Abstract
PURPOSE
Because of expanding interoperability requirements, structured patient data are increasingly available in electronic health records. Many oncology data elements (eg, staging, biomarkers, documentation of adverse events and cancer outcomes) remain challenging. The Minimal Common Oncology Data Elements (mCODE) project is a consensus data standard created to facilitate transmission of data of patients with cancer.
METHODS
In 2018, mCODE was developed through a work group convened by ASCO, including oncologists, informaticians, researchers, and experts in terminologies and standards. The mCODE specification is organized by 6 high-level domains: patient, laboratory/vital, disease, genomics, treatment, and outcome. In total, 23 mCODE profiles are composed of 90 data elements.
RESULTS
A conceptual model was published for public comment in January 2019 and, after additional refinement, the first public version of the mCODE (version 0.9.1) Fast Healthcare Interoperability Resources (FHIR) implementation guide (IG) was presented at the ASCO Annual Meeting in June 2019. The specification was approved for balloting by Health Level 7 International (HL7) in August 2019. mCODE passed the HL7 ballot in September 2019 with 86.5% approval. The mCODE IG authors worked with HL7 reviewers to resolve all negative comments, leading to a modest expansion in the number of data elements and tighter alignment with FHIR and other HL7 conventions. The mCODE version 1.0 FHIR IG Standard for Trial Use was formally published on March 18, 2020.
CONCLUSION
The mCODE project has the potential to offer tremendous benefits to cancer care delivery and research by creating an infrastructure to better share patient data. mCODE is available free from www.mCODEinitiative.org. Pilot implementations are underway, and a robust community of stakeholders has been assembled across the oncology ecosystem.
Context
Key Objective
To determine how the Minimal Common Oncology Data Elements (mCODE) project will improve sharing data of patients with cancer between electronic health records and promote enhanced cancer care delivery.
Knowledge Generated
The mCODE project was created in 2018 as a consensus data standard to ease the sharing of data of patients with cancer between different electronic health records. Health Level Seven International formally published the mCODE version 1.0 Fast Healthcare Interoperability Resources implementation guide Standard for Trial Use on March 18, 2020.
Relevance
A large group of diverse stakeholders has contributed to the mCODE project and created a framework focused on interoperability of data of patients with cancer. Six high-level domains organize mCODE: patient, laboratory/vital, disease, genomics, treatment, and outcome. Twenty-three mCODE profiles are organized within the domains and are composed of 90 data elements in total. mCODE has the potential to benefit cancer care delivery and research. Multiple pilot implementations are underway.
INTRODUCTION
Cancer Data Landscape
Improved outcomes in cancer care can optimally be achieved by accelerating data sharing and aggregation and by the creation of a learning health system in which routine patient care data seamlessly inform scientific discovery, and, reciprocally, research informs practice.1 However, the current state of oncology data interoperability is far from optimal. Many foundational types of oncology data, such as cancer staging, biomarkers, and the documentation of adverse events and cancer outcomes, are captured in the electronic health record (EHR) primarily in noncomputable form in notes and other unstructured documents. Research-grade data are usually found only for the small percentage of patients participating in clinical trials, and expensive and unsustainable manual data abstraction is otherwise required to extract data from unstructured sources. In addition, today’s EHRs are not well designed to support modern oncology practice, with deficiencies in data entry, practice workflows, and a common library of oncology-specific discrete data elements.2 These shortcomings are particularly amplified in the case of genomic data, because most EHRs poorly support the needs of precision oncology, and next-generation sequencing (NGS) results are usually reported in the EHR as noncomputable PDF files.3
ASCO and Data Standards Development
ASCO has long had an interest in developing oncology data standards as a way of facilitating cancer care quality—one of the pillars of its organizational mission. In 2005, ASCO began to develop medical oncology treatment plan and summary (TPS) data templates and elements.4 Subsequently, in 2009, a collaboration between ASCO, the National Cancer Institute, and the National Community Cancer Center Program led to the Clinical Oncology Requirements for the EHR (CORE) project, which sought to develop a functional requirements document, data element specifications, and interoperability requirements of an EHR to support oncologists providing patient-centered care.5 In 2013, the ASCO Health Information Technology Work Group created a new oncology-specific data standard entitled the electronic Clinical Oncology Treatment Plan and Summary (eCOTPS), derived from the paper-based ASCO Breast Cancer Adjuvant TPS. The eCOTPS was developed to be congruent with the Health Level Seven International (HL7) Clinical Document Architecture Release 2 (CDA-R2) standard. The concepts in the parent Breast Cancer Adjuvant TPS were mapped to standard terminologies and modeled to conform to or extend existing CDA-R2 templates, and some new templates were added. The eCOTPS implementation guide was balloted through HL7 and published as a Draft Standard for Trial Use (DSTU) in October 2013, the first oncology-specific CDA standard that achieved the DSTU designation.6 The eCOTPS was extended to encompass templates for colon cancer adjuvant therapy and general cancer survivorship care.7
Oncology Data Standards: Challenges and New Federal Requirements
Although there were early adopters of the ASCO-developed eCOTPS, it was not widely adopted by EHR vendors or used by cancer clinicians. Historically, the uptake of health care–specific data standards has been frustratingly slow, and the field of oncology does not have a history of many successes in this area. Likely contributing factors include a lack of focus on clinical workflows during standards development, the failure to center standards development on specific use cases, a focus on the exchange of data rather than the data themselves, the lack of EHR vendor engagement in the development process, and, for example, of the eCOTPS, the fact that the emerging HL7 standard Fast Healthcare Interoperability Resources (FHIR) was beginning to eclipse the CDA standard shortly after the eCOTPS was released.
The 21st Century Cures Act, passed by the US Congress and signed into law in December 2016, provided a needed stimulus for the facilitation of wider health care data sharing.8 The law required the US Department of Health and Human Services (HHS) and the Office of the National Coordinator for Health Information Technology (ONC) to improve the overall interoperability of health information through the development of data standards and application programming interfaces (APIs), with a particular focus on patient access to their own health care data “without special effort.”9
In early 2020, ONC and the HHS Centers for Medicare and Medicaid Services (CMS) announced a pair of final rules for full implementation of some of the provisions of the 21st Century Cures Act, known respectively as the ONC Cures Act Final Rule10 and CMS Interoperability and Patient Access Final Rule.11 A key provision of these rules was that health information technology vendors, to maintain their ONC-certified status, must implement an API using HL7 FHIR Release 4.0.1 for patients to be able to access their claims, encounter information, and all data covered by the US Core Data for Interoperability (USCDI)12 data elements through a third-party application of their choice. In addition, data exchange between payers would have to use the USCDI content standard. It is reasonable to expect that a fortunate byproduct of these regulations is that patients will experience lower barriers to accessing their own data, because EHR vendors will be required to provide FHIR APIs for access, and this may spur the incorporation of FHIR-based standards like mCODE into source systems.
CONCEIVING MCODE
ASCO’s CancerLinQ and the Origins of the Minimal Common Oncology Data Elements Standard
To begin to realize the vision of the oncology learning health system, ASCO, through its wholly owned nonprofit subsidiary CancerLinQ LLC, created the health technology platform CancerLinQ in 2014.13 CancerLinQ extracts data from EHRs and other sources, such as cancer registries, via direct software connections, and then aggregates, harmonizes, and normalizes the data in a cloud-based environment where it is used by the contributing centers for quality improvement purposes and clinical care. Aggregated de-identified data also can be used for data exploration, insights, and secondary research.14 As of April 2020, the CancerLinQ database contains data on ≥ 2.5 million total patients, ≥ 1.5 million of whom have a primary diagnosis of a malignant neoplasm.
Although CancerLinQ has fully operationalized oncology data extraction, aggregation, and processing at scale, supporting a learning health system with such a platform remains a daunting task because of the nature of the broader EHR ecosystem and sheer scope and variability of data entry by clinicians. Even highly discrete and well-defined concepts such as patient tobacco-use assessment are beset by extreme variability in data capture, rendering aggregation into broad clinical categories challenging. Although CancerLinQ incorporates > 180,000 software rules to normalize inbound data into its common data model, it became clear that the most effective solution to improve data quality would be to collect the data in the EHR in a more standardized format, because postprocessing manual review using trained human curators was not a sustainable solution.15
MCODE DEVELOPMENT
In 2018, ASCO and a group of collaborators, including oncologists, informaticians, researchers, and experts in terminologies and standards, came together to create the Minimal Common Oncology Data Elements (mCODE) initiative to develop and maintain standard computable oncology data formats. In addition to ASCO, founding collaborators included a federally funded research and development center (MITRE, Bedford, MA), a National Cancer Institute–sponsored clinical trials research consortium (Alliance for Clinical Trials in Oncology), and the US Food and Drug Administration. The group agreed on the following guiding principles for the initiative: mCODE incorporates a use case development model; mCODE development is iterative; mCODE maintenance is reductionist and parsimonious; mCODE is developed and maintained by its users and is highly collaborative and noncommercial. The current mCODE governance structure is shown in Figure 1.
FIG 1.
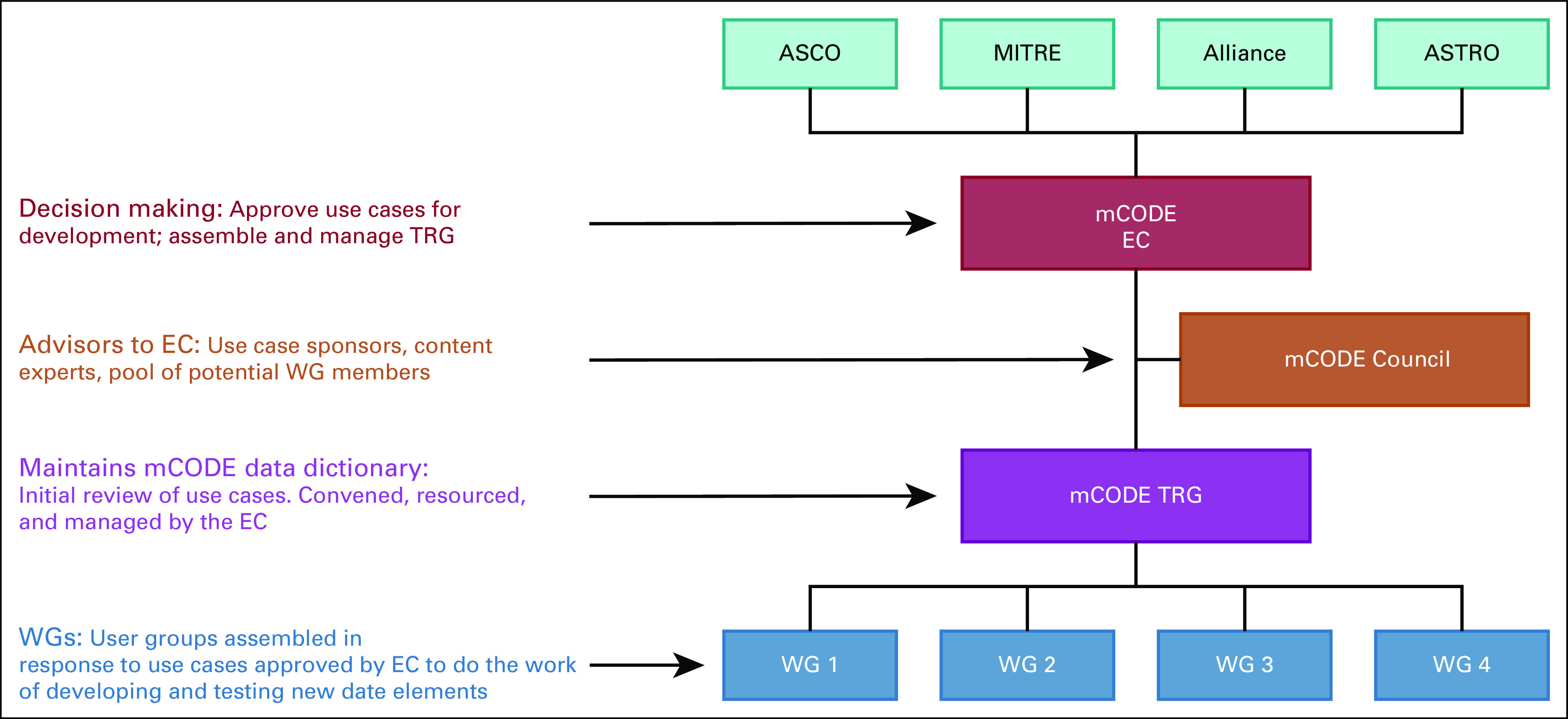
Minimal Common Oncology Data Elements (mCODE) governance structure (as of May 15, 2020). Alliance, Alliance for Clinical Trials in Oncology; ASTRO, American Society for Radiation Oncology; EC, Executive Committee; MITRE, the MITRE Corporation; TRG, Technical Review Group; WG, working group.
Doug Blaney, MD (Stanford University) and James Chen, MD (The Ohio State University) chaired the mCODE work group. The group agreed on the listed set of principles for the project, which shaped its evolution toward the goal of developing and maintaining a standard computable data format to achieve data interoperability and enable progress in clinical-care quality initiatives, clinical research, and health care policy development. The work group drove discussions and development by focusing on two hypothetical clinical cases.
Case 1: Newly Diagnosed Stage IV Non–Small Cell Lung Cancer
The first example case was that of a 58-year-old man who presented with dyspnea and pleuritic chest pain and who had multiple pulmonary masses and boney lesions on imaging that were suspicious for metastatic cancer. A magnetic resonance imaging scan showed 5 brain metastases, all < 1 cm, with no resulting edema. From a computed tomography (CT)-guided core needle biopsy specimen, diagnosis was confirmed of adenocarcinoma consistent with lung primary. The biopsy specimen was sent for programmed death ligand 1 (PD-L1) testing and NGS. The patient has not yet begun treatment.
Case 2: Newly Diagnosed Stage IV Clear-Cell Renal Cell Carcinoma
The second example case was that of a 68-year-old man with a history of hypertension, obesity, and 40 pack-years of smoking cigarettes. He presented with multiple episodes of painless hematuria, decreased, appetite, and weight loss. After negative cystoscopy, a CT scan of the abdomen showed a 9-cm hypervascular left renal mass, several low-density hepatic lesions ≤ 2.5 cm, and multiple lower-lung nodules ≤ 2 cm. Laboratory evaluation showed anemia. Examination of a renal biopsy specimen revealed clear-cell carcinoma, WHO/International Society of Urologic Pathologists grade 3, without sarcomatoid features. The patient discussed preference for oral treatment, if available, with his medical oncologist.
Case 1 highlighted several domains that the work group would need to address within mCODE, including tumor staging, disease status, biomarkers such as PD-L1, and advanced laboratory testing such as NGS. Case 2 demonstrated the complexity of comorbidities, social history, laboratory testing, prognostic scoring, and treatment selection.
Even with the 6 guiding principles and the clinical cases to drive discussion, creating the set of data elements was arduous. One central discussion point was whether implementation challenges should influence the inclusion or exclusion of data elements. There was clear agreement that if EHR vendors implemented mCODE by requiring additional data entry by users, the project would fail. The group, however, did not want to exclude key data elements if those were routinely collected. This led to inclusion of elements such as demographics, vital signs, and laboratory tests, which are routinely collected in the EHR as structured data and have underlying standards. Other elements such as disease status, social history, social determinants of health, and several outcome variables such as progression-free survival dominated discussions during the months spent creating the initial proposal. The formula for shaping these discussions centered around maintaining mCODE as a minimal set, acknowledging that the first version would not meet all potential use cases, and using existing standards when possible.
The work group agreed on a first draft of mCODE elements in December 2018, and the first, high-level, conceptual model was published for public comment in January 2019. The work group reviewed public comments and iteratively refined the model, after which 2 pilot implementations began. The first pilot used Compass, an open-source, mCODE-based FHIR implementation and Substitutable Medical Applications and Reusable Technologies–on–FHIR application developed by MITRE.15 This pilot was implemented at Intermountain Healthcare and aimed to connect the local EHR to CancerLinQ, allowing providers and patients to make real-time, informed, shared, data-driven decisions by showing similar patients treatments and outcomes from CancerLinQ. The Integrating Clinical Trials and Real-World Endpoints (ICARE) was the second mCODE pilot.15 The ICAREdata study is a partnership with the Alliance for Clinical Trials in Oncology to demonstrate using real-world data to improve clinical research without sacrificing the quality as traditional clinical trials.
The modeling effort was led by MITRE and the first public version of the mCODE (version 0.9.1) FHIR implementation guide (IG) was presented at the ASCO Annual Meeting in 2019.16
MCODE DATA SPECIFICATION
After work-group deliberation, public comment, refinement, and feedback from HL7 working groups and members, the final data standard (version 1.0) included six primary domains (Fig 2): patient, disease, lab/vital, genomics, treatment, and outcome. Each domain is organized into several concepts, which then have associated data elements.17 For example, the treatment domain includes concepts for cancer-related medication statements and cancer-related procedure statements (for surgery and radiation). These concepts are referred to as profiles. Cancer-related medications contains data elements of treatment intent (eg, curative, palliative), medication start and end date, and reason for termination.
FIG 2.
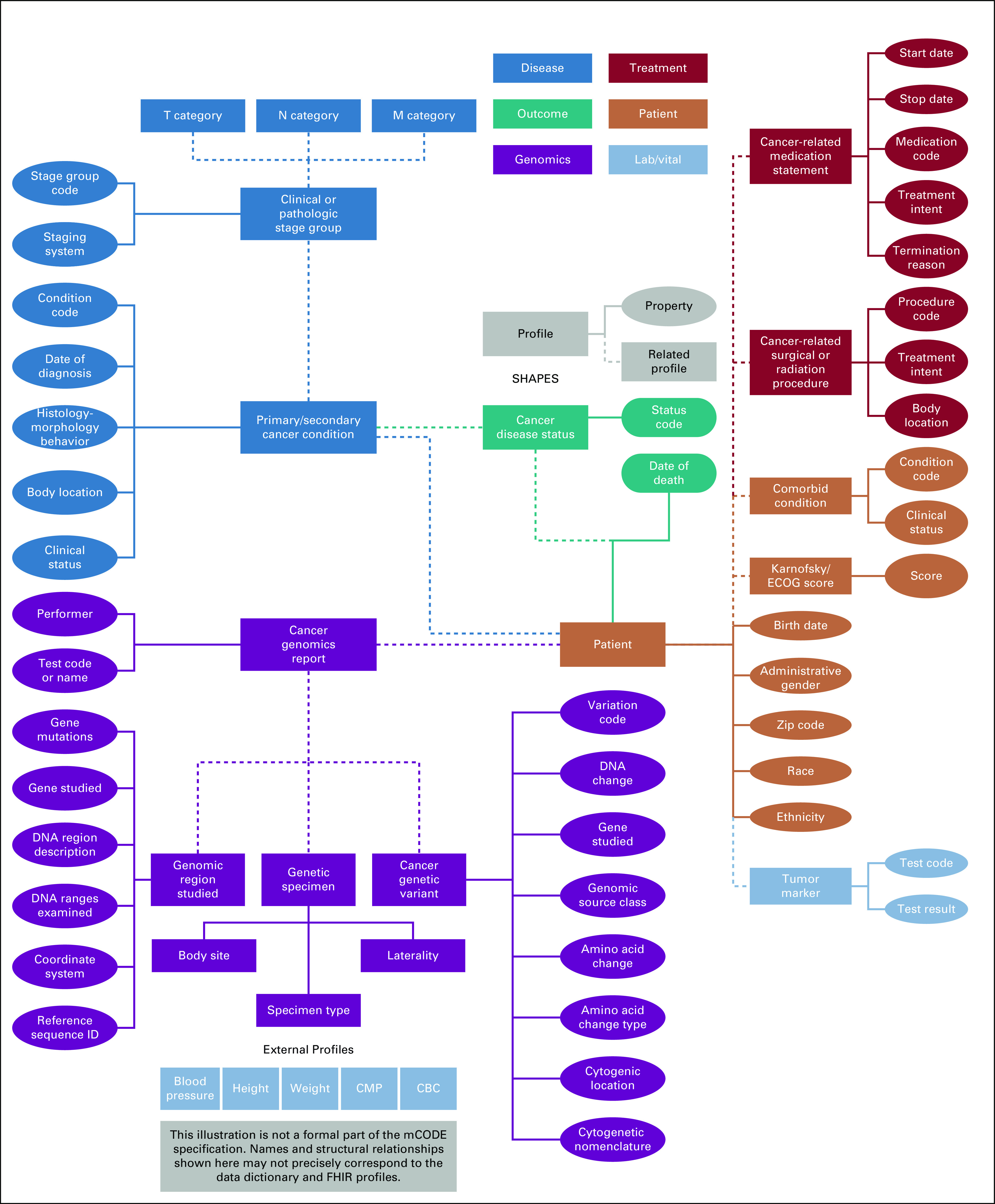
Minimal Common Oncology Data Elements (mCODE), version 1.0, high-level conceptual model describing domains, profiles, and properties. CMP, comprehensive metabolic panel; ECOG, Eastern Cooperative Oncology Group; FHIR, Fast Healthcare Interoperability Resources; Lab/Vital, laboratory test and vital signs values.
The data elements are modeled onto value sets that define the possible values of a given data element. A value set, when possible, is based on at least one terminology; however, it may also be an enumerated list, or data type. The example of “medication,” in the domain “treatment” under the profile of “cancer-related medication statement,” references the set of FHIR USCoreMedicationCodes.18 This reference standard is built on a subset of the RxNorm19 vocabulary and maintained by HL7. In total, 90 data elements exist across mCODE’s 23 profiles. These data elements are linked to standard coding systems such as American Join Committee on Cancer,20 ClinVar,21 Human Genome Variation Society,22 Human Genome Organization Gene Nomenclature Committee,23 International Classification of Diseases for Oncology (ICD), third edition,24 ICD-10, Clinical Modification,25 ICD-10 Procedure Coding System,26 Logical Observation Identifiers Names and Codes,27 and Systematized Nomenclature of Medicine, Clinical Terms.28
mCODE Organization and Future Development
The mCODE Executive Committee (EC) comprises four to seven representatives appointed from the stakeholder groups (currently ASCO, Alliance, MITRE, and ASTRO; Fig 1). The EC is responsible for adding data elements to the standard, considering implementation pilots, and approving public statements regarding mCODE. Members of a multidisciplinary advisory body, the mCODE Council, serve as external content experts and use case sponsors. The EC relies on the mCODE Technical Review Group (TRG) to provide recommendations on any updates on the data standard, including additions, subtractions, and mapping changes. The TRG comprises a diverse group of oncology subspecialists, informaticians, and standards experts. Although the TRG may recommend changes to the EC directly, additions to the standard will more likely be reviewed by the TRG after external pilot implementations to address new uses cases, which is in line with the project’s guiding principles. To assist with organizing projects to expand mCODE to include new use cases, MITRE helped to create the CodeX HL7 FHIR Accelerator (CodeX).29 CodeX is a collection of providers and health systems, researchers, regulators, EHR vendors, and information technology innovators working to improve data from patients with cancer through individual projects that may ultimately expand the scope of mCODE.
MCODE STANDARDS PUBLICATION PROCESS
As previously described, the mCODE specification was in development in late 2018, and there was an early public release in the spring of 2019. Although the IG conformed to an early version of FHIR, it was not approved by HL7 at the time. Rather, to accelerate the progress of mCODE specification development, the process of HL7’s approval for conformance to a FHIR standard was done concurrently with the iterative process outlined by the mCODE work group. Figure 3 illustrates mCODE data modeling activities aligned with the HL7 publication timeline.
FIG 3.
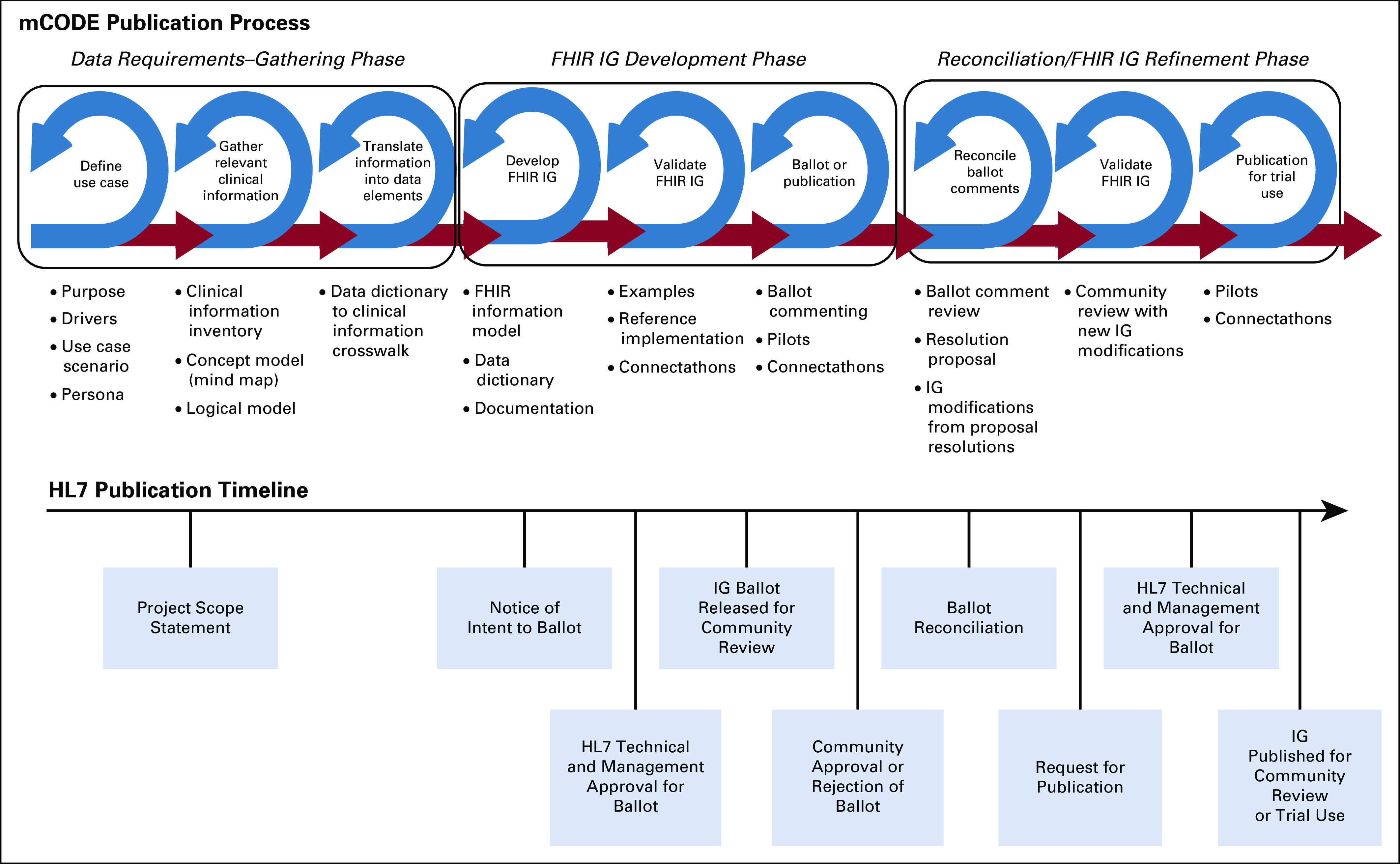
Minimal Common Oncology Data Elements (mCODE) timeline for Health Level Seven International (HL7) publication (source: MITRE). FHIR, Fast Healthcare Interoperability Resources; IG, Implementation Guide.
The mCODE modeling process was split into three phases of activities: (1) data requirements gathering; (2) modeling and implementation guide development; and (3) reconciliation and model refinement.
Data Requirements Gathering
Clinical subject matter experts (SMEs) in oncology used terms relevant to the clinical oncology community and the use cases described previously to determine the key clinical content. Clinical SMEs worked closely with non-SME informaticians to map equivalent clinical terms identified in the initial use case scenarios with terms from the more technical data model in the mCODE FHIR specification.
In January 2019, the mCODE modeling team engaged HL7 in the standardization process. A Project Scope Statement (PSS) was proposed to the HL7 Clinical Interoperability Council, a working group willing to sponsor the project and oversee mCODE through the publication process.30 The PSS identified HL7 FHIR as the specification standard, with an intent to ballot the first IG as a Standard for Trial Use (STU), and a project time frame to run through May 2022.
Model Development
The model development phase occurred between January and August 2019 and involved multiple iterations of technical analysis, modeling, and internal mCODE work group reviews. The data elements identified in the requirements gathering phase were analyzed to find structural and semantic mappings to the appropriate FHIR resources. In FHIR, any customization to a base FHIR resource by either extending the resource with new data elements or specifying more narrow constraints to existing elements required the creation of a FHIR profile. A data dictionary summarized the list of profiles, data elements within the profiles, and terminology value sets bound to the elements.
After several rounds of review and approval by HL7 committees and management teams, the balloted mCODE IG was announced on the HL7 ballot page for review by the entire community in August 2019. Although commenting is open to all, only HL7 members who sign up in advance can vote on the IG ballot approval. mCODE passed ballot in September 2019 with 86.5% approval.
The mCODE team also hosted a Cancer Interoperability track for participants to test the mCODE model and IG specification at the September 2019 FHIR Connectathon. Eight organizations representing oncology EHR vendors, standards developers, and research organizations participated in this track. Participants shared their use cases and tested FHIR API calls to an FHIR server loaded with mCODE FHIR structure definitions and sample data.
Ballot Reconciliation and IG Refinement
mCODE received 142 reviewer comments that ranged from positive comments on the organization of the IG to negative comments requiring changes to the model. Because all negative comments are required to be withdrawn by the submitter before publication, the IG authors worked with each submitter on an agreeable resolution. Some of the resolutions resulted in major changes to the originally balloted FHIR IG. Custom mCODE profiles for vital signs, CBC count, and comprehensive metabolic panel were replaced with documentation specifying use of equivalent USCDI profiles. Clinical genomics profiles were expanded to better align with the HL7 Clinical Genomics Reporting IG. These changes were presented to the mCODE TRG and approved.
Once all comments were reconciled and IG changes applied to address negative comments, the IG authors were required to reach out to each person who voted negative to the overall ballot and have them withdraw their negative vote. mCODE created two rounds of STU prerelease candidate versions of the specification for review. After withdrawal of the negative votes, an STU publication request was then submitted and followed by another round of reviews by the FHIR work groups.
The mCODE version 1.0 FHIR IG STU1 was formally published on March 18, 2020. This publication is now recognized by the overall HL7 community for trial use implementation. Trial implementers can submit issues, enhancements, and other feedback through an HL7 issue tracker, which is visible to the entire HL7 community. Nonsubstantive changes result in the release of a technical errata, whereas substantive changes would follow the process of a new STU, which would repeat the FHIR IG development and refinement phases, as noted in Figure 3. This iterative process of development, refinement, and trial implementation in an organized way improves and matures the IG, thereby increasing the level of confidence and adoption.
IMPACT AND FUTURE
The mCODE project offers potential benefits to clinical care delivery and research by creating an infrastructure to share patient data between health care systems. Although this sort of interoperability is not novel, the success of mCODE to date in part has been due to its approach. By engaging diverse stakeholders, leveraging existing data standards, and using use cases to drive its creation, mCODE has transformed from concept to a standard approved for test use in < 2 years, a remarkably short development cycle for a consensus data standard. Like any standard, adoption will be the key to mCODE’s ultimate success. Clinical and research use cases will likely drive the adoption and potential expansion of mCODE going forward.
A pivotal aspect of the mCODE development process was the decision made by the mCODE EC, shortly after the first version of the specification was approved in mid 2019, to ballot and publish the standard through HL7, as described in detail in the previous section. This demonstrates to external stakeholders, especially EHR vendors and regulatory agencies, the stability of the standard, an essential requirement for its incorporation into software or federal regulations, respectively. By contrast, had mCODE only been supported and published by clinical organizations like ASCO or Alliance, software vendors might have viewed it more as a project and not a product, and consequently they may not have had the confidence that the standard was stable enough to invest in for production systems. However, reconciling the ballot comments did result in numerous changes to the original version and perhaps less clinical flexibility, because strict adherence to FHIR and other HL7 standards was required. As a result, the total number of data elements increased from 73 to 90 going from version 0.9.1 to version 1.0. Managing mCODE scope creep and keeping mCODE minimal remains a challenge.
In summary, mCODE is a consensus data standard for oncology that specifies a computable set of data elements based on clinical use cases. This standard, published through HL7, is free and intended to provide a stable platform for implementers along with flexibility to expand data elements in the future through processes described in this article.31 Currently, ASCO, Alliance, MITRE, and ASTRO oversee development through representation on the EC, but anyone can participate in mCODE development via the mCODE Council or CodeX. mCODE’s potential ultimately will be realized by the development of other applications that use the standard or extend it to improve care and discovery, and some currently under consideration include matching patients to clinical trials, cancer registry reporting, and facilitating the use of oncology clinical pathways. Most important, better cancer data interoperability stands to benefit patients the most.
ACKNOWLEDGMENT
The authors thank Dr Karen Hagerty, Andre Quina, and Mark Kramer for their thoughtful review of and input to the manuscript.
SUPPORT
Supported by National Institutes of Health, National Cancer Institute Grant No. P30CA068485 (T.J.O.).
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Collection and assembly of data: All authors
Data analysis and interpretation: Travis J. Osterman, Robert S. Miller
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Travis J. Osterman
Stock and Other Ownership Interests: Infostratix
Consulting or Advisory Role: eHealth, AstraZeneca, Outcomes Insights, Biodesix, MD Outlook, GenomOncology, Cota Healthcare
Research Funding: GE Healthcare, Microsoft
Travel, Accommodations, Expenses: GE Healthcare
No other potential conflicts of interest were reported.
REFERENCES
- 1.Abernethy AP, Etheredge LM, Ganz PA, et al. Rapid-learning system for cancer care. J Clin Oncol. 2010;28:4268–4274. doi: 10.1200/JCO.2010.28.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patt D, Stella P, Bosserman L. Clinical challenges and opportunities with current electronic health records: Practicing oncologists’ perspective. J Oncol Pract. 2018;14:577–579. doi: 10.1200/JOP.18.00306. [DOI] [PubMed] [Google Scholar]
- 3.Conway JR, Warner JL, Rubinstein WS, et al. doi: 10.1200/PO.19.00232. Next-generation sequencing and the clinical oncology workflow: Data challenges, proposed solutions, and a call to action. Precision Oncol 2019. https://ascopubs.org/doi/10.1200/PO.19.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Developing the medical oncology treatment plan and summary. J Oncol Pract. 2006;2:95–96. doi: 10.1200/jop.2006.2.2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. https://www.asco.org/sites/new-www.asco.org/files/content-files/practice-and-guidelines/documents/2009-asco-nci-core-white-paper.pdf American Society of Clinical Oncology: Clinical Oncology Requirements for the EHR (CORE). 2009.
- 6.Warner J. L., Maddux SE, Hughes KS, et al. Development, implementation, and initial evaluation of a foundational open interoperability standard for oncology treatment planning and summarization. J Am Med Inform Assoc. 2015;22:577–586. doi: 10.1093/jamia/ocu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. https://www.hl7.org/implement/standards/product_brief.cfm?product_id=327 HL7 International: Section 1a: Clinical Document Architecture (CDA).
- 8. https://www.congress.gov/bill/114th-congress/house-bill/34 114th Congress: H.R.34 - 21st Century Cures Act. 2016.
- 9.Rucker D. https://www.healthaffairs.org/do/10.1377/hblog20180618.138568/full/ Achieving the interoperability promise of 21st century cures. Health Aff. June 19, 2018.
- 10. https://www.healthit.gov/curesrule/ Office of the National Coordinator for Health Information Technology: ONC’s Cures Act Final Rule. HealthIT.gov website.
- 11. https://www.cms.gov/Regulations-and-Guidance/Guidance/Interoperability/index Centers for Medicare and Medicaid Services: CMS Interoperability and Patient Access final rule.
- 12. https://www.healthit.gov/isa/united-states-core-data-interoperability-uscdi Office of the National Coordinator for Health Information Technology: United States Core Data for Interoperability (USCDI).
- 13.Schilsky RL, Michels DL, Kearbey AH, et al. Building a rapid learning health care system for oncology: The regulatory framework of CancerLinQ. J Clin Oncol. 2014;32:2373–2379. doi: 10.1200/JCO.2014.56.2124. [DOI] [PubMed] [Google Scholar]
- 14. doi: 10.2217/fon-2017-0521. https://www.futuremedicine.com/doi/10.2217/fon-2017-0521 Miller RS, Wong JL: Using oncology real-world evidence for quality improvement and discovery: The case for ASCO’s CancerLinQ. Future Medicine. 2017. [DOI] [PubMed]
- 15.Bertagnolli MM, Anderson B, Norsworthy K, et al. Status update on data required to build a learning health system. J Clin Oncol. 2020;38:1602–1607. doi: 10.1200/JCO.19.03094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chambers DA, Amir E, Saleh RR, et al. The impact of big data research on practice, policy, and cancer care. Am Soc Clin Oncol Educ Book. 2019;39:e167–e175. doi: 10.1200/EDBK_238057. [DOI] [PubMed] [Google Scholar]
- 17. http://hl7.org/fhir/us/mcode/ HL7 International: HL7 FHIR Implementation Guide: minimal Common Oncology Data Elements.
- 18. http://hl7.org/fhir/us/core/STU3.1/ValueSet-us-core-medication-codes.html HL7 International: HL7 FHIR US Core Implementation Guide STU3 Release 3.1.0.
- 19. https://www.nlm.nih.gov/research/umls/rxnorm/ National Library of Medicine: Unified Medical Language System. RxNorm.
- 20.American Joint Committee on Cancer Cancer staginghttps://cancerstaging.org/Pages/default.aspx
- 21. https://www.ncbi.nlm.nih.gov/clinvar/ National Center for Biotechnology Information. ClinVar.
- 22.Human Genome Variation Society About the society [homepage]http://www.hgvs.org/
- 23.HUGO Gene Nomenclature Committee [HUGO homepage]https://www.genenames.org/
- 24. https://seer.cancer.gov/icd-o-3/index.html National Cancer Institute, Surveillance, Epidemiology, and End Results Program: ICD-O-3 coding materials.
- 25. https://www.cms.gov/Medicare/Coding/ICD10/2020-ICD-10-CM Centers for Medicare & Medicaid Services: 2020 ICD-10-CM. [PubMed]
- 26. https://www.cms.gov/Medicare/Coding/ICD10/2020-ICD-10-PCS Centers for Medicare & Medicaid Services: 2020 ICD-10-PCS. [PubMed]
- 27. https://loinc.org/ Regenstrief Institute: LOINC.
- 28. http://www.snomed.org/ SNOMED International: [Homepage].
- 29. https://confluence.hl7.org/display/COD/CodeX+Home HL7 International: Code X home.
- 30. http://www.hl7.org/Special/committees/cic/overview.cfm HL7 International: Clinical Interoperability Council.
- 31. https://mcodeinitiative.org American Society of Clinical Oncology: mCODE: Minimal Common Oncology Data Elements.


