Abstract
The gut microbiome is fundamental in neurogenesis processes. Alterations in microbial constituents promote inflammation and immunosuppression. Recently, in immune-oncology, specific microbial taxa have been described to enhance the effects of therapeutic modalities. However, the effects of microbial dysbiosis on glioma are still unknown. The aim of this study was to explore the effects of glioma development and Temozolomide (TMZ) on fecal microbiome in mice and humans. C57BL/6 mice were implanted with GL261/Sham and given TMZ/Saline. Fecal samples were collected longitudinally and analyzed by 16S rRNA sequencing. Fecal samples were collected from healthy controls as well as glioma patients at diagnosis, before and after chemoradiation. Compared to healthy controls, mice and glioma patients demonstrated significant differences in beta diversity, Firmicutes/Bacteroides (F/B) ratio, and increase of Verrucomicrobia phylum and Akkermansia genus. These changes were not observed following TMZ in mice. TMZ treatment in the non-tumor bearing mouse-model diminished the F/B ratio, increase Muribaculaceae family and decrease Ruminococcaceae family. Nevertheless, there were no changes in Verrucomicrobia/Akkermansia. Glioma development leads to gut dysbiosis in a mouse-model, which was not observed in the setting of TMZ. These findings seem translational to humans and warrant further study.
Subject terms: CNS cancer, Cancer in the nervous system
Introduction
Glioma is the most common primary malignant tumor of the Central Nervous System (CNS), with 50% of patients presenting with the most aggressive type, glioblastoma (GBM)1. In spite of recent advances in chemoradiation and surgical approaches, the median survival continues to be uniformly poor2,3. Despite progress in basic and clinical research, the overall survival (OS) of GBM has not significantly increased for the past decades3. The rapid proliferation, infiltrating nature, molecular heterogeneity, and challenges to deliver drugs across the blood–brain barrier have limited the effectiveness of therapies4. Development of new therapeutic strategies through the study of molecular and genomic characteristics, tumor microenvironment, and tumor-host interactions has become of great interest in the need to develop new potential therapies.
The human gastrointestinal microbiome is composed of several microorganisms with diverse natures and functions that are dependent on genetic and environmental factors5–8. In recent years, a growing body of evidence supports the role of the gut microbiome (GM) in human physiology and biological activity under homeostatic and pathological conditions6,9. Evidence now suggests that some microbes may confer susceptibility to certain cancers and may influence response to therapies10–13. Gut dysbiosis, a disequilibrium in the bacterial ecosystem, leads to overrepresentation of some bacteria and favors chronic inflammation and immunosuppression14, as recently described in several CNS diseases, including stroke, Parkinson’s disease, amyotrophic lateral sclerosis (ALS), and multiple sclerosis (MS)5,15–19. However, the relationship between the gut-brain axis, glioma development, and how the GM responds and interacts with Temozolomide (TMZ) has not been investigated. The goals of this study were (1) to explore the GM changes following glioma growth and after oral TMZ in a glioma mouse-model and (2) to identify glioma induced GM changes and the effects of TMZ in a prospective glioma patient cohort.
Results
Glioma leads to dysbiosis in the gut microbiome in mice
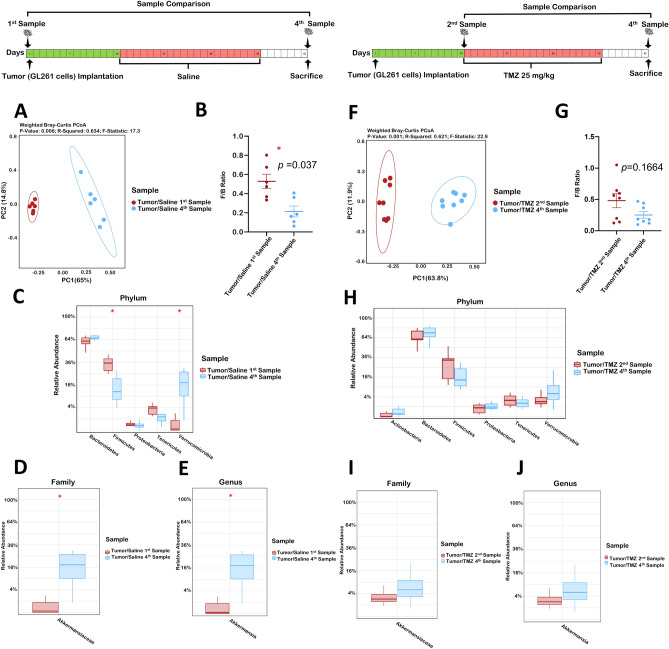
First, we performed a comparison of samples before tumor implantation (1st sample-baseline) vs. sacrifice (4th sample) to evaluate if glioma growth changes the GM. This comparison revealed that the operational taxonomic units (OTUs) were significantly different (p = 0.0049) while the Shannon index showed similar bacterial community richness and diversity (p = 0.065, Supplementary Fig. S1A). Further assessment using the Principal coordinate analysis (PCoA) of weighted pairwise Bray–Curtis distances demonstrated differential clustering patterns (p = 0.006), Fig. 1A. There was a significant decrease in the Firmicutes to Bacteroides (F/B) ratio suggesting dysbiosis following tumor-growth (p = 0.037, Fig. 1B). The relative abundance after tumor development demonstrated decreased Firmicutes (p = 0.03) and increased Verrucomicrobia phyla (p = 0.03, Fig. 1C).
Figure 1.
(A–E) Glioma induces dysbiosis in the mouse gut microbiome. Comparison of Tumor/Saline 1st and 4th samples (n = 6). (A) Beta diversity Weighted Bray–Curtis PCoA. (B) Firmicutes to Bacteroides ratio. (C) Relative abundance taxa summary at the Phylum level. (D) Relative abundance of Akkermansiaceae at the family level. (E) Relative abundance of Akkermansia at the genus level. (F–J) Temozolomide reshapes the Glioma-induced Dysbiosis. Comparison of Tumor/TMZ 2nd and 4th samples (n = 8). (F) Beta diversity Weighted Bray–Curtis PCoA. (G) Firmicutes to Bacteroides ratio. (H) Relative abundance taxa summary at the Phylum level. (I) Relative abundance of Akkermansiaceae at the family level. (J) Relative abundance of Akkermansia at the genus level.
Akkermansiaceae family, the dominant family in the phylum Verrucomicrobia20 and its genus, Akkermansia were evaluated, demonstrating an increased relative abundance between the 1st and 4th sample in tumor-bearing mice (p = 0.03 and 0.035, respectively, Fig. 1D,E).
To confirm that the observed changes were secondary to tumor growth rather than weight loss, we performed a similar analysis between the 1st and 3rd samples (collected prior to bodyweight loss). Results demonstrated similar changes in alpha diversity, F/B ratio, PCoA, Verrucomicrobia, Akkermansiaceae and Akkermansia (Supplementary Fig. S2). Furthermore, the 1st and 4th samples comparison in the Sham/Saline group did not display changes in Verrucomicrobia, Akkermansiaceae, or Akkermansia (Supplementary Fig. S3).
Since GBM typically occurs within the 5–7th decade21, and aging alters the microbiome composition22; we implanted GL261 cells into 18–20-month-old mice. Alpha diversity indices were comparable to young mice at baseline (Supplementary Fig. S4A). Consistent with previous findings22, the F/B ratio in aged mice was increased at baseline, suggestive of underlying dysbiosis (Supplementary Fig. S4B). PCoA exhibited separate clusters in weighted Bray–Curtis distances between young and aged mice (Supplementary Fig. S4C). The aged microbiome differed from young in the relative abundance of Bacteroidetes, Firmicutes, Tenericutes, and Actinobacteria phyla (Supplementary Fig. S4D). However, the relative abundance of Verrucomicrobia, Akkermansiaceae, and Akkermansia remained unchanged with aging (Supplementary Fig. S4D–F). Following tumor implantation, there was a similar pattern of microbiome alteration in aged and young mice (Supplementary Fig. S5). Overall our results demonstrated that glioma development leads to a GM dysbiosis in mice.
Oral temozolomide prevented the glioma-induced dysbiosis
Then, we investigated the TMZ (oral chemotherapy) effect on glioma-induced dysbiosis, which is the standard of care chemotherapy for GBM. Mice were gavaged with TMZ (25 mg/kg) for three-weeks after tumor implantation. Stool analysis was performed between the 2nd (before treatment) and 4th samples (before sacrifice). Our results demonstrated no differences in OTUs or Shannon index (Supplementary Fig. S1B). Although the fecal samples (FS) in the 2nd and 4th samples in the Tumor/TMZ group clustered separately (p = 0.001, Fig. 1F) there was no difference in the F/B ratio (p = 0.17, Fig. 1G). In addition, our results did not demonstrate changes at the Firmicutes and Verrucomicrobia phyla or Akkermansiaceae family and Akkermansia genus (Fig. 1H–J). Similar results were observed in aged mice (Supplementary Fig. S5). Collectively, our results in mice, suggest that the glioma-induced microbiome changes were not observed in the TMZ setting.
Effect of oral temozolomide on non-tumor bearing mice microbiome
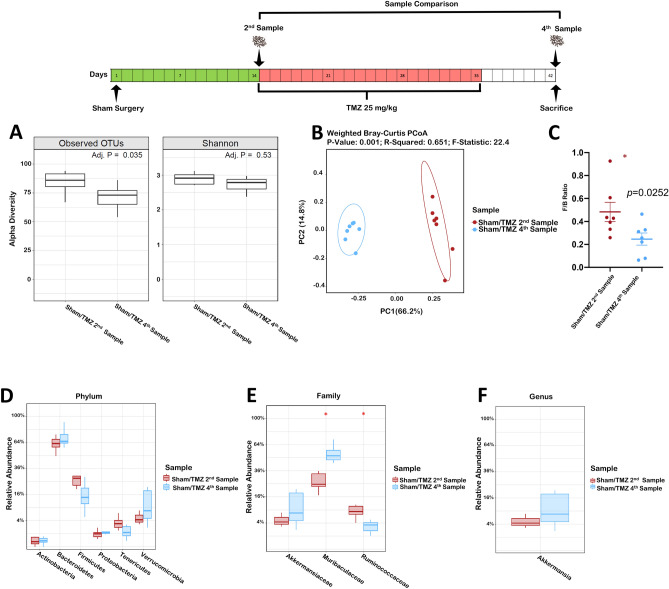
After demonstrating a glioma induced GM dysbiosis and that this was not observed in the setting of TMZ, we aimed to determine the TMZ effect on mouse GM. We evaluated this in non-tumor bearing mice (Sham/TMZ group). Samples were compared prior (2nd Sample) and after TMZ therapy (4th Sample/sacrifice), Fig. 2. Observed OTUs were different between samples (p = 0.035), however, the Shannon index showed comparable microbiome community richness and diversity (p = 0.53, Fig. 2A). TMZ administration for three-weeks resulted in a clear separation of the microbiome beta diversity (p = 0.001, Fig. 2B). In addition, a decrease in the F/B (p = 0.02, Fig. 2C) was observed. However, there were no significant differences in Verrucomicrobia, Akkermansiaceae, or Akkermansia (Fig. 2D–F). Importantly, there was no weight loss following TMZ administration (pre-TMZ 27 ± 0.97 vs. post-TMZ 27.29 ± 0.94, p = 0.27). Muribaculaceae and Ruminococcaceae families increased and decreased following TMZ, respectively (p = 0.01 and 0.04, Fig. 2E). In addition, TMZ altered gut permeability in aged mice as measured by the dextran assay (p < 0.05, Supplementary Fig. S6). Altogether, these results show that TMZ itself causes dysbiosis in the GM, but without affecting Verrucomicrobia or Akkermansia abundance.
Figure 2.
(A–F) Temozolomide alters the mouse gut microbiome. Comparison of Sham/TMZ 2nd and 4th samples (n = 7). (A) Observed OTU and the Shannon alpha diversity index. (B) Beta diversity Weighted Bray–Curtis PCoA. (C) Firmicutes to Bacteroides ratio. (D) Relative abundance taxa summary at the Phylum level. (E) Relative abundance taxa abundance at the family level. (F) Relative abundance of Akkermansia at the genus level.
Effect of glioma and temozolomide in patients with glioma
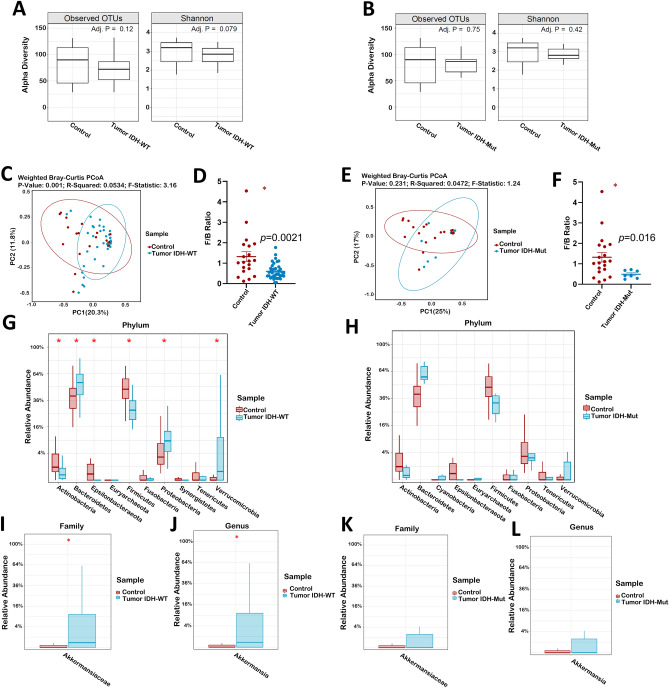
After our mice experiments demonstrating the effects of glioma development and TMZ in the GM of mice, we evaluated if this effects were also observed in glioma patients compared to healthy controls. Eighty-three stool samples from 53-glioma patients were prospectively collected. 47-samples at diagnosis (39 IDH-wildtype and 8 IDH-mutant), and 18 matching pairs before and after chemoradiation. Tumors were classified according to the WHO 2016 classification23. Median age was 55.5-years and 64% were males. Median pre-operative Karnofsky Performance Status (KPS) was 70, and most patients (n = 39, 74%) were treated with the Stupp protocol24, Table 1. Median follow-up was 7.0 months. No differences were observed in OTUs and Shannon index between controls and IDH-WT (Fig. 3A) or IDH-Mut (Fig. 3B) glioma at diagnosis. The PCoA plot displayed separate clustering between controls and glioma IDH-WT (p = 0.001, Fig. 3C), but not in IDH-Mut patients (p = 0.23, Fig. 3E). F/B ratio demonstrated significant differences between IDH-WT and IDH-Mut patients compared to controls (p = 0.0021 and p = 0.016, respectively, Fig. 3D–F). Interestingly, marked differences were observed at the phylum level in IDH-WT patients compared to controls, with increased Bacteroidetes, Proteobacteria, and Verrucomicrobia (p = 0.02, 0.02 and 0.01, respectively, Fig. 3G), however, these were not observed in IDH-Mut patients (Fig. 3H). Akkermansiaceae (p = 0.035, Fig. 3I), and Akkermansia (p = 0.037, Fig. 3J) were significantly higher in IDH-WT patients compared to controls. In IDH-Mut patients, there was no difference in Akkermansiaceae (p = 1.0, Fig. 3K), and Akkermansia (p = 1.0, Fig. 3L). Twenty-one control samples were analyzed and compared to samples from glioma patients at initial diagnosis. The control samples demographic characteristics are demonstrated in Supplementary Table S1.
Table 1.
Clinical Characteristics and Demographics of Glioma Patients (N = 53).
| Variable | N (%) |
|---|---|
| Age group at diagnosis | |
| 18–54 | 26 (49) |
| > 55 | 27 (51) |
| Sex | |
| Male | 34 (64) |
| Female | 19 (36) |
| Race/ethnicity | |
| White/Caucasian | 26 (49) |
| African American | 6 (12) |
| Hispanic | 15 (28) |
| Asian/Pacific Islander | 5 (9) |
| Others | 1 (2) |
| Pre-operative KPS | |
| 70 or more | 31 (58) |
| < 70 | 22 (42) |
| BMI | |
| < 18.5 | 3 (6) |
| 18.5–24.9 | 12 (23) |
| 25–29.9 | 21 (40) |
| 30–34.9 | 9 (17) |
| 35–39.9 | 4 (7) |
| > 40 | 4 (7) |
| Tumor location* | |
| Frontal | 24 (45) |
| Parietal | 16 (30) |
| Temporal | 25 (47) |
| Occipital | 8 (15) |
| Insular/Paralimbic | 21 (40) |
| Brainstem, thalamus, and basal ganglia | 8 (15) |
| Intraventricular | 4 (7) |
| Butterfly | 4 (7) |
| Multifocal | 4 (7) |
| Diagnosis | |
| Glioblastoma IDH-wild type | 40 (75) |
| Glioblastoma IDH-mutant | 6 (12) |
| Anaplastic astrocytoma IDH-wild type | 1 (2.5) |
| Anaplastic oligodendroglioma IDH-mutant 1p19q co-deleted | 1 (2.5) |
| Diffuse astrocytoma IDH-wild type | 2 (4) |
| Diffuse astrocytoma IDH-mutant | 3 (6) |
| Tumor volume (cc) ** | |
| 16.4 -113.3 | 14 (26) |
| 113.3—178.9 | 13 (24.67) |
| 178.9 -219.3 | 13 (24.67) |
| 219.3–374.4 | 13 (24.67) |
| Radiological extent of resection (1st surgery) | |
| GTR | 19 (36) |
| NTR | 12 (23) |
| STR | 18 (34) |
| Biopsy | 4 (7) |
| Temozolomide | |
| Yes | 39 (74) |
| No | 11 (20) |
| Not available | 3 (6) |
| Radiotherapy | |
| Yes | 39 (74) |
| No | 11 (20) |
| Not available | 3 (6) |
| Other treatments*** | |
| Yes | 10 (19) |
| No | 42 (81) |
| Progression | |
| Yes | 20 (38) |
| No | 33 (62) |
| Survival status | |
| Death | 5 (9) |
| Alive | 48 (91) |
KPS Karnofsky Performance Score, BMI body mass index, IDH isocitrate dehydrogenase, GTR gross-total resection, NTR near-total resection, STR Sub-total resection.
*More than one location was listed if the tumor involved more than one lobe.
**Volumetric analysis included the enhancing tumor, necrosis, and T2-Flair changes.
***Other treatments included: Depatuxizumab Mafodotin n = 5 (Clinical Trial), Bevacizumab n = 3, Novo TTF (Tumor Treating Fields) n = 3, Immunotherapy n = 2, CPT-11 n = 1, Laser Interstitial Thermal Therapy—LITT n = 1.
Figure 3.
IDH-WT and IDH-Mut gliomas display different effects on the human gut microbiome. Comparison of patients' gut microbiome at diagnosis, based on IDH gene status (IDH-WT, n = 39 and IDH-Mut, n = 8) against controls (n = 21). (A,B) Observed OTU and the Shannon alpha diversity index between controls and IDH-WT or IDH-Mut gliomas patients. (C) Beta diversity Weighted Bray–Curtis PCoA of IDH-WT gliomas. (D) Firmicutes to Bacteroides ratio of IDH-WT gliomas. (E) Beta diversity Weighted Bray–Curtis PCoA of IDH-Mut gliomas. (F) Firmicutes to Bacteroides ratio of IDH-Mut gliomas. (G,H) Relative abundance taxa summary at the Phylum level of IDH-WT and IDH-Mut gliomas. (I) Relative abundance of IDH-WT glioma in Akkermansiaceae at the family level (J) Relative abundance of IDH-WT glioma in Akkermansia at the genus level. (K) Relative abundance of IDH-Mut glioma in Akkermansiaceae at the family level. (L) Relative abundance of IDH-WT glioma in Akkermansia at the genus level.
TMZ effect on the microbiome was determined by comparing samples before and after TMZ in 18-patients (14-IDH-WT and 4-IDH-Mut gliomas). Our results showed that Verrucomicrobia, Akkermansiaceae, and Akkermansia abundance trend to decrease following TMZ administration, however, no alpha diversity, F/B ratio, and PCoA changes were observed (Supplementary Fig. S7).
Preliminary progression-free survival (PFS) analysis in Akkermansia positive (12) vs Akkermansia negative (14) GBM patients demonstrated a non-significant difference between groups (8.8 months vs 6.3 months, p = 0.842), respectively.
Discussion
In the present study, we investigated GM changes in mice following both tumor development and TMZ chemotherapy, as well as in a prospective cohort of newly diagnosed glioma patients. Our results demonstrate, significant glioma-induced change in the F/B in young mice following glioma development. Furthermore, glioma development was associated with a significant decrease in Firmicutes and an increase in the Verrucomicrobia phylum of which the predominant genus is Akkermansia20. The GM of aged mice differed at baseline from young mice, including changes in Firmicutes, Bacteroidetes, Actinobacteria, and Tenericutes. Although, Firmicutes and Bacteroidetes changes have been previously demonstrated to occur with age22, little is known about the changes of Actinobacteria and Tenericutes with aging. Despite these differences, similar results were observed in both young and aged mice following tumor development and TMZ treatment.
Our findings in both young and aged were consistent, including a clear clustering in the weighted Bray–Curtis distances of the GM, decreased F/B ratio, and the relative increased of the phylum Verrucomicrobia with a resulting increase in the genus Akkermansia and its more common species Akkermansia muciniphila (AM).
AM a mucin-degrading bacteria has been found to be inversely associated with inflammation, diabetes, obesity, and cardio-metabolic diseases and has been described as the next-generation beneficial probiotic5,15,25. AM converts mucin to short-chain fatty acids, including acetate and propionate, which may mediate its immunoregulatory effects, conferring both regulatory and inflammatory properties via interactions with the host immune system26.
Recent studies have demonstrated that A. muciniphila is increased in stroke, Parkinson’s disease, and untreated MS15–19. These reports suggest that Akkermansia abundance correlates with interferon signaling, T-cell and monocyte gene expression, namely NF-kB, that have been implicated in disease pathogenesis17. Furthermore, Akkermansia abundance has been positively correlated to pro-inflammatory pathways including upregulation of genes involved in antigen-presentation, B and T-cell receptor signaling, activation of complement, and coagulation cascades17,26. These pro-inflammatory features may be related to its ability to degrade mucus, leading to gut barrier breakdown and increased exposure of resident immune cells to microbial antigens26. Similar signaling pathway (IFN-ƴ and IL-2) alterations have been implicated in CNS toxicity, blood–brain barrier (BBB) disruption, and vasogenic brain edema in glioma27.
In our study, the Akkermansia population significantly increased following glioma development both in mice and in IDH-WT glioma patients. The similarity of findings across humans and mice do not come as a surprise as GL261 cells used in this study are IDH-WT28. Considering that IDH-WT and IDH-Mut gliomas are distinct molecular subgroups with IDH-WT being more aggressive and having a worse prognosis29, it was not surprising to observe dysbiosis particularly in the IDH-WT human cohort.
Recent studies demonstrate how the GM composition in cancer patients influences the response to ICI. Importantly, A. muciniphila abundance correlated with clinical response to PD-1/PD-L1 therapy12. Despite ICI success in many solid cancers, little efficacy has been seen in GBM30,31. Studies evaluating if certain GM taxa, including Akkermansia, may enhance specific therapies previously failed in GBM are warranted. Moreover, the potential of therapeutic modulation of the microbiome by probiotics/ fecal transplant to expand therapeutic options for glioma deserves further study.
TMZ, an oral agent with a unique pharmacological profile, is considered the standard of care for adjuvant therapy, in combination with radiotherapy24,32. Prior studies have evaluated the effects of chemotherapeutic agents on the GM33. Cyclophosphamide has been found to cause disruption of the intestinal barrier, translocation of commensal bacteria, and small intestine microbiome alterations34. Despite its well-established safety and ease of administration, the TMZ effects on the GM remain unknown.
Our mice results demonstrate that TMZ alters the GM with clear separation in weighted Bray–Curtis distances and a significant decrease in the F/B following oral TMZ. In addition, levels of , Muribaculaceae and Ruminococcaceae increased and decreased respectively. The later, has been associated with maintenance of gut health35. A higher relative abundance of Ruminococcaceae has been found in GM of melanoma responders versus non-responders to PD-1 immunotherapy. Furthermore, higher effector CD4+ and CD8+ T-cells frequency and a preserved cytokine response coincided with higher Ruminococcaceae levels in anti-PD-1 responders, while those who were non-responders displayed higher Bacteroidales levels13.
Muribaculaceae abundance has been correlated with increased short-chain fatty acids (SCFAs) levels36, which affect microglial density, morphology, and maturity5,36. Additional studies of SCFA producing bacteria, fecal metabolites, neurotransmitters and their interactions with the immune system may allow for potential microbiome manipulations in glioma patients.
Verrucomicrobia and Akkermansia increased abundance following tumor implantation in mice were not observed following TMZ treatment. Interestingly, TMZ by itself (Sham/TMZ) did not alter Verrucomicrobia or Akkermansia levels in mice. We hypothesize that the observed Verrucomicrobia/Akkermansia reduction in the tumor/TMZ group is the result of tumor control. Similar to our findings in mice, Akkermansia increased abundance in glioma patients demonstrated a decreasing trend following TMZ treatment (Supplementary Fig. S7). Understanding the interactions between glioma and the GM is vital as a particular microbiome composition may enhance the effects of existing therapies (chemotherapies, targeted therapies, immunotherapies), which have not yet been effective in glioma30,31,37.
Despite our robust findings, there are several limitations to our study. First, we did not find a relationship between the GM, clinical factors, and survival in mice or humans, which could be secondary to the limited number of patients and short follow-up in this preliminary study. Future studies with larger cohorts and longer follow-up are needed to investigate possible clinical associations. Second, TMZ effects in humans may be influenced by cohort-specific cofounders, small sample size, and radiotherapy treatment32,38. Third, glioma patients received dexamethasone prior to surgery, which could potentially affect the GM39. Lastly, while our microbiome analysis using 16S rRNA is useful for providing taxonomic information, shotgun metagenomics, metatranscriptomic and metabolomic profiling of FS may reveal changes that would complement phylogenic and taxonomic data. Additionally, further experiments are needed to evaluate the effect of other intracranial tumors on the GM to assess if the findings in our study are specific to glioma development. In addition, the short follow-up of this preliminary study might preclude us to identify specific microbial taxa that correlated with survival. Even though our experiments fell short of establishing a relationship between the microbiome and clinical parameters, there have been numerous studies that have shown a correlation between Akkermansia and pro-inflammatory genes in macrophages and T-cells with downstream effects in IFN-ƴ and IL-2; which in turn cause BBB disruption and cerebral edema in glioma patients. This may be a plausible explanation for the increased level of Akkermansia in our mouse-model and in IDH-WT glioma patients.
Conclusions
This study demonstrates for the first time the relationship between the gut-brain axis, glioma development, and TMZ in both mice and humans. We demonstrated that Verrucomicrobia phylum and Akkermansia genus abundance increase after glioma growth in a mouse-tumor model, which was not observed in the setting of TMZ treatment. These findings appear to be translational, as humans with IDH-WT gliomas also show similar microbiome changes when compared to controls. Further studies to evaluate the relationship between the gut-brain axis and glioma are needed to address the role that the GM, especially the genus Akkermansia plays in this disease.
Methods
Animals
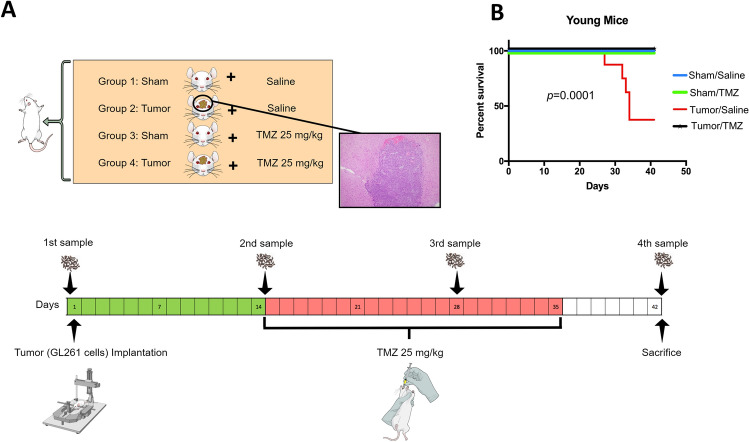
Six- to eight-week-old male C57BL/6 mice from the Jackson Laboratory (Bar Harbor, ME, USA) and 20-month-old male C57BL/6 mice from the National Institute on Aging were used. Thirty young mice were assigned to four groups: G1 Sham/Saline (n = 7), G2 Tumor/Saline (n = 8), G3 Sham/TMZ (n = 7), and G4 Tumor/TMZ (n = 8). Tumor implantation was performed on day 0. On day 1440–42, mice were orally gavaged with TMZ (25 mg/kg) or saline daily, 5 days per week for 3 weeks. FS were collected and stored in sterile tubes at -80 °C prior to tumor implantation (1st sample), before and after the initiation of TMZ/Saline treatment (2nd and 3rd sample), and at sacrifice, on day 42 or at mice death if this was prior to sacrifice (4th sample) (Fig. 4)43. Brains implanted with GL261 cells were examined to confirm tumor growth44. Gut permeability assay was performed as previously described45. All experimental work was approved by the Laboratory Animal Medicine and Care at the University of Texas Health Science Center at Houston institutional review board (IRB) in compliance with the Guide for the Care and Use of Laboratory Animals.
Figure 4.
Experimental design and stool sample collection. (A) Twenty-eight male C57BL/6 mice were randomly divided into four groups: Sham/ Saline (N = 7), Tumor/ Saline (N = 6), Sham/ TMZ (N = 7), and Tumor/ TMZ (N = 8). The tumor was implanted using GL261 mouse cells. Two weeks post tumor implantation, mice were treated orally by either saline or TMZ 25 mg/kg for three weeks, five days per week. Stool samples were collected periodically as depicted in the timeline. (B) Kaplan–Meier survival curve using the Log-rank test of the 4 groups, in which mortality was significantly higher in the tumor/saline group (p < 0.001) compared to the other three groups.
Twelve aged mice were assigned to two groups: G1Tumor/Saline (n = 6) and G2 Tumor/TMZ (n = 6). However, both group's numbers were reduced due to mortality after tumor implantation (G1 n = 3, G2 n = 4). Tumor implantation, drug dosing, and stool collection followed the same protocol as young mice.
Microbial DNA extraction and the 16S rRNA gene sequencing
16S rRNA gene sequencing methods were adapted from the methods developed for the Earth Microbiome Project and NIH-Human Microbiome Project46–49. Subsequent analysis steps were performed at the Alkek Center for Metagenomics and Microbiome Research (CMMR) at Baylor College of Medicine. 16Sv4rRNA sequences were clustered into Operational Taxonomic Units (OTUs) at a similarity cutoff value of 97% using the UPARSE algorithm50 and mapped to an optimized version of the SILVA Database51 containing only sequences from the v4 region of the 16S rRNA gene to determine taxonomies. A custom script constructs an OTU table from the output files for downstream analyses using a visualization toolkit also developed at the CMMR named ATIMA (Agile Toolkit for Incisive Microbial Analyses).
Patients’ characteristics and fecal sample collection
The prospective study was conducted from January 2018-July 2019. Patients with newly diagnosed glioma who provided a pre-surgical FS were included. Patients with recent antibiotic exposure (30-days), < 18-years, other cancers, and gastrointestinal diseases were excluded. Patients’ demographics, clinical, histological, and follow-up information, were obtained from the electronic medical record and stored in a REDCap database, which was later exported for analysis (Table 1). Patients underwent biopsy or maximum safe tumor resection at the discretion of the treating neurosurgeon followed by chemoradiotherapy24. IDH1 p.R13H was assessed by immunohistochemistry. FS were collected from patients at three-time points: before resection (Pre-Sx) / prior to cephalosporin per-protocol; before and after 6-weeks of chemoradiation (Pre-Tx and Post-Tx). To minimize confounding factors known to impinge on the human microbiome composition32,38, twenty-one FS were utilized as controls. FS for DNA extraction and microbiome sequencing were collected using microbial collection and stabilization kits (OMNIgene Gut; DNA Genotek). This prospective human study was approved by our institution IRB and it was in accordance with the 1964 Helsinki Declaration and its later amendments.
Statistical analysis
The ATIMA software was used to perform the downstream analyses. The significance of categorical variables was determined using the Mann–Whitney/Kruskal–Wallis test. All p-values were two-sides and adjusted for multiple comparisons with the FDR-algorithm.
The F/B was calculated by Mann–Whitney/paired t-test. This was evaluated as F/B represents the two most common phyla in the GM. Additionally, F/B ratio has been known to affect outcome in neurological diseases including stroke22. PFS of patients with 6-months or longer follow-up was plotted using the Kaplan–Meier method in GraphPad Prism (version 8.2.1 for Mac, GraphPad, CA, USA). Analysis in the aged-mice was performed using paired t-test, prior confirmation of normal distribution. Mice were randomly assigned to treatment groups. All analyses were performed by a blinded investigator. For a complete methodology description, see Supplementary Methodology.
Ethical approval
All procedures performed in this prospective human study were approved by the institutional review board of The University of Texas Health Science Center at Houston and Memorial Hermann Hospital, Houston, TX and it was in accordance with the 1964 Helsinki Declaration and its later amendments.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Supplementary information
Acknowledgments
We would like to acknowledge our patients and family members for participating in the study. We also greatly appreciate the help of Memorial Hermann Hospital nurses.
Author contributions
A.P., S.Z., Y.O., J.S. Y.E, performed experiments. A.P., S.Z., B.K., L.D.M., L.Y.B., Y.E., designed experiments. A.P., A.D., S.Z., G.H., T.T., N.H., O.A., Y.E., gather human samples and information. A.P., A.D., S.Z., G.H., T.T., YE analyzed and recollection of data. A.P., A.D., S.Z., Y.E., wrote the paper. A.P., A.D., S.Z., H.A.C., J.S., N.T., B.P.G., L.D.M., L.Y.B., Y.E., participated in editing and provided conceptual advice. All authors approved the final manuscript.
Funding
This work was supported by the Center for Clinical and Translational Sciences (YE), which is funded by National Institutes of Health Clinical and Translational Award UL1 TR003167 from the National Center for Advancing Translational Sciences. Also, the research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number K08CA241651 (LYB). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Anthony Patrizz, Antonio Dono and Soheil Zorofchian.
Contributor Information
Leomar Y. Ballester, Email: Leomar.Y.Ballester@uth.tmc.edu
Yoshua Esquenazi, Email: Yoshua.EsquenaziLevy@uth.tmc.edu.
Supplementary information
is available for this paper at 10.1038/s41598-020-77919-w.
References
- 1.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 2.Esquenazi Y, et al. The survival advantage of “supratotal” resection of glioblastoma using selective cortical mapping and the subpial technique. Neurosurgery. 2017;81:275–288. doi: 10.1093/neuros/nyw174. [DOI] [PubMed] [Google Scholar]
- 3.Ostrom QT, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro. Oncol. 2018;20:1–86. doi: 10.1093/neuonc/noy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delgado-López PD, Corrales-García EM. Survival in glioblastoma: a review on the impact of treatment modalities. Clin. Transl. Oncol. 2016;18:1062–1071. doi: 10.1007/s12094-016-1497-x. [DOI] [PubMed] [Google Scholar]
- 5.Cryan JF, et al. The microbiota-gut-brain axis. Physiol. Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 6.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, G. J. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 7.Velagapudi VR, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res. 2010;51:1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaser MJ. The microbiome revolution. J. Clin. Invest. 2014;124:4162–4165. doi: 10.1172/JCI78366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selgrad M, et al. The role of viral and bacterial pathogens in gastrointestinal cancer. J. Cell. Physiol. 2008;216:378–388. doi: 10.1002/jcp.21427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat. Med. 2019;25:377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 12.Routy B, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 13.Gopalakrishnan V, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajagopala SV, et al. The human microbiome and cancer. Cancer Prev. Res. 2017;10:226–234. doi: 10.1158/1940-6207.CAPR-16-0249. [DOI] [PubMed] [Google Scholar]
- 15.Stanley D, Moore RJ, Wong CHY. An insight into intestinal mucosal microbiota disruption after stroke. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li N, et al. Change of intestinal microbiota in cerebral ischemic stroke patients. BMC Microbiol. 2019;19:1–8. doi: 10.1186/s12866-018-1372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jangi S, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016;7:1–11. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unger MM, et al. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Park. Relat. Disord. 2016;32:66–72. doi: 10.1016/j.parkreldis.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 19.Blacher E, et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019;572:474–480. doi: 10.1038/s41586-019-1443-5. [DOI] [PubMed] [Google Scholar]
- 20.Geerlings S, Kostopoulos I, de Vos W, Belzer C. Akkermansia muciniphila in the human gastrointestinal tract: when, where, and how? Microorganisms. 2018;6:75. doi: 10.3390/microorganisms6030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu P, Du XL, Zhu J-J, Esquenazi Y. Improved survival of glioblastoma patients treated at academic and high-volume facilities: a hospital-based study from the National Cancer Database. J. Neurosurg. 2019 doi: 10.3171/2018.10.JNS182247. [DOI] [PubMed] [Google Scholar]
- 22.Spychala MS, et al. Age-related changes in the gut microbiota influence systemic inflammation and stroke outcome. Ann. Neurol. 2018;84:23–36. doi: 10.1002/ana.25250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louis DN, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 24.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 25.Cani PD, de Vos WM. Next-generation beneficial microbes: The case of Akkermansia muciniphila. Front. Microbiol. 2017;8:1–8. doi: 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derrien M, et al. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Microbiol. 2011;2:1–14. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tjuvajev J, et al. RG-2 glioma growth attenuation and severe brain edema caused by local production of interleukin-2 and interferon-γ. Cancer Res. 1995;55:1902–1910. [PubMed] [Google Scholar]
- 28.Bunse L, et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 2018;24:1192–1203. doi: 10.1038/s41591-018-0095-6. [DOI] [PubMed] [Google Scholar]
- 29.Yan H, et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fecci PE, Sampson JH. The current state of immunotherapy for gliomas: an eye toward the future. J. Neurosurg. 2019;131:657–666. doi: 10.3171/2019.5.JNS181762. [DOI] [PubMed] [Google Scholar]
- 31.Caccese M, Indraccolo S, Zagonel V, Lombardi G. PD-1/PD-L1 immune-checkpoint inhibitors in glioblastoma: a concise review. Crit. Rev. Oncol. Hematol. 2019;135:128–134. doi: 10.1016/j.critrevonc.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez JM, et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015;26:1–17. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panebianco C, Andriulli A, Pazienza V. Pharmacomicrobiomics: exploiting the drug-microbiota interactions in anticancer therapies. Microbiome. 2018;6:92. doi: 10.1186/s40168-018-0483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;32:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the genetic basis of fibrolytic specialization by lachnospiraceae and ruminococcaceae in diverse gut communities. Diversity. 2013;5:627–640. doi: 10.3390/d5030627. [DOI] [Google Scholar]
- 36.Erny D, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lassman AB, et al. Safety and efficacy of depatuxizumab mafodotin + temozolomide in patients with EGFR -amplified, recurrent glioblastoma: results from an international phase I multicenter trial. Neuro. Oncol. 2019;21:106–114. doi: 10.1093/neuonc/noy091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 39.Zhao H, Jiang X, Chu W. Shifts in the gut microbiota of mice in response to dexamethasone administration. Int. Microbiol. 2020 doi: 10.1007/s10123-020-00129-x. [DOI] [PubMed] [Google Scholar]
- 40.Liaw K, et al. Dendrimer size effects on the selective brain tumor targeting in orthotopic tumor models upon systemic administration. Bioeng. Transl. Med. 2020;5:e10160. doi: 10.1002/btm2.10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh T, et al. Immunocompetent murine models for the study of glioblastoma immunotherapy. J. Transl. Med. 2014;12:1–10. doi: 10.1186/1479-5876-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zagzag D, et al. Vascular apoptosis and involution in gliomas precede neovascularization: a novel concept for glioma growth and angiogenesis. Lab. Invest. 2000;80:837–849. doi: 10.1038/labinvest.3780088. [DOI] [PubMed] [Google Scholar]
- 43.Lee J, et al. Young versus aged microbiota transplants to germ-free mice: increased short-chain fatty acids and improved cognitive performance. Gut Microbes. 2020;35:1–14. doi: 10.1080/19490976.2020.1814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoo JY, et al. Oncolytic HSV therapy increases trametinib access to brain tumors and sensitizes them in vivo. Neuro. Oncol. 2019;21:1131–1140. doi: 10.1093/neuonc/noz079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crapser J, et al. Ischemic stroke induces gut permeability and enhances bacterial translocation leading to sepsis in aged mice. Aging. 2016;8:1049–1063. doi: 10.18632/aging.100952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huttenhower C, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Methé BA, et al. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 50.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 51.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.