INTRODUCTION
Utilization of checkpoint inhibition therapy in the management of malignant diseases is increasing. However, response to these therapies is highly variable, and robust predictive markers remain elusive. Identification of predictive biomarkers is crucial considering the toxicities and high financial cost associated with this approach. Here, we report an adolescent with relapsed clear cell adenocarcinoma of the cervix (CCAC) who experienced a complete response to checkpoint blockade despite not exhibiting positive PD-L1 immunohistochemical (IHC) staining, microsatellite instability (MSI), or a high tumor mutational burden (TMB). Using whole-genome sequencing (WGS) and RNA sequencing (RNA-seq), we identified a high neoantigen burden and an APOBEC mutational signature that we suggest may explain her exceptional response.
Although the standard of care for relapsed cervical carcinoma is conventional cytotoxic chemotherapy (cisplatin and paclitaxel) with bevacizumab, this therapy yields dismal outcomes in adults, with a median overall survival time of 17 months.1 In the CheckMate 358 trial evaluating nivolumab, five of 19 patients with advanced gynecologic cancers had an objective response.2 More favorable responses were achieved with nivolumab plus ipilimumab at shorter intervals.3 Intriguingly, results were similar among women with and without PD-L1–positive disease. Therefore, we elected to treat the patient with nivolumab (3 mg/kg every 2 weeks) instead of cytotoxic chemotherapy despite the absence of biomarkers believed to predict favorable responses in other cancers.
CASE REPORT
The patient is a previously healthy 15-year-old girl who presented with vaginal discharge. Biopsy of an exophytic mass protruding into the vagina confirmed a human papillomavirus–/p16-negative CCAC (Fig 1). Family history was notable for paternal clear cell renal carcinoma. The patient underwent radical hysterectomy, bilateral salpingectomy, oophoropexy, and lymph node dissection, followed by cisplatin and 45 Gy of radiation to the pelvis. Sixteen months after entering remission, surveillance positron emission tomography (PET)–computed tomography (CT) revealed new hypermetabolic disease, including a new right upper lobe (RUL) pulmonary nodule, enlarged prevascular right internal mammary, and right paratracheal nodes (Fig 2A) and a left supraclavicular node. Biopsy of the supraclavicular node confirmed recurrence of CCAC (Fig 3).
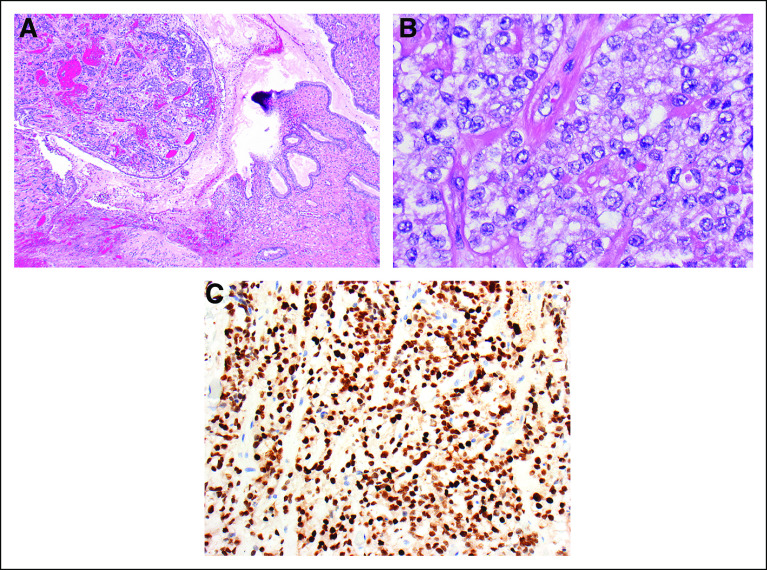
FIG 1.
Pathologic confirmation of clear cell carcinoma of the cervix. (A) Primary clear cell carcinoma of the cervix, showing hyperchromatic tumor cell nests in the stroma at left and normal endocervical glandular epithelium at the right (×40). (B) The tumor cells grow in small nests and trabeculae and have vesicular nuclei with prominent nucleoli. Mitotic figures are sparse, as is typical of this tumor type. The cells have abundant amphophilic to clear cytoplasm (×400). (C) An immunohistochemical stain for hepatocyte nuclear factor 1-β, which is a marker of clear cell carcinoma, shows strong positive nuclear staining in the tumor cells, a positive test result (×200).
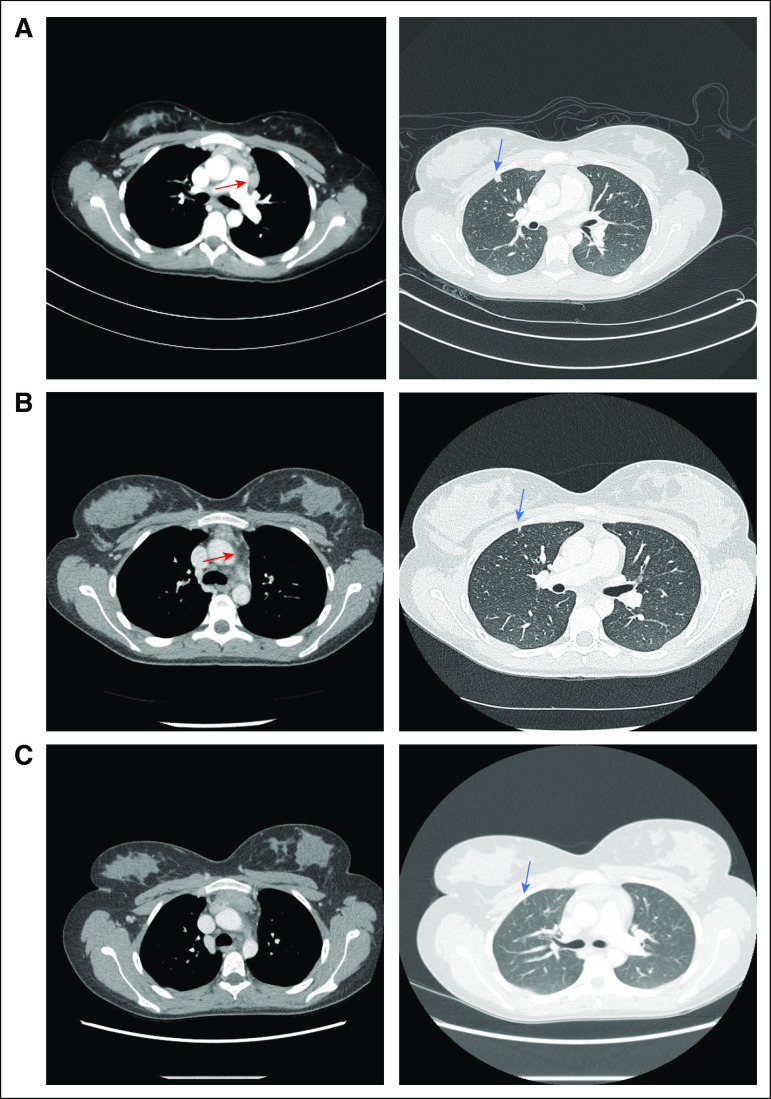
FIG 2.
Radiographic response to checkpoint blockade. (A) Patient developed a new hypermetabolic pulmonary nodule in the right upper lobe (blue arrow), along with prominent hypermetabolic prevascular nodes (red arrow). (B) After initial nivolumab therapy, patient’s right upper lobe nodule decreased in size (blue arrow), and the prevascular node had nearly resolved (red arrow). (C) Approximately 16 months after initial recurrent disease, the pulmonary nodule has nearly completely resolved (blue arrow), and no new pathologic nodes have developed.
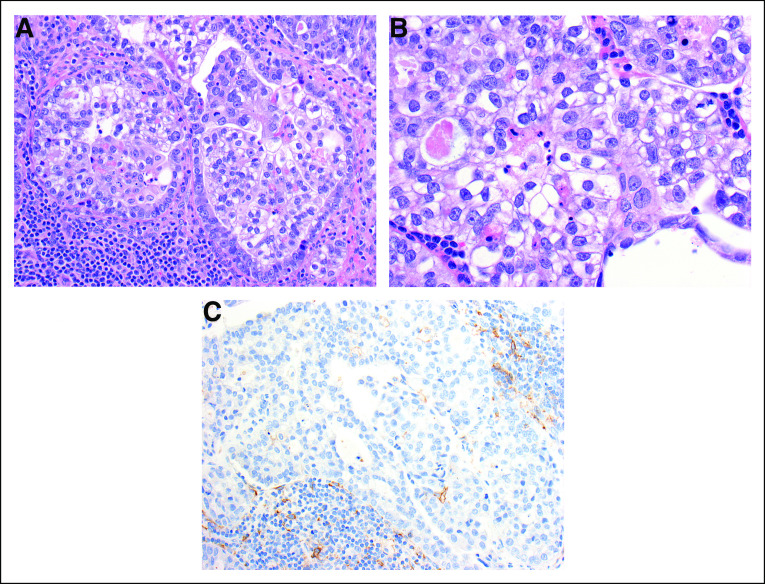
FIG 3.
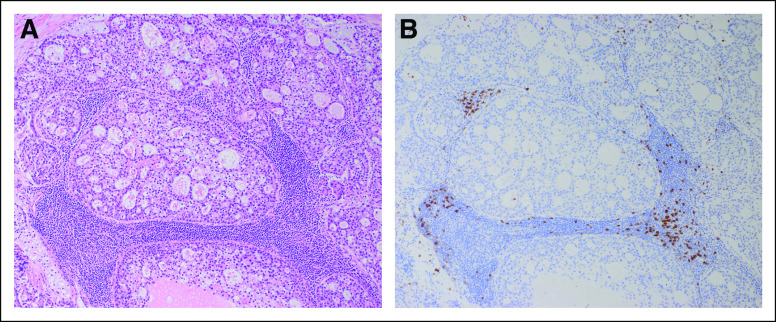
Biopsy of supraclavicular node confirms metastatic disease. (A) Metastatic clear cell carcinoma in a supraclavicular lymph node. The tumor cells line glands or grow in nests and are surrounded by lymphocytes, seen at the lower left (×200). (B) Metastatic clear cell carcinoma in a supraclavicular lymph node. The tumor cells are mainly polygonal with abundant clear or amphophilic cytoplasm and vesicular nuclei with prominent nucleoli. The appearance is similar to that seen in the cervical primary (×400). (C) Metastatic clear cell carcinoma in a supraclavicular lymph node. An immunohistochemical stain for PD-L1 shows minimal staining in the tumor cells and staining of scattered lymphoid cells around the tumor (×200).
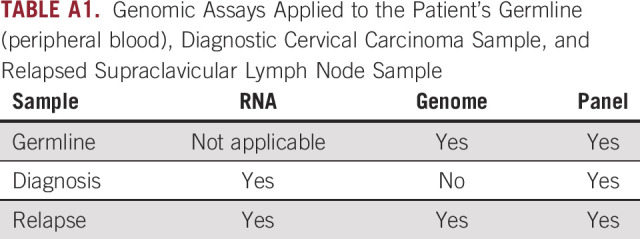
Her relapsed tumor exhibited 0% staining for PD-L1 by IHC, microsatellite stability (1,149 of 1,157 tested microsatellite regions were stable), and a low TMB (1.9 single nucleotide variants per megabase on WGS, less than the pediatric threshold of 2; Appendix). CD8+ lymphocytes were absent (Appendix Fig A1). Genomic assays performed included targeted DNA sequencing, WGS, and RNA-seq (Appendix Table A1).
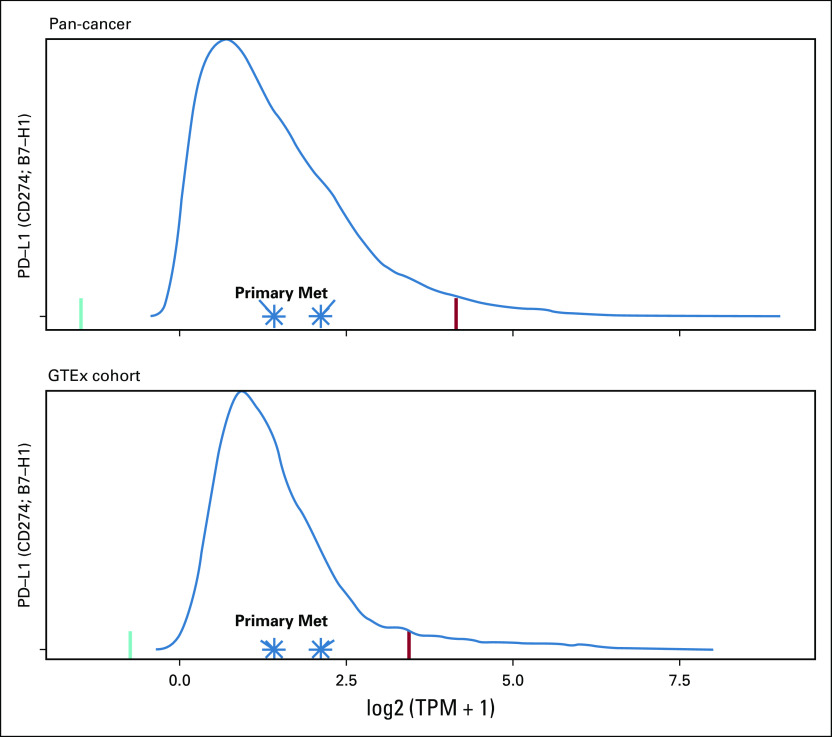
A targeted institutional DNA-seq panel assaying 479 cancer-relevant genes4 showed that both diagnostic and relapsed tumors had a broad approximately 11-fold amplification on chromosome 20 containing Aurora kinase A as a putative driver.5 The diagnostic sample also displayed a chromosome 1p deletion containing ARID1A, a gene frequently mutated in cervical cancer,6 which was absent at relapse. No pathogenic point mutations, insertions/deletions (indels), or germline alterations were detected on this panel at diagnosis or relapse. Findings from WGS and targeted sequencing were concordant (Appendix Fig A2). RNA-seq demonstrated normal expression of PD-L1 at diagnosis and relapse (Appendix Fig A3) and also detected the chromosome 20 amplification and 1p deletion at diagnosis.
After four doses of nivolumab monotherapy, PET-CT scan showed decreased size and hypermetabolism of the RUL pulmonary nodule and the prevascular lymph node (Fig 2B). The other mediastinal lymph nodes decreased in size, although hypermetabolism was not evaluable as a result of development of an intense background of hypermetabolic brown fat. Nivolumab was continued as monotherapy for 11 doses, with only the prevascular node and small RUL pulmonary nodule persisting. Ipilimumab (1 mg/kg every 3 weeks), a monoclonal antibody against CTLA-4, was then added based on superior outcomes with the combination in adults with melanoma.7 After one combined dose, CT demonstrated continued interval decrease in the size of the prevascular node and complete resolution of the pulmonary nodule, but therapy was discontinued as a result of inflammatory colitis. One month later, nivolumab monotherapy was restarted, and the patient achieved a complete radiographic remission, which has persisted for 18 months (Fig 2C).
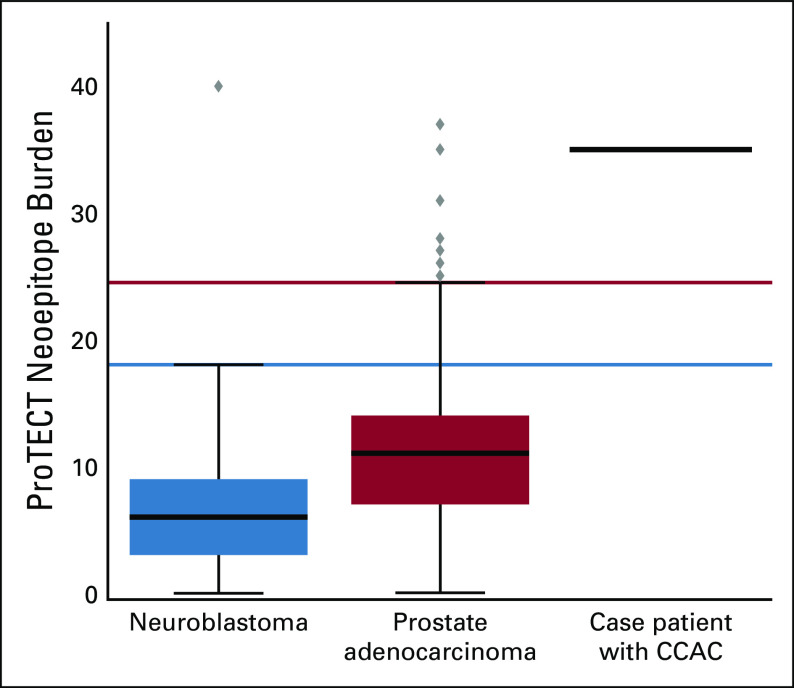
We used the epitope prediction algorithm Prediction of T-Cell Epitopes for Cancer Therapy (ProTECT)8 to evaluate the relapsed tumor, yielding a neoepitope burden of 35 high-affinity neoepitopes (of which 14 had a variant allele frequency > 0.4, suggesting that a large fraction of tumor cells express these neoepitopes). In contrast, the mean neoepitope burdens for available control cohorts, including neuroblastoma and prostate, were only 6.7 (95% CI, 5.6 to 7.7 neoepitopes) and 12.7 (95% CI, 10.6 to 14.9 neoepitopes), respectively.9 The tumor’s neoepitope burden was a statistical outlier relative to these cohorts, crossing the outlier thresholds for both (Fig 4).
FIG 4.
High neoepitope burden compared with pediatric neuroblastoma and adult prostate adenocarcinoma cohorts using Prediction of T-Cell Epitopes for Cancer Therapy (ProTECT) neoepitope analysis. Box-and-whisker plots display the interquartile range (box) and outlier thresholds (whiskers) for the Therapeutically Applicable Research to Generate Effective Treatments neuroblastoma and The Cancer Genome Atlas prostate adenocarcinoma data sets. A solid black line represents the patient. CACC, clear cell adenocarcinoma of the cervix.
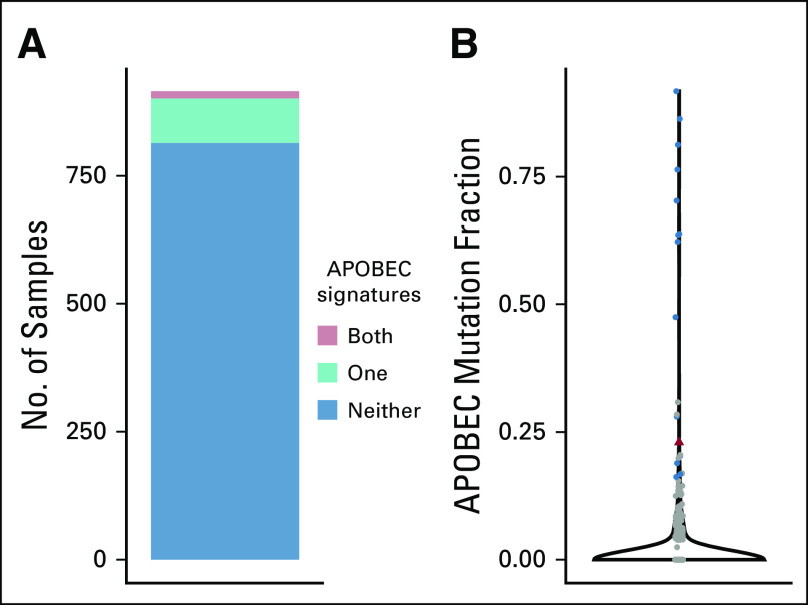
We next sought to evaluate the mutational signatures at relapse. Somatic mutations in cancer genomes are caused by mutational processes including DNA damage, modification, repair, and replication, which generate characteristic mutational signatures.10 Single base substitution mutational signature analysis of WGS data identified five active signatures out of 30 (Fig 5), including the AID/APOBEC family of enzymes.11 Although the APOBEC mutational signature is common in adult cancers, it is rare in pediatric cancers.11 Using pediatric cancer genomic and transcriptomic data from 915 tumor samples published by Ma et al,11 we determined that 98 (10.7%) of 915 had signature 2, 17 (1.9%) had signature 13, and only 14 (1.5%) had both APOBEC signatures (Fig 6).
FIG 5.
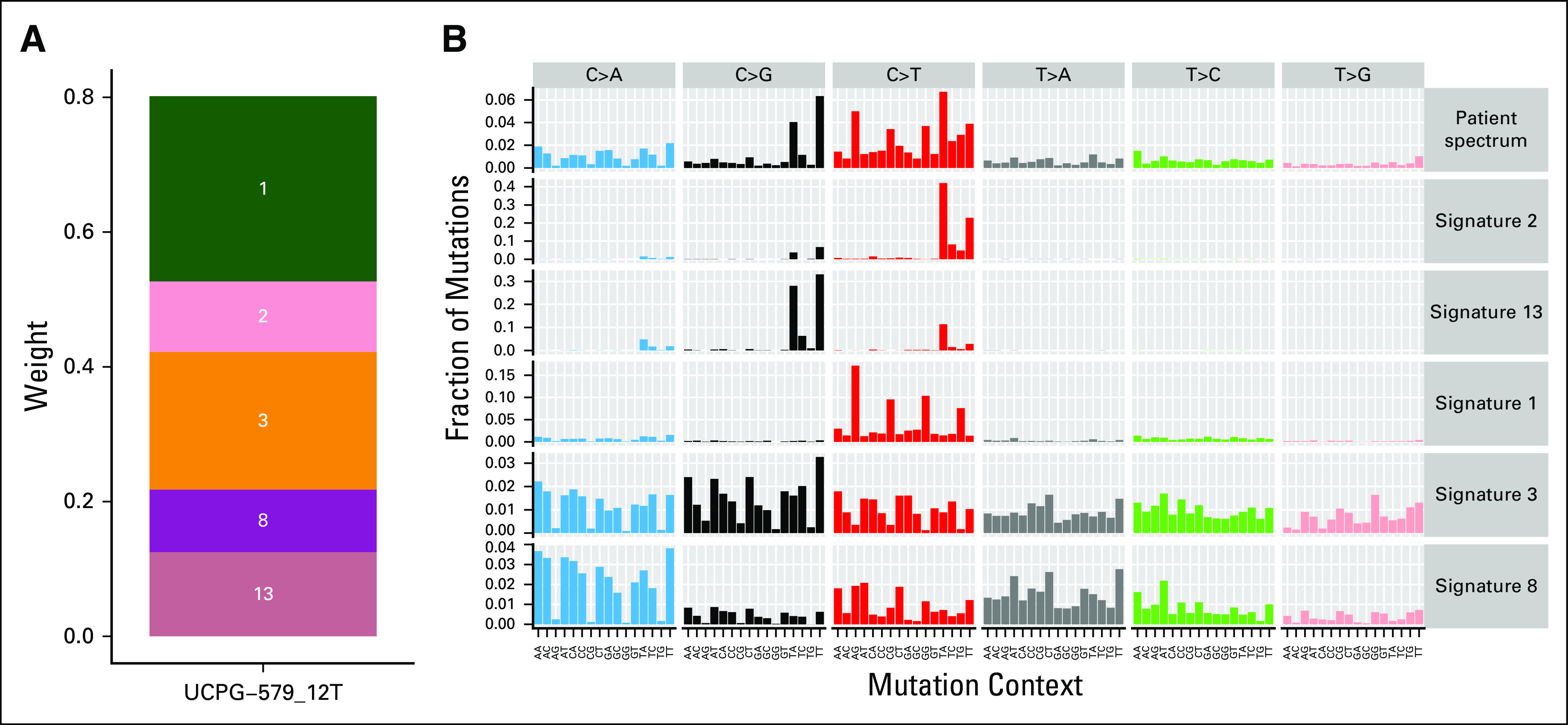
Mutational signature analysis identifies five active signatures. (A) Each signature is shown as a segment on the plot, with the names of the signatures overlaid. The size of the segment corresponds to its weight. The greater the weight, the more active the signature is (ie, the more mutations it has caused). (B) Plot of the mutational spectrum of the patient’s diagnostic sample alongside the five active signatures.
FIG 6.
APOBEC mutational signature frequency in a large pediatric cancer cohort. (A) Numbers of pediatric cancer samples with one or both APOBEC signatures (signatures 2 and 13). Fourteen (1.5%) of 915 samples from Ma et al11 display both signatures. (B) Violin plot of the fraction of mutations in each sample attributed to the combined APOBEC signatures. Samples with both signatures are shown in blue, samples with one or neither signature are shown in gray, and the patient sample is shown in red.
DISCUSSION
CCAC, which is historically linked to prenatal exposure to diethylstilbestrol, represents only 4% to 9% of cervical adenocarcinomas.12 A recent multi-institutional review of CCAC identified 34 patients with CCAC, of whom two had CCAC associated with diethylstilbestrol. The median age was 53 years, and only three patients were < 30 years of age,13 making the patient presented here one of the youngest patients ever reported with idiopathic CCAC. The genomic landscape of CCAC has not been published, although among 24 patients in a 1996 analysis, more than half had evidence of MSI,14 suggesting a possible role for checkpoint inhibition. An isolated case of POLE-mutated DES-associated CCAC with high PD-L1 expression and elevated tumor-infiltrating lymphocytes has also been reported.15
Known biomarkers for response to checkpoint inhibition are IHC staining for PD-L1, presence of MSI, and high TMB. This patient, who achieved a complete and durable remission from checkpoint inhibition without any of these markers, highlights the need for additional biomarkers. We sought to analyze the genomics of her tumors to explain her dramatic response.
Somatic variations that give rise to amino acid substitutions in tumors yield neoepitopes, or mutated, tumor-specific peptides on the surface of cancer cells that can serve as neoantigens for the adaptive immune system, even if they are not oncogenic. Determinants of neoantigen fitness are the likelihood of their presentation by the major histocompatibility complex and of subsequent T-cell recognition.16 The number of neoepitopes per tumor can be more functionally relevant than the TMB. This idea is supported by the fact that renal cell cancers (including clear cell carcinoma) respond well to checkpoint inhibition despite having low TMB but display an exceptionally high frequency of immunogenic indel-derived tumor-specific neoantigens.17 In fact, renal cell carcinomas have the highest pan-cancer number of indels.17
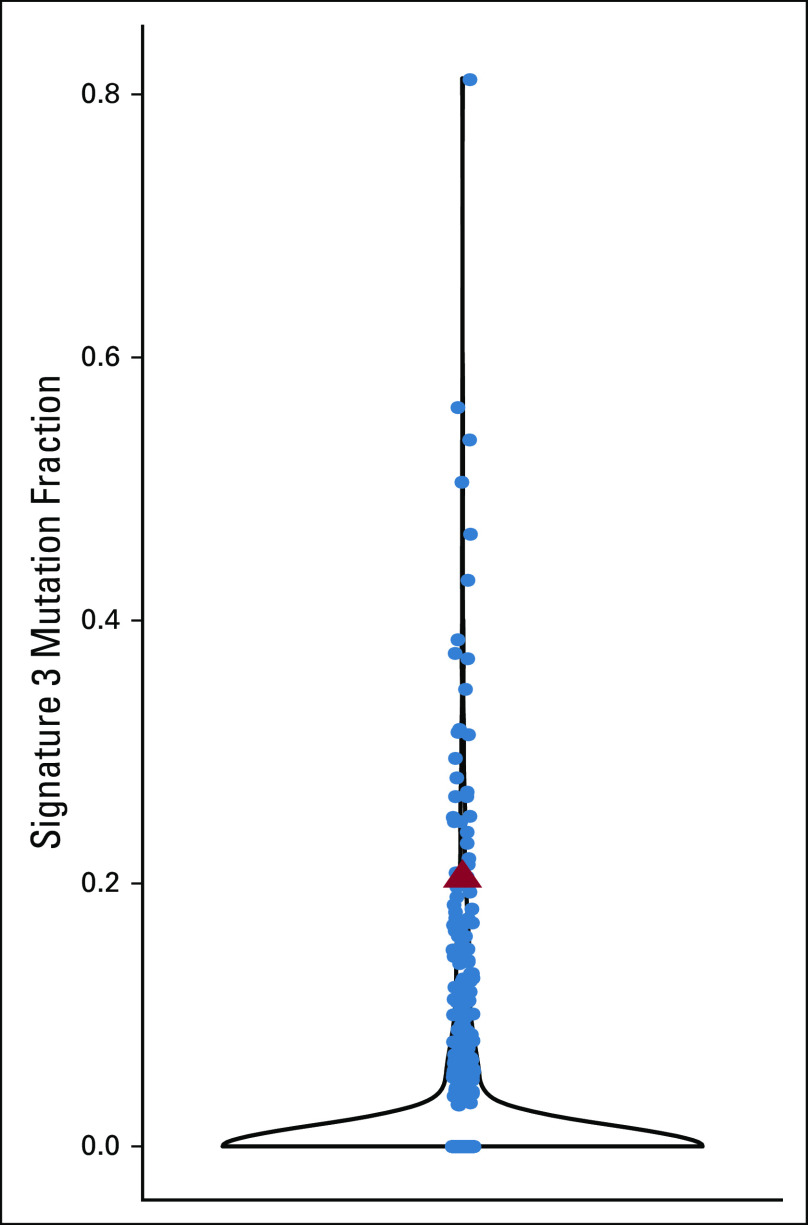
The ProTECT epitope prediction algorithm yielded a neoepitope burden of 35 high-affinity neoepitopes in our patient’s relapsed tumor, higher than that of available (although biologically distinct) comparison cohorts, supporting the idea that high neoepitope burden may predict favorable response to immunotherapy. Single base substitution mutational signature analysis of WGS data identified five active signatures. Of these, signatures 1 (aging) and 3 (homologous recombination deficiency; Appendix Fig A4) are commonly found in cancer.18,19 Signatures 2 and 13 represent activity of the AID/APOBEC family of enzymes.18,19
APOBEC, a family of zinc-coordinating enzymes that convert cytosines to uracils, has been implicated in mutagenesis of non–small-cell lung cancer. Systematic cancer genomic and transcriptomic association studies have shown that overexpression of one of the family members, APOBEC3B, is associated with expression of immune genes and known immunotherapy response biomarkers such as PD-L1.20 A positive correlation was recently documented between strength of the APOBEC signature and the neoepitope burden in many cancers, including cervical.21 The authors also found that this mutational signature corresponded to increased abundance of tumor-infiltrating lymphocytes in some cancers,21 although these were absent in this case. Clinically, the APOBEC mutational signature is enriched in patients with durable clinical benefit after immunotherapy, and an APOBEC signature may be better than TMB in predicting immunotherapy response.22,23 Finally, there was a small contribution from signature 8 of uncertain etiology (it is perhaps related to nucleotide excision repair deficiency).24
The present patient is of interest for several reasons. This patient is one of the youngest reported individuals with non–diethylstilbestrol-associated CCAC. Upon relapse, she demonstrated a complete response to anti–PD-1 and anti–CTLA-4 checkpoint blockade, despite the fact that her tumor did not exhibit PD-L1 staining by IHC and had a low TMB and MSI. The case highlights the promise of checkpoint blockade in relapsed cervical cancer and the need for more comprehensive biomarker development in the field of immunotherapy. Although there may be other factors that contributed to the patient’s dramatic response, this case supports emerging evidence that a high neoepitope burden and an APOBEC mutational signature are potentially actionable biomarkers of response to checkpoint blockade. Finally, these findings support the routine use of larger-footprint sequencing panels or, where possible, WGS in children with advanced cancer to detect potentially actionable mutational signatures.
ACKNOWLEDGMENT
We thank the patient and her family for their willingness to share their experience using an unconventional treatment approach. Written consent to publish the information contained in this article was obtained from the patient and her mother.
APPENDIX
Tissue Source, Processing, and Analysis
The cervical resection from the patient was formalin fixed and paraffin embedded (FFPE). The lymph node from the patient was snap frozen and FFPE into optimal cutting temperature (OCT) medium. All tissues were sectioned to a depth of 5 μm and stained with hematoxylin and eosin. Samples were evaluated for tumor content by a certified pathologist. For FFPE samples, sections were deparaffinized with xylene and nucleic acid extracted using the Allprep DNA/RNA FFPE micro kit 80234 (QIAGEN, Hilden, Germany).
For FFPE samples, tumor was macrodissected from the OCT block to a depth of up to 5 mm, disrupted with a mortar and pestle under liquid nitrogen, and homogenized with a QIAshredder 79654 (QIAGEN). Nucleic acids were extracted using the AllPrep DNA/RNA micro kit 80204 (QIAGEN). RNA integrity was quantified with the High Sensitivity RNA kit DNF-472 (Advanced Analytical Technologies, Orangeburg, NY) on the Fragment Analyzer (Advanced Analytical Technologies). Immunohistochemical PD-L1 staining was performed on tumor cells with the US Food and Drug Administration–approved PD-L1 28-8 pharmDx antibody for OPDIVO (NeoGenomics Laboratories, Fort Myers, FL).
Whole blood was collected during lymph node metastasectomy in a 4-mL EDTA vacutainer. Germline DNA was extracted using the DNeasy Blood and Tissue kit 69504 (QIAGEN).
DNA was quantified using the Nanodrop 2000 (Thermo Fisher, Waltham, MA) and QuBit High Sensitivity dsDNA assay Q32851 (Thermo Fisher) and integrity quantified using the High Sensitivity gDNA kit DNF488 (Advanced Analytical Technologies) on the Fragment Analyzer (Advanced Analytical Technologies). RNA was quantified using the Nanodrop 2000 and integrity quantified with the High Sensitivity RNA kit DNF-472 (Advanced Analytical Technologies) on the Fragment Analyzer. RNA proceeded on to library generation if the RIN was > 7 or DV200 > 30. DNA proceeded if the DIN was > 7.
RNA Sequencing
Preparation.
Libraries from FFPE samples were made using the TruSeq RNA Access kit (RS-301-2001; Illumina, San Diego, CA) with an input of 100 ng in accordance with manufacturer’s instructions. Libraries from FFPE samples were made using the TruSeq Stranded mRNA kit (RS-122-2101; Illumina) with an input of 400 ng in accordance with manufacturer’s instructions. All manufacturer’s controls were used in preparation. Libraries were quantified using the High Sensitivity NGS kit (DNF-474; Advanced Analytical Technologies) on the Fragment Analyzer. Libraries from FFPE and FFPE samples were sequenced on the Illumina NovaSeq using chemistry for 2 × 150 bp reads and on the Illumina HiSEquation 400 using chemistry for 2 × 100 bp reads, respectively.
Analysis.
All external cancer data were downloaded from the University of California, Santa Cruz (UCSC) treehouse childhood cancer initiative public portal site (https://treehousegenomics.soe.ucsc.edu/), version v11 (Vaske OM, et al: JAMA Netw Open 2:e1913968, 2019). The Genotype-Tissue Expression (GTEx) data set was downloaded directly from the xena browser (UCSC Genomic Institute). To compare the two RNA sequencing (RNA-seq) samples, we derived transcripts per million (TPM) counts using the same UCSC rnaseq-cgl-pipeline (https://github.com/BD2KGenomics/toil-rnaseq/tree/master/docker), docker version 6d5313d0706 (Vivian J, et al: Nat Biotechnol 35:314-316, 2017). This pipeline aligns the reads using STAR (hg38) and RSEM for the final TPM counts. Thereafter, gene counts were aggregated with summation and transformation with log2 (X + 1). By producing TPM with this method, we are ensuring that the TPM counts were harmonized with the two external data sets. To calculate outliers, we plotted expression against either the entire cancer cohort or normal GTEx cohort. Outliers (ie, under- or overexpression) were identified using Tukey outlier definition.
Targeted DNA Sequencing
An institutional DNA sequencing panel assaying 479 cancer-related genes was used.4 As per Kline et al,4 genomic DNA was extracted from peripheral blood and tumor tissue microdissected from FFPE blocks. Capture-based next-generation sequencing (NGS) was performed at the University of California, San Francisco (UCSF) Clinical Cancer Genomics Laboratory, using an assay targeting the coding regions of these genes, TERT promoter, select introns from 40 genes (for detection of gene fusions and other structural variants), and intergenic regions at regular intervals along each chromosome (for chromosomal copy number assessment), altogether with a total sequencing footprint of 2.8 Mb (UCSF500 Cancer Gene Panel, Appendix Fig A2). Sequencing libraries were prepared from genomic DNA with target enrichment performed by hybrid capture using a custom oligonucleotide library. Sequencing was performed on an Illumina HiSEquation 2500. Duplicate sequencing reads were removed computationally to allow for accurate allele frequency determination and copy number estimates. The analysis was based on the human reference sequence UCSC build hg19 (NCBI build 37). Single nucleotide variants and small insertions/deletions (indels) were visualized and verified using Integrated Genome Viewer. A list of genes included can be found at the following URL (although some genes may have been added or deleted since this patient’s analysis in 2017): https://genomics.ucsf.edu/content/ucsf-500-cancer-gene-panel-test-ucsf500-uc500.
Whole-Genome Sequencing
Libraries were generated using the TruSeq Nano kit FC-121-4001 (Illumina) with a 350-bp insert, per manufacturer's instructions. Libraries were made using 200-ng input gDNA, and then quantified on the TapeStation 4200 (Agilent, Santa Clara, CA) and sequenced on the HiSeq X Ten (Illumina) using 2 × 150 bp paired-end reads. One germline library and two sets of somatic libraries were sequenced for 30× and 60× coverage, respectively. Whole-genome sequencing (WGS) data were aligned to the GRCh38 reference genome using bwa-mem (Li H: https://arxiv.org/abs/1303.3997) and variants called using MuTect2 (Cibulskis K, et al: Nat Biotechnol 31:213-219, 2013). Tumor mutation burden was calculated as the total number of single nucleotide variants over the length of the female reference genome (in megabases). Nonsynonymous variants were annotated using Variant Effect Predictor (McLaren W, et al: Genome Biol 17:122, 2016).
Predicted Tumor Neoepitope Analysis
We used the Prediction of T-cell Epitopes for Cancer Therapy (ProTECT) pipeline v 2.6.18 to investigate the patient’s neoepitope burden. ProTECT is built on a scalable pipeline management system and uses state-of-the-art mutation calling and MHC binding prediction algorithms to identify and rank neoepitopes. We identified neoepitopes using tumor WGS and RNA-seq data generated from our patient’s biopsied metastasis and normal WGS data. We used the hg38_references under the AWS S3 bucket protect-data. The GRCh38 reference sequence and the GENCODE v25 reference annotation were used to generate the indexes and reference files. Default ProTECT parameters were used for all tools, except the Transgene tool was updated to version 2.6.1. Neoepitopes derived from fusion genes were manually filtered to remove fusions associated with mitochondrial and immunoglobulin genes as well as transcription read-through events. We compared the patient’s tumor mutation and neoepitope burden to publicly available ProTECT data for the Therapeutically Applicable Research to Generate Effective Treatments neuroblastoma (PMID: 23334666) and The Cancer Genome Atlas prostate adenocarcinoma data sets (PMID: 26544944) and assessed for significance using Tukey’s outlier definition.
APOBEC Signature Analysis
Single base substitution mutational signatures were extracted from the sample’s WGS data using deconstructSigs (Rosenthal R, et al: Genome Biol 17:31, 2016), with default parameters. The COSMIC V2 Mutational Signature Set19 was used as the reference signature set.
FIG A1.
Few CD8+ lymphocytes are present within and around the patient’s relapsed tumor. (A) Hematoxylin and eosin stain at ×10 magnification. (B) CD8 stain at ×10 magnification shows rare intratumoral CD8+ lymphocytes without convincing peritumoral condensation of CD8+ lymphocytes, which are confined to the interfollicular zones of the background lymph node.
FIG A2.
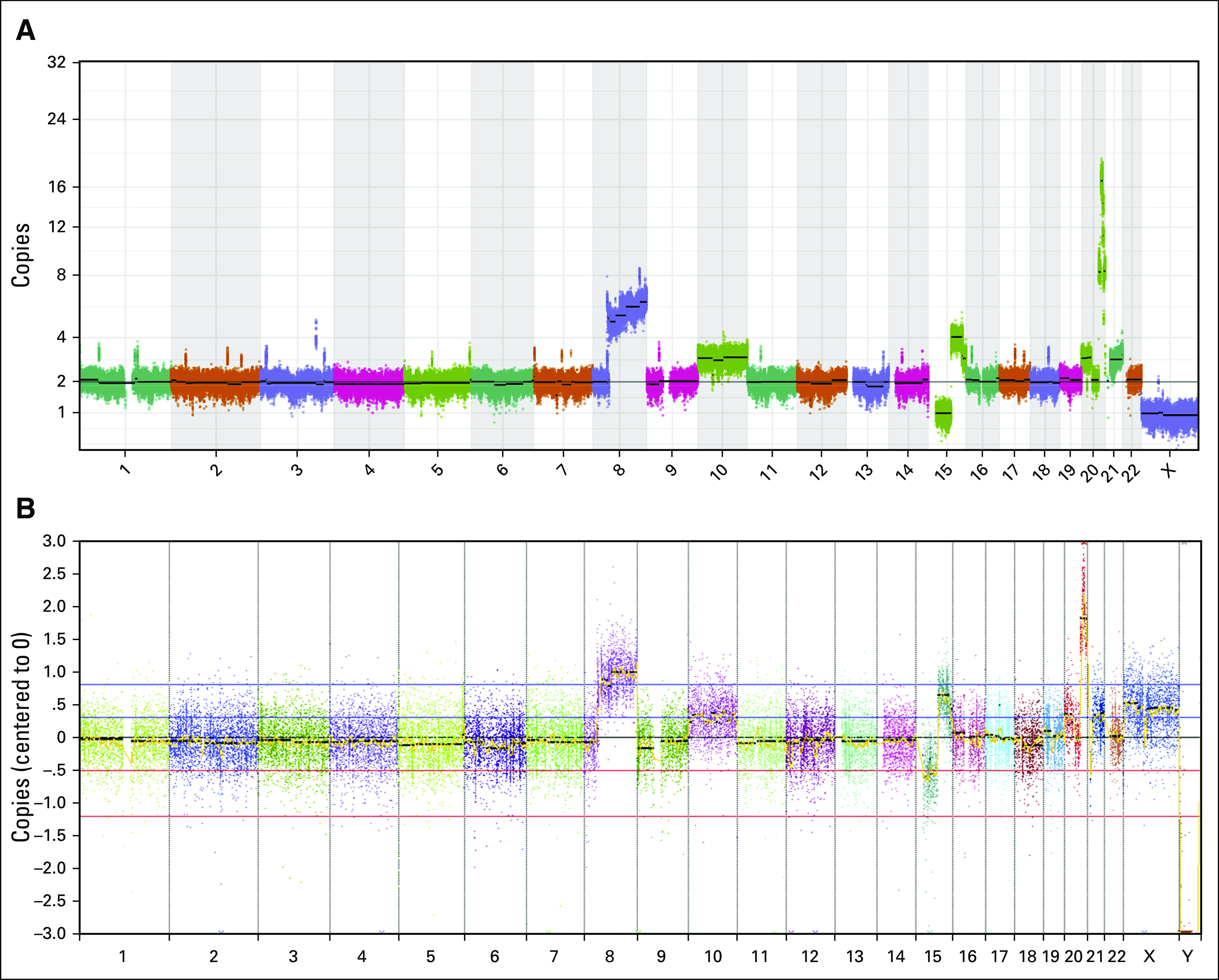
Copy number variant (CNV) analyses are concordant between targeted sequencing and whole-genome sequencing (WGS). (A) CNV plot from WGS. (B) CNV plot from targeted sequencing panel.
FIG A3.
The patient’s primary and relapsed samples have normal PD-L1 expression on RNA sequencing (RNA-seq) when compared with cancer and normal tissue cohorts. Both primary and relapsed or metastatic (Met) samples have normal PD-L1 expression on RNA-seq. Transcripts per million (TPM) were plotted against (A) a cancer cohort (n = 12,748) and (B) Genotype-Tissue Expression (GTEx; normal tissue; n = 7,795) cohorts. Green and red lines represent the lower and upper Tukey outlier bounds, respectively.
FIG A4.
The patient’s sample is notable for a signature 3 (homologous recombination deficiency [HRD]) signature. Violin plot of the fraction of mutations in each sample from Ma et al11 attributed to signature 3. The patient’s sample is represented as a red triangle.
TABLE A1.
Genomic Assays Applied to the Patient’s Germline (peripheral blood), Diagnostic Cervical Carcinoma Sample, and Relapsed Supraclavicular Lymph Node Sample
EQUAL CONTRIBUTION
E.A.S.-C. and E.S. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Anya Levinson, Jacob Pfeil, Inge Behroozfard, E. Alejandro Sweet-Cordero, Elliot Stieglitz
Financial support: Elliot Stieglitz, E. Alejandro Sweet-Cordero
Administrative support: Elliot Stieglitz, E. Alejandro Sweet-Cordero
Provision of study materials or patients: Jessica Van Ziffle, M. Dwight Chen, Elliot Stieglitz
Collection and assembly of data: Anya Levinson, Marcus R. Breese, Charles Zaloudek, Jessica Van Ziffle, Stanley G. Leung, M. Dwight Chen, Lee-may Chen, Jacob Pfeil, Nicholas R. Ladwig, Avanthi Tayi Shah, E. Alejandro Sweet-Cordero
Data analysis and interpretation: Anya Levinson, Alex G. Lee, Henry J. Martell, Marcus R. Breese, Charles Zaloudek, Benjamin Laguna, Jacob Pfeil, Nicholas R. Ladwig, Arjun Arkal Rao, Sofie R. Salama, E. Alejandro Sweet-Cordero, Elliot Stieglitz
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jessica Van Ziffle
Employment: Adaptive Biotechnologies (I)
Stock and Other Ownership Interests: Adaptive Biotechnologies (I)
Patents, Royalties, Other Intellectual Property: Adaptive Biotechnologies (I)
Benjamin Laguna
Leadership: Sira Medical
Stock and Other Ownership Interests: Sira Medical
Stanley G. Leung
Stock and Other Ownership Interests: Illumina, Pfizer, Thermo Fisher
Jacob Pfeil
Employment: AbbVie
No other potential conflicts of interest were reported.
REFERENCES
- 1.Tewari KS, Sill MW, Long HJ, III, et al. Improved survival with bevacizumab in advanced cervical cancer. N Engl J Med. 2014;370:734–743. doi: 10.1056/NEJMoa1309748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naumann RW, Hollebecque A, Meyer T, et al. Safety and efficacy of nivolumab monotherapy in recurrent or metastatic cervical, vaginal, or vulvar carcinoma: Results from the phase I/II CheckMate 358 trial. J Clin Oncol. 2019;37:2825–2834. doi: 10.1200/JCO.19.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naumann RW: Nivolumab-ipilimumab combination benefits women with advanced cervical cancer. European Society for Medical Oncology Meeting, Barcelona, Spain, September 27-October 1, 2019 (abstr LBA62) [Google Scholar]
- 4.Kline CN, Joseph NM, Grenert JP, et al. Targeted next-generation sequencing of pediatric neuro-oncology patients improves diagnosis, identifies pathogenic germline mutations, and directs targeted therapy. Neuro Oncol. 2017;19:699–709. doi: 10.1093/neuonc/now254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glasspool RM, McNeish IA. Clear cell carcinoma of ovary and uterus. Curr Oncol Rep. 2013;15:566–572. doi: 10.1007/s11912-013-0346-0. [DOI] [PubMed] [Google Scholar]
- 6.Cho H, Kim JS, Chung H, et al. Loss of ARID1A/BAF250a expression is linked to tumor progression and adverse prognosis in cervical cancer. Hum Pathol. 2013;44:1365–1374. doi: 10.1016/j.humpath.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Wolchok JD, Chiarion-Sileni V, Gonzalez R, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377:1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. doi: 10.3389/fimmu.2020.483296. Rao AA, Madejska AA, Pfeil J, et al: ProTECT: Prediction of T-cell epitopes for cancer therapy. https://www.biorxiv.org/content/10.1101/696526v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toor JS, Rao AA, McShan AC, et al. A recurrent mutation in anaplastic lymphoma kinase with distinct neoepitope conformations. Front Immunol. 2018;9:99. doi: 10.3389/fimmu.2018.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma X, Liu Y, Liu Y, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018;555:371–376. doi: 10.1038/nature25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reich O, Tamussino K, Lahousen M, et al. Clear cell carcinoma of the uterine cervix: Pathology and prognosis in surgically treated stage IB-IIB disease in women not exposed in utero to diethylstilbestrol. Gynecol Oncol. 2000;76:331–335. doi: 10.1006/gyno.1999.5700. [DOI] [PubMed] [Google Scholar]
- 13.Thomas MB, Wright JD, Leiser AL, et al. Clear cell carcinoma of the cervix: A multi-institutional review in the post-DES era. Gynecol Oncol. 2008;109:335–339. doi: 10.1016/j.ygyno.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd J, Takahashi H, Waggoner SE, et al. Molecular genetic analysis of clear cell adenocarcinomas of the vagina and cervix associated and unassociated with diethylstilbestrol exposure in utero. Cancer. 1996;77:507–513. doi: 10.1002/(SICI)1097-0142(19960201)77:3<507::AID-CNCR12>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 15.Lee EK, Lindeman NI, Matulonis UA, et al. POLE-mutated clear cell cervical cancer associated with in-utero diethylstilbestrol exposure. Gynecol Oncol Rep. 2019;28:15–17. doi: 10.1016/j.gore.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Łuksza M, Riaz N, Makarov V, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551:517–520. doi: 10.1038/nature24473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turajlic S, Litchfield K, Xu H, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: A pan-cancer analysis. Lancet Oncol. 2017;18:1009–1021. doi: 10.1016/S1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 18.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tate JG, Bamford S, Jubb HC, et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Jia M, He Z, et al. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene. 2018;37:3924–3936. doi: 10.1038/s41388-018-0245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, Wen W, Bao J, et al. Integrative genomic analyses of APOBEC-mutational signature, expression and germline deletion of APOBEC3 genes, and immunogenicity in multiple cancer types. BMC Med Genomics. 2019;12:131. doi: 10.1186/s12920-019-0579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen PL, Roh W, Reuben A, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6:827–837. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boichard A, Tsigelny IF, Kurzrock R. High expression of PD-1 ligands is associated with kataegis mutational signature and APOBEC3 alterations. Oncoimmunology. 2017;6:e1284719. doi: 10.1080/2162402X.2017.1284719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jager M, Blokzijl F, Kuijk E, et al. Deficiency of nucleotide excision repair is associated with mutational signature observed in cancer. Genome Res. 2019;29:1067–1077. doi: 10.1101/gr.246223.118. [DOI] [PMC free article] [PubMed] [Google Scholar]