mL Eppendorf tube containing 300 µL CTAB buffer (CTAB 2% (w/v), NaCl 1.4 M, Tris-HCl 100 mM, pH 8.0; EDTA 20 mM, b-mercaptoethanol 0.2% (v/v)) and about 80 mg of a silica mixture (silica gel H, Merck, Darmstadt, Germany / Celite 545, Biotec, São Paulo, SP, Brazil, 2:1, w/w). Cells were grinded manually with a sterile pestle for approximately 5 min. Subsequently, 200 µL CTAB buffer was added; the mixture was vortexed and incubated for 10 min at 65 °C. After the addition of 500 µL 24:1 chloroform:isoamylalcohol, the solution was mixed and centrifuged for 5 min at 20,500× g and the supernatant was transferred to a new tube with two volumes of ice-cold 96% ethanol. DNA was allowed to precipitate for 30 min at −20 °C and then centrifuged again for 5 min at 20,500× g. Subsequently, the pellet was washed with cold 70% ethanol. After drying at room temperature, it was resuspended in 100 µL in ultrapure water.
Amplification of the internal transcribed region (ITS) was performed as previously described [12]. Amplicons were subjected to direct sequencing, as follows: 95 °C for 1 min, followed by 30 cycles consisting of 95 °C for 10 s, 50 °C for 5 s, and 60 °C. The sequences obtained were aligned using Mega 7 software and compared to the Isham Barcoding Database (http://its.mycologylab.org/) and GenBank Blast (NCBI https://blast.ncbi.nlm.nih.gov/Blast.cgi).
3. Results
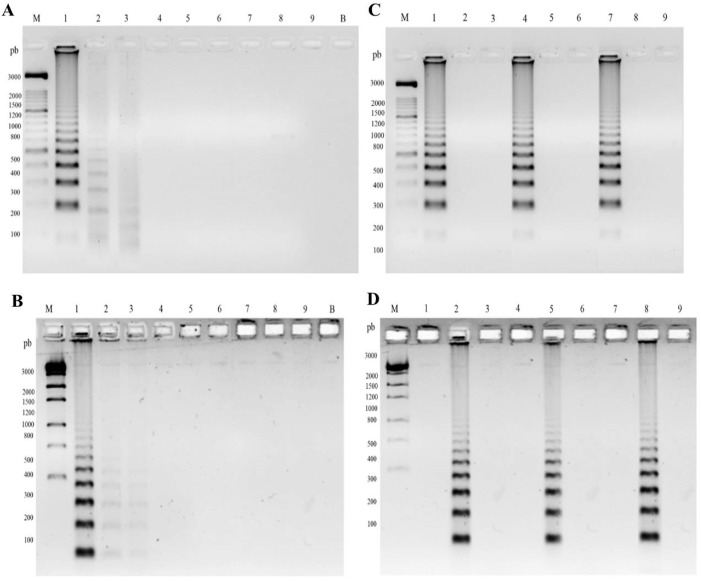
The sensitivity of the RCA padlock probes FOP and FOM was determined using serial dilutions of the F. pedrosoi (CBS 271.37) and F. monophora (CBS 269.37) DNA as templates, respectively. The oligonucleotides ITS1 and ITS4 showed a product of approximately 644 bp. Fonsecaea pedrosoi and F. monophora were detected at 2.88 × 107 copies of purified DNA (Figure 2A,B). A decrease in the signal intensity was observed at lower concentrations for both probes.
Figure 2.
Specificity and limit of detection of padlock probes FOP and FOM by Rolling Circle Amplification (RCA). (A) Sensibility of padlock probe FOP amplified by RCA in internal transcribed region (ITS) amplicons of F. pedrosoi (CBS 271.37). (B) Sensibility of padlock probe FOM amplified by RCA in ITS amplicons of F. monophora (CBS 269.37). M, molecular marker 1 Kb; B, blank; 1 to 9, 2.88 × 109, 2.88 × 108, 2.88 × 107, 2.88 × 106, 2.88 × 105, 2.88 × 104, 2.88 × 103, and 2.88 × 102, 2.88 × 101 copies per tube, respectively. (C,D) Specificity of padlock probe FOP and FOM, respectively, amplified by RCA in plant and soil samples. In vitro specificity analysis: 1–3, total DNA of plant Mimosa pudica; 4–6, total DNA of soil, F. pedrosoi, F. monophora, and Fonsecaea erecta, respectively; 7–9, in vivo specificity analysis of Bactris gasipaes in a vessel after injury with F. pedrosoi, F. monophora, and F. erecta, respectively.
The in vitro specificity analysis demonstrated that the FOP and FOM probe species remained without cross-reaction with closely related species F. nubica, F. erecta, and F. pugnacius, as well as reference DNAs of Candida albicans, Penicillium citrinum, and Aspergillus nidulans (Table 1). The presence of plant material, soil, and other components of total DNA did not interfere with the specificity of the probes and RCA amplification, as demonstrated by in vitro and in vivo analyses (Figure 2C,D). The used RCA padlock probes (FOP and FOM) [25] were shown to be a specific tool for the environmental detection of F. pedrosoi and F. monophora, with no cross-reaction being observed. In vitro and in vivo assays confirmed the specificity and applicability of the method for environmental sample screening (Figure 2C,D).
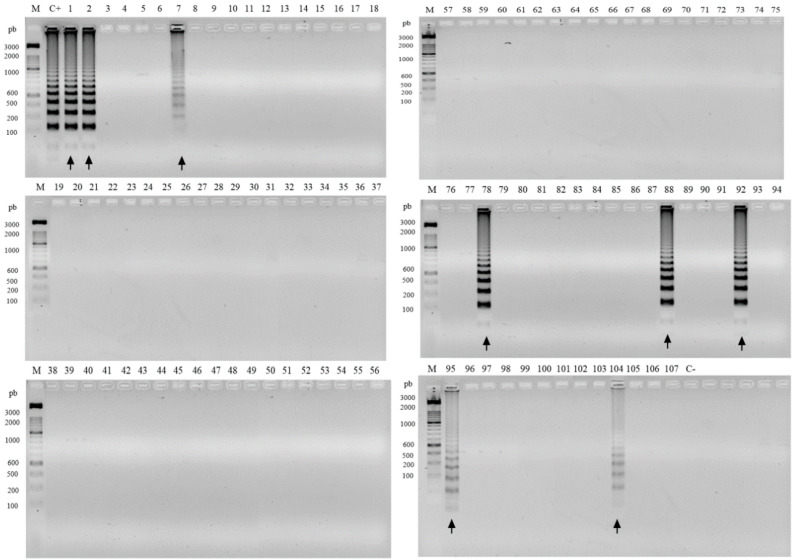
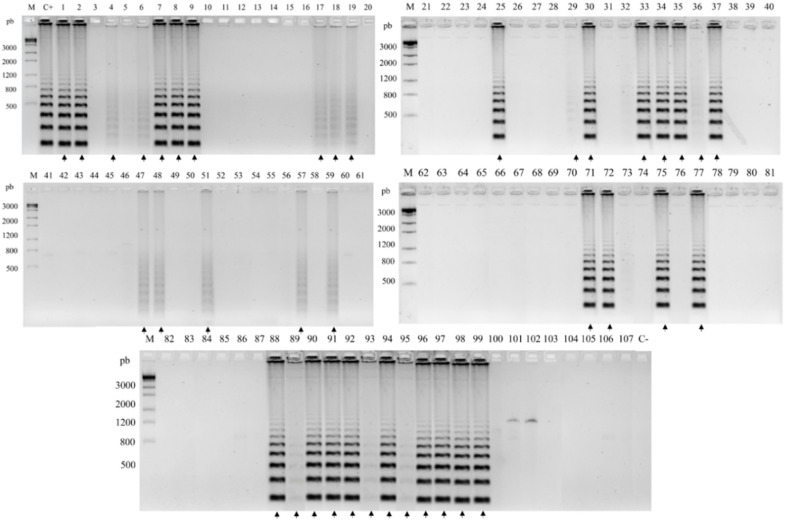
A total of 107 environmental samples were analyzed by RCA padlock probes (Table S1). Eight (7.48%) of the samples were positive for F. pedrosoi, including four samples from shells of babassu coconuts, two from plant debris, one from Solanum paniculatum (Jurubeba tree), and one from soil (Figure 3). The FOM padlock probe amplified forty-two (39.25%) of the samples with twelve samples of babassu coconut shell; seven samples of debris plant; twenty samples of plant material of Murraya paniculata (Murta tree), Astrocaryum vulgare (Tucum tree), Scoparia dulcis (Vassourinha tree), and Platonia insignis (Bacuri tree); and four samples of soil (Figure 4).
Figure 3.
Positive environmental samples for FOP by RCA screening. Molecular marker 1 kb. C+, positive control F. pedrosoi (CBS 271.37); C-, negative control F. erecta (CBS125760). Bacabeira: 1–5, decomposing plant material; 6–10, leaf; 11–15, thorns; 16–20, steam; 68–72, soil. São Benedito do Rio Preto: 21–25, decomposing plant material; 26–30, leaf; 31–32, thorn; 33–37, steam; 73–77, soil. Pinheiro: 58–62, decomposing plant material; 63–67, leaf; 48–52, steam; 78–82, soil. Nina Rodrigues: 53–57, decomposing plant material; 43–47, leaf; 38–42, steam; 83–87, soil; 88–107, shell of babassu coconut. Positive samples are indicated by arrows.
Figure 4.
Positive environmental samples for FOM by RCA screening. Molecular marker 1 kb. C+, positive control F. monophora (CBS 269.37); C-, negative control F. erecta (CBS125760). Bacabeira: 1–5, decomposing plant material; 6–10, leaf; 11–15, thorns; 16–20, steam; 68–72, soil. São Benedito do Rio Preto: 21–25, decomposing plant material; 26–30, leaf; 31–32, thorn; 33–37, steam; 73–77, soil. Pinheiro: 58–62, decomposing plant material; 63–67, leaf; 48–52, steam; 78–82, soil. Nina Rodrigues: 53–57, decomposing plant material; 43–47, leaf; 38–42, steam; 83–87, soil; 88–107, shell of babassu coconut. Positive samples are indicated by arrows.
Positive samples for RCA padlock probes were submitted to isolation by oil-mineral flotation. Sequencing of the acquired isolates showed that they belonged to species other than F. pedrosoi and F. monophora. Judging from the ITS sequencing results, the isolates were affiliated to the order Chaetothyriales in families Trichomeriaceae, Herpotrichiellaceae, and Cyphellophoraceae, and order Capnodiales, families Mycosphaerellaceae and Cladosporiaceae (Table 2).
Table 2.
Fungi isolates from positive samples by RCA padlock probes (FOP and FOM).
| Positive Sample | Substrate | Padlock Probe Positive | N. Isolates | CMRP | Molecular ID. | GenBank Accession |
|---|---|---|---|---|---|---|
| 1 | Decomposing Material | FOP; FOM | 4 | CMRP2566 | Melanoctona tectonae | MT075634 |
| CMRP2821 | Melanoctona tectonae | MT075635 | ||||
| CMRP2840 | Cladosporium sp. | MT075636 | ||||
| CMRP2863 | Cyphellophora sp. | MT075637 | ||||
| 2 | Decomposing Material | FOP; FOM | 2 | CMRP2617 | Strelitziana sp. | MT080291 |
| CMRP2859 | Cyphellophora ambigua | MT075638 | ||||
| 4 | Decomposing Material | FOM | 5 | CMRP2619 | Cladosporium sp. | MT075639 |
| CMRP2594 | Cladosporium sp. | MT075640 | ||||
| CMRP2598 | Mycosphaerellaceae | MT080292 | ||||
| CMRP2826 | Exophiala alcalophila | MT075641 | ||||
| CMRP2822 | Exophiala spinifera | MT075642 | ||||
| 6 | Leaf, A. vulgare | FOM | 2 | CMRP2601 | Hyalocladosporiella cannae | MT075643 |
| CMRP2850 | Exophiala spinifera | MT075644 | ||||
| 7 | Leaf, S. paniculatum | FOP; FOM | 14 | CMRP2560 | Hyalocladosporiella cannae | MT075645 |
| CMRP2848 | Hyalocladosporiella cannae | MT075646 | ||||
| CMRP2564 | Hyalocladosporiella cannae | MT075647 | ||||
| CMRP2567 | Hyalocladosporiella cannae | MT075648 | ||||
| CMRP2557 | Hyalocladosporiella cannae | MT075649 | ||||
| CMRP2589 | Nigrograna obliqua | MT075650 | ||||
| CMRP2591 | Hyalocladosporiella cannae | MT075651 | ||||
| CMRP2609 | Hyalocladosporiella cannae | MT075652 | ||||
| CMRP2615 | Hyalocladosporiella cannae | MT075653 | ||||
| CMRP2851 | Hyalocladosporiella cannae | MT075654 | ||||
| CMRP2852 | Hyalocladosporiella cannae | MT075655 | ||||
| CMRP3098 | Hyalocladosporiella cannae | MT075656 | ||||
| CMRP3094 | Hyalocladosporiella cannae | MT075657 | ||||
| CMRP2867 | Hyalocladosporiella cannae | MT075658 | ||||
| 8 | Leaf, S. dulcis | FOM | 1 | CMRP2868 | Hyalocladosporiella cannae | MT075659 |
| 17 | Stalk, S. paniculatum | FOM | 11 | CMRP2562 | Chaetothyriales | MT080293 |
| CMRP2569 | Cyphellophora sp. | MT075660 | ||||
| CMRP2620 | Cladosporium sp. | MT075661 | ||||
| CMRP2568 | Hyalocladosporiella cannae | MT075662 | ||||
| CMRP2586 | Hyalocladosporiella cannae | MT075663 | ||||
| CMRP2622 | Mycosphaerellaceae | MT080294 | ||||
| CMRP2614 | Hyalocladosporiella cannae | MT075664 | ||||
| CMRP2828 | Strelitziana sp. | MT080295 | ||||
| CMRP2837 | Cyphellophora oxyspora | MT075665 | ||||
| CMRP2839 | Teratosphaeria sp. | MT080296 | ||||
| CMRP3086 | Chaetothyriales | MT080297 | ||||
| 35 | Stalk, A. vulgare | FOM | 1 | CMRP2624 | Strelitziana sp. | MT080298 |
| 47 | Leaf, S. dulcis | FOM | 1 | CMRP3082 | Mycosphaerellaceae | MT080299 |
| 51 | Stalk, S. dulcis | FOM | 6 | CMRP3116 | Cladosporium sp. | MT075666 |
| CMRP3114 | Ochroconis sp. | MT075667 | ||||
| CMRP3113 | Cladosporium sp. | MT075668 | ||||
| CMRP3001 | Pyriculariaceae | MT080300 | ||||
| CMRP3074 | Chaetothyriales | MT080301 | ||||
| CMRP2985 | Mycosphaerellaceae | MT080302 | ||||
| 57 | Decomposing Material | FOM | 6 | CMRP2986 | Mycosphaerellaceae | MT080303 |
| CMRP2998 | Fonsecaea brasiliensis | MT075669 | ||||
| CMRP3104 | Mycosphaerellaceae | MT080304 | ||||
| CMRP3085 | Mycosphaerellaceae | MT080305 | ||||
| CMRP3107 | Mycosphaerellaceae | MT080306 | ||||
| CMRP3088 | Chaetothyriales | MT080307 | ||||
| 59 | Decomposing Material | FOM | 8 | CMRP2855 | Chaetothyriales | MT080308 |
| CMRP2856 | Chaetothyriales | MT080309 | ||||
| CMRP2865 | Chaetothyriales | MT080310 | ||||
| CMRP2869 | Chaetothyriales | MT080311 | ||||
| CMRP2874 | Chaetothyriales | MT080312 | ||||
| CMRP3002 | Chaetothyriales | MT080313 | ||||
| CMRP3109 | Fonsecaea brasiliensis | MT075670 | ||||
| CMRP2582 | Cladosporium sp. | MT075671 | ||||
| 71 | Soil | FOM | 1 | CMRP2561 | Exophiala spinifera | MT075672 |
| 72 | Soil | FOM | 5 | CMRP2580 | Mycosphaerellaceae | MT080314 |
| CMRP2602 | Exophiala spinifera | MT075673 | ||||
| CMRP2605 | Exophiala spinifera | MT075674 | ||||
| CMRP2610 | Trichomeriaceae | MT080315 | ||||
| CMRP3117 | Cyphellophora oxyspora | MT075675 | ||||
| Total | 67 | |||||
N, number of isolates; ID, identification; FOP, F. pedrosoi padlock probe; FOM, F. monophora padlock probe; CMRP, Coleções Microbiológicas da Rede Paranaense.
4. Discussion
The epidemiology of chromoblastomycosis suggests that the etiological agents of the disease are present in the environment and the infection is accidental. However, why only a small selection of the apparently saprobic fungi have repeatedly been found in humans, while they theoretically all have a comparative chance of being inoculated, has remained unexplained. Isolates from cactus thorns obtained near a house of a CBM patient had a similar morphology to the common CBM agent Cladophialophora carrionii, but by molecular methods, they were identified as Cladophialophora yegresii, which has never been found in human hosts [15]. Isolates of both species were demonstrated to be able to form muriform cells when inoculated into cactus plants. Sibling species of CBM agents in Fonsecaea, i.e., F. erecta and F. minima, were commonly associated with plants, but did not have any relationship with chromoblastomycosis [11]. In order to investigate the environmental sources of F. pedrosoi and F. monophora, substrates reported as possible sources of CBM infection in epidemic areas in Brazil [13,26] were collected, including living plants (leaf, stem, and thorns) and babassu coconut shells.
The RCA probe was previously applied to other causal agents of fungal infection, such as Sporothrix spp. [31], Fusarium graminearum [32], and Histoplasma capsulatum [33]. The limit of detection of the method was 2.88 × 107 copies of DNA for both F. pedrosoi and F. monophora. A decrease in intensity of the signal amplification was observed when low concentrations of DNA were available, as also reported for Histoplasma spp. [33], although this was not observed in Sporothrix spp. [31]. The probes for Fonsecaea spp. seem to have a higher detection limit, which can be useful for environmental screening.
The isolation of Fonsecaea spp. from environmental substrates by oil flotation was reported in several studies [4,11,13,16,26,34,35]. These studies demonstrate that, despite sophisticated selective efforts, the recovery of pathogenic environmental strains is limited. Environmental inoculation is the most parsimonious hypothesis for the onset of chromoblastomycosis, since some patients have presented fragments of plant material in tissue [6,36,37]. Our study shows that the agents of CBM are commonly present in the environment, as are the strictly saprobes species, with some plant material detection unclear. Even though few pathogenic strains are recovered from live plants by conventional isolation methods, these agents of chromoblastomycosis are able to survive in plant tissue after inoculation [38]. The method applied in the present study allowed the detection of DNA of chromoblastomycosis agents in living plants. The presence of F. pedrosoi was detected in the leaves of S. paniculatum (Jurubeba), in decomposing material, soil, and shells of babassu coconut (Orbygnia phalerata) (Figure 3). Fonsecaea monophora tests were positive in stems of M. paniculata (Murta tree), leaves and stems of A. vulgare (Tucum tree), leaves and stems of S. dulcis (Vassourinha tree), and leaves of P. insignis (Bacuri tree), decomposing material, soil, and shell of babassu coconut (O. phalerata) (Figure 4). The isolation by mineral oil flotation was performed in positive samples for RCA, but the chromoblastomycosis agents F. pedrosoi and F. monophora were not isolated. The babassu coconut has been suggested as a source of agents in Maranhão state of the Brazilian Amazon rainforest [26,27]. Nascimento et al. (2017) [2] isolated numerous chaetothyrialean fungi from this source, but no CBM agents. Our study showed the presence of CBM agents in the DNA from the same study.
The environmental screening of Fonsecaea spp. by RCA padlock probes, when compared with selective isolation, demonstrated that CBM agents must be present in environmental samples, even though they were not previously detected [4,11,16,34,35,39,40]. RCA was positive for F. pedrosoi and F. monophora in most of the substrates analyzed, showing that living plants provide a habitat for the agents, such as S. paniculatum, M. paniculata, A. vulgare, S. dulcis, and P. insignis (Table S1). This may be explained by the high sensitivity of the probe to detect small concentrations of fungal DNA, even when among competing saprobes. Alternatively, and perhaps more likely, is an explanation by a larger preference for the CBM agents to grow in habitats with similarity to animal tissue; such factors have not yet been revealed. Our results demonstrate that the hypothesis of a route of infection via plant material could be considered, as suggested in clinical reports.
Similar results have been published for environmental pathogens (i.e., pathogens with a double, environment/host alternating life cycle), such as Histoplasma and Paracoccidioides. Furthermore, these fungi were difficult to isolate from environmental samples, despite positivity with molecular methods [41,42]. The RCA padlock approach applied in this study represents an important method of pathogen detection in environmental samples, contributing to understanding routes of infection. The difference in habitat preference between strict saprobes and opportunistic saprobes remains enigmatic.
5. Conclusions
In conclusion, the data obtained in this study showed that the RCA padlock probe represents an efficient, sensitive, and reproducible molecular tool for the environmental screening of opportunistic fungi related to chromoblastomycosis in natural substrates, such as babassu coconut, plants, soil, and decomposing plant material. The use of the padlock probe contributes to new insights into the environmental occurrence and infection route of these agents.
Acknowledgments
The authors are grateful for the support from the Federal University of Paraná (UFPR) and Federal University of Maranhão (UFMA). We are grateful to Jiufeng Sun for his contribution to previous work on the design of the probe.
Supplementary Materials
The following are available online at https://www.mdpi.com/2309-608X/6/4/290/s1, Table S1: Environmental samples from Maranhão state, Brazil, analyzed by RCA padlock probes.
Author Contributions
M.F.V., V.A.V., G.S.d.H., and R.R.G. conceived and designed the experiments. M.F.V., B.J.F.d.S.L., G.F., A.B., I.C.L.d.S., and F.d.F.C. performed the experiments. C.d.M.P.e.S.d.A. collected the samples. M.F.V., V.A.V., and R.R.G. analyzed the data. V.A.V., C.d.M.P.e.S.d.A., and G.F. contributed reagents, materials, and analysis tools. M.F.V., V.A.V., B.J.F.d.S.L., F.d.F.C., B.P.R.L., and S.H. contributed to preparing the manuscript and revising it critically. M.F.V., F.d.F.C., I.C.L.d.S., and V.A.V. contributed to preparation, creation, and/or presentation of the tables, graphics, and figures. V.A.V., C.d.M.P.e.S.d.A., L.V.A. and M.J.N. offered strains and/or substantial contributions to the work. M.F.V., V.A.V., and G.S.d.H. conceived and designed the work, wrote the manuscript, and revised the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Brazilian Federal Agency for Support and Evaluation of Graduate Education: National Council for Scientific and Technological Development (CNPq), Brasilia, Brazil (http://cnpq.br/) and Education Coordination for the Improvement of Higher Education Personnel—CAPES– Finance Code 001 (www.capes.gov.br). Vânia Aparecida Vicente received fellowships from CNPq (grant number 312811/2018–17), Brasilia, Brazil and the Institutional Program of Internationalization CAPES/PrInt n. 8887.311835/2018-00-AUXPE-2796/2018.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Deng S., Tsui C.K.M., van den Ende A.H.G.G., Yang L., Najafzadeh M.J., Badali H., Li R., Hagen F., Meis J.F., Sun J., et al. Global spread of human chromoblastomycosis is driven by recombinant Cladophialophora carrionii and predominantly clonal Fonsecaea species. PLoS Negl. Trop. Dis. 2015;9:e0004004. doi: 10.1371/journal.pntd.0004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nascimento M.M.F., Vicente V.A., Bittencourt J.V.M., Gelinski J.M.L., Prenafa-Boldúe F., Romero-Guizae M., Fornari G., Gomes R.R., Santos G.D., Van Den Ende A.H.G., et al. Diversity of opportunistic black fungi on babassu coconut shells, a rich source of esters and hydrocarbons. Fungal Biol. 2017;121:488–500. doi: 10.1016/j.funbio.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Teixeira M.M., Moreno L.F., Stielow B.J., Muszewska A., Hainaut M., Gonzaga L., Abouelleil A., Patané J.S.L., Priest M., Souza R., et al. Exploring the genomic diversity of black yeasts and relatives (Chaetothyriales, Ascomycota) Stud. Mycol. 2017;86:1–28. doi: 10.1016/j.simyco.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vicente V.A., Ribeiro O., Najafzadeh M.J., Sun J., Guerra R.S., Miesch S., Ostrensky A., Meis J.F., Klaassen C.H., de Hoog G.S., et al. Black yeast-like fungi associated with lethargic crab disease (LCD) in the mangrove-land crab, Ucides cordatus (Ocypodidae) Vet. Microbiol. 2012;158:109–122. doi: 10.1016/j.vetmic.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 5.Seyedmousavi S., Netea G.M., Mouton J.W., Melchers W.J.G., Verweij P.E., de Hoog G.S. Black yeasts and their filamentous relatives: Principles of pathogenesis and host defense. Clin. Microbiol. Rev. 2014;27:527–542. doi: 10.1128/CMR.00093-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Queiroz-Telles F., de Hoog G.S., Santos D.C., Salgado C.G., Vicente V.A., Bonifaz A., Roilides E., Xi L., Azevedo A.M.P., Silva M.B., et al. Cromoblastomycosis: A neglected global disease. Clin. Microbiol. Rev. 2017;30:233–276. doi: 10.1128/CMR.00032-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Queiroz-Telles F. Chromoblastomycosis, a neglected tropical disease. Rev. Inst. Med. Trop. São Paulo. 2015;57:46–50. doi: 10.1590/S0036-46652015000700009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esterre P., Andriantsimahavandy A., Ramarcel E.R., Pecarrere J.L. Forty years of chromoblastomycosis in Madagascar: A review. Am. J. Trop. Med. Hyg. 1996;55:45–47. doi: 10.4269/ajtmh.1996.55.45. [DOI] [PubMed] [Google Scholar]
- 9.Silva J.P., de Souza W., Rozental S. Chromoblastomycosis: A retrospective study of 325 cases on Amazonic Region (Brazil) Mycopathologia. 1999;143:171–175. doi: 10.1023/A:1006957415346. [DOI] [PubMed] [Google Scholar]
- 10.Pérez-Blanco M., Hernández Valles R., García-Humbría L., Yegres F. Chromoblastomycosis in children and adolescents in the endemic area of the Falcón State, Venezuela. Med. Mycol. 2006;44:467–471. doi: 10.1080/13693780500543238. [DOI] [PubMed] [Google Scholar]
- 11.Vicente V.A., Najafzadeh M.J., Sun J., Romes R.R., Robl D., Marques S.G., Azevedo C.M.P.S., de Hoog G.S. Environmental siblings of black agents of human chromoblastomycosis. Fungal Divers. 2013;65:47–63. doi: 10.1007/s13225-013-0246-5. [DOI] [Google Scholar]
- 12.Gomes R.R., Vicente V.A., Azevedo C.M.P.S., Salgado C.G., Silva M.B., Queiroz-Telles F., Marques S.G., Santos D.W.C.L., Andrade T.S., Takagi E.H., et al. Molecular epidemiology of agents of human chromoblastomycosis in Brazil with the description of two novel species. PLoS Negl. Trop. Dis. 2016;11:e0005315. doi: 10.1371/journal.pntd.0005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salgado C.G., Silva J.P., Diniz J.A.P., Silva M.B., Costa P.F., Teixeira C., Salgado U.I. Isolation of Fonsecaea pedrosoi from thorns of Mimosa pudica, a probable natural source of chromoblastomycosis. Rev. Inst. Med. Trop. Sao Paulo. 2004;46:33–36. doi: 10.1590/S0036-46652004000100006. [DOI] [PubMed] [Google Scholar]
- 14.Sudhadham M., de Hoog G.S., Menken S.B.J., van den Ende A.H.G.G., Sihanonth P. Rapid screening for genotypes as possible markers of virulence in the neurotropic black yeast Exophiala dermatitidis using PCR-RFLP. J. Microbiol. Methods. 2010;80:138–142. doi: 10.1016/j.mimet.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 15.De Hoog G.S., Nishikaku A.S., Fernandez-Zeppenfeldt G., Padín-González C., Burger E., Badali H., Richard-Yegres N., van den Ende A.H.G.G. Molecular analysis and pathogenicity of the Cladophialophora carrionii complex, with the description of a novel species. Stud. Mycol. 2007;58:219–234. doi: 10.3114/sim.2007.58.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vicente V.A., Attili-Agelis D., Pie M.R., Queiroz-Telles F., Cruz M., Najafzadeh M.J., de Hoog G.S., Zhao J., Pizzirani-Kleiner A. Environmental isolation of black yeast-like fungi involved in human infection. Stud. Mycol. 2008;61:137–144. doi: 10.3114/sim.2008.61.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vicente V.A., Weiss V.A., Bombassaro A., Moreno L.F., Costa F.F., Raittz R.T., Leão A.C., Gomes R.R., Bocca A.L., Fornari G., et al. Comparative genomics of sibling species of Fonsecaea associated with human chromoblastomycosis. Front. Microbiol. 2017;8:1924. doi: 10.3389/fmicb.2017.01924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J., Najafzadh M.J., Vicente V.A., Xi L., de Hoog G.S. Rapid detection of pathogenic fungi using loop-mediated isothermal amplification, exemplified by Fonsecaea agents of chromoblastomycosis. J. Microbiol. Methods. 2010;80:19–24. doi: 10.1016/j.mimet.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Van Elsas J.D., Boersma F.G.H. A review of molecular methods to study the microbiota of soil and the mycosphere. Eur. J. Soil Biol. 2011;47:77–87. doi: 10.1016/j.ejsobi.2010.11.010. [DOI] [Google Scholar]
- 20.Najafzadeh M.J., Dolatabadi S., Saradeghi Keisari M., Naseri A., Feng P., de Hoog G.S. Detection and identification of opportunistic Exophiala species using the rolling circle amplification of ribosomal internal transcribed spacers. J. Microbiol. Methods. 2013;94:338–342. doi: 10.1016/j.mimet.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 21.Irinyi L., Lackner M., de Hoog G.S., Meyer W. DNA barcoding of fungi causing infections in humans and animals. Fungal Biol. 2016;120:125–136. doi: 10.1016/j.funbio.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Tsui C.K.M., Woodhall J., Chen W., Levesque C.A., Lau A., Schoens C.D., Bachiens C., Najafzadeh M.J., de Hoog G.S. Molecular techniques for pathogen identification and fungus detection in the environment. IMA Fungus. 2011;2:177–189. doi: 10.5598/imafungus.2011.02.02.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilson M., Malmgren H., Samiotaki M., Kwiatkowski M., Chowdhary B.P., Landegren U. Padlock probes: Circularizing oligonucleotides for localized DNA detection. Science. 1994;265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 24.Atkins S.D., Clarck I.M. Fungal molecular diagnostics: A mini review. J. Appl. Genet. 2004;45:3–15. [PubMed] [Google Scholar]
- 25.Najafzadeh M.J., Sun J., Vicente V.A., de Hoog G.S. Rapid detection and identification of fungal pathogens by rolling circle amplification (RCA) using Fonsecaea as a model. Mycoses. 2011;54:577–582. doi: 10.1111/j.1439-0507.2010.01995.x. [DOI] [PubMed] [Google Scholar]
- 26.Marques S.G., Silva C.M.P., Saldanha P.C., Rezende M.A., Vicente V.A., Queiroz-Telles F., Costa J.M.L. Isolation of Fonsecaeae from the shell of the babassu coconut (Orbygnia phalerata Martius) in the Amazon region of Maranhão Brazil. Jpn. J. Med Mycol. 2006;47:305–311. doi: 10.3314/jjmm.47.305. [DOI] [PubMed] [Google Scholar]
- 27.Silva C.M., Da Rocha R.M., Moreno J.S., Silva R.R., Marques S.G., Costa J.M. The coconut babaçu (Orbignya phalerata martins) as a probable risk of human infection by the agent of chromoblastomycosis in the State of Maranhão. Rev. Soc. Bras. Med. Trop. 1995;28:49–52. doi: 10.1590/S0037-86821995000100009. [DOI] [PubMed] [Google Scholar]
- 28.Ferreira M.E., Grattapaglia D. Introdução ao Uso de Marcadores Moleculares em Análise Genética. 3rd ed. Embrapa Recursos genéticos e Biotecnologia; Brasília, Brazil: 1998. [Google Scholar]
- 29.White T.J., Bruns T., Lee S., Taylor J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to the Methods and Applications. Academic Press; New York, NY, USA: 1990. pp. 315–322. [Google Scholar]
- 30.Iwatsu T., Miyaji M., Okamoto S. Isolation of Phialophora verrucosa and Fonsecaea pedrosoi from nature in Japan. Mycopathologia. 1981;75:149–158. doi: 10.1007/BF00482809. [DOI] [Google Scholar]
- 31.Rodrigues A.M., Najafzadeh M.J., de Hoog G.S., De Camargo Z.P. Rapid identification of emerging human-pathogenic Sporothrix species with rolling circle amplification. Front. Microbiol. 2015;6:1385. doi: 10.3389/fmicb.2015.01385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davari M., van Diepeningen A.D., Badai-Ahari A., Arzanlou M., Najafzadeh M.J., Van Der Lee T.A.J., de Hoog G.S. Rapid identification of Fusarium graminearum species complex using Rolling Circle Amplification (RCA) J. Microbiol. Methods. 2012;89:63–70. doi: 10.1016/j.mimet.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 33.Furuie J.L., Sun J., Do Nascimento M.F., Gomes R.R., Waculicz-Andrade C.E., Sessegolo G.C., Rodrigues A.M., Galvão-Dias M.A., De Camargo Z.P., Queiroz-Telles F., et al. Molecular identification of Histoplasma capsulatum using rolling circle amplification. Mycoses. 2016;59:12–19. doi: 10.1111/myc.12426. [DOI] [PubMed] [Google Scholar]
- 34.Vicente V.A., Attili-Agelis D., Queiroz-Telles F., Pizzirani-Kleiner A.A. Isolation of herpotrichiellacious fungi from the environment. Braz. J. Microbiol. 2001;32:47–51. doi: 10.1590/S1517-83822001000100011. [DOI] [Google Scholar]
- 35.Lima B.J.F.D.S., Voidaleski M.F., Gomes R.R., Fornari G., Barbosa Soares J.M., Bombassaro A., Schneider G.X., Soley B.D.S., Azevedo C.D.M.P.E.S.D., Menezes C., et al. Selective isolation of agents of chromoblastomycosis from insect-associated environmental sources. Fungal Biol. 2020;124:194–204. doi: 10.1016/j.funbio.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Heidrich D., González G.M., Paganic D.M., Ramírez-Castrillónc M., Scrofernekera M.L. Chromoblastomycosis caused by Rhinocladiella similis: Case report. Med. Mycol. Case Rep. 2017;16:25–27. doi: 10.1016/j.mmcr.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal R., Singh G., Ghosh A., Verma K.K., Pandey M., Xess I. Chromoblastomycosis in India: Review of 169 cases. PLoS Negl. Trop. Dis. 2017;11:e0005534. doi: 10.1371/journal.pntd.0005534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fornari G., Gomes R.R., Degenhardt-Goldbach J., Lima B.J.S., Voidaleski M.F., Santos S.S., Almeida S.R., Muro M.D., Bonna C., Scola R.H., et al. A Model for Trans-Kingdom Pathogenicity in Fonsecaea Agents of Human Chromoblastomycosis. Front. Microbiol. 2018;9:2211. doi: 10.3389/fmicb.2018.02211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satow M.M., Attili-Angelis D., de Hoog G.S., Angelis D.F., Vicente V.A. Selective factors involved in oil flotation isolation of black yeasts from the environment. Stud. Mycol. 2008;61:157–163. doi: 10.3114/sim.2008.61.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerra R.S., Nascimento M.M.F., Miesch S., Najafzadeh M.J., Ribeiro R.O., Ostrensky A., de Hoog G.S., Vicente V.A., Boeger W.A. Black Yeast Biota in the Mangrove, in Search of the Origin of the Lethargic Crab Disease (LCD) Mycopathologia. 2013;75:421–430. doi: 10.1007/s11046-013-9636-1. [DOI] [PubMed] [Google Scholar]
- 41.Arantes T.D., Theodoro R.C., Macoris S.A.G., Bagagli E. Detection of Paracoccidioides spp. in environmental aerosol samples. Med. Mycol. 2013;51:83–92. doi: 10.3109/13693786.2012.698444. [DOI] [PubMed] [Google Scholar]
- 42.Norkaew T., Ohno H., Sriburee P., Tanabe K., Tharavichitkul P., Takarn P., Puengchan T., Bumrungsri S., Miyazake Y. Detection of environmental sources of Histoplasma capsulatum in Chiang Mai, Thailand, by Nested PCR. Mycopathologia. 2013;6:395–402. doi: 10.1007/s11046-013-9701-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.