Abstract
Extracellular vesicles (EVs) containing specific cargo molecules from the cell of origin are naturally secreted from bacteria. EVs play significant roles in protecting the bacterium, which can contribute to their survival in the presence of antibiotics. Herein, we isolated EVs from methicillin-resistant Staphylococcus aureus (MRSA) in an environment with or without stressor by adding ampicillin at a lower concentration than the minimum inhibitory concentration (MIC). We investigated whether EVs from MRSA under stress condition or normal condition could defend susceptible bacteria in the presence of several β-lactam antibiotics, and directly degrade the antibiotics. A comparative proteomic approach was carried out in both types of EVs to investigate β-lactam resistant determinants. The secretion of EVs from MRSA under antibiotic stressed conditions was increased by 22.4-fold compared with that of EVs without stress. Proteins related to the degradation of β-lactam antibiotics were abundant in EVs released from the stressed condition. Taken together, the present data reveal that EVs from MRSA play a crucial role in the survival of β-lactam susceptible bacteria by acting as the first line of defense against β-lactam antibiotics, and antibiotic stress leads to release EVs with high defense activity.
Subject terms: Antibiotics, Antimicrobial resistance
Introduction
Although the discovery of antibiotics has prolonged human life and helped develop a variety of health-related technologies, the overuse of antibiotics has led to the emergence of "superbugs" as bacteria have evolved the means of resisting traditional antibiotics. The increase in superbug strains is believed to be a serious threat facing public health around the world. Damage caused by antimicrobial resistance is expected to reach a total gross domestic product (GDP) loss of $ 100 trillion worldwide by 2050, at which point it could kill 10 million people annually1. To combat the spread of superbugs globally, it is important to understand all possible bacterial protection mechanisms against antibiotics.
The multidrug-resistant (MDR) pathogenic bacterium, as the main cause of recurrent opportunistic infections, is methicillin-resistant Staphylococcus aureus (MRSA). Since MRSA was reported in 19612, the incidence of this strain has spread worldwide. MRSA is a major Gram-positive bacterial pathogen that causes a range of illnesses including pneumonia, meningitis, osteomyelitis, endocarditis, and septicemia, which leads to high mortality and is expensive to treat3. Numerous cases of infections due to MRSA have been reported in companion, diverse domesticated, and livestock animals4,5. The transmission between humans and animals indicated that both directions of humanosis and zoonosis are possible5,6. Therefore, emerging MRSA infections are no longer primarily a human healthcare-related threat, but a global community-associated problem.
Bacteria secret proteins, polysaccharides and diverse molecules to their extracellular milieu to communicate and to coordinate population behaviors7,8. Among them, extracellular vesicles (EVs) are known to possess diverse cellular factors and have a variety of functions to aid bacteria survival7–11. EVs are defined as spherical, and bilayered proteolipids which form lumen-containing spheres with an average diameter of 20–200 nm, and composed of proteins from various cellular origins, unique lipids, enzymes, toxins and nucleic acids7,8,12,13. Packed within vesicles, the cargo molecules can be transported over long distances away from dilution and degradation, which might explain the effective interaction among bacteria14. These vesicles have been elucidated to play diverse roles7–9,12,15, thus EVs are considered the powerful intercellular and interspecies ‘communicasomes’ in the microbial ecosystem.
Although several studies suggested that EVs can defend the bacteria against the effects of several antibiotics by acting as decoys9,16, either through horizontal transfer of antibiotic resistance genes15,17, or degradation/sequestration of antibiotics7,9,18,19, the mechanism of EVs against antibiotics remain uncharacterized. Previous studies have reported that exposure to some physiological or environmental stressors such as antibiotic treatment, oxidative stress, and temperature influenced the level of vesicles secretion, and stressors could cause alterations in the composition of vesicles11,20,21. Based on these studies, we hypothesize that a physiological stressor such as sub-lethal antibiotics can trigger bacteria secreting EVs with significant changes in the proteome to adapt and survive in the antibiotic stressed environment.
In the present study, we compared the protein constituents of EVs from MRSA cultured under normal conditions (EVNor) and cultured in the presence of sub-lethal concentrations of ampicillin (EVStrs). The results implicated that EVStrs, which have more proteins that can degrade antibiotics than EVNor, can offer protection to the β-lactam-susceptible S. aureus against lethal β-lactam antibiotic concentrations better than EVNor.
Results
Physical characterization of EVs
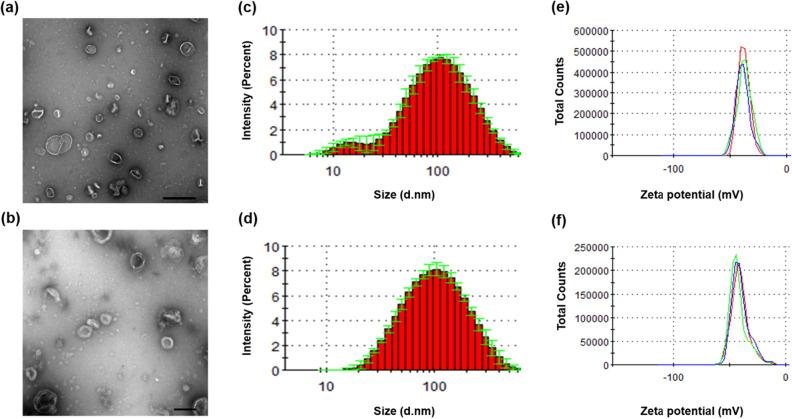
EVs of MRSA ST692 cells were isolated and designated as EVNor and EVStrs with respect to the culture conditions. Transmission electron microscopy (TEM) analysis showed bi-layered spherical EVs (Fig. 1a,b). Dynamic light scattering (DLS) revealed the average diameter of EVStrs (78.22 ± 0.81 nm) and EVNor (86.84 ± 0.25 nm) (Fig. 1c,d, and see Supplementary Table S1 online). EVStrs has more vesicles of 10–20 nm size than EVNor, but the size distribution except for 10–20 nm is almost identical with EVNor. Their polydispersity index (PDI) were measured below 0.3, indicating that the arrangements were monodispersed (Supplementary Table S1). Their zeta potentials were more negative than -30 mV, implying that there were no considerable differences in the cohesion of the vesicles (Fig. 1e,f, and Supplementary Table S1).
Figure 1.
Physical characterizations of EVs derived from stressed and normal ST692 cells. TEM image of EVs derived from stressed ST692 (a) (scale bar: 500 nm) and normal ST692 (b) (scale bar: 100 nm) cells. The size distribution EVs released from stressed ST692 (c) and normal ST692 (d) cells. Three independent analyses were performed; means are shown with ± standard deviation (error bars) of the percentage intensity. The zeta potential of EVs from stressed ST692 (e) and normal ST692 (f) cells.
Vesiculation enhances upon physiological stress
To understand further how ampicillin stress affects EVs production, ST692 was exposed to incremental concentrations of ampicillin and each of the respective EVs was purified (Fig. 2). The quantity of secreted EVs increased dose-dependently according to the amount of ampicillin added to the culture of ST692 cells. In particular, the production of EVStrs (64 μg/mL of ampicillin treated) increased by 22.4-fold compared to EVNor, the untreated control. Even when only 1 μg/mL of ampicillin was added, the yield was 2.5-fold higher than EVNor.
Figure 2.
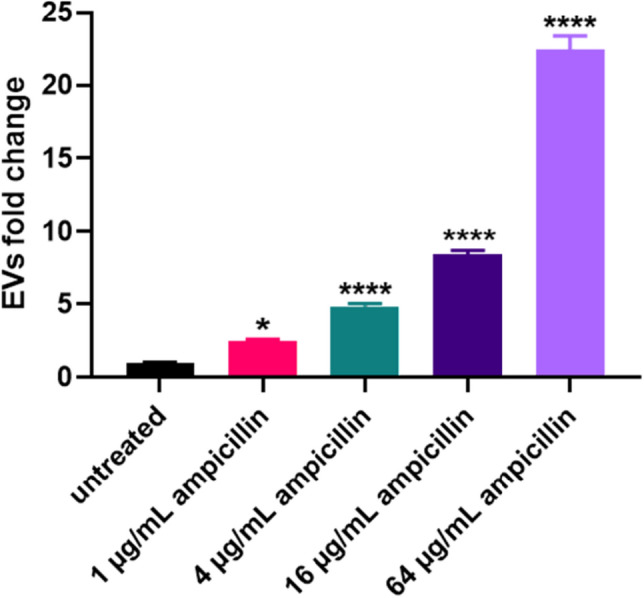
Induction of EVs production by treatments with stressors. (a) EVs were purified and quantified from cultures of ST692 cells treated with 64, 16, 4, 1, or 0 μg/mL ampicillin. EVs yields were averaged and normalized to untreated controls to adjust fold change. One way ANOVA was used for analyses and data were presented as mean ± standard deviations (SD). *P < 0.05, ****P < 0.0001.
EVs defend β-lactam susceptible S. aureus cells against β-lactam antibiotics
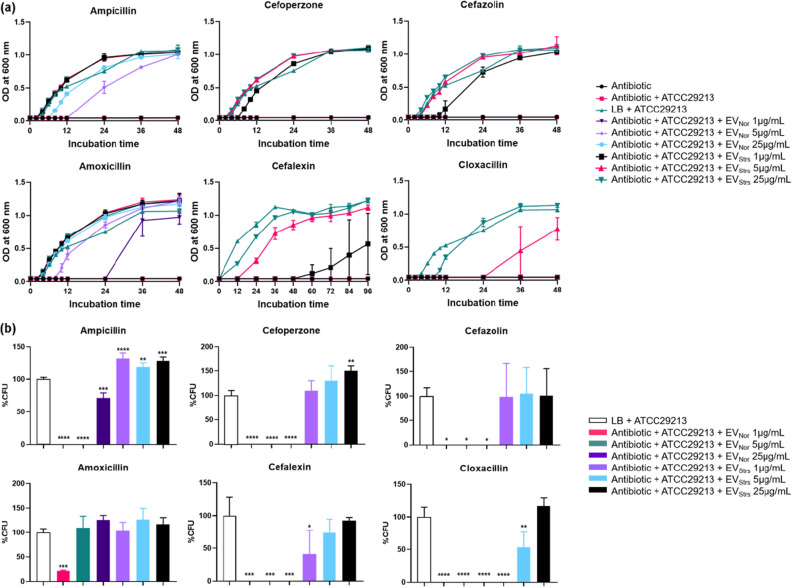
Minimum inhibitory concentrations (MICs) of the MRSA strain ST692 and the susceptible bacterium S. aureus ATCC29213 were measured to determine whether the different concentrations of EVStrs and EVNor from ST692 could protect S. aureus ATCC29213 against several antibiotics (Table 1). The growth kinetics presented in Fig. 3a showed that EVs can protect susceptible bacteria against each antibiotic at a higher concentration than the MIC of S. aureus ATCC29213. For the six β-lactam antibiotics, EVStrs dose-dependently protected S. aureus ATCC29213, allowing it to tolerate antibiotic exposures above the MICs. EVNor also dose-dependently defended ATCC29213 against ampicillin, and amoxicillin, but did not protect the susceptible bacteria from the other antibiotics tested over the incubation time. Since the equivalent amount of EVStrs protects susceptible bacteria from antibiotics more strongly than that of EVNor, the susceptible strains grew much faster in the EVStrs group. One microgram per milliliter of EVStrs appears to be more protective than 25 μg/mL of EVNor. However, neither EVStrs nor EVNor protected the susceptible bacteria against antibiotics other than the six mentioned above (cefotaxime, imipenem, methicillin, chloramphenicol, gentamicin, kanamycin, streptomycin, and tetracycline) (data not shown).
Table 1.
The MIC of several antibiotics against the β-lactam-susceptible Staphylococcus aureus ATCC29213 and β-lactam-resistant S. aureus ST692.
| Class | Antibiotics | MIC (μg/mL) | |
|---|---|---|---|
| ATCC29213 | ST692 | ||
| β-Lactams | Ampicillin | 2 | 256 |
| Amoxicillin | 4 | 256 | |
| Cefalexin | 1 | 32 | |
| Cefazolin | 0.25 | 32 | |
| Cefoperazone | 4 | 64 | |
| Cefotaxime | 1 | 32 | |
| Cloxacillin | 0.25 | < 0.5 | |
| Imipenem | 8 | > 1024 | |
| Methicillin | 1 | 32 | |
| Chloramphenicol | Chloramphenicol | 8 | 16 |
| Aminoglycosides | Gentamicin | 4 | 4 |
| Kanamycin | 16 | 32 | |
| Streptomycin | 32 | > 1024 | |
| Tetracyclines | Tetracycline | 0.5 | 64 |
MIC minimum inhibitory concentration.
Figure 3.
EVs from methicillin-resistant Staphylococcus aureus (MRSA) can defend β-lactam-susceptible S. aureus and fully protect them from β-lactam antibiotic-induced growth inhibition. (a) Representative growth profiles of β-lactam-susceptible S. aureus cells in the presence of growth-inhibiting concentrations of β-lactam antibiotics. The growth-inhibiting concentrations of antibiotics were: ampicillin, 40 μg/mL; cefoperazone, 8 μg/mL; cefazolin, 1.25 μg/mL; amoxicillin, 40 μg/mL; cefalexin, 4 μg/mL; and cloxacillin, 1.25 μg/mL. The data were presented as means and SEMs of at least three independent experiments. (b) The survival percentages of β-lactam-susceptible S. aureus cells in the presence of the above-listed growth-inhibiting concentrations of antibiotics and EVs from stress condition or normal condition were calculated by bacterial counts of cultures at a certain time points (ampicillin, 12 h; cefoperazone, 12 h; cefazolin, 24 h; amoxicillin, 36 h; cefalexin, 96 h; and cloxacillin, 48 h). CFU of S. aureus cells in medium without any antibiotics were used as a positive control and taken as 100%, and the corresponding CFU of samples was computed. The data were presented as means and SEMs of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Bacterial protection percentage of EVStrs and EVNor from antibiotics
To determine the degree of bacterial protection of each EV against antibiotics, we performed quantitative plate assays based on growth kinetics (Fig. 3b). Notably, the viability of the susceptible cells inoculated with EVStrs at 25 μg/mL in the presence of respective antibiotics showed no loss in viability compared to the culture of susceptible cells without antibiotics. In contrast, the viability of the susceptible cells with EVNor at 25 μg/mL in the presence of cefoperazone, cefazolin, cefalexin, and cloxacillin showed no viability, which can be explained by the rapid killing by the respective antibiotic concentrations.
Specific molecules of EVs are important for the protection of bacteria
After the growth curve experiment, all samples were plated on TSA agar with or without respective antibiotics in the same concentration as was used in the growth curve experiment (see Supplementary Fig. S1 online). All samples that grew in the above experiment were grown in TSA agar but not in TSA agar with respective antibiotics. These results suggested that the enhanced survival rates were not due to a mutation of the β-lactam resistant genetic materials of EVs transferred to susceptible S. aureus, but rather from the molecules owned by EVs that protected the bacteria from the β-lactam antibiotic environment. In addition, colonies grown in TSA agar were identified as S. aureus at the species level using MALDI-TOF MS (data not shown).
EVs protect different genera of Gram-negative bacteria against ampicillin
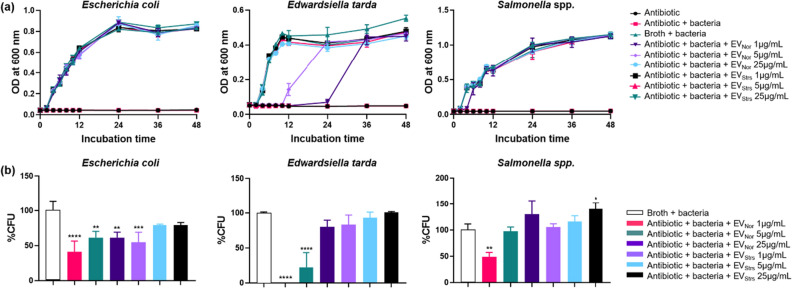
To determine whether the EVs isolated from the Gram-positive MRSA can protect Gram-negative bacteria, which belong to different genera, growth curve profiles in the presence of growth-inhibiting concentrations of ampicillin and qualitative plate assays were carried out. The MICs of ampicillin against Escherichia coli (RC85), Edwardsiella tarda (ED45), and Salmonella spp. (Sal26B) were 8 μg/mL, 4 μg/mL, and 8 μg/mL, respectively9. Both EVStrs and EVNor defended RC85, ED45, and Sal26B against ampicillin (Fig. 4a). ED45 cells treated with 5 μg/mL EVNor exhibited growth at 12 h, whereas both RC85 and Sal26B cells supplemented with 5 μg/mL exhibited growth at 2 h. When EVNor 1 μg/mL was added to each strain, RC85 exhibited growth at 2 h, ED45 at 24 h, and Sal26B at 4 h. Since the MIC of ED45 was lower than that of RC85 or Sal26B, the protective effect caused by EVs molecules appears to be slower. Quantitative plate assays were investigated by counting the CFU based on growth kinetics (Fig. 4b). Both EVStrs and EVNor dose-dependently protected each susceptible strains against the bactericidal effect of ampicillin.
Figure 4.
EVs from methicillin-resistant Staphylococcus aureus (MRSA) can protect other bacterial species from ampicillin-induced growth inhibition. (a) Representative growth profiles of Escherichia coli (RC85), Edwardsiella tarda (ED45), and Salmonella spp. (Sal26B) cells in the presence of a growth-inhibiting concentration of ampicillin (30 μg/mL) plus increasing amounts of EVs from stress conditions or normal conditions. The data were presented as means and SEMs of at least three independent experiments. (b) The survival percentages of RC85, ED45, and Sal26B cells were calculated by counting CFUs at specific time points (12 h) from cultures grown with 30 μg/mL ampicillin and increasing quantities of EVs from stress condition or normal condition. CFU of respective bacterial cells in medium without ampicillin was used as a positive control and taken as 100%, and the corresponding CFU of samples was calculated. The data were presented as means and SEMs of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
EVs enable degradation of β-lactam antibiotics
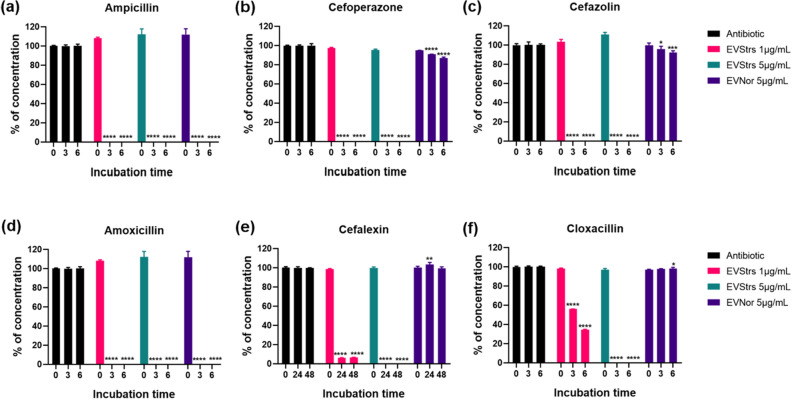
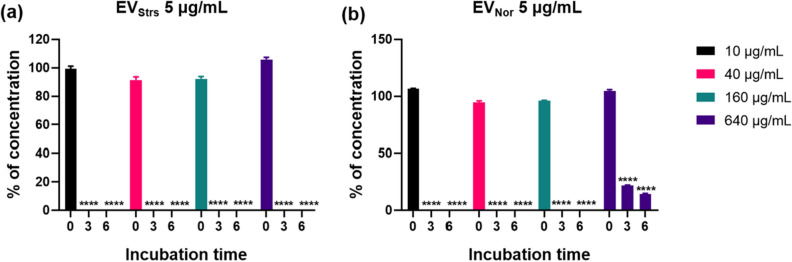
To explain the reason why EVs were capable of protecting bacteria in the β-lactam antibiotic environment, LC-ESI-QQQ analysis was carried out to measure concentrations of antibiotics after treatment of EVs in the presence of antibiotics in a cell-free system (Fig. 5). The concentration of six antibiotics was dramatically decreased in samples treated with 5 μg/mL of EVStrs for 3 h (five antibiotics except for cefalexin) or 24 h (cefalexin) compared with respective antibiotics without EVs. Samples treated with 1 μg/mL of EVStrs could hydrolyze six antibiotics, but the capacity appears to be lower than that of 5 μg/mL. Moreover, 5 μg/mL of EVNor also completely decomposed ampicillin and amoxicillin in 3 h, but the activity against cefoperazone and cefazolin was very weak, and against cefalexin and cloxacillin was not detected. The extent of the capacity of EVStrs and EVNor to hydrolyze ampicillin for 6 h was compared by increasing the dose of ampicillin after EVs concentrations were fixed to a certain amount (Fig. 6). EVStrs degraded 640 μg/mL of ampicillin in 3 h, but EVNor could not degrade the equivalent concentration of antibiotics for 6 h.
Figure 5.
LC-QQQ-based evaluation of the concentration of β-lactam antibiotics following incubation with different doses of EVs from stress conditions or normal conditions in a cell-free system. The original concentrations were as follows: ampicillin, 40 μg/mL (a); cefoperazone, 8 μg/mL (b); cefazolin, 1.25 μg/mL (c); amoxicillin, 40 μg/mL (d); cefalexin, 4 μg/mL (e); cloxacillin, 1.25 μg/mL (f). One microgram per milliliter or 5 μg/mL of EVs from stress condition or 5 μg/mL of EVs from a normal conditions in sterilized PBS were mixed with respective antibiotics. Each of the antibiotic without EVs was averaged and normalized as 100%, and the corresponding concentrations of antibiotics in samples treated with EVs were analyzed. The concentrations of antibiotics were registered at 3-h intervals for 6 h (a–d,f) or 24-h intervals for 48 h (e) in triplicate. Bars indicate standard deviations. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Figure 6.
Consumptive ampicillin titration of EVs from stress condition and normal condition using LC-QQQ-based assessment in a cell-free system. The concentration of each EVs was fixed at 5 μg/mL, and the concentration of ampicillin was changed to 10, 40, 160, or 640 μg/mL. Ampicillin concentration was determined at an interval of 3 h up to 6 h. Ampicillin without EVs was used as a positive control and taken as 100%, and the corresponding concentrations of ampicillin in samples treated with EVStrs and EVNor were analyzed. ****P < 0.0001.
Proteomic characterization of EVStrs and EVNor
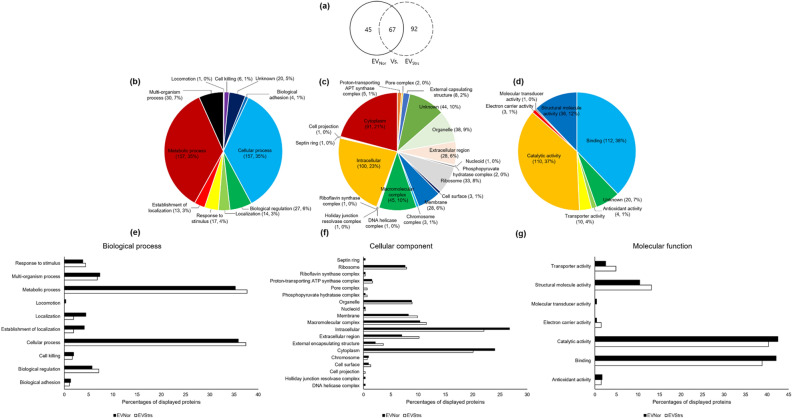
We compared the protein constituents of EVStrs and EVNor using LC–MS/MS analysis because delineation of the biological role of the protein components of EVs is important in understanding their relevance to antibiotic resistance. A total of 204 proteins were identified using the uni_bacteria database (Fig. 7a, See Dataset 1 online). EVStrs alone has 159 proteins, of which 67 proteins overlapped with EVNor. All proteins possessed by EVStrs and EVNor were classified according to these categories: relevant biological processes, cellular components, and molecular functions (Fig. 7b–d). Each of the proteins of EVStrs and EVNor was also classified with the same categories (Fig. 7e–g). The introduction of the LC–MS/MS data into the Staphylococcus aureus USA300 strain database revealed that there are several different proteins identified which are not included in the uni_bacteria database. So we re-examined the proteins under the categories, biological processes, cellular components, and molecular functions of both EVStrs and EVNor, separately and combined (see Supplementary Fig. S2 and Dataset 2 online). The results obtained here implied that the majority of the protein composition of EVs was altered and some of the proteins are more abundant which might be the result of the reaction of the host bacteria to the exposure of sub-MICs of ampicillin.
Figure 7.
Venn diagrams show the entire proteins identified from EVNor and EVStrs searched against the uni_bacteria database while the bar graphs classify the proteins which showed differential expression in EVNor and EVStrs. (a) A total of 204 proteins were established in EVNor (112 Proteins) and EVStrs (159 Proteins) together. Entire proteins were classified depending on the related biological process (b), cellular component (c), and molecular function (d). The percentages of displayed EVNor (black) and EVStrs (white) proteins were compared with respect to the consistent biological process (e), cellular component (f), and molecular function (g).
Comparative analysis of EVs protein concerned with resistance to a β-lactam antibiotic
Among the EVs proteins analyzed from the two databases (Dataset 1 and Dataset 2), we examined the candidate proteins related to the consumption of β-lactam antibiotics (Table 2). The β-lactamase proteins encoded by blaZ and SA1529 and native PBPs such as PBP1, -2, -3 were upregulated when compared to the protein compositions of EVStrs versus EVNor.
Table 2.
Quantitative candidate protein profiling involved in the consumption of β-lactam antibiotics by EVs.
| Identified proteins | Accession number | Alternate ID | Quantitative value (normalized total spectra) | |
|---|---|---|---|---|
| EVNor | EVStrs | |||
| Beta-lactamase | BLAC_STAAU | blaZ | 111.86 | 607.48 |
| UPF0173 metal-dependent hydrolase | Y1529_STAAN (+ 12) | SA1529 | 0 | 4.4352 |
| Penicillin-binding protein 1 | A0A0H2XJZ5_STAA3 | pbpA | 0 | 25.135 |
| Penicillin binding protein 2 | A0A0H2XI32_STAA3 | pbp2 | 16.733 | 119.2 |
| Penicillin-binding protein 3 | A0A0H2XJ39_STAA3 | pbp3 | 12.932 | 122.17 |
β-lactamase content in EVs was regulated by ampicillin stress
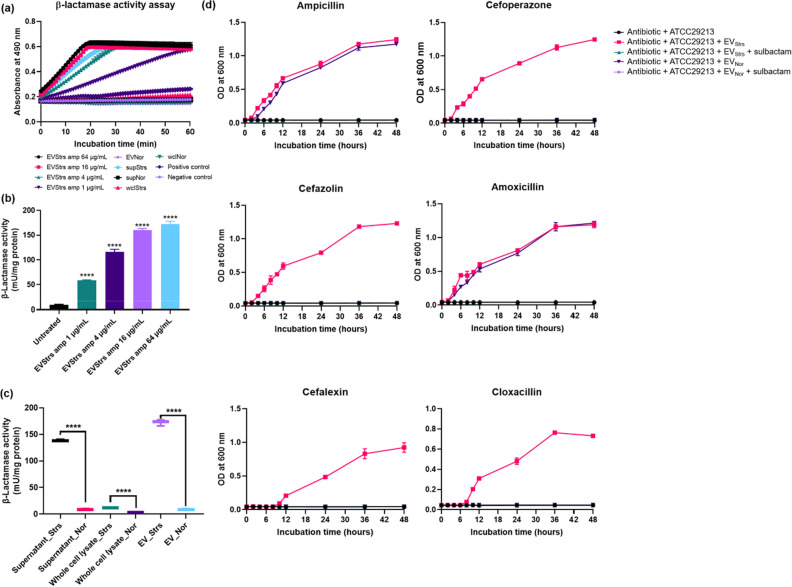
To further elaborate on the relevance of ampicillin stress in the total production of β-lactamase in EVs of the host bacteria, we then analyzed the β-lactamase activity associated with the degradation of various β-lactam antibiotics. All samples of EVs from cultures treated with increasing concentrations of ampicillin showed higher absorbance than the positive control, other samples had higher absorbance than negative control but lower than the positive control (Fig. 8a). To quantify the β-lactamase activity of EVs was presented as milliunit per milligram of protein (Fig. 8b). The activity of EVs isolated from bacteria exposed to increasing concentration of ampicillin in sub-MICs increased gradually, wherein, EVs treated with 1 μg/mL exhibited more β-lactamase activity than corresponding EVNor, by 6.5-fold. When comparing the samples with β-lactamase activities per mg, the EVStrs (treated with 64 μg/mL of ampicillin) was the highest, followed by the supernatant from stress condition (Fig. 8c). All of the samples subjected to ampicillin stress showed significantly increased levels as opposed to those in the stress-free environment, specifically in the supernatant by 15.8-fold, whole-cell lysate by 4.0-fold, and EVs by 19.1-fold.
Figure 8.
Comparison and analysis of β-lactamase activity of supernatant (sup), whole-cell lysate (wcl), and EVs isolated from stressed strain and normal strain. ‘S’ means stressed and ‘N’ means normal. (a) β-Lactamase activity profiles of respective samples, as examined by measuring absorbance at 490 nm in kinetic mode. (b) EVs from cultures of ST692 cells treated with 64, 16, 4, 1 μg/mL ampicillin, or untreated were assayed for β-lactamase activity. (c) β-Lactamase activities expressed per milligram of protein. (d) The effects of the β-lactamase inhibitor, sulbactam, were examined by growth curve experiments of β-lactam-susceptible Staphylococcus aureus treated with six antibiotics (ampicillin, 40 μg/mL; cefoperazone, 8 μg/mL; cefazolin, 1.25 μg/mL; amoxicillin, 40 μg/mL; cefalexin, 4 μg/mL; cloxacillin 1.25 μg/mL) plus sulbactam in the presence of 25 μg/mL EVStrs or EVNor. All data were presented as means and SEMs of three independent experiments. ****P < 0.0001.
The function of EVs mixed with a β-lactamase inhibitor
To determine whether β-lactamase is involved in the antibiotic degradation capacity of EVs, we observed the growth kinetics after the treatment of EVs and β-lactamase inhibitors (sulbactam) in ATCC29213 cells in the respective antibiotic environment (Fig. 8d). EVStrs degraded each β-lactam antibiotics more than EVNor, allowing susceptible cells to grow in an antibiotic environment, but no susceptible cells grew within 48 h in the respective EVs added with sulbactam.
Ampicillin stress affects β-lactam antibiotic tolerance of bacteria
The MICs of each antibiotic against stressed and normal ST692 cells were compared (Table 3). Treatment of ampicillin lower than MICs (64 μg/mL) increased the MICs from 2 to 4 times for β-lactam antibiotics except for cefotaxime, imipenem, and methicillin, but when the stressed ST692 cells were sub-cultured in antibiotic-free agar they reverted to their normal susceptibility to a β-lactam antibiotic. The MICs of stressed ST692 cells showed no change in all other antibiotics except for β-lactam antibiotics.
Table 3.
Alteration in MIC of several antibiotics against Staphylococcus aureus ST692 and ST692 stressed on ampicillin.
| Class | Antibiotics | MIC (μg/mL) | Fold increase in MIC (stress/normal) | |
|---|---|---|---|---|
| Normal ST692 | Stressed ST692 | |||
| β-Lactams | Ampicillin | 256 | 1024 | 4 |
| Amoxicillin | 256 | 1024 | 4 | |
| Cefalexin | 32 | 64 | 2 | |
| Cefazolin | 32 | 64 | 2 | |
| Cefoperazone | 64 | 128 | 2 | |
| Cefotaxime | 32 | 32 | 1 | |
| Cloxacillin | < 0.5 | 1 | –a | |
| Imipenem | > 1024 | > 1024 | –a | |
| Methicillin | 32 | 32 | 1 | |
| Chloramphenicol | Chloramphenicol | 16 | 16 | 1 |
| Aminoglycosides | Gentamicin | 4 | 4 | 1 |
| Kanamycin | 32 | 32 | 1 | |
| Streptomycin | > 1024 | > 1024 | –a | |
| Tetracyclines | Tetracycline | 64 | 64 | 1 |
MIC minimum inhibitory concentration.
aIndicates not exactly known because the range of MIC is greater than the maximum value or less than a minimum value.
Discussion
In this study, the potential function in β-lactam resistance of EVs released by bacteria in stressed conditions was examined. EVs from MRSA cultured with or without sub-MICs of ampicillin were purified, and their capability to degrade several β-lactam antibiotics investigated, and their proteomics compared to identified β-lactam antibiotic resistance-related protein constituents. It was found that the production of MRSA EVs increased dose-dependently by ampicillin treatment, and EVs from cultures treated with ampicillin became more potent in degrading β-lactam antibiotics.
Several factors that trigger vesicle production including genetic background of the strains, and factors such as temperature, iron and oxygen availability, media composition, and growth phase10,22. In addition, bacterial vesiculation could be triggered more intensely when bacteria were challenged with certain antibiotics at sub-lethal concentrations. A previous study showed that the treatment of β-lactam antibiotics creates holes in the peptidoglycan layer of Gram-positive bacteria which cause protrusion of cytoplasmic membrane material into the extracellular area and more EVs are generated10,23–25. There is no significant difference in the size of EVs isolated from MRSA treated w/ and w/o ampicillin by the analysis of TEM and DLS (Fig. 1 and Supplementary Table S1 in manuscript). However, the total protein amount of respective EVs was different in BCA protein assay. In addition, we have confirmed that the production of EVs augmented when ampicillin or gentamicin was treated with different MRSA strains named ST541 in sub-inhibitory concentration (data not shown).
Several types of research have shown that vesicles from bacteria can affect various antibiotics or antimicrobial peptides by hydrolysis/sequestration and acting as decoys. For instance, the vesicles can protect the bacteria for some antibiotics16,24,26 such as the case of colistin18, melittin18, polymyxin B26, and daptomycin24 thereby assisting the survival of the bacterium both in vitro and ex vivo24. It has been demonstrated in the past that vesicles can defense bacteria by degrading some β-lactam antibiotics, like amoxicillin, ampicillin, cefoperazone, and cefotaxime through their own proteins which are related to β-lactam resistance7,9,19. Packed into vesicles, substances related to antibiotic resistance can be delivered over long distances, unharmed by dilution and degradation14; such packaging poses a great benefit for the antibiotic resistance mechanism of the bacteria.
β-lactams are one of the most widely utilized groups of antibiotics available for the treatment of several bacterial infections27. β-lactam antibiotics attach to transpeptidase enzymes, also referred to penicillin-binding proteins (PBPs), interfering them from forming a peptidoglycan layer that produces the cell wall. Due to differences in cell wall organization, Gram-positive bacteria are bounded by a single membrane making them more sensitive to the bactericidal activity of β-lactam antibiotics than Gram-negative which are surrounded by an outer membrane that can protect exposure to antibiotics28. These bacteria have been exposed to naturally occurring antibiotic compounds for at least one billion years, and the evolutionary advantages they have adapted have led to the rapid development of resistance mechanisms for survival against antibiotics which are currently being used for medical applications29. The major mechanisms by which Gram-positive bacteria have developed to avoid the inhibitory effect of β-lactam antibiotics are as follows30: (1) β-lactamases are secreted into the extracellular space to degrade β-lactam antibiotics before they reach the cell wall; (2) Expression of mutated PBPs that are still capable of synthesizing the cell wall but are incapable of binding to β-lactam antibiotics. Therefore, EVs that have the ability to resist the bactericidal effect of β-lactam antibiotics as newly observed in Gram-positive strains are more important than those from the less susceptible Gram-negative strains.
The β-lactamase enzyme7 and metallo β-lactamase superfamily protein31 hydrolytically destroy β-lactam antibiotics and both proteins were significantly upregulated in EVStrs compared with EVNor (Table 2 and see Dataset 1 and Dataset 2 online). It is assumed that the β-lactamase may be bound to the membrane of EVs32, therefore the EVs secreted from Gram-positive S. aureus exhibit β-lactamase activity (Fig. 8) and degrade antibiotics in the extracellular environment (Fig. 5). The number of PBPs located on the extracellular surface of the cytoplasmic membrane varies between bacterial species33,34. Cell wall synthesis in S. aureus is intrinsically controlled by four native PBPs, PBP1 to 4, and β-lactam resistance of MRSA is determined by the production of one non-native PBP, PBP2a33. Once β-lactam covalently bound to native PBPs, this stable covalent adduct could not be removed by neutral buffers, acids or detergents35,36, therefore we assumed that the native PBPs which are more abundant in EVStrs than EVNor (Table 2) are capable of holding β-lactam antibiotics. This study hypothesized that EVStrs are more efficient in degrading β-lactam antibiotics than EVNor, and this capability can be due to the proteins produced in abundance since bacteria have a mechanism to selectively package-specific proteins and concentrate them into the vesicles to resist stressors37,38.
We confirmed that EVs did not affect the growth of strains in the presence of the other tested antibiotics (data not shown) which include imipenem, cefotaxime, and methicillin. These three antibiotics belong in the group of β-lactam antibiotics but were not degraded by the EVs containing β-lactamases due to their resistance to β-lactamase39–41. Therefore, although the activity of β-lactamase was increased fourfold by ampicillin stress in ST692 cells (Fig. 8c), these three mentioned antibiotics were not influenced in terms of their MICs by the stressed ST692 (Table 3).
Ampicillin below concentrations of MICs value intensifies the production of chromosomal β-lactamase42,43 and this phenomenon was confirmed in Fig. 8C and Table 3. These findings suggested that MRSA temporarily facilitates the intrinsic ability of bacterial defense mechanism to over-produce β-lactamase as a reaction after exposure to high levels of ampicillin drug, thereby causing an increased hyposensitivity to β-lactam antibiotics. Thus, for an appropriate antibiotic challenge, it might require an increased dose of antibiotics to treat bacteria.
Actually, S. aureus has been identified as one of the microbial infections in many polymicrobial infections44. Studies proving ‘cooperative interaction’ between different strains of bacteria had been discussed in the past. For example, Candida albicans and S. aureus colonized human mucosal surfaces with commensals and cause enhanced disease severity during biofilm-related coinfections44–46. Haemophilus influenzae and S. aureus showed cooperative interactions and both colonized in nasopharynx, instances, and genital tract44,47. S. aureus co-colonized together with Enterococcus faecalis in the intestinal tracts44, and conjugation between the two strains causes a horizontal transfer of the vanA gene, resulting in multidrug-resistant staphylococci48,49. Schaar et al. also indicated that vesicles including β-lactamase help to protect producer bacteria as well as co-occurring organisms in the human19,50. The results of Fig. 4 further imply that the EVs secreted from MRSA can indeed protect co-existing bacterial communities against β-lactam antibiotics.
To summarize, global protein modulation of EVs by ampicillin stress response of MRSA involves processes that are directly related to β-lactam antibiotic resistance. EVs naturally secreted from MRSA possess β-lactam-resistant proteins, which can help bacteria to survive in the antibiotic environment by hydrolyzing the capacity of antibiotics. Inducing stress on MRSA with a sub-lethal dose of ampicillin could stimulate production of EVs enhancing the ability to consume antibiotic compared with EVs released in no-stress condition. Therefore, proper drug treatment is necessary to impede the progeny of MRSA because indiscriminate abuse of β-lactam antibiotics may create an opportunity for bacteria to be more resistant to β-lactam antibiotics. Besides, this is a novel mechanism not related to PBP2a-based, the major resistance mechanism of β-lactam antibiotics of MRSA strains known up to this point. This information equips us with a new perspective on how to lessen (if not eradicate) the impact of multi-drug resistant bacteria which impose a grave threat to global public health.
Methods
Bacterial strains
Methicillin-resistant Staphylococcus aureus C-S03-S237 strain of ST69251 isolated from the chicken was provided from Animal and Plant Quarantine Agency, Korea and β-lactam-sensitive S. aureus ATCC29213 strain was purchased from ATCC. Luria–Bertani (LB; Oxoid) broth or tryptone soya agar (TSA; Oxoid, United Kingdom) were used to grow both cells at 37 °C. Ampicillin-sensitive bacteria, Escherichia coli RC859, and Edwardsiella tarda ED4552 were cultured on TSA and Salmonella spp. Sal26B53 was incubated on brain heart infusion (BHI; Oxoid) agar at 37 °C.
Determination of minimum inhibitory concentrations
Nine β-lactam antibiotics known to confer bactericidal effects by inhibiting cell wall biosynthesis, namely ampicillin, amoxicillin, cefalexin, cefazolin, cefoperazone, cefotaxime, cloxacillin, imipenem, and methicillin (Sigma-Aldrich, USA) and five other class antibiotics, such as chloramphenicol, gentamicin, kanamycin, streptomycin, and tetracycline (Sigma-Aldrich) were selected. The minimum inhibitory concentration (MIC) of each antimicrobial agent was determined in ST692 and ATCC29213 cells using the broth-dilution method in 96-well plates9,54 according to Clinical and Laboratory Standards Institute (CLSI) guidelines, except that cation-adjusted Muller Hinton broth was substituted with LB. The MIC values were measured from three independent experiments.
EVs isolation and characterization
EVs from ST692 cells were purified from bacterial culture supernatant as described previously7, with some modifications. Briefly, when isolating EVNor, the strain was cultured in nutrient broth (NB; Difco) without any antibiotic addition. To determine the change in the production of EVs, the ampicillin dose was treated at 1, 4, 16, or 64 μg/mL (EVStrs means EVs treated with 64 μg/mL of ampicillin when bacteria are cultured). Each culture medium was centrifuged at 6,000×g for 15 min and concentrated by QuixStand Benchtop system (GE Healthcare, Sweden). Each supernatant was centrifuged at 150,000×g at 4 °C for 3 h. Further purification was performed by a continuous sucrose density gradient followed by ultracentrifugation. The final EV pellet was resuspended in 10 mM Tris–HCl (pH 8.0) (Biopure, Korea) and filtered through a 0.2 μm filter (Thermo Fisher Scientific, IL). The protein yields of EVs samples were measured using a Pierce BCA protein assay kit (Thermo Fisher Scientific, USA). Transmission electron microscopy (TEM) of EVs was performed as previously described9 using a Tecnai G2 Spirit Twin TEM system (FEI, USA). Dynamic light scattering (DLS) of EVs for particle size distribution and measurement of zeta potential was performed as described previously9 using a Nano ZS instrument (Malvern Instruments, Malvern, UK) and the Zetasizer software (version 7.11; Malvern Instruments).
Proteome analysis by liquid chromatography combined with tandem mass spectrometry (LC–MS/MS)
Each of ST692 EVs was mixed with sample buffer and separated by Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) according to Laemmli’s method55 and in-gel digestion was performed as previously described9. Each obtained peptide mixture was resuspended in 0.1% (v/v) TFA and passed through an analytical column (Zorbax 300SB-C18 75 μm i.d. × 15 cm column; Agilent, Germany) via a trap column (Zorbax 300SB-C18 300 μm i.d. × 5 mm column; Agilent). The peptides were separated from acetonitrile gradient of buffer A (0.1% (v/v) formic acid in water) and buffer B (0.1% (v/v) formic acid in pure acetonitrile) at a constant flow rate of 0.2 μl/min using the Agilent 100 series nano HPLC system coupled on-line to a LTQ ion-trap mass spectrometer (Thermo Fisher Scientific). The gradient started linearly with 5% B and rose linearly to 40% B over 100 min, increased to 80% B over 1 min, and then increased to 80% B isocratically over 15 min. Full-scan mode (m/z 350–1600) was enabled and three MS/MS scans were performed with a 30-s dynamic exclusion option set for each survey MS scan.
Quantitative protein profiling, statistics and database searching
Peptide peaks were detected with an average peak width of 1 min and matched with a mass accuracy of at least 0.6 Da. Differentially expressed proteins were defined to exhibit more than twofold or greater increase/decrease in comparable intensity or the complete appearance/disappearance of the spot. After alignment of the retention times of the chromatogram, normalization was carried out with the measured intensity distribution and the proteome was quantified with the peak intensity ratio. The MS/MS spectra of the peptide peaks were searched against a SwissProt uni_bacteria database or Staphylococcus aureus USA300 strain database using Mascot 2.3 (Matrix Science, London, UK). The obtained LC/MS data were analyzed with the DeCyder MS software (version 2.0; GE Healthcare, Uppsala, Sweden). For quantitative profiling, proteins identified by multiple peptides with significant Mascot scores were selected (p < 0.05).
In silicon analysis of functional associations
Gene ontology (GO) terms such as biological process, cellular component, and molecular function are derived from differentially expressed proteins obtained using a software tool for researching annotations of proteins (STRAP) version 1.5 (Boston University School of Medicine, USA).
Effect of EVs on the growth of bacteria in the presence of β-lactam antibiotics
The effects of EVNor and EVStrs on the cytotoxicity of β-lactam antibiotics were monitored by assessing the growth curves of EVs-treated Staphylococcus aureus (ATCC29213) cells as previously described with slight modifications9. The following antibiotics were used at the growth-inhibiting concentrations: ampicillin, 40 μg/mL; cefoperazone, 8 μg/mL; cefazolin, 1.25 μg/mL; amoxicillin, 40 μg/mL; cefalexin, 4 μg/mL; and cloxacillin, 1.25 μg/mL. Cultured ATCC29213 cells were separately inoculated in a medium containing each antibiotic and 1, 5, 25 μg/mL of EVNor or EVStrs. To test whether EVs can affect different genera of bacteria, Gram-negative bacteria such as RC85, ED45, and Sal26B were cultured in LB, TSB, and BHI, respectively, and same concentrations of EVNor or EVStrs were treated in the medium with respective cultured bacteria in the presence of 30 μg/mL of ampicillin. The bacterial growth curves at OD600 were recorded at 2 h intervals up to 12 h, and then at 12 h intervals up to 48 h or 12 h intervals for 96 h using an xMark microplate spectrophotometer (Bio-Rad). The bacterial cultures since the last measurement time were streaked on TSA with or without the respective same concentrations of antibiotics to test whether the EVs could confer resistance to antibiotic-susceptible bacteria. The samples collected at certain time points (ampicillin, 12 h; cefoperazone, 12 h; cefazolin, 24 h; amoxicillin, 36 h; cefalexin, 96 h; and cloxacillin, 48 h) and quantitative plate assays were carried out9. The count obtained in the absence of antibiotics was taken as 100%, and the corresponding counts in the presence of different concentrations EVNor or EVStrs plus the respective β-lactam antibiotics were calculated. Colonies from each cultured sample (n = 5, colonies per sample) were randomly selected and identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS)56 to check for contamination.
Measurement of antibiotic concentrations
To evaluate whether EVs could directly degrade β-lactam antibiotics, the effects of EVNor and EVStrs on the concentrations of six antibiotics in a cell-free system were analyzed by liquid chromatography/electrospray ionization mass spectrometry (LC-ESI-QQQ-MS/MS; 6420 Triple Quad LC/MS; Agilent) as previously described with slight modifications9. One microgram per milliliter or 5 μg/mL of EVStrs or 5 μg/mL of EVNor in PBS were mixed with ampicillin (40 μg/mL), cefoperazone (8 μg/mL), cefazolin (1.25 μg/mL), amoxicillin (40 μg/mL), cefalexin (4 μg/mL) or cloxacillin (1.25 μg/mL). Filtered PBS containing the respective antibiotics without EVs was used as a positive control. In addition, to compare the degradability of EVNor and EVStrs against ampicillin, 5 μg/mL of each EVs was added to various concentrations of ampicillin. The concentrations of antibiotics were recorded at 3-h intervals for 6 h or at 24-h intervals for 48 h in triplicate.
Quantification of β-lactamase activity
To test for differences in β-lactamase activity between whole-cell lysates, supernatants, and EVs from the stressed condition and normal condition, a colorimetric β-lactamase activity assay kit (BioVision, Canada) was used according to the manufacturer’s instructions. The assay involves the hydrolysis of nitrocefin which produces a colored product that is measured by spectrophotometry (OD490). A Bradford assay kit (Thermo Fisher Scientific) was used to determine the protein concentrations of samples. Equivalent concentrations of each sample were dispensed to the wells of a clear flat-bottomed 96-well, and the provided nitrocefin and buffer were added. The OD490 was immediately measured in kinetic mode.
Changes in the effect of EVs on β-lactam antibiotics by β-lactamase inhibitor
The effects of the β-lactamase inhibitor, sulbactam (Abcam, United Kingdom), were investigated by growth curve experiments. The β-lactamase inhibitors were added at the previously reported and fixed concentrations9, the final concentration of 25 μg/mL was set in each case. Briefly, the growth curves of ATCC29213 treated with antibiotics (ampicillin, 40 μg/mL; cefoperazone, 8 μg/mL; cefazolin, 1.25 μg/mL; amoxicillin, 40 μg/mL; cefalexin, 4 μg/mL; or cloxacillin, 1.25 μg/mL) plus sulbactam were determined in the presence of 25 μg/mL EVStrs or EVNor.
Statistical analysis
Statistical analyses were carried out using Graphpad Prism, version 8.1.1. (GraphPad, CA, USA). Significant differences were determined by Student’s t-test, One-way Analysis of Variance (ANOVA), Two-way ANOVA, or Turkey’s multiple comparison test. All data were expressed as means ± standard deviations (SD). Differences were considered statistically significant at P < 0.05.
Supplementary information
Acknowledgements
This research was supported by a Grant (19162MFDS563) from Ministry of Food and Drug Safety in 2020 and by a Korea Research Foundation Grant (NRF-2018R1A2B2005505).
Author contributions
S.W.K. and S.B.P. designed the study; S.W.K., J.S.S., A.R.L., J.H.K., and J.W.S. prepared samples and performed experiments; S.W.K., J.S.L., J.W.J., J.H.C., and M.W.H. processed and analyzed data; S.W.K. and S.B.P. wrote the original draft; S.W.K., J.M.S.L., J.K., C.F., W.Z., and S.M.P. reviewed and edited the manuscript; M.J., and T.S.J. supervised the study. All authors read and approved this manuscript.
Data availability
All data generated or analyzed during this study are included in this published article and its supplemental material files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-78121-8.
References
- 1.O’Neill, J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Drug Resist. Updat. 1–16 (2014).
- 2.Jevons MP. “Celbenin”-resistant staphylococci. Br. Med. J. 1961;1:124. doi: 10.1136/bmj.1.5219.124-a. [DOI] [Google Scholar]
- 3.Cosgrove SE, et al. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: A meta-analysis. Clin. Infect. Dis. 2003;36:53–59. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 4.Leonard F, et al. Methicillin-resistant Staphylococcus aureus isolated from a veterinary surgeon and five dogs in one practice. Vet. Rec. 2006;158:155–159. doi: 10.1136/vr.158.5.155. [DOI] [PubMed] [Google Scholar]
- 5.Morgan M. Methicillin-resistant Staphylococcus aureus and animals: ZOONOSIS or humanosis? J. Antimicrob. Chemother. 2008;62:1181–1187. doi: 10.1093/jac/dkn405. [DOI] [PubMed] [Google Scholar]
- 6.Springer B, et al. Methicillin-resistant Staphylococcus aureus: A new zoonotic agent? Wien Klin Wochenschr. 2009;121:86–90. doi: 10.1007/s00508-008-1126-y. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, et al. Staphylococcus aureus extracellular vesicles carry biologically active β-lactamase. Antimicrob. Agents Chemother. 2013;57:2589–2595. doi: 10.1128/AAC.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015;13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SW, et al. Outer membrane vesicles from β-lactam-resistant Escherichia coli enable the survival of β-lactam-susceptible E. coli in the presence of β-lactam antibiotics. Sci. Rep. 2018;8:5402. doi: 10.1038/s41598-018-23656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toyofuku M, Nomura N, Eberl L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019;17:13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 11.Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: A novel mechanism of enzyme secretion. J. Bacteriol. 1995;177:3998–4008. doi: 10.1128/JB.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claßen L, et al. Extracellular vesicles mediate intercellular communication: Transfer of functionally active microRNAs by microvesicles into phagocytes. Eur. J. Immunol. 2017;47:1535–1549. doi: 10.1002/eji.201646595. [DOI] [PubMed] [Google Scholar]
- 13.Maas SL, Breakefield XO, Weaver AM. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell. Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quan K, et al. Escherichia coli-derived outer membrane vesicles deliver galactose-1-phosphate uridyltransferase and yield partial protection against Actinobacillus pleuropneumoniae in mice. J. Microbiol. Biotechnol. 2018;28:2095–2105. doi: 10.4014/jmb.1809.09004. [DOI] [PubMed] [Google Scholar]
- 15.Fulsundar S, et al. Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl. Environ. Microbiol. 2014;80:3469–3483. doi: 10.1128/AEM.04248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015;13:605. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rumbo C, et al. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: A new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011;55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulkarni HM, Nagaraj R, Jagannadham MV. Protective role of E. coli outer membrane vesicles against antibiotics. Microbiol. Res. 2015;181:1–7. doi: 10.1016/j.micres.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Schaar V, Nordström T, Mörgelin M, Riesbeck K. Moraxella catarrhalis outer membrane vesicles carry β-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob. Agents Chemother. 2011;55:3845–3853. doi: 10.1128/AAC.01772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauwens A, Kunsmann L, Karch H, Mellmann A, Bielaszewska M. Antibiotic-mediated modulations of outer membrane vesicles in enterohemorrhagic Escherichia coli O104: H4 and O157: H7. Antimicrob.. Agents Chemother. 2017;61:e00937–e1917. doi: 10.1128/AAC.00937-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacDonald IA, Kuehn MJ. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. J. Bacteriol. 2013;195:2971–2981. doi: 10.1128/JB.02267-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orench-Rivera N, Kuehn MJ. Environmentally controlled bacterial vesicle-mediated export. Cell. Microbiol. 2016;18:1525–1536. doi: 10.1111/cmi.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biagini M, et al. The human pathogen Streptococcus pyogenes releases lipoproteins as lipoprotein-rich membrane vesicles. Mol. Cell. Proteom. 2015;14:2138–2149. doi: 10.1074/mcp.M114.045880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andreoni F, et al. Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage-dependent and-independent fashions and via different routes. Antimicrob. Agents Chemother. 2019;63:e01439–e11418. doi: 10.1128/AAC.01439-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devos S, et al. The effect of imipenem and diffusible signaling factors on the secretion of outer membrane vesicles and associated Ax21 proteins in Stenotrophomonas maltophilia. Front. Microbiol. 2015;6:298. doi: 10.3389/fmicb.2015.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manning AJ, Kuehn MJ. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011;11:258. doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta RS. Origin of diderm (Gram-negative) bacteria: Antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes. Antonie Van Leeuwenhoek. 2011;100:171–182. doi: 10.1007/s10482-011-9616-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez JL. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. Biol. Sci. 2009;276:2521–2530. doi: 10.1098/rspb.2009.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munita JM, Bayer AS, Arias CA. Evolving resistance among Gram-positive pathogens. Clin. Infect. Dis. 2015;61:S48–S57. doi: 10.1093/cid/civ523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.31Scherr, T. D. Staphylococcus aureus biofilms interfere with macrophage antimicrobial responses through differential gene regulation, toxin production, and purine metabolism. Theses & Dissertations145 (2016).
- 32.Wilke MS, Lovering AL, Strynadka NC. β-Lactam antibiotic resistance: A current structural perspective. Curr. Opin. Microbiol. 2005;8:525–533. doi: 10.1016/j.mib.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 33.Georgopapadakou NH, Liu FY. Penicillin-binding proteins in bacteria. Antimicrob. Agents Chemother. 1980;18:148–157. doi: 10.1128/AAC.18.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuda C, Fisher J, Mobashery S. β-Lactam resistance in Staphylococcus aureus: the adaptive resistance of a plastic genome. Cell. Mol. Life Sci. 2005;62:2617. doi: 10.1007/s00018-005-5148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zervosen A, Sauvage E, Frère J-M, Charlier P, Luxen A. Development of new drugs for an old target—the penicillin binding proteins. Molecules. 2012;17:12478–12505. doi: 10.3390/molecules171112478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong KF, Schneper L, Mathee K. Beta-lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS. 2010;118:1–36. doi: 10.1111/j.1600-0463.2009.02563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haurat MF, et al. Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 2011;286:1269–1276. doi: 10.1074/jbc.M110.185744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonnington K, Kuehn M. Protein selection and export via outer membrane vesicles. Biochim. Biophys. Acta Mol. Cell. Res. 2014;1843:1612–1619. doi: 10.1016/j.bbamcr.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clissold SP, Todd PA, Campoli-Richards DM. Imipenem/cilastatin. Drugs. 1987;33:183–241. doi: 10.2165/00003495-198733030-00001. [DOI] [PubMed] [Google Scholar]
- 40.Van TTH, Nguyen HNK, Smooker PM, Coloe PJ. The antibiotic resistance characteristics of non-typhoidal Salmonella enterica isolated from food-producing animals, retail meat and humans in South East Asia. Int. J. Food Microbiol. 2012;154:98–106. doi: 10.1016/j.ijfoodmicro.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 41.Chang Y-M, et al. Structural study of TcaR and its complexes with multiple antibiotics from Staphylococcus epidermidis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8617–8622. doi: 10.1073/pnas.0913302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moosdeen F, Keeble J, Williams J. Induction/inhibition of chromosomal β-lactamases by β-lactamase inhibitors. Rev. Infect. Dis. 1986;8:S562–S568. doi: 10.1093/clinids/8.Supplement_5.S562. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y, Livermore D, Williams RJ. Chromosomal β-lactamase expression and antibiotic resistance in Enterobacter cloacae. J. Med. Microbiol. 1988;25:227–233. doi: 10.1099/00222615-25-3-227. [DOI] [PubMed] [Google Scholar]
- 44.Nair N, Biswas R, Götz F, Biswas L. Impact of Staphylococcus aureus on pathogenesis in polymicrobial infections. Infect. Immun. 2014;82:2162–2169. doi: 10.1128/IAI.00059-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters BM, Noverr MC. Candida albicans-Staphylococcus aureus polymicrobial peritonitis modulates host innate immunity. Infect. Immun. 2013;81:2178–2189. doi: 10.1128/IAI.00265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kojic EM, Darouiche RO. Candida infections of medical devices. Clin. Microbiol. Rev. 2004;17:255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Margolis E, Yates A, Levin BR. The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: The role of competition and interactions with host's immune response. BMC Microbiol. 2010;10:59. doi: 10.1186/1471-2180-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark NC, Weigel LM, Patel JB, Tenover FC. Comparison of Tn1546-like elements in vancomycin-resistant Staphylococcus aureus isolates from Michigan and Pennsylvania. Antimicrob. Agents Chemother. 2005;49:470–472. doi: 10.1128/AAC.49.1.470-472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Périchon B, Courvalin P. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009;53:4580–4587. doi: 10.1128/AAC.00346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaar V, Uddbäck I, Nordström T, Riesbeck K. Group A streptococci are protected from amoxicillin-mediated killing by vesicles containing β-lactamase derived from Haemophilus influenzae. J. Antimicrob. Chemother. 2013;69:117–120. doi: 10.1093/jac/dkt307. [DOI] [PubMed] [Google Scholar]
- 51.Lim S-K, et al. Prevalence and characterization of methicillin-resistant Staphylococcus aureus in raw meat in Korea. J. Microbiol. Biotechnol. 2010;20:775–778. [PubMed] [Google Scholar]
- 52.Park SB, et al. Outer membrane vesicles as a candidate vaccine against edwardsiellosis. PLoS ONE. 2011;6:e17629. doi: 10.1371/journal.pone.0017629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bae D-H, et al. Prevalence and characteristics of Salmonella spp. isolated from poultry slaughterhouses in Korea. J. Vet. Med. Sci. 2013;75:1193–1200. doi: 10.1292/jvms.13-0093. [DOI] [PubMed] [Google Scholar]
- 54.Andrews JM. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 55.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 56.Kim SW, et al. Comparison of proteome typing and serotyping of Streptococcus parauberis isolates from olive flounder (Paralichthys olivaceus) J. Microbiol. Methods. 2015;118:168–172. doi: 10.1016/j.mimet.2015.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplemental material files.