Abstract
PURPOSE
To detect alterations in DNA damage repair (DDR) genes, measure homologous recombination deficiency (HRD), and correlate these findings with clinical outcome in patients with leiomyosarcoma (LMS).
PATIENTS AND METHODS
Patients with LMS treated at Memorial Sloan Kettering (MSK) Cancer Center who consented to prospective targeted next-generation sequencing with MSK-IMPACT were screened for oncogenic somatic variants in one of 33 DDR genes; where feasible, an experimental HRD score was calculated from IMPACT data. Progression-free survival (PFS) and overall survival (OS) were estimated after stratifying patients by DDR gene alteration status and HRD score.
RESULTS
Of 211 patients with LMS, 20% had an oncogenic DDR gene alteration. Univariable analysis of PFS in 117 patients who received standard frontline chemotherapy in the metastatic setting found that an altered homologous recombination pathway gene was significantly associated with shorter PFS (hazard ratio [HR], 1.79; 95% CI, 1.04 to 3.07; P = .035). Non-BRCA homologous recombination gene alteration was associated with shorter PFS (HR, 2.61; 95% CI, 1.35 to 5.04; P = .004) compared with BRCA-altered and wild-type homologous recombination genes. Univariable analysis of OS from diagnosis in the entire cohort of 211 patients found that age, tumor size, number of metastatic sites, localized disease, and non-BRCA homologous recombination gene alteration were significantly associated with OS. On multivariable analysis, non-BRCA homologous recombination pathway gene alteration remained significant (HR, 4.91; 95% CI, 2.47 to 9.76; P < .001). High HRD score was not associated with a different PFS or OS.
CONCLUSION
Patients with LMS with homologous recombination pathway gene alterations have poor clinical outcomes, particularly those with non-BRCA gene alterations. HRD score calculated from a targeted exome panel did not discern disparate clinical outcomes.
INTRODUCTION
Leiomyosarcoma (LMS) is a mesenchymal malignancy that arises from differentiated smooth muscle cells. It most commonly originates in the uterus, retroperitoneum, or extremities.1 LMS is one of the most common subtypes of sarcoma, estimated to comprise up to 20% of newly diagnosed soft tissue sarcoma cases.2 Although surgical resection of localized disease can be curative, LMS has a high risk of recurrence, and patients with unresectable or metastatic disease have a poor prognosis.3,4
CONTEXT
Key Objective
Leiomyosarcomas (LMSs) have a relatively frequent aberration of DNA damage repair (DDR) pathways, but data are limited with regard to the impact of these gene alterations on clinical outcome. This retrospective study used real-word data to determine whether DDR gene alterations or an experimental homologous recombination deficiency (HRD) score calculated from a targeted next-generation sequencing panel is associated with differential clinical outcome in patients with LMS.
Knowledge Generated
Patients with LMS and somatic alteration of a homologous recombination pathway gene have significantly worse clinical outcomes, particularly those with altered non-BRCA genes within this pathway. HRD score was associated with homologous recombination gene and overall DNA copy number alteration but not with clinical outcome.
Relevance
Patients with a homologous recombination gene alteration have a more aggressive disease course and may represent a targetable subset of patients with LMS. Additional study and refinement of HRD score calculated from targeted gene panels are likely necessary to increase the score’s clinical utility in this patient population.
LMS typically has a complex karyotype characterized by recurrent alterations in the tumor suppressor genes TP53, RB1, and PTEN, often accompanied by widespread chromosomal structural damage. Genomic studies of LMS have reported frequent alterations in BRCA2 and other DNA damage repair (DDR) pathway genes, including ATM, ATR, and CHEK2.5-7 Patients with LMS and somatic loss of BRCA2 have dedifferentiated tumors with a high mitotic index, signifying a potentially aggressive phenotype.8
The presence of a deficiency in the homologous recombination pathway, one of many pathways critical in the repair of damaged DNA, has become especially relevant in the age of poly (ADP-ribose) polymerase (PARP) inhibitors, which have transformed the treatment of BRCA1 or BRCA2 (BRCA)-associated cancers.9-12 Synthetically lethal approaches may also be efficacious in cancers with homologous recombination deficiency (HRD) and wild-type (WT) BRCA. For example, breast and ovarian cancers with high HRD scores and WT BRCA have higher response rates to platinum chemotherapy and PARP inhibition than those without evidence of HRD.11,13 The HRD score is an unweighted sum of three distinct measures of genomic scarring: telomeric allelic imbalance (NtAI), large-scale state transitions (LSTs), and loss of heterozygosity (HRD-LOH).13-16 A pan-cancer analysis of DDR pathway alterations across The Cancer Genome Atlas (TCGA) found that soft tissue sarcomas have relatively high HRD scores compared with other cancers, and patients with high HRD scores trend toward worse clinical outcomes.17
Aside from homologous recombination pathway deficiencies, other DDR pathways, such as the base excision repair and DNA damage sensor pathways, have also been susceptible to PARP inhibitors in preclinical and clinical studies.12,18 For instance, pancreatic adenocarcinoma and urothelial carcinoma with alterations in DDR genes have improved outcomes after platinum chemotherapy.19,20 It is unknown whether oncogenic DDR gene alterations or high HRD scores are prognostic or predictive in LMS.
We hypothesized that patients with LMS with BRCA loss would have improved clinical outcomes compared with patients with WT BRCA, as has been demonstrated in ovarian cancer.21,22 Similarly, we hypothesized that patients with somatic alteration in any DDR pathway gene would have improved survival compared with patients with WT DDR, also as a result of synthetic lethality after DNA-damaging cytotoxic chemotherapy. We used targeted genomic sequencing of tumor samples to identify the frequency of DDR gene alterations and quantified genomic scarring indicative of HRD to generate an HRD score in a cohort of patients with LMS treated at Memorial Sloan Kettering (MSK).
PATIENTS AND METHODS
Patient Selection and Study Design
This study was approved by the MSK institutional review board. Patients with histologically confirmed LMS who had targeted genetic sequencing of their tumor with MSK-Integrated Molecular Profiling of Actionable Cancer Targets (IMPACT) between March 2014 and October 2018 and had clinical data available for review were included in this analysis. The objectives of this study were to determine the incidence of DDR gene alterations in patients with LMS and the prognostic and predictive potential of DDR gene, homologous recombination pathway gene, and BRCA gene status, respectively, on clinical outcomes. We sought to measure HRD with a copy number signature modeled after those reported in the literature that combined NtAI, LSTs, and HRD-LOH23 and to correlate this measure with clinical outcome.
Demographic, pathologic, and clinical information were retrieved from the medical record for each patient. The following variables were included: age, sex, race, date of diagnosis, tumor size at diagnosis, site of primary disease, presence of locally invasive or metastatic disease at diagnosis, number of metastatic sites at diagnosis, use of neoadjuvant and adjuvant systemic therapy, first-line chemotherapy start and end dates, reason for cessation of first-line chemotherapy, and date of death or last follow-up. The cutoff date for clinical follow-up was May 15, 2019.
Classification of DDR Gene Alterations and Calculation of an HRD Score
All patients included in this study provided informed written consent to participate in a prospective tumor sequencing initiative at MSK using the MSK-IMPACT assay. The IMPACT assay has been described in detail elsewhere.24,25 It is a hybridization capture–based next-generation sequencing platform of 341-468 exons and select introns, depending on the assay version. We selected 33 genes from the IMPACT panel demonstrated in the medical literature to be involved in at least one DNA damage response pathway19 (Data Supplement). Any nonsense, frameshift, or splice site mutation predicted to lead to loss of function of the encoded protein or homozygous deletion of a DDR gene was considered deleterious. In addition, missense mutations annotated as oncogenic by OncoKB were considered deleterious.26 To calculate the HRD score, FACETS was used to generate copy number profiles27 from IMPACT data, from which the HRD scores, which consisted of the sum of scores from measures of genomic scars (LSTs, NtAI, and HRD-LOH), were estimated. Patients with at least one alteration in a DDR pathway gene were labeled DDR altered; those with an alteration in at least one homologous recombination pathway gene were labeled homologous recombination pathway altered; those with BRCA loss were labeled BRCA altered; and those with an alteration in a non-BRCA homologous recombination pathway were labeled non-BRCA homologous recombination pathway altered.
Statistical Analyses
Patients were divided into subgroups on the basis of the detection of a deleterious DDR pathway alteration, homologous recombination pathway alteration, BRCA gene, or non-BRCA homologous recombination pathway alteration in their tumor. The HRD score was tested as a continuous variable using the median as a cutoff. Survival analyses were performed on two overlapping sets of patients. The first set analyzed the progression-free survival (PFS) of patients who received standard-of-care chemotherapy (either doxorubicin- or gemcitabine-based treatment) in the first-line metastatic setting. Patients who received systemic chemotherapy in the neoadjuvant or adjuvant settings were excluded. PFS was defined as the time from the first dose of chemotherapy in the metastatic setting until the date of progression as determined by the treating clinician, the date of treatment cessation because of toxicity, or death; patients who stopped chemotherapy for any reason other than clinical or radiologic progression, toxicity, or death were censored. A second patient set included all patients with LMS who had IMPACT testing and an analyzed overall survival (OS). OS was defined as the time from the date of diagnosis until the date of death or last contact; patients who were alive at the time of last contact were censored.
Summary statistics, medians, and ranges, were used to describe continuous variables, and counts and percentages were used for categorical variables. To compare variables across groups, Fisher’s exact test was used for categorical variables and Wilcoxon rank sum test for continuous variables. Correlation was assessed with the Spearman test. Kaplan-Meier survival analysis was performed, and log-rank P values are reported. Univariable and multivariable analyses were performed with Cox proportional hazard regression models. Multivariable models were selected using backward selection with inclusion criteria being significant at P < .10 in the univariable analysis. SAS 9.4 statistical software (SAS Institute, Cary, NC) was used for outcome analyses. All tests were two sided, and P < .05 was considered significant.
RESULTS
Patient and Tumor Characteristics
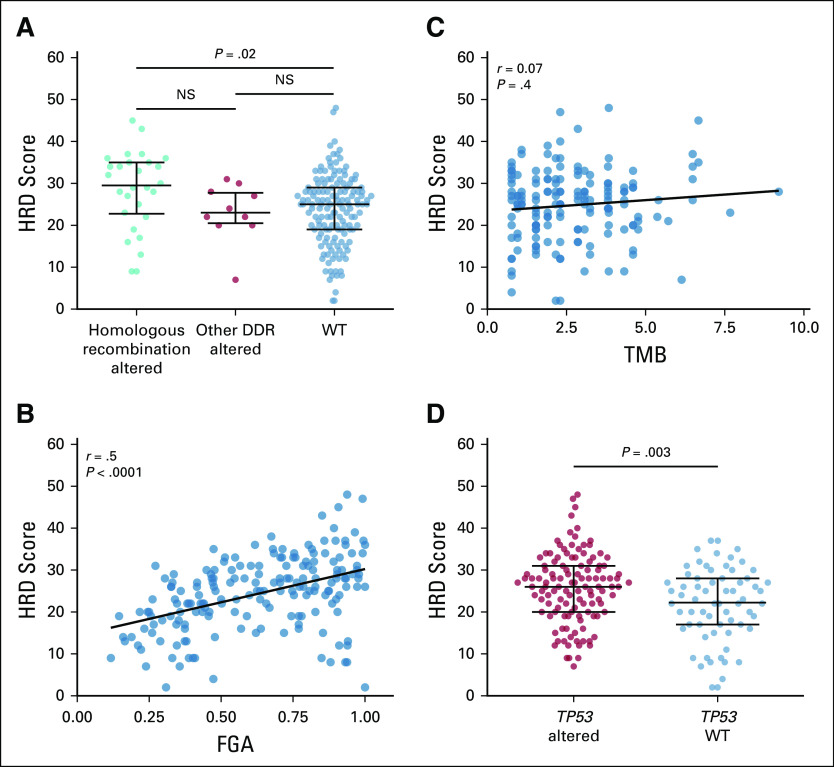
A total of 211 patients with LMS consented to IMPACT testing of their tumors and were included in this analysis: 121 patients (57%) had LMS of the uterus (uLMS), and 90 (43%) had LMS of other soft tissue sites (stLMS). Patient demographic and tumor characteristics are listed in Table 1. The median age was 54 years (range, 20-79 years), and 83% of the study population was female. Among patients with stLMS, 55 were female (60%). The most common extrauterine primary sites were the abdomen, pelvis, and retroperitoneum. Thirty-five percent of patients had locally invasive or metastatic disease at the time of diagnosis; 47% of IMPACT samples were from primary tumors, and the remaining were from metastatic foci. Sixty-one percent of samples were obtained before initiation of chemotherapy or radiation therapy.
TABLE 1.
Patient Demographic and Clinical Characteristics
Frequency of DDR Gene Alterations in LMSs
Forty-three patients (20%) had a somatic putatively oncogenic DDR gene alteration, 72% of whom had an alteration in the homologous recombination pathway. The most frequently altered DDR genes were BRCA2 (n = 14; 7%), RAD51B (n = 8; 4%), and ERCC5 (n = 4; 2%; Fig 1). Seventy-seven percent of the alterations in our cohort were homozygous deletions, while the remainder were putatively loss-of-function single nucleotide variants or indels. Patients with uLMS had more DDR alterations than those with stLMS (25% v 14%, respectively; P = .084).
FIG 1.
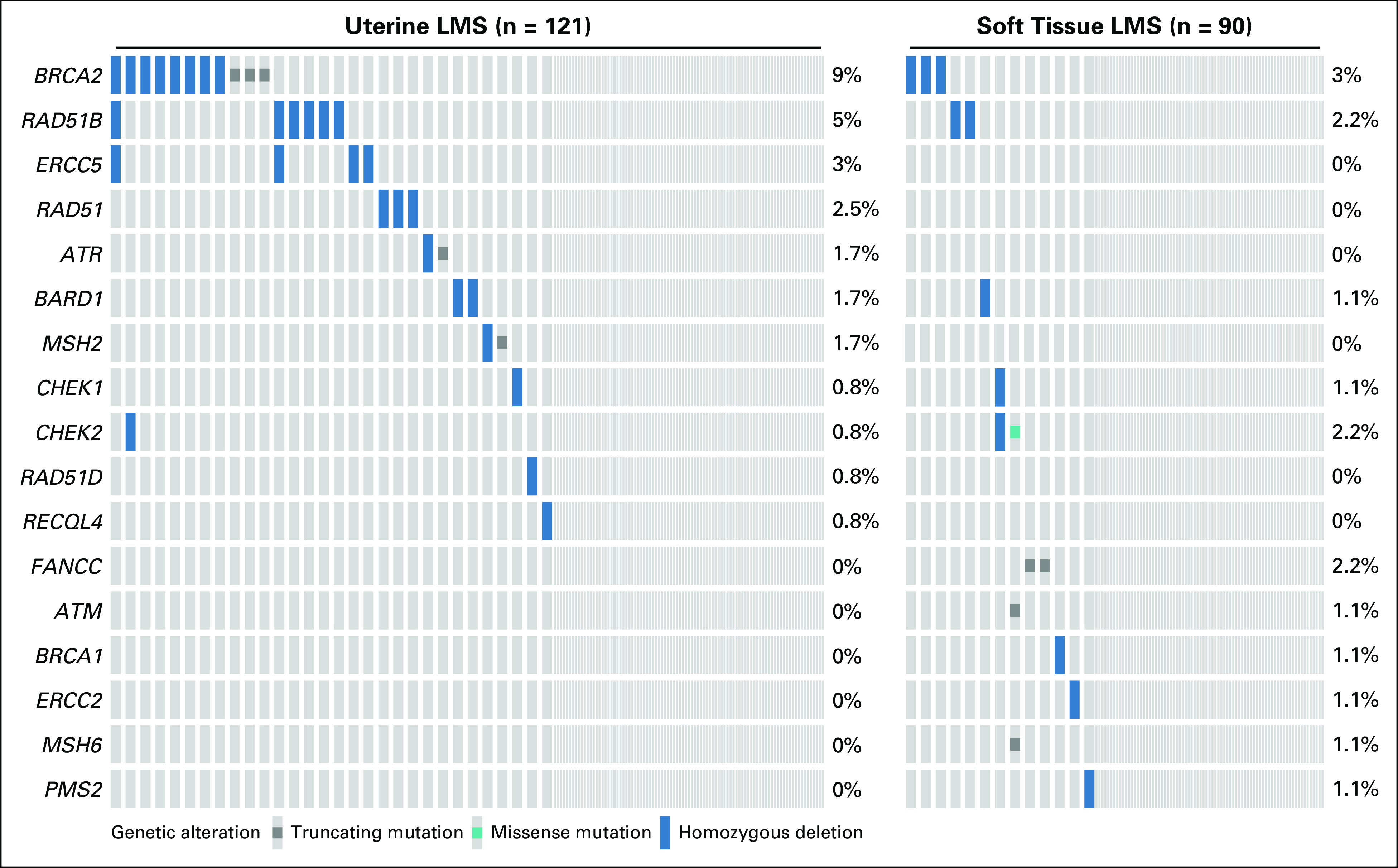
Altered DNA damage repair (DDR) genes across 211 patients with leiomyosarcoma (LMS) by subtype. DDR genes with no detectable oncogenic or likely oncogenic alterations in this study population are excluded from this figure.
Association Between Genomic Scarring and Homologous Recombination Pathway Gene Alterations
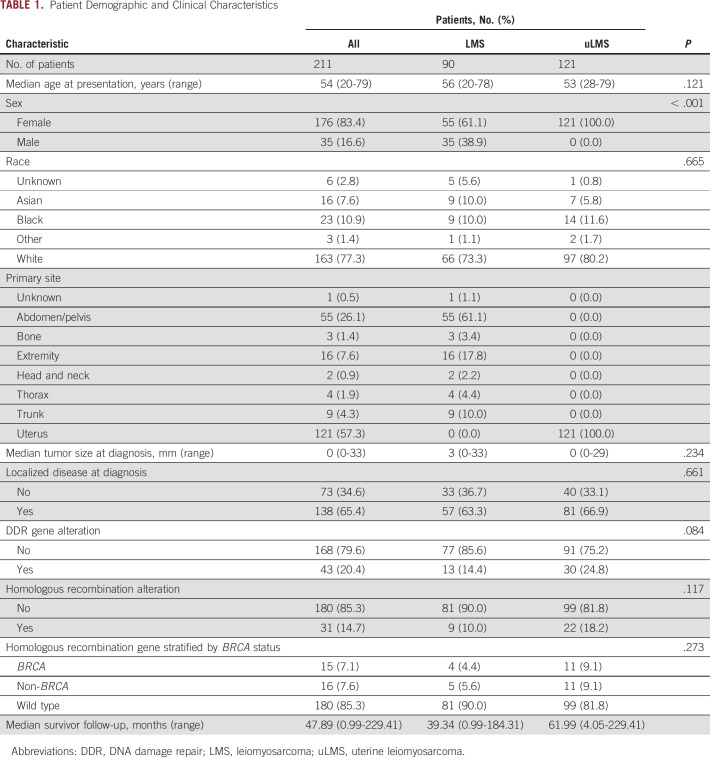
An HRD score was calculated from the patients for whom a copy number profile was available (n = 185; 88%). The median HRD score was 25 (interquartile range, 19-30). HRD score was associated with homologous recombination pathway gene loss of function (P = .004) but not with other DDR pathway gene alterations (P = .616; Fig 2A). HRD score did not significantly differ between patients with uLMS and stLMS (median, 26 and 24, respectively; P = .181) or between those who had IMPACT testing before or after treatment (median, 25 and 27, respectively; P = .130). Higher HRD score was correlated with the fraction genome altered, defined as the percentage of the genome affected by copy number gains or losses (Spearman’s r = 0.5; P ≤ .001). Tumor mutation burden and HRD score had a low correlation (Spearman’s r = 0.07; P = .391), likely reflecting the low overall mutation burden in LMS and the higher frequency of copy number alterations rather than mutations in DDR pathways (Figs 2B and 2C). Because HRD score calculated from TCGA data across cancer types is associated with altered TP53 status,28 we analyzed the association between TP53 alteration status in our cohort with the HRD score calculated from IMPACT. In total, 135 patients (64%) had a loss-of-function alteration in TP53, and these patients had a significantly higher median HRD score compared with patients with TP53 WT (median, 26 and 22.5, respectively; P = .009; Fig 2D).
FIG 2.
Homologous recombination deficiency (HRD) score calculated from targeted genome sequencing measures genomic scarring. (A) Median HRD score and interquartile range (whiskers) by homologous recombination and DNA damage repair (DDR) gene alteration status. Correlation between (B) HRD score and fraction genome altered (FGA) and (C) tumor mutation burden (TMB). (D) Median HRD score by TP53 alteration status. NS, not significant; WT, wild type.
DDR Gene Status, HRD Score, and Response to Chemotherapy in the Metastatic Setting
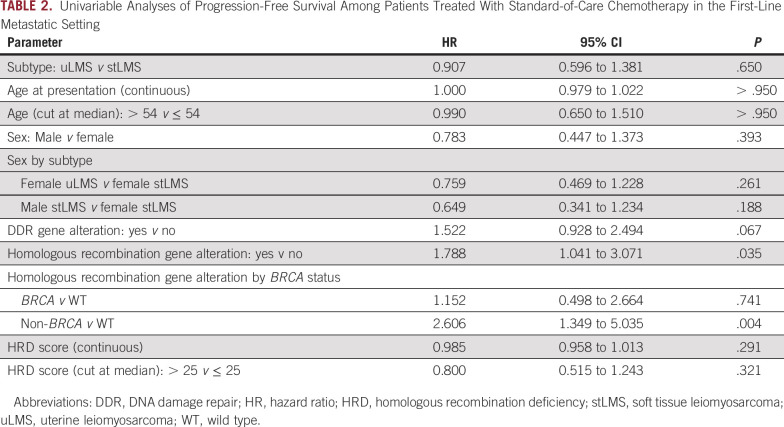
To determine whether somatic alterations in DDR genes predict response to cytotoxic chemotherapy, we analyzed the PFS of 117 patients with metastatic LMS who received standard-of-care first-line therapy. Twenty-seven patients (23%) received doxorubicin-based therapy, and 90 (77%) received gemcitabine-based treatment (Data Supplement). When stratified by the presence or absence of any DDR gene alteration, PFS did not significantly differ between groups (hazard ratio [HR], 1.52; 95% CI, 0.93 to 2.49; P = .096; Table 2). By contrast, the presence of a homologous recombination pathway alteration was associated with shorter PFS (HR, 1.79; 95% CI, 1.04 to 3.07; P = .035). The median PFS of patients with homologous recombination pathway alterations was 6.0 months (95% CI, 2.0 to 9.2 months) compared with 9.3 months (95% CI, 6.9 to 11.4 months) in patients with WT (P = .032; Fig 3A). HRD score, age, and sex did not associate with a significantly different PFS.
TABLE 2.
Univariable Analyses of Progression-Free Survival Among Patients Treated With Standard-of-Care Chemotherapy in the First-Line Metastatic Setting
FIG 3.
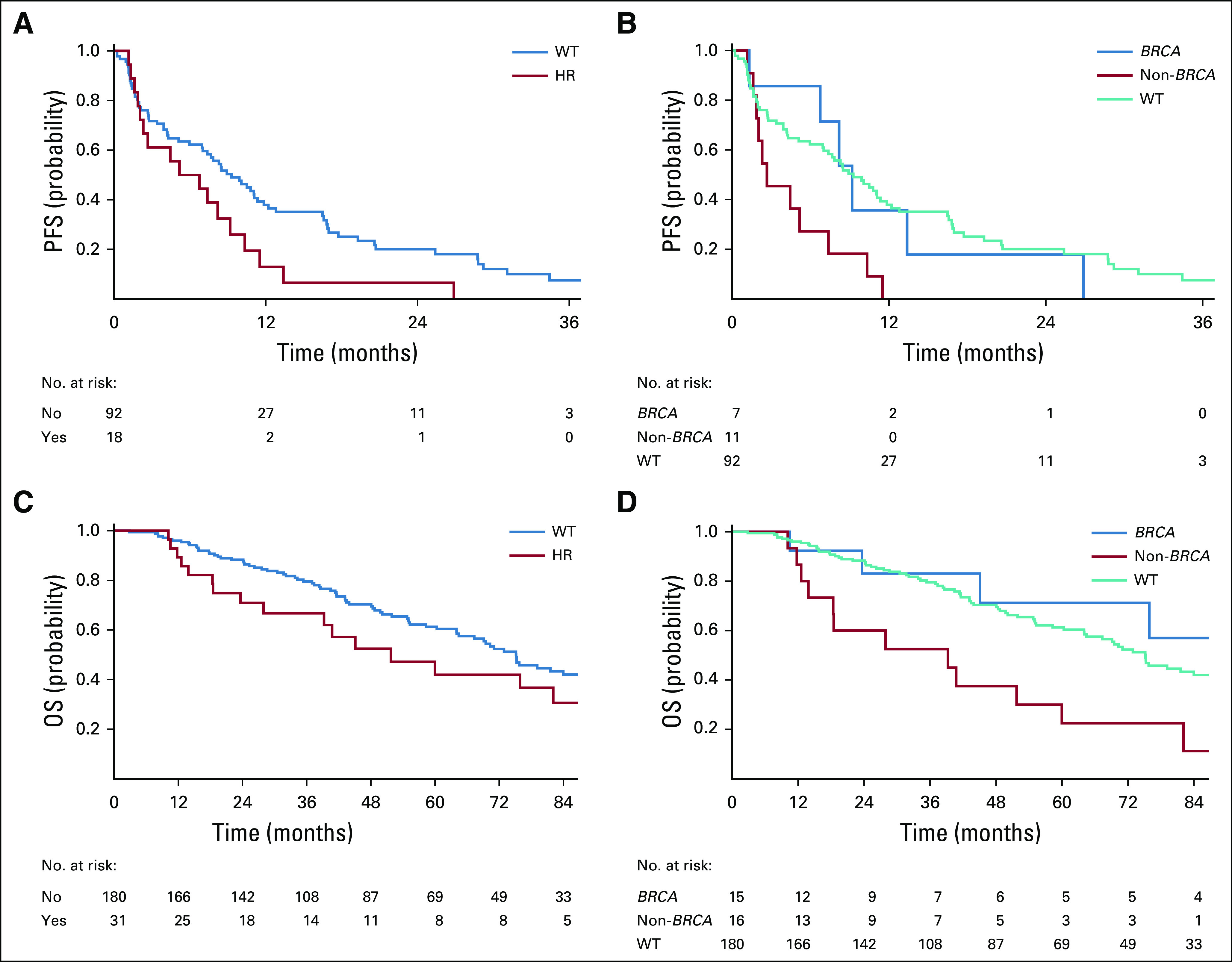
Clinical outcome after infusion of first-line chemotherapy. Kaplan-Meier curves for (A) progression-free survival (PFS) of patients with homologous recombination pathway alterations compared with those with wild type (WT). (B) PFS of patients with homologous recombination pathway alterations compared with those with WT stratified by BRCA status. (C) Overall survival (OS) of patients with homologous recombination pathway alterations compared with those with WT. (D) OS of patients with homologous recombination pathway alterations compared with those with WT stratified by BRCA status. HR, hazard ratio.
In a three-category comparison estimating PFS, homologous recombination pathway gene alteration status was stratified by BRCA and non-BRCA alterations and compared with patients with WT homologous recombination. Compared with WT, patients with non-BRCA homologous recombination pathway alterations had a significantly shorter PFS (HR, 2.61; 95% CI, 1.35 to 5.04; P = .004), while those with BRCA alterations did not (HR, 1.15; 95% CI, 0.50 to 2.66; P = .741). Median PFS was 9.2 months (95% CI, 1.3 to 26.9 months) in the BRCA-altered group, 2.7 months (95% CI, 1.6 to 7.4 months) in the non-BRCA homologous recombination pathway–altered group, and 9.3 months (95% CI, 6.9 to 11.4 months) in the WT group (P = .012; Fig 3B). These findings remained consistent when uLMS and stLMS were analyzed separately: Patients with non-BRCA homologous recombination pathway alterations had a significantly shorter PFS (Data Supplement). The same three-category comparison analyzing OS from the date of chemotherapy initiation again found that the non-BRCA homologous recombination pathway–altered group had a significantly shorter survival (Data Supplement).
DDR Gene Status, HRD Score, and OS From Diagnosis
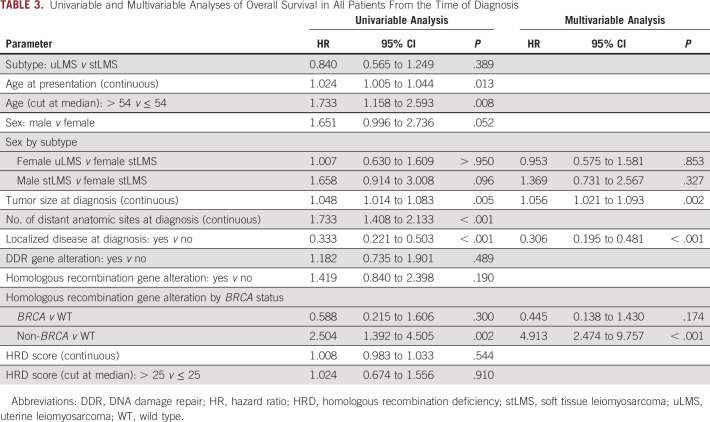
Among patients with localized disease at diagnosis (n = 138), median OS was 75.9 months (95% CI, 39.2 months to not reached) in the homologous recombination pathway–altered group and 91.4 months (95% CI, 72.3 to 158.1 months) in the WT group (P = .524). In those with advanced disease at diagnosis (n = 73), median OS was 16.9 months (95% CI, 10.1 to 45.1 months) in the homologous recombination pathway–altered group and 43.2 months (95% CI, 32.2 to 64.1 months) in the WT group (P = .013).
Median OS from the time of diagnosis in the entire cohort of 211 patients independent of treatment modality was 51.7 months (95% CI, 27.9 to 163.9 months) in the homologous recombination pathway–altered group and 75.2 months (95% CI, 64.1 to 89.9 months) in the WT group (P = .188). Upon further stratification by BRCA status, patients with non-BRCA homologous recombination pathway alterations had a median OS of 39.2 months (95% CI, 12.6 to 60.0 months), compared with not reached (95% CI, 23.6 months to not reached) and 75.2 months (95% CI, 64.1 to 89.9 months) in patients with BRCA alterations and WT, respectively (P = .003; Figs 3C and 3D).
On multivariable analysis, larger tumor size (HR, 1.06; 95% CI, 1.02 to 1.09; P = .002), presence of localized disease at diagnosis (HR, 0.31; 95% CI, 0.20 to 0.48; P < .001), and non-BRCA homologous recombination pathway–altered status (HR, 4.91; 95% CI, 2.47 to 9.76; P < .001) remained significantly associated with OS (Table 3). A detectable non-BRCA homologous recombination pathway gene alteration remained significantly associated with shorter survival when patients with uLMS and stLMS were analyzed separately (Data Supplement).
TABLE 3.
Univariable and Multivariable Analyses of Overall Survival in All Patients From the Time of Diagnosis
DISCUSSION
In a large cohort of patients with LMS who underwent targeted genomic sequencing, one fifth had at least one known or likely oncogenic alteration in a DDR gene, most commonly in the homologous recombination pathway. BRCA2 was the most frequently altered gene and was more commonly lost in uLMS. Patients with deleterious homologous recombination pathway gene alterations had a shorter PFS on frontline chemotherapy, which suggests that these tumors have a distinct underlying biology that results in aggressive behavior. Patients with deleterious alterations in a non-BRCA homologous recombination pathway gene had consistently poor clinical outcomes, irrespective of LMS subtype or treatment modality.
We hypothesized that patients with BRCA alterations would have improved clinical outcomes compared with those with BRCA WT. The survival of patients with BRCA loss was longer than those with non-BRCA homologous recombination alterations or WT, although the difference was not statistically significant. This may be because platinum chemotherapy is not generally used in LMS, and hence, BRCA status may not have affected outcomes to the current standardly used agents. Alternatively, the biology of BRCA-altered tumors in LMS may differ from other BRCA-associated cancers with altered BRCA, especially those that frequently harbor germline mutations in BRCA. Recently published data indicate that the phenotype of patients with BRCA loss is lineage dependent.29
Our data consistently demonstrated that patients with non-BRCA homologous recombination pathway gene alterations had a poor prognosis. Given the small number of patients with these alterations overall, these findings need to be replicated in a larger data set. Similar findings were reported in breast cancer, where BRCA gene alterations did not affect survival, but co-occurrence of a mutated DDR gene, such as RAD51B with BRCA1, increased resistance to chemotherapy as reflected in a worse relapse-free survival and OS.30 In ovarian cancer, detection of gene breakage in tumor suppressor genes, such as RAD51B, was associated with development of resistance to chemotherapy.31 Additional study is needed to better understand why alteration in these genes confers a worse prognosis.
In our analysis, patients with LMS and an homologous recombination pathway gene alteration had slightly higher HRD scores calculated from IMPACT samples compared with patients with WT. Use of an HRD score as a potential biomarker of response in LMS is of interest because it could increase the number of patients, above and beyond those with BRCA alterations, amenable to targeted treatment approaches. In support of using HRD score as a biomarker in this disease is the frequent loss of BRCA2, RAD51B, and TP53 in LMS, three genes that were found to significantly contribute to increased HRD scores across the pan-cancer landscape.17 Although there were indications that our HRD score successfully measured genomic scars, it did not have discriminatory capacity in terms of clinical outcome. Because targeted panels are based on sequencing of limited genomic regions, this score is likely less sensitive than the HRD score measured from larger sequencing panels. Use of validated measures of HRD that incorporate genomic data from single nucleotide polymorphisms across the whole genome32 may have more accuracy in identifying patients with LMS with HRD.
This study has several limitations, first among which is the retrospective nature of this work. Analyzing a time-dependent end point such as PFS outside the context of a clinical trial that uses objective criteria is challenging and depends on subjective determination of progression by the treating clinician. In addition, while the IMPACT panel is a prospective sequencing effort, many samples were obtained in the metastatic setting in patients who were pretreated with chemotherapy or radiation. These treatments may have introduced a selective pressure among tumors to develop alterations in DDR pathways. Treatment history notwithstanding, an alteration in a homologous recombination pathway gene and an elevated HRD score are inherently limited measures that may not reflect in vivo function of the homologous recombination pathway.33 In addition, many patients in this cohort did not provide consent for germline genetic testing, and thus, their germline DDR or homologous recombination pathway gene alteration status were unknown at the time of this analysis.
There has only been one sarcoma-specific clinical trial of PARP inhibition reported to date: the phase Ib TOMAS trial of trabectedin plus olaparib in patients with advanced sarcomas.34 This trial enrolled 15 patients with LMS, both uterine and nonuterine, five of whom had prolonged clinical benefit of > 6 months with the treatment combination. It is challenging to interpret the added benefit of olaparib above and beyond that of trabectedin, which is a known active agent in LMS.35 Whether responses between those with uterine or nonuterine disease differed was not reported.
Future studies are needed to confirm our finding that homologous recombination pathway gene alterations confer a worse prognosis, particularly non-BRCA alterations, and to determine the predictive potential of homologous recombination pathway gene alterations in LMS. Additional work to incorporate germline genomic sequencing efforts are warranted, as are studies to explore the molecular mechanism behind those who have better or worse clinical outcomes. Prospective studies of targeted agents like PARP inhibitors to exploit homologous recombination pathway deficiencies are needed to ultimately determine whether the homologous recombination pathway can serve as a biomarker predictive of response.
ACKNOWLEDGMENT
We thank the patients and their families for participating in this clinical research.
EQUAL CONTRIBUTION
E.R. and P.J. contributed equally to this work.
PRIOR PRESENTATION
Presented at the American Society of Clinical Oncology 2019 Annual Meeting, Chicago, IL, May 31-June 4, 2019.
SUPPORT
Supported by grant funds from the Sarcoma Foundation of America (S.P.D. and W.D.T.). The Memorial Sloan Kettering Cancer Center is supported in part by National Cancer Institute core grant P30 CA008748.
AUTHOR CONTRIBUTIONS
Conception and design: Evan Rosenbaum, Philip Jonsson, Kenneth Seier, Li-Xuan Qin, Martee L. Hensley, Sandra P. D’Angelo, William D. Tap
Provision of study material or patients: All authors
Collection and assembly of data: Evan Rosenbaum, Philip Jonsson, Ping Chi, Mark Dickson, Mary L. Keohan, Samuel Singer, Marc Ladanyi, Cristina R. Antonescu, Sandra P. D’Angelo, William D. Tap
Data analysis and interpretation: Evan Rosenbaum, Philip Jonsson, Kenneth Seier, Li-Xuan Qin, Ping Chi, Mark Dickson, Mrinal Gounder, Ciara Kelly, Mary L. Keohan, Benjamin Nacev, Mark T. A. Donoghue, Sarah Chiang, Marc Ladanyi, Cristina R. Antonescu, Sujana Movva, Sandra P. D’Angelo, William D. Tap
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Li-Xuan Qin
Employment: MedImmune (I), VielaBio (I)
Leadership: VielaBio (I)
Stock and Other Ownership Interests: VielaBio (I)
Ping Chi
Consulting or Advisory Role: Deciphera, Exelixis, Merck (I)
Research Funding: Deciphera (Inst), Array BioPharma (Inst)
Mark Dickson
Consulting or Advisory Role: Celgene
Research Funding: Eli Lilly (Inst), AADi (Inst)
Mrinal Gounder
Honoraria: Bayer AG, Flatiron Health, PER, Medscape, SpringWorks Therapeutics, Guidepoint Global
Consulting or Advisory Role: Daiichi Sankyo, Karyopharm Therapeutics, Epizyme, Bayer AG, SpringWorks Therapeutics, Boehringer Ingelheim
Speakers’ Bureau: Amgen
Patents, Royalties, Other Intellectual Property: UpToDate
Travel, Accommodations, Expenses: Epizyme
Other Relationship: Desmoid Tumor Research Foundation
Uncompensated Relationships: Foundation Medicine, Rain Therapeutics, Boehringer Ingelheim, Athenex
Open Payments Link: https://openpaymentsdata.cms.gov/physician/459583
Ciara Kelly
Research Funding: AGIOS (Inst), Amgen (Inst), Merck (Inst), Incyte (Inst), Kartos (Inst), Exicure (Inst)
Benjamin Nacev
Uncompensated Relationships: Delfi, Rapafusyn
Marc Ladanyi
Consulting or Advisory Role: Bristol Myers Squibb, Bayer AG
Research Funding: Loxo (Inst), Helsinn Therapeutics, Merus NV (Inst), Elevation Oncology (Inst)
Martee L. Hensley
Employment: Sanofi (I), Sanofi (I)
Consulting or Advisory Role: Eli Lilly, Janssen Pharmaceuticals, Tesaro, Research to Practice, GOG Foundation, Merck, GlaxoSmithKline
Research Funding: Bristol Myers Squibb (Inst)
Patents, Royalties, Other Intellectual Property: Up to Date
Travel, Accommodations, Expenses: Eli Lilly
Sujana Movva
Consulting or Advisory Role: Genmab
Research Funding: Novartis (Inst), Takeda Pharmaceuticals (Inst)
Sandra P. D’Angelo
Consulting or Advisory Role: EMD Serono, Amgen, Nektar, Immune Design, GlaxoSmithKline, Incyte, Merck, Adaptimmune, Immunocore
Research Funding: EMD Serono (Inst), Amgen (Inst), Merck (Inst), Incyte (Inst), Nektar (Inst), Bristol Myers Squibb (Inst), Deciphera (Inst)
Travel, Accommodations, Expenses: Adaptimmune, EMD Serono, Nektar
William D. Tap
Leadership: Certis Oncology Solutions, Atropos Pharmaceuticals
Stock and Other Ownership Interests: Certis Oncology Solutions, Atropos
Consulting or Advisory Role: EMD Serono, Eli Lilly, Daiichi Sankyo, Eisai, Blueprint Medicines, Agios, GlaxoSmithKline, NanoCarrier, Deciphera
Research Funding: Novartis, Eli Lilly, Plexxikon, Daiichi Sankyo, TRACON Pharma, Blueprint Medicines, Immune Design, BioAtla, Deciphera
Patents, Royalties, Other Intellectual Property: Companion diagnostic for CDK4 inhibitors-14/854,329
No other potential conflicts of interest were reported.
REFERENCES
- 1.Hernando E, Charytonowicz E, Dudas ME, et al. The AKT-mTOR pathway plays a critical role in the development of leiomyosarcomas. Nat Med. 2007;13:748–753. doi: 10.1038/nm1560. [DOI] [PubMed] [Google Scholar]
- 2.George S, Serrano C, Hensley ML, et al. Soft tissue and uterine leiomyosarcoma. J Clin Oncol. 2018;36:144–150. doi: 10.1200/JCO.2017.75.9845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gladdy RA, Qin LX, Moraco N, et al. Predictors of survival and recurrence in primary leiomyosarcoma. Ann Surg Oncol. 2013;20:1851–1857. doi: 10.1245/s10434-013-2876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoushtari AN, Landa J, Kuk D, et al. Overall survival and response to systemic therapy in metastatic extrauterine leiomyosarcoma. Sarcoma. 2016;2016:3547497. doi: 10.1155/2016/3547497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Movva S, Wen W, Chen W, et al. Multi-platform profiling of over 2000 sarcomas: Identification of biomarkers and novel therapeutic targets. Oncotarget. 2015;6:12234–12247. doi: 10.18632/oncotarget.3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuppens T, Moisse M, Depreeuw J, et al. Integrated genome analysis of uterine leiomyosarcoma to identify novel driver genes and targetable pathways. Int J Cancer. 2018;142:1230–1243. doi: 10.1002/ijc.31129. [DOI] [PubMed] [Google Scholar]
- 7.Chudasama P, Mughal SS, Sanders MA, et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat Commun. 2018;9:144. doi: 10.1038/s41467-017-02602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. doi: 10.1634/theoncologist.2018-0448. Seligson ND, Kautto EA, Passen EN, et al: BRCA1/2 functional loss defines a targetable subset in leiomyosarcoma. Oncologist 24:973-979, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 10.Golan T, Hammel P, Reni M, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirza MR, Monk BJ, Herrstedt J, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 12.Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Telli ML, Timms KM, Reid J, et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birkbak NJ, Wang ZC, Kim JY, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2:366–375. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popova T, Manié E, Rieunier G, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72:5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 16.Abkevich V, Timms KM, Hennessy BT, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knijnenburg TA, Wang L, Zimmermann MT, et al: Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep 23:239-254.e6, 2018. [DOI] [PMC free article] [PubMed]
- 18.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teo MY, Bambury RM, Zabor EC, et al. DNA damage response and repair gene alterations are associated with improved survival in patients with platinum-treated advanced urothelial carcinoma. Clin Cancer Res. 2017;23:3610–3618. doi: 10.1158/1078-0432.CCR-16-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sehdev A, Gbolahan O, Hancock BA, et al. Germline and somatic DNA damage repair gene mutations and overall survival in metastatic pancreatic adenocarcinoma patients treated with FOLFIRINOX. Clin Cancer Res. 2018;24:6204–6211. doi: 10.1158/1078-0432.CCR-18-1472. [DOI] [PubMed] [Google Scholar]
- 21.Hyman DM, Zhou Q, Iasonos A, et al. Improved survival for BRCA2-associated serous ovarian cancer compared with both BRCA-negative and BRCA1-associated serous ovarian cancer. Cancer. 2012;118:3703–3709. doi: 10.1002/cncr.26655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang D, Khan S, Sun Y, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306:1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sztupinszki Z, Diossy M, Krzystanek M, et al. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer. 2018;4:16. doi: 10.1038/s41523-018-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. doi: 10.1038/nm.4333. Zehir A, Benayed R, Shah RH, et al: Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23:703-713, 2017 [Erratum: Nat Med 23:1004, 2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chakravarty D, Gao J, Phillips S, et al: OncoKB: A precision oncology knowledge base. JCO Precis Oncol doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed]
- 27.Shen R, Seshan VE. FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marquard AM, Eklund AC, Joshi T, et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark Res. 2015;3:9. doi: 10.1186/s40364-015-0033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. doi: 10.1038/s41586-019-1382-1. Jonsson P, Bandlamudi C, Cheng ML, et al: Tumour lineage shapes BRCA-mediated phenotypes. Nature 571:576-579, 2019 [Erratum: Nature 577:E1, 2020] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takada M, Nagai S, Haruta M, et al. BRCA1 alterations with additional defects in DNA damage response genes may confer chemoresistance to BRCA-like breast cancers treated with neoadjuvant chemotherapy. Genes Chromosomes Cancer. 2017;56:405–420. doi: 10.1002/gcc.22445. [DOI] [PubMed] [Google Scholar]
- 31. doi: 10.1038/nature14410. Patch AM, Christie EL, Etemadmoghadam D, et al: Whole-genome characterization of chemoresistant ovarian cancer. Nature 521:489-494, 2015 [Erratum: Nature 527:398, 2015] [DOI] [PubMed] [Google Scholar]
- 32.Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:1949–1961. doi: 10.1016/S0140-6736(17)32440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. doi: 10.1158/1078-0432.CCR-18-0063. Meijer TG, Verkaik NS, Sieuwerts AM, et al: Functional ex vivo assay reveals homologous recombination deficiency in breast cancer beyond BRCA gene defects. Clin Cancer Res 24:6277-6287, 2018 [Erratum: Clin Cancer Res 25:2935, 2019] [DOI] [PubMed] [Google Scholar]
- 34.Grignani G, D’Ambrosio L, Pignochino Y, et al. Trabectedin and olaparib in patients with advanced and non-resectable bone and soft-tissue sarcomas (TOMAS): An open-label, phase 1b study from the Italian Sarcoma Group. Lancet Oncol. 2018;19:1360–1371. doi: 10.1016/S1470-2045(18)30438-8. [DOI] [PubMed] [Google Scholar]
- 35.Demetri GD, von Mehren M, Jones RL, et al. Efficacy and safety of trabectedin or dacarbazine for metastatic liposarcoma or leiomyosarcoma after failure of conventional chemotherapy: Results of a phase III randomized multicenter clinical trial. J Clin Oncol. 2016;34:786–793. doi: 10.1200/JCO.2015.62.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]