Abstract
SARS-CoV-2, the virus causing the current COVID-19 pandemic, primarily targets the airway epithelium and in lungs can lead to acute respiratory distress syndrome. Clinical studies in recent months have revealed that COVID-19 is a multi-organ disease causing characteristic complications. Stem cell models of various organ systems—most prominently, lung, gut, heart, and brain—are at the forefront of studies aimed at understanding the role of direct infection in COVID-19 multi-organ dysfunction.
Simoneau and Ott discuss various stem-cell-derived models used to elucidate susceptibility of different organ systems to SARS-CoV-2, exploring their permissiveness to viral infection, consequent innate immune responses, and the disruption of essential cellular functions.
Introduction
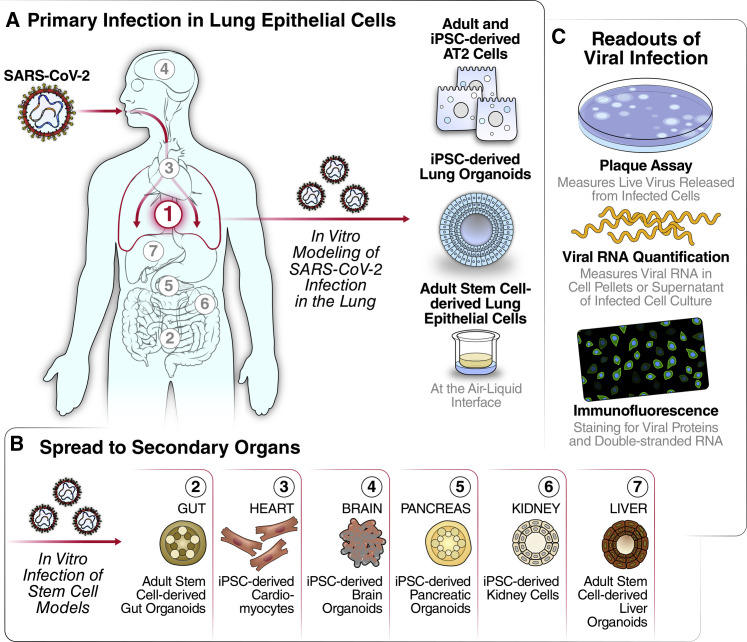
SARS-CoV-2 is a single-stranded RNA virus in the Coronaviridae family, which includes the highly pathogenic SARS-CoV and MERS-CoV viruses, as well as the circulating human coronaviruses OC43, NL63, HKU1, and 229E that cause mild seasonal respiratory illness (Fung and Liu, 2019). SARS-CoV-2 emerged in human populations in December 2019 as the causative agent of the current COVID-19 pandemic. COVID-19 primarily manifests as a respiratory illness due to upper and lower airway infection, viral pneumonia, and, in severe cases, acute respiratory distress syndrome (ARDS). Along with the primary lung and upper airway infections and respiratory symptoms, COVID-19 patients can experience neurological, cardiac, pancreatic, digestive, and renal symptoms, and SARS-CoV-2 RNA has been detected in various organ systems (Figures 1A and 1B) (Wiersinga et al., 2020; Puelles et al., 2020). Respiratory symptoms, as well as sequelae in secondary organs, reportedly last long after the virus has been cleared and can cause significant impairment in the quality of life for COVID-19 survivors (Marshall, 2020). Therefore, it is important to understand how SARS-CoV-2 affects lung epithelia and other organs. Specifically, whether the symptoms observed in the secondary organs reflect a direct infection by SARS-CoV-2 or systemic immune overactivation remains an open question. To investigate the possibility that SARS-CoV-2 infects multiple organs, many groups have turned to stem-cell-derived culture systems. The purpose of this review is to describe the contribution of these model systems to our understanding of SARS-CoV-2 pathogenesis (Figures 1A and 1B).
Figure 1.
Stem-Cell-Derived Models for Studying SARS-CoV-2 Complications in Multiple Organ Systems
(A) SARS-CoV-2 enters through the mouth and nasal cavity and spreads down through the upper airway into the alveoli. Lung infection of SARS-CoV-2 has been modeled through iPSC-derived lung organoids, stem-cell-derived AT2 cells, and adult stem-cell-derived upper airway cells.
(B) Direct infection of SARS-CoV-2 may occur in other organs during COVID-19 disease. Gut, heart, brain, pancreatic, kidney, and liver infection have been modeled through stem-cell-derived models.
(C) Understanding of viral replication dynamics in in vitro models is quantifiable through plaque assay, viral RNA quantification, and immunofluorescence
Cancer cell lines have elucidated many aspects of virology but often have a dysregulated proteome and an impaired immune response, are homogeneous, and lack polarity (Ramani et al., 2018). In contrast, differentiated stem cell models, especially cultured as 3D organoids, maintain polarity, have functional immune signaling, and generally include multiple cell types. In 2D or 3D configurations, they have emerged as excellent models for studying viral pathogenesis (Bar-Ephraim et al., 2020; Ramani et al., 2018). They can be derived from induced pluripotent stem cells (iPSCs), embryonic stem cells, or adult organ-derived progenitor cells and model a variety of organ systems including upper airway cells, alveolar type II cells, enterocytes, colonoids, cardiomyocytes, brain organoids, pancreatic organoids, kidney cells, and liver cells (Yang et al., 2020; Sachs et al., 2019; Ramani et al., 2018). In their undifferentiated “stem-like” state, these models can be maintained for many passages in a laboratory, until differentiated into non-proliferating parenchymal cells. The adult stem-cell-derived organoids in both the proliferating state, such as the alveolar type II (AT2) and basal cells in the lung, and the differentiated state can be used to elucidate essential biology of viral infection. Here, we will discuss various stem-cell-derived models used to elucidate susceptibility of different organ systems to SARS-CoV-2, exploring their permissiveness to viral infection, consequent innate immune responses, and the disruption of essential cellular functions (Table 1 ).
Table 1.
Summary of In Vitro Stem Cell Models for SARS-CoV-2 in Various Organ Systems
| Cell Types | Stem Cell Model | Culture Method | SARS-CoV-2 Susceptibility | Cell-Type-Specific Effects | Immune Response | Relevant Citations |
|---|---|---|---|---|---|---|
| Lung | ||||||
| Alveolar type 2 cells | Adult stem cells | 2D | Yes | Decreased expression of surfactant protein, cell death | Type I response in both studies with an MOI of 1 and ISG stimulation | Youk et al., 2020; Katsura et al., 2020; Tindle et al., 2020 |
| Alveolar type 2 cells | iPSC derived | 3D | Yes | Decreased expression of surfactant proteins, cell death | Delayed type I interferon response with an MOI of 5 and ISG stimulation | Huang et al., 2020 |
| Airway cells | Adult stem cells | Pseudo-stratified layer | Yes | Cell death | Increase in interferon genes 72 h post infection with an MOI of 0.1 | Lamers et al., 2020; Tindle et al., 2020; Purkayastha et al., 2020 |
| Airway cells | iPSC derived | 3D | Yes | Not discussed | Not discussed | Samuel et al., 2020; Duan et al., 2020 |
| Gut | ||||||
| Ileal organoid | Adult stem cells | 2D/3D | Yes | Cell death observed | Types I and III IFN responses after 24 h with an MOI of 0.1 | Zang et al., 2020; Lamers et al., 2020 |
| Colonoids | iPSC derived | 3D | Yes | Not discussed | TNF and IL-17 signatures reported after 24 h with an MOI of 0.1 | Han et al., 2020 |
| Heart | ||||||
| Cardiomyocytes | iPSC derived | 2D | Yes | Impairment of contraction, fragmentation of troponin T, cell death | Type I interferon response and ISG stimulation at MOI 0.01 | Pérez-Bermejo et al., 2020, Samuel et al., 2020; Bojkova et al., 2020; Sharma et al., 2020; Yang et al., 2020 |
| Brain | ||||||
| Neurons | iPSC derived | 2D/3D | Conflicting evidence | Abnormally localized Tau protein, cell death | Not discussed | Ramani et al., 2020; Zhang et al., 2020; Jacob et al., 2020; Pellegrini et al., 2020 |
| Astrocytes | iPSC derived | 2D | Low | N/A | N/A | Jacob et al., 2020 |
| Microglia | iPSC derived | 2D | Low | N/A | N/A | Yang et al., 2020; Jacob et al., 2020 |
| Choroid plexus | iPSC derived | 3D | Yes | Disruption of ion transport and cell adhesion pathways | Inflammatory cytokines induced at MOI ~0.1 after 24 h | Pellegrini et al., 2020; Jacob et al., 2020 |
| Pancreas | ||||||
| Endocrine cells | iPSC derived | 3D | Yes | Upregulation of insulin resistance and cell death pathways | Chemokine induction at 24 h after MOI 0.01 | Yang et al., 2020 |
| Kidney | ||||||
| Capillary organoids | iPSC derived | 3D | Yes | Not discussed | Not discussed | Monteil et al., 2020 |
| Liver | ||||||
| Hepatocytes | Adult stem cells | 3D | Yes | Metabolism pathways downregulated | Chemokine, IL-17, TNF and NFkB signaling at MOI 0.1 at 24 h | Yang et al., 2020 |
| Cholangiocytes | Adult stem cells | 3D | Yes | Not discussed | Chemokine and IL-17 signaling pathway activated at MOI 0.1 at 24 h | Yang et al., 2020 |
SARS-CoV-2 Virology 101
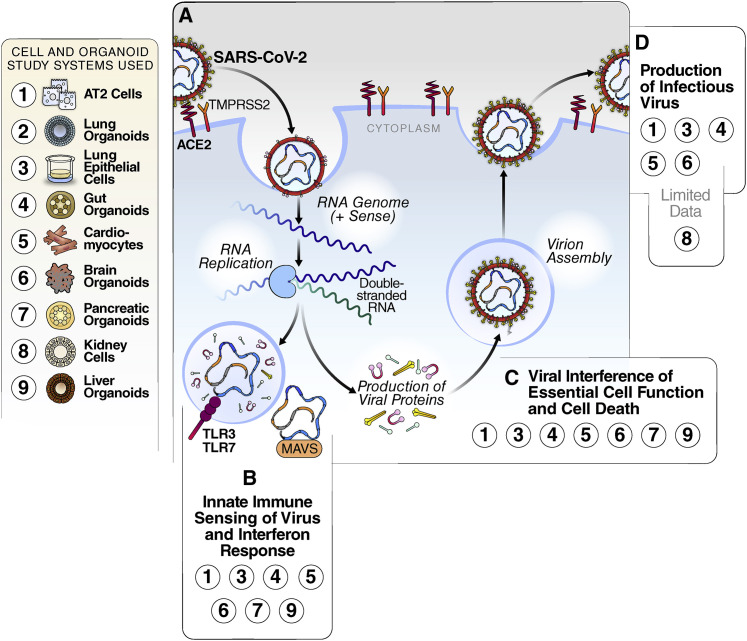
The SARS-CoV-2 life cycle is intricately connected with its host cell. The single-stranded RNA genome of SARS-CoV-2 is approximately 30 kb in length and encodes 16 nonstructural proteins and four structural proteins. Entry into the cell occurs when the viral Spike (S) protein on the surface of the virion binds to angiotensin-converting enzyme 2 (ACE2) on the apical exterior of the target cell (Shang et al., 2020) (Figure 2 A). Studies that focus specifically on the entry of the virus into cells can use pseudotyped viruses, which only reproduce the entry and fusion steps of SARS-CoV-2 but do not reproduce the full viral life cycle. Pseudotyped viruses are generally built of lentiviral or vesiculo-stomatitis virus-based vectors carrying the SARS-CoV-2 S protein (Crawford et al., 2020). After processing of the S protein by host proteases—most prominently, transmembrane serine protease 2 (TMPRSS2)—and successful uptake and membrane fusion, the viral genome is released into the host cell and translated by the host ribosomes. This produces a large polyprotein of nonstructural proteins, prominently the viral polymerase and two viral protease complexes that allow viral RNA replication to begin. Replication occurs via a labile negative RNA step transiently creating a double-stranded RNA (dsRNA) intermediate (Figure 2A) (de Wilde et al., 2018). The structural proteins, including spike and the viral nuclear capsid (N) protein, as well as several laterally acquired ORFs encoded at the 3′ end of the viral RNA genome, are transcribed and translated separately via subgenomic RNA transcription and translation (Fehr and Perlman, 2015).
Figure 2.
SARS-CoV-2 Replication and Interaction with Host Cells
(A) Simplified schematic of the viral replication cycle. The virus enters the cell through binding of the ACE2 receptor and is internalized. Inside of the cell, viral RNA is produced, and structural and non-structural proteins are translated. New virions are assembled and released from the cell, resulting in cell lysis.
(B) Interferon pathway activation by SARS-CoV-2. Innate immune sensors recognize the virus, and transcription of interferon is turned on, leading to the production of interferon-stimulated genes. Stem-cell-derived models in which this has been detected are highlighted in the box.
(C) Disruption of cellular processes by SARS-CoV-2 and cell death. Through the hijacking of host cell proteins and pathways, SARS-CoV-2 can disrupt the essential functions of cells. Stem-cell-derived models in which this has been detected are highlighted in the box.
(D) Viral production. When cells are productively infected, SARS-CoV-2 virions are released at the end of the viral life cycle, causing cell lysis and death. Stem-cell-derived models in which this has been detected are highlighted in the box.
The SARS-CoV-2 dsRNA, as well as other pathogen-associated molecular patterns (PAMPs) produced by the virus, can be recognized by host immune sensors such as the RIG-I receptors in the cytoplasm or Toll-like receptors in the endosome (Lei et al., 2020). Immune response pathways are activated in the infected cells, stimulating an innate antiviral immune response indicated by interferon production and consequent activation of interferon-stimulated genes (ISGs) (Busnadiego et al., 2020). In parallel, structural and nonstructural viral proteins interact with numerous host proteins to support viral replication, interfere with host pathways, and diminish antiviral immune responses (Gordon et al., 2020; Lei et al., 2020). SARS-CoV-2 viral replication causes apoptosis and cytopathic effects (CPEs), releasing virions into the surrounding environment (de Wilde et al., 2018). While the basic principles of SARS-CoV-2 viral replication are likely similar between different organ systems, cell-type-specific differences in host factors supporting viral replication may exist, underscoring the need for a broad spectrum of cell systems to study the pathogenesis of SARS-CoV-2.
Quantification of SARS-CoV-2 Viral Infection: A How-To Guide
The viral life cycle offers multiple parameters for quantification of viral infection. Essential for reproducible in vitro infection of cell culture models is knowing how many infectious viral particles were added per cell during an experimental infection, a ratio known as multiplicity of infection (MOI). Using a low MOI (<1) allows the infection to spread in culture if infectious viral particles can be produced in the host cell and mimics the natural course of SARS-CoV-2 infection. MOIs can be increased (>1) if the cell type is minimally permissive to infection or if a high infection rate is desired for molecular studies. Notably, in organoid cultures, MOIs are difficult to standardize due to the variability in cell numbers per organoid, and MOIs are sometimes—erroneously—reported as a ratio of virions to organoids or simply as a number of virions added. One caveat is that viral titers and MOIs are usually calculated based on results in highly susceptible Vero cells, a long-cultured African green monkey kidney cell line that is highly susceptible to viral infections and may not accurately reflect the infectivity of a virus in primary human cells.
Quantification of SARS-CoV-2 viral loads in infected cultures through a so-called plaque assay is the gold standard (Figure 1C). This method quantifies the amount of infectious viral particles produced into the supernatant of a culture system by measuring the CPE the particles exert in a secondary culture. Briefly, supernatants of infected cells are serially diluted and transferred onto highly susceptible cells overlaid with a layer of agar to concentrate the virus to the areas of initial infection. Infectious virus is quantified as plaque-forming units (PFUs), which represent the number of areas where infected cells have died and left a macroscopic blank spot—a plaque—on a plate.
The number of SARS-CoV-2-infected cells in a culture can also be determined by immunofluorescence microscopy using antibodies against the structural proteins, usually N or S, or against dsRNA intermediates. Human cells do not harbor dsRNA, and antibodies against it have been widely used to identify cells infected with positive stranded RNA viruses (Figure 1C) (Weber et al., 2006). While N and S antibodies determine the percentage of cells harboring virus, antibodies against dsRNA further reveal whether cells are actively replicating viral RNA as these antibodies detect the duplex formed of positive and negative RNA strands during active RNA replication. By incorporating viral stains with co-stains, information about the cells’ viability and the subcellular localization of viral proteins or RNA replication centers can simultaneously be assessed with the number of infected cells. Finally, viral RNA counts can also be assessed by quantitative RT-PCR in cells or in the cell supernatant (Figure 1C). While this is the most sensitive method to detect viral production, in SARS-CoV-2 infection, it also captures non-infectious viral RNA species, including subgenomic RNAs, that are produced in the infected cells but can be released into the supernatant after CPE. In combination, the readouts of viral titer, immunofluorescence and RT-qPCR fully capture the infectious state of a target cell in different cell systems.
Upper and Lower Airway: The Primary Target
Expression of the ACE2 protein in the respiratory tract is highest in the nasal cavity and decreases through the upper airway and into the alveoli, where it is described as lowly or variably expressed between different donors. (Hou et al., 2020; Ortiz et al., 2020). Whether ACE2 mRNA and protein levels correlate is not clear, but low mRNA levels have also been measured in the alveoli (Ziegler et al., 2020; Ortiz et al., 2020). More definitive data at the protein level are needed to quantify ACE2 expression in the respiratory tract. Consistent with the ACE2 expression pattern, SARS-CoV-2 virus is found in few airway cells upon autopsy, raising the possibility that (1) only a small percentage of lung epithelial cells are susceptible to infection or (2) infection is more widespread in the early days of infection but has been cleared from most parts of the lung by the time of autopsy (Schaefer et al., 2020). Evidence points to ciliated cells as the main target of the virus in the upper airway and to AT2 cells as the target in the alveoli (Hou et al., 2020; Schaefer et al., 2020). While the virus replicates actively in the nasal epithelium and the upper airway, lung pathology occurs when the virus reaches the alveoli and has the potential to evolve into ARDS.
Both iPSC and adult stem cell models of upper airways in organoid form have been previously established and characterized (Huang et al., 2014; Sachs et al., 2019). The adult stem cell organoid model was successfully infected after differentiation at the air-liquid interface (ALI), a specific procedure where cells are plated on a thin layer of Matrigel in a transwell system with the basolateral surface of the cells in contact with differentiation media while the apical side is exposed to air (Lamers et al., 2020). At an MOI of 0.1, the differentiated cells produce infectious virus, quantified through plaque assay as early as 24 h after infection (Lamers et al., 2020) (Figure 2D). Similar results were obtained when basal cells isolated from donors were directly differentiated at the ALI, with virus detected 2 days post infection. At an MOI of 0.1, less than 5% of cells were positive for SARS-CoV-2 72 h post infection, and single-cell RNA sequencing revealed upregulation of interferon-related genes (Purkayastha et al., 2020). Notably, this model was also used to investigate the effect of cigarette smoke on SARS-CoV-2 infection, showing that smoke increased the number of SARS-CoV-2-positive cells (Purkayastha et al., 2020). iPSC-derived models of airway cells have also successfully been infected with SARS-CoV-2 and used to model drug treatment, although the dynamics of infection in this model must be explored further (Samuel et al., 2020).
Although infection of AT2 cells in the lung is recognized as the primary driver of ARDS, in vitro models of AT2 cells have multiple challenges, including rapid dedifferentiation and the need for feeder cells such as fibroblasts. The SARS-CoV-2 pandemic and central role of AT2 cells in severe disease has spurred development of new culture systems for AT2 cells, through either successful differentiation of iPSCs or maintenance of primary AT2 cells, aligning research in stem cell biology and SARS-CoV-2 infection. Three groups have successfully infected these new models with SARS-CoV-2, two using novel culture systems for primary AT2 cells and one using the iPSC model (Katsura et al., 2020; Huang et al., 2020; Youk et al., 2020). While primary AT2 cell cultures were successfully trans-differentiated into AT1 cells, the role of SARS-CoV-2 infection in the trans-differentiated cells has not been explored (Youk et al., 2020; Katsura et al., 2020). AT2 cells, grown in 2D or 3D culture conditions, were characterized by surfactant protein-C (SFTPC) expression, a protein exclusively produced in AT2 cells. ACE2 and TMPRSS2 localized to the apical surface in these models, consistent with entry of the virus from the airways in vivo. MOIs varied in these studies, but a high MOI (>1) was necessary to achieve infection of more than 20% of cells early after infection, pointing to relatively low permissiveness of these cells, at least in culture (Huang et al., 2020).
Bulk RNA sequencing in all three studies demonstrated the ability of AT2 cells to produce type I interferon and upregulate ISGs upon infection (Youk et al., 2020; Katsura et al., 2020; Huang et al., 2020). While all studies reported an upregulation of ISGs by at least 96 h of infection, the kinetics of the immune response varied. In the iPSC-derived model, the immune response did not appear until 4 days post infection and has up to 33% of reads mapped to the viral genome as early as 1 day post infection (Huang et al., 2020). In contrast, in the primary AT2 model, interferon response was upregulated at 48 h post infection and only 4.7% of transcripts mapped to the SARS-CoV-2 genome (Katsura et al., 2020). Whether this difference is due to viral load or differences in the cell culture system remains an open question. Single-cell RNA sequencing revealed two distinct patterns of interferon response: cells expressing high levels of ISGs and cells with low ISG expression but upregulated apoptotic pathways (Youk et al., 2020). However, correlation with viral load was not discussed. Genes related to cell death, cell adhesion, and surfactant proteins were also upregulated during infection in the AT2 cells, potentially providing a window into the mechanisms of lung epithelium dysfunction during COVID-19 (Figure 2B).
Because the pathology of severe COVID-19 disease is caused by a dysregulated immune response, more complex models have been applied to studying lung pathogenesis of SARS-CoV-2 infection. A system derived from adult stem cells and containing a mixture of upper airway and lung cells has been used to probe the interplay between airway and alveolar cells (Tindle et al., 2020). Mimicking what has been seen in vivo, SARS-CoV-2 infection was more widespread in airway cells than in the alveolar cells. This culture system generated an immune gene signature very similar to that seen in COVID-19 patient lungs when assessed by bulk RNA sequencing, whereas the epithelial cell types cultured alone did not. The similarity to in vivo results suggests that this platform could be essential for testing interventions that aim to decrease viral replication and overactive immune signaling (Tindle et al., 2020).
A previously established differentiated iPSC system that contained ciliated, goblet, and alveolar cells was infected with SARS-CoV-2 in the presence of autologous iPSC-derived M1 and M2 macrophages (Huang et al., 2014; Duan et al., 2020). The SARS-CoV-2 N protein was detectable via immunofluorescence in all types of airway epithelial cells, and the addition of macrophages decreased the expression of N protein (though this effect was not quantified), indicating that macrophages can recognize and remove infected cells. Notably, N protein was also detected in cocultured macrophages, in contrast to previous reports showing no infection of isolated macrophage cultures (Duan et al., 2020; Yang et al., 2020). One possibility is that macrophages in proximity to infected lung epithelial cells may be rendered permissive for infection. Another option is that N+ macrophages phagocytosed infected epithelial cells without themselves becoming actively infected.
Further studies are necessary to evaluate SARS-CoV-2 infection of complex lung cell models. It is critical to understand the dynamics of innate immune responses in these models as the timing of infiltration of innate immune cells such as macrophages is a recognized determinant of the pathology of COVID-19 (Merad and Martin, 2020). These models could serve as unique drug-testing platforms of the cytokine storm associated with ARDS. They will also shed light on how the upper and lower airways can be the major site of pathogenesis and damage despite their low ACE2 expression and relatively low permissiveness to infection in monocultures.
Primary or Secondary Infection in the Gut?
The presence of viral RNA in the gut is widely recognized, and the detection of viral RNA in stool samples, along with gastrointestinal symptoms, occurs in up to 20% of symptomatic carriers during COVID-19 (Chen et al., 2020b). However, whether virus found in the gut is infectious remains controversial: the few studies that looked for infectious virus in the feces rarely found any (Zang et al., 2020). In culture, primary gut cells are highly permissive to SARS-CoV-2 and capable of producing infectious virions. Stem-cell-derived intestinal organoids are well characterized and widely used to study viral infection—notably, norovirus infection (Sato et al., 2011; Ettayebi et al., 2016). Because these organoids are widely used in research and support high levels of viral replication, they are poised to become an essential model system for studying the gut pathogenesis of SARS-CoV-2.
Intestinal organoids are readily grown from adult stem cells isolated from both large and small intestine biopsies (Sato et al., 2011). Upon differentiation, these organoids contain mature enterocytes, goblet cells, enteroendocrine cells, and Paneth cells. ACE2 is expressed robustly in the small intestine along with the serine protease TMPRSS2, both in vivo and in the associated stem cell models. In contrast, colonic organoids have low ACE2 receptor expression (Zang et al., 2020). TMPRSS4 can perform the same function as TMPRSS2 in intestinal organoids, supporting entry of the virus (Zang et al., 2020). As in other organoid systems, ACE2 receptors, which are expressed on the apical side of organoids cells, face inward; thus, manipulation of the organoids is generally necessary for successful infection. This can be achieved by dissociating the organoids prior to infection. Alternatively, intestinal cell cultures can be grown in 2D in transwell systems.
Two groups have shown that ileal organoids—organoids modeling the small intestine— can be infected by SARS-CoV-2 and produce live virus (Lamers et al., 2020; Zang et al., 2020) (Figure 2D). In these organoids, SARS-CoV-2, similar to its close cousin SARS-CoV, infects primarily mature enterocytes and cells that are undergoing cell division (Lamers et al., 2020; Zang et al., 2020). Bulk RNA sequencing analysis shows that SARS-CoV-2 generates a robust interferon response as early as 24 h post infection and even stronger by 60 h, exceeding the response triggered by SARS-CoV (Figure 2B) (Lamers et al., 2020). Caspase-3, a marker of apoptotic cell death, is upregulated in infected cell cultures, indicating gastrointestinal cell damage caused by the virus. However, alterations of ilium-specific cellular pathways have not yet been reported after infection. There is a brief report of successful SARS-CoV-2 infection of iPSC-derived colonoids, leading to an upregulation of cytokine signaling and apoptotic pathways, and the difference in infection between the two models should be further explored (Han et al., 2020).
Ileal organoids in culture produce infectious virions, as confirmed by plaque assays (Lamers et al., 2020). However, whether the virus survives in the gut in vivo remains a question. When SARS-CoV-2 virus was incubated with gastric fluids from the large and small intestine, it rapidly lost infectivity, suggesting that while infection in the gut may occur transiently, the fecal-oral route may not be a major driver of viral transmission (Zang et al., 2020).
Widespread Damage in the Heart
Cardiac complications are increasingly taking center stage during COVID-19 illness. Increased levels of troponin, a marker of cardiac dysfunction, occur in over 25% of hospitalized patients (Marshall, 2020; Bonow et al., 2020), and COVID-19 survivors often experience myocarditis (Marshall, 2020). The role of direct infection of heart cells by SARS-CoV-2 remains unclear, but viral RNA has been detected in the heart in a small number of case studies (Puelles et al., 2020). In vitro, multiple groups have demonstrated that iPSC-derived cardiomyocytes are highly susceptible to SARS-CoV-2 infection and to ensuing functional damage.
iPSC-derived cardiomyocytes express ACE2 but only low to undetectable levels of TMPRSS2. Infection of cardiomyocytes is not abrogated by the addition of Camostat, a TMPRSS2 inhibitor, suggesting that TMPRSS2 is not required for SARS-CoV-2 infection in cardiomyocytes. Instead, the cathepsins CTSL and CTSB, which are expressed in iPSC-derived cardiomyocytes, appear sufficient to allow SARS-CoV-2 entry (Pérez-Bermejo et al., 2020). Direct infection has been confirmed with both S-protein pseudotyped viruses, which only model entry and infectious viruses (Yang et al., 2020; Bojkova et al., 2020; Sharma et al., 2020). Multiple virological assays—detection of dsRNA, RT-qPCR in the culture supernatant S protein staining and plaque assays—have all confirmed their permissiveness of iPSC-derived cardiomyocytes to SARS-CoV-2 infection (Figure 2D) (Sharma et al., 2020; Bojkova et al., 2020; Pérez-Bermejo et al., 2020). Primary heart tissue is also successfully infected by SARS-CoV-2 ex vivo, underscoring the validity of the stem cell model to recapitulate the in vivo situation (Bojkova et al., 2020). In vitro infection of cardiomyocytes induces widespread CPE and a strong interferon response as demonstrated by multiple groups via bulk RNA sequencing (Figure 2B) (Pérez-Bermejo et al., 2020; Sharma et al., 2020; Bojkova et al., 2020).
Direct infection of cultured cardiomyocytes and reports of myocarditis in infected individuals raise the specter of long-term cardiovascular impairment in COVID-19 survivors. SARS-CoV-2 infection impairs the in vitro ability of cardiomyocytes to beat, because of either viral cytopathic effects or specific disruption of contractility (Figure 2C) (Sharma et al., 2020). Fragmentation of troponin T filaments observed during SARS-CoV-2 infection may explain the disruption of cardiomyocyte contractility (Pérez-Bermejo et al., 2020). Overall, the highly permissive cardiomyocyte model offers not only plenty of opportunities to study the pathogenesis of cardiac complications during COVID-19 but also a solid model for in vitro infection of the virus.
Brain: A Not-So-Innocent Bystander?
A variety of neurological symptoms—most commonly, loss of smell and taste, headache, and dizziness—are common among symptomatic SARS-CoV-2 carriers, while severe symptoms, such as cerebrovascular events or seizures, remain rare (Chen et al., 2020). Loss of taste was reported in up to 50% of patients and headache in up to 20% (Chen et al., 2020). Here, too, the question of whether direct infection or systemic immune responses cause organ damage remains open. Individual cases of SARS-CoV-2 RNA detection in cerebrospinal fluid (CSF) have been reported, but overall, viral RNA is found in less than 1% of tested CSF samples (Destras et al., 2020).
A number of iPSC-derived systems have been used to identify potential target cells of SARS-CoV-2 in the central nervous system, but results remain controversial. Variation in differentiation protocol, length of infection, or differences in MOI could explain the discrepancy between reported results. For example, infection of iPSC-derived cortical neurons alone, either with S-protein pseudotyped or infectious virus at a high MOI, resulted in minimal infection (<1%), suggesting that neurons are not a primary target of viral infection (Jacob et al., 2020; Yang et al., 2020). However, when a model of brain organoids containing both cortical neurons and neural progenitor cells (NPCs) was infected, SARS-CoV-2 antigen was found in the neurons (though not quantified) and not the NPCs, and the supernatant was reported to contain 103 PFU/mL of released virus (Figure 2D) (Zhang et al., 2020). In a third study of brain organoids, no live virus was recovered from the supernatant, but antigen was found in the neurons, suggesting that the virus had entered but not replicated in cortical neurons in this case (Ramani et al., 2020). However, this abrogated entry of live virus alone dysregulated neuronal functions, as shown by abnormally localized Tau protein and neuronal cell death (Ramani et al., 2020). This observation indicates that abortive infection of neurons may contribute to some of the reported neurological symptoms (Figure 2C).
While iPSC-derived brain organoids representing the cortical, hippocampal, hypothalamic, and midbrain regions of the brain reportedly show low infection rates, organoids representing choroid plexus epithelial cells adjacent to the hippocampus were infected at rates of 10%–20%, depending on the donor (Jacob et al., 2020; Pellegrini et al., 2020). These cells produce most of the cerebrospinal fluid and thus could contribute to widespread distribution of virions in the brain and spinal cord. Choroid plexus organoids produced infectious virus as early as 24 h post infection, even when infected with very low MOIs. Bulk RNA sequencing indicated that infection occurred despite a vigorous innate immune response, resulting in expression of interferon and ISGs (Jacob et al., 2020) (Figure 2B). Notably, the expression of genes related to ion transport and cell adhesion was altered, showing that viral infection could directly interfere with the function of these cells. Decline of tight junction proteins during SARS-CoV-2 infection was also confirmed by microscopy by others (Pellegrini et al., 2020).
Overall, in vivo infection of the nervous system remains a matter of debate, and further evidence from autopsies of COVD-19 deaths is required in the search for direct viral targets. Understanding the signature of direct infection in vitro will suggest areas of investigation in vivo and possibly lead to neuroprotective treatments in the future.
Kidney, Liver, and Pancreas: Too Little Attention Yet
Emerging evidence shows that kidney, liver, and pancreas are affected by SARS-CoV-2, but these organs remain understudied in in vitro models so far (Puelles et al., 2020; Wang et al., 2020b; Rubino et al., 2020). All three organs express the viral ACE2 receptor, and stem-cell-derived models of the liver and pancreas have shown that viral entry is possible in vitro (Yang et al., 2020). In the pancreas, the endocrine α and β cells are the major targets of in vitro SARS-CoV-2 infection. Apoptotic and insulin resistance pathways activated upon infection of pancreatic β cells (Yang et al., 2020). The direct infection of β cells demonstrated in vitro could explain the increased incidence of type I diabetes in asymptomatic SARS-CoV-2 carriers (Rubino et al., 2020). In the liver, infection of adult stem-cell-derived cholangiocyte and hepatocyte organoids with SARS-CoV-2 led to high percentages of S+ cells (40% and 80%, respectively), leading to an increase in chemokine production and IL-17 signaling (Yang et al., 2020). Some people hospitalized for COVID-19 show increased levels of liver enzymes, a sign of temporary liver damage, and acute kidney injury requiring dialysis has been reported in otherwise healthy individuals. An iPSC-derived model of human kidney organoids has been used to test soluble ACE2 as a protective treatment against SARS-CoV-2, but viral infection in this system has not been characterized (Monteil et al., 2020). More clinical and molecular studies are needed to understand the impact of SARS-CoV-2 infection on these organs.
Conclusion
The advance in stem cell biology and resulting differentiated cell models has significantly accelerated research of SARS-CoV-2 pathogenesis in different organs. Completion of the viral life cycle of SARS-CoV-2 has been shown in airway, alveolar, gut, cardiac, and pancreatic stem-cell-derived systems, with significant questions remaining in the brain, kidney, and liver models. Many reported similarities between these cell culture systems point to universal mechanisms of viral infection and the innate immune response. While MOI and kinetics of infection vary, all reported infections eventually lead to an interferon or cytokine response and cell death. However, the virus diminishes the function of these cells it infects in various ways, from abrogating the beating of cardiomyocytes to weakening the tight junctions between choroid plexus cells in the brain. As the long-term symptoms arising in various organ systems are naturally different, understanding these differences and developing treatments to target these organ-specific pathways are essential. Nevertheless, the basics of viral replication remain consistent across cell types, with some variability in the viral entry process.
We are still in the early stages of SARS-CoV-2 research and susceptibility or resistance to SARS-CoV-2 in these models must be confirmed with rigorous clinical studies. In vitro studies have obvious limitations, lacking immune cells and interplay with other organ systems. While great strides have been made in improving and refining cellular differentiation protocols, stem-cell-derived models can have significant differences in gene expression as compared to primary organ-derived cells and may not fully recapitulate the cellular response in primary cells. iPSC models, in particular, lack genomic stability and may not recapitulate the full maturity of primary cells (Doss and Sachinidis, 2019). As research on SARS-CoV-2 moves forward, these models will need to be reassessed, improved, and used for new questions. For example, these studies have focused on the role of ACE2 as the necessary factor for entry into the cell. Recently, the role of other factors in entry have been identified, and the expression of these factors in the models discussed in this review will have to be addressed (Bestle et al., 2020; Clausen et al., 2020; Cantuti-Castelvetri et al., 2020).
While the wave of first reports discussed here demonstrate more or less successful infection and offer important first insight into the mechanisms of organ involvement, more questions have arisen from these studies. For example, why are airway cells relatively resistant to viral infection in vitro while other organ systems like the heart and the gut are widely infectible? How is infection spreading in vivo from the upper and lower airways to secondary organs despite the very low levels of virus reported in the blood (Wang et al., 2020a)? The stem-cell-derived models focus on the terminally differentiated cells as targets of SARS-CoV-2, but the role of tissue-resident stem cells remains largely unaddressed. In particular, the question of possible hit-and-run mechanisms affecting stem cells is becoming a pressing question in light of clinical “long haulers,” COVID survivors suffering from long-term clinical symptoms after clearance of the virus. While iPSCs and some tissue-resident progenitor cells are resistant to viral infections due to an abnormal interferon response, the question of direct infection by SARS-CoV-2 or bystander damage remains open (Wu et al., 2018). Current stem cell technologies offer unique opportunities to study this and other pressing issues, including the role of genomic variation, age, and pre-existing conditions in donors. Future work incorporating these factors and more complex immune cell populations into stem-cell-derived models will be critical to understanding the complete pathology of multi-organ involvement in COVID-19.
Acknowledgments
We thank Dr. Françoise Chanut for editorial assistance, Veronica Fonseca for administrative assistance, and John C.W. Carroll for graphic design. Figures were based on Biorender.com templates and edited by John C.W. Carroll. M.O. was supported by NIH 5DP1DA038043, NIDDK 1R01DK123746-01, and NIAID R01AI097552-01A1 and acknowledges support through gifts from the Roddenberry Foundation and Atlantic Philanthropies.
References
- Bar-Ephraim Y.E., Kretzschmar K., Clevers H. Organoids in immunological research. Nat. Rev. Immunol. 2020;20:279–293. doi: 10.1038/s41577-019-0248-y. [DOI] [PubMed] [Google Scholar]
- Bestle D., Heindl M.R., Limburg H., Van Lam van T., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020;3:e202000786. doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Wagner J.U.G., Shumliakivska M., Aslan G.S., Saleem U., Hansen A., Luxán G., Günther S., Pham M.D., Krishnan J. SARS-CoV-2 infects and induces cytotoxic effects in human cardiomyocytes. Cardiovasc. Res. 2020:cvaa267. doi: 10.1093/cvr/cvaa267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonow R.O., Fonarow G.C., O’Gara P.T., Yancy C.W. Association of Coronavirus Disease 2019 (COVID-19) With Myocardial Injury and Mortality. JAMA Cardiol. 2020;5:751–753. doi: 10.1001/jamacardio.2020.1105. [DOI] [PubMed] [Google Scholar]
- Busnadiego I., Fernbach S., Pohl M.O., Karakus U., Huber M., Trkola A., Stertz S., Hale B.G. Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2. MBio. 2020;11:11. doi: 10.1128/mBio.01928-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Chen X., Laurent S., Onur O.A., Kleineberg N.N., Fink G.R., Schweitzer F., Warnke C. A systematic review of neurological symptoms and complications of COVID-19. J. Neurol. 2020:1–11. doi: 10.1007/s00415-020-10067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183:1043–1057. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12:12. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Snijder E.J., Kikkert M., van Hemert M.J. Host Factors in Coronavirus Replication. Curr. Top. Microbiol. Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destras G., Bal A., Escuret V., Morfin F., Lina B., Josset L., COVID-Diagnosis HCL Study Group Systematic SARS-CoV-2 screening in cerebrospinal fluid during the COVID-19 pandemic. Lancet Microbe. 2020;1:e149. doi: 10.1016/S2666-5247(20)30066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss M.X., Sachinidis A. Current Challenges of iPSC-Based Disease Modeling and Therapeutic Implications. Cells. 2019;8:8. doi: 10.3390/cells8050403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan F., Guo L., Yang L., Han Y., Thakur A., Nilsson-Payant B.E., Wang P., Zhang Z., Ma C.Y., Zhou X. Modeling COVID-19 with Human Pluripotent Stem Cell-Derived Cells Reveals Synergistic Effects of Anti-inflammatory Macrophages with ACE2 Inhibition Against SARS-CoV-2. Res. Sq. 2020 doi: 10.21203/rs.3.rs-62758/v1. [DOI] [Google Scholar]
- Ettayebi K., Crawford S.E., Murakami K., Broughman J.R., Karandikar U., Tenge V.R., Neill F.H., Blutt S.E., Zeng X.L., Qu L. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S. Identification of SARS-CoV-2 Inhibitors using Lung and Colonic Organoids. Nature. 2020 doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.X., Islam M.N., O’Neill J., Hu Z., Yang Y.G., Chen Y.W., Mumau M., Green M.D., Vunjak-Novakovic G., Bhattacharya J., Snoeck H.W. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Hume A.J., Abo K.M., Werder R.B., Villacorta-Martin C., Alysandratos K.D., Beermann M.L., Simone-Roach C., Lindstrom-Vautrin J., Olejnik J. SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response. Cell Stem Cell. 2020;27:962–973. doi: 10.1016/j.stem.2020.09.013. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Pather S.R., Huang W.K., Zhang F., Wong S.Z.H., Zhou H., Cubitt B., Fan W., Chen C.Z., Xu M. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell. 2020;27:937–950. doi: 10.1016/j.stem.2020.09.016. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H., Sontake V., Tata A., Kobayashi Y., Edwards C.E., Heaton B.E., Konkimalla A., Asakura T., Mikami Y., Fritch E.J. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2 mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell. 2020;27:890–904. doi: 10.1016/j.stem.2020.10.005. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. The lasting misery of coronavirus long-haulers. Nature. 2020;585:339–341. doi: 10.1038/d41586-020-02598-6. [DOI] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M.E., Thurman A., Pezzulo A.A., Leidinger M.R., Klesney-Tait J.A., Karp P.H., Tan P., Wohlford-Lenane C., McCray P.B., Jr., Meyerholz D.K. Heterogeneous expression of the SARS-Coronavirus-2 receptor ACE2 in the human respiratory tract. EBioMedicine. 2020;60:102976. doi: 10.1016/j.ebiom.2020.102976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Albecka A., Mallery D.L., Kellner M.J., Paul D., Carter A.P., James L.C., Lancaster M.A. SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids. Cell Stem Cell. 2020;27:951–961. doi: 10.1016/j.stem.2020.10.001. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Bermejo J.A., Kang S., Rockwood S.J., Simoneau C.R., Joy D.A., Ramadoss G.N., Silva A.C., Flanigan W.R., Li H., Nakamura K. SARS-CoV-2 infection of human iPSC-derived cardiac cells predicts novel cytopathic features in hearts of COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.08.25.265561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A., Sen C., Garcia G., Langerman J., Vijayaraj P., Shia D.W., Meneses L.K., Rickabaugh T.M., Mulay A., Konda B. Direct exposure to SARS-CoV-2 and cigarette smoke increases infection severity and alters the stem cell-derived airway repair response. Cell Stem Cell. 2020;27:869–875. doi: 10.1016/j.stem.2020.11.010. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani S., Crawford S.E., Blutt S.E., Estes M.K. Human organoid cultures: transformative new tools for human virus studies. Curr. Opin. Virol. 2018;29:79–86. doi: 10.1016/j.coviro.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A., Müller L., Ostermann P.N., Gabriel E., Abida-Islam P., Müller-Schiffmann A., Mariappan A., Goureau O., Gruell H., Walker A. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020;39:e106230. doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino F., Amiel S.A., Zimmet P., Alberti G., Bornstein S., Eckel R.H., Mingrone G., Boehm B., Cooper M.E., Chai Z. New-Onset Diabetes in Covid-19. N. Engl. J. Med. 2020;383:789–790. doi: 10.1056/NEJMc2018688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N., Papaspyropoulos A., Zomer-van Ommen D.D., Heo I., Böttinger L., Klay D., Weeber F., Huelsz-Prince G., Iakobachvili N., Amatngalim G.D. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38:38. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel R., Majd H., Richter M., Ghazizadeh Z., Zekavat S.M., Navickas A., Ramirez J., Asgharian H., Simoneau C.R., Bonser L. Androgen Regulates SARS-CoV-2 Receptor Levels and Is Associated with Severe COVID-19 Symptoms in Men. Cell Stem Cell. 2020;27:876–889. doi: 10.1016/j.stem.2020.11.009. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Schaefer I.M., Padera R.F., Solomon I.H., Kanjilal S., Hammer M.M., Hornick J.L., Sholl L.M. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod. Pathol. 2020;33:2104–2114. doi: 10.1038/s41379-020-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Garcia G., Jr., Wang Y., Plummer J.T., Morizono K., Arumugaswami V., Svendsen C.N. Human iPSC-Derived Cardiomyocytes Are Susceptible to SARS-CoV-2 Infection. Cell Rep Med. 2020;1:100052. doi: 10.1016/j.xcrm.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindle C., Fuller M., Fonseca A., Taheri S., Ibeawuchi S.R., Beutler N., Claire A., Castillo V., Hernandez M., Russo H. Adult Stem Cell-derived Complete Lung Organoid Models Emulate Lung Disease in COVID-19. bioRxiv. 2020 doi: 10.1101/2020.10.17.344002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu S., Liu H., Li W., Lin F., Jiang L., Li X., Xu P., Zhang L., Zhao L. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J. Hepatol. 2020;73:807–816. doi: 10.1016/j.jhep.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Wagner V., Rasmussen S.B., Hartmann R., Paludan S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Wu X., Dao Thi V.L., Huang Y., Billerbeck E., Saha D., Hoffmann H.H., Wang Y., Silva L.A.V., Sarbanes S., Sun T. Intrinsic Immunity Shapes Viral Resistance of Stem Cells. Cell. 2018;172:423–438.e25. doi: 10.1016/j.cell.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffré F. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youk J., Kim T., Evans K.V., Jeong Y.I., Hur Y., Hong S.P., Kim J.H., Yi K., Kim S.Y., Na K.J. Three-Dimensional Human Alveolar Stem Cell Culture Models Reveal Infection Response to SARS-CoV-2. Cell Stem Cell. 2020;27:905–919. doi: 10.1016/j.stem.2020.10.004. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:5. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.Z., Chu H., Han S., Shuai H., Deng J., Hu Y.F., Gong H.R., Lee A.C., Zou Z., Yau T. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30:928–931. doi: 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., HCA Lung Biological Network. Electronic address: lung-network@humancellatlas.org. HCA Lung Biological Network SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]