Abstract
Mutations in isocitrate dehydrogenase 1 (IDH1) and IDH2 are found frequently in malignant gliomas and are likely involved in early gliomagenesis. To understand the prevalence of these mutations and their relationship to other genetic alterations and impact on prognosis for Japanese glioma patients, we analyzed 250 glioma cases. Mutations of IDH1 and IDH2 were found in 73 (29%) and 2 (1%) cases, respectively. All detected mutations were heterozygous, and most mutations were an Arg132His (G395A) substitution. IDH mutations were frequent in oligodendroglial tumors (37/52, 71%) and diffuse astrocytomas (17/29, 59%), and were less frequent in anaplastic astrocytomas (8/29, 28%) and glioblastomas (13/125, 10%). The pilocytic astrocytomas and gangliogliomas did not have either mutation. Notably, 28 of 30 oligodendroglial tumors harboring the 1p/19q co‐deletion also had an IDH mutation, and these alterations were significantly correlated (P < 0.001). The association between TP53 and IDH mutation was significant in diffuse astrocytomas (P = 0.0018). MGMT promoter methylation was significantly associated with IDH mutation in grade 2 (P < 0.001) and grade 3 (P = 0.02) gliomas. IDH mutation and 1p/19q co‐deletion were independent favorable prognostic factors for patients with grade 3 gliomas. For patients with grade 3 gliomas and without 1p/19q co‐deletion, IDH mutation was strongly associated with increased progression‐free survival (P < 0.0001) and overall survival (P < 0.0001), but no such marked correlation was observed with grade 2 gliomas or glioblastomas. Therefore, IDH mutation would be most useful when assessing prognosis of patients with grade 3 glioma with intact 1p/19q; anaplastic astrocytomas account for most of these grade 3 gliomas. (Cancer Sci 2012; 103: 587–592)
Gliomas are among the most common and formidable brain tumors.( 1 ) Despite intensive treatment, most patients die within 2‐10 years. Therefore, development of novel therapeutic strategies based on greater understanding of tumor characteristics is needed. Recently, a comprehensive sequence analysis of human GBM that included most human genes revealed frequent mutations in IDH1.( 2 ) Subsequent analyses revealed that these mutations occur more frequently in low‐grade glioma than in GBM, with a rate of IDH1 mutation as high as 59–90%.( 3 , 4 , 5 , 6 , 7 ) The IDH gene mutation is currently believed to occur in the early stage of gliomagenesis( 4 , 6 ) and to play a critical role in tumor development.
The IDH genes encode redox enzymes; these enzymes convert isocitrate to alpha‐ketoglutarate, use NAD(P)+ as a co‐enzyme, and function in energy metabolism. There are three IDH genes in humans, and only mutations in IDH1 are frequently found in gliomas; the IDH1 enzyme resides in the cytosol and peroxisomes.( 2 , 3 , 4 , 5 , 6 , 7 ) Mutations in IDH2 are rare in gliomas; IDH2 localizes to mitochondria and functions in the Krebs (citric acid) cycle. To date, no IDH3 mutation has been reported. Most IDH1 mutations in gliomas are missense mutations at amino acid 132, which is in the catalytic domain and binds to substrate. Similarly, IDH2 mutations in gliomas are substitutions at amino acid 172, which is functionally equivalent to amino acid 132 of IDH1. IDH1 and IDH2 mutations in the catalytic domain are also found in 8–23% of acute myeloid leukemias.( 8 , 9 ) Mutations in IDH genes are rarely found in other tumors.( 7 , 10 )
In general, a tumor with an IDH mutation has either an IDH1 or IDH2 mutation, and the mutation is heterozygous with a wild‐type allele.( 2 , 4 , 6 , 7 ) This observation led to the notion that mutated IDH1/IDH2 genes gain novel functions and are oncogenes, and that the wild‐type IDH genes are not tumor suppressor genes. In fact, mutant IDH1 has novel enzymatic activity; it converts alpha‐ketoglutarate to 2‐HG, and accumulated 2‐HG is presumed to contribute to tumorigenesis as an “onco‐metabolite”.( 9 , 11 , 12 , 13 )
Malignant gliomas categorized as WHO grade 4( 2 , 5 , 7 , 14 , 15 ) or grade 3( 5 , 7 , 15 , 16 , 17 ) with an IDH mutation were reportedly associated with higher PFS( 15 , 16 , 17 ) and OS( 2 , 5 , 7 , 14 , 15 , 16 , 17 ) than those without an IDH mutation. However, for grade 2 gliomas, the relation between the presence of an IDH mutation and prognosis is controversial.( 15 , 18 , 19 , 20 )
The IDH mutations apparently have an important role in many aspects of glioma, including gliomagenesis, patient prognosis, and development of therapeutic strategies. However, information on IDH mutations in gliomas, such as prevalence, relation to other genetic alterations, and prognostic value, is still limited, particularly for Asian populations,( 21 , 22 ) including Japanese patients.( 5 , 17 ) Thus, to further clarify the significance of IDH mutations with regard to proper diagnosis and optimized treatment of malignant gliomas, we sought basic data on a large number of Japanese glioma patients for IDH1 and IDH2 mutations and other genetic and epigenetic alterations frequently found in gliomas, specifically 1p/19q LOH, TP53 mutation, and MGMT promoter methylation.
Materials and Methods
Tumor specimens. Tumor samples and paired blood samples were obtained following surgery. Of 250 gliomas, 168 tumors were collected at the University of Tokyo hospital (Tokyo, Japan) and 82 gliomas were collected at collaborating hospitals. The study was approved by the Ethics Committee of the University of Tokyo and all patients gave written informed consent. Histological diagnoses were made on formalin‐fixed, paraffin‐embedded tissues following the WHO classification( 1 ) by a neuropathologist (J.S.) for samples from the University of Tokyo hospital and consensus diagnoses were made by four neuropathologists for samples from other hospitals as reported previously.( 23 ) Genomic DNA was extracted for genetic analyses. Patients with the same grade (2, 3, or 4) glioma were treated similarly with surgical resection followed by radiotherapy and alkylating agent chemotherapy.
Genetic analysis. For IDH gene mutations, the genomic regions spanning the catalytic domain of IDH1, including codon 132, and of IDH2, including codon 172, were analyzed by direct sequencing using the Genetic Analyzer 310 (Applied Biosystems, Foster City, CA, USA). An aliquot of DNA was amplified by PCR using AmpliTaq Gold (Applied Biosystems) with annealing temperature at 55°C. The primers 5′‐TGCCACCAACGACCAAGTCA and 5′‐TGTGTTGAGATGGACGCCTATTTG were used for IDH1 amplification and sequencing, as reported previously.( 15 ) Amplification of IDH2 was carried out using the primers 5′‐CTCTGTCCTCACAGAGTTCAAGC and 5′‐CCACTCCTTGACACCACTGCC, and the IDH2 sequencing reactions were carried out using the primers 5′‐AAGTCCCAATGGAACTATCCG and 5′‐TCTGTGGCCTTGTACTGCAGAG.
Loss of heterozygosity on chromosomes 1p and 19q was determined using microsatellite analysis as described previously.( 23 ) When tumors had no available paired blood DNA or when the LOH assay was ambiguous because of non‐informative microsatellite markers, MLPA assay was carried out using the SALSA MLPA kit P088 (MRC Holland, Amsterdam, the Netherlands) following the manufacturer’s instructions. TP53 gene mutation was determined by direct sequencing following PCR‐SSCP screening of exons 5–8 of TP53, as described previously.( 24 )
Methylation‐specific PCR. Genomic DNA samples (250 ng each) were used for bisulfite reactions using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocol. DNA methylation status of the MGMT promoter was then determined by methylation‐specific PCR as described by Esteller et al. ( 25 )
Statistical analysis. Fisher’s exact test was used to compare the genotype distributions. Overall survival was defined as the time between initial surgery and death or last follow‐up. Progression‐free survival was defined as the time between initial surgery and recurrence or last follow‐up. Both OS and PFS were calculated according to the Kaplan–Meier method, and differences among patient subsets were evaluated using the log–rank test. Statistical calculations were carried out using JMP 9 (SAS Institute, Cary, NC, USA).
Results
Frequency and characteristics of IDH mutations in glioma samples from Japanese patients. We analyzed 250 human glioma samples obtained following surgery; these tumors consisted of 125 GBM, 29 AA, 29 DA, 52 oligodendroglial tumors, 9 PA, and 6 GGL. Mutations of IDH1 and IDH2 were found in 73 (29%) and 2 (1%) tumors, respectively. All detected mutations were heterozygous, missense mutations. Among the 73 IDH1 mutations, the G395A (R132H) substitution was the most frequent mutation (occurring in 70/73 cases, 96%), C394A (R132S) substitutions occurred in two cases, and a C394T (R132C) substitution occurred in one case. Of the two IDH2 mutations, one was a G515A (R172K) substitution, and the other was an A514T (R172W) substitution. IDH mutations (IDH1 or IDH2) were found in 13 (10%) of 125 GBM, 8 (28%) AA, 17 (59%) DA, and in 37 (71%) oligodendroglial tumors (Table 1). In the 52 oligodendroglial tumors, IDH mutations were found in 19/25 (76%) OG, 4/7 (57%) OA, 10/15 (67%) AOG, and 4/5 (80%) AOA. No mutation was detected in any case of PA or GGL. A higher rate of IDH mutation was found in secondary GBM (6/13, 46%) than primary GBM (6/109, 6%). In the three GBMO cases, there was only one IDH1 mutation.
Table 1.
IDH mutation and common genetic and epigenetic alterations in gliomas from Japanese patients
| Tumor pathology (WHO grade) IDH1 or IDH2 status | No. of patients | Frequency of IDH1 or IDH2 mutation | Median age, years | Male sex (%) | 1p/19q co‐deletion | TP53 mutation | Methylated MGMT promoter (%) |
|---|---|---|---|---|---|---|---|
| GBM primary (Gr. 4) | 109 | 6/109 (6%) | 62 | 61 | 1 | 23 (22%) | 27/57 (47) |
| Mutant | 6 | 43 | 50 | 1 | 3 n.s. | 1/1 (100) | |
| Wild‐type | 103 | 62 | 61 | 0 | 20 | 26/56 (46) | |
| GBM secondary (Gr. 4) | 13 | 6/13 (46%) | 47 | 69 | 0 | 2 (15%) | 7/9 (78) |
| Mutant | 6 | 52 | 50 | 0 | 2 n.s. | 4/5 (80) | |
| Wild‐type | 7 | 43 | 86 | 0 | 0 | 3/4 (75) | |
| GBMO (Gr.4) | 3 | 1/3 (33%) | 80 | 67 | 1 | 0 | 3/3 (100) |
| Mutant | 1 | 62 | 100 | 1 | 0 | 1/1 (100) | |
| Wild‐type | 2 | 80 | 50 | 0 | 0 | 2/2 (100) | |
| Anaplastic astrocytoma (Gr. 3) | 29 | 8/29 (28%) | 57 | 55 | 1 | 10 (34%) | 5/18 (28) |
| Mutant | 8 | 46 | 50 | 1 | 5 n.s. | 2/5 (40) | |
| Wild‐type | 22 | 60 | 57 | 0 | 5 | 3/13 (23) | |
| Anaplastic oligoastrocytoma (Gr. 3) | 5 | 4/5 (80%) | 43 | 40 | 0 | 4 (80%) | 2/3 (66) |
| Mutant | 4 | 48 | 25 | 0 | 3 n.s. | 2/2 (100) | |
| Wild‐type | 1 | 11 | 100 | 0 | 1 | 0/1 (0) | |
| Anaplastic oligodendroglioma (Gr. 3) | 15 | 10/15 (67%) | 62 | 56 | 10 (67%) | 0 | 6/7 (86) |
| Mutant | 10 | 49 | 43 | 9* | 0 | 5/5 (100) | |
| Wild‐type | 5 | 66 | 100 | 1 | 0 | 1/2 (50) | |
| Diffuse astrocytoma (Gr. 2) | 29 | 17/29 (59%) | 32 | 61 | 3 | 13 (45%) | 10/17 (59) |
| Mutant | 17 | 33 | 59 | 2 | 12*** | 10/10 (100) | |
| Wild‐type | 12 | 30 | 64 | 1 | 1 | 0/7 (0) | |
| Oligoastrocytoma (Gr. 2) | 7 | 4/7 (57%) | 44 | 71 | 1 | 1 | 5/6 (83) |
| Mutant | 4 | 37 | 100 | 1 | 1 | 3/3 (100) | |
| Wild‐type | 3 | 53 | 33 | 0 | 0 | 2/3 (67) | |
| Oligodendroglioma (Gr. 2) | 25 | 19/25 (76%) | 46 | 52 | 19 (76%) | 3 | 8/12 (66) |
| Mutant | 19 | 47 | 53 | 18** | 1 | 7/10 (70) | |
| Wild‐type | 6 | 34 | 50 | 1 | 2 | 1/2 (50) | |
| Pilocytic astrocytoma (Gr. 1) | 9 | 0% | 12 | 56 | 0 | 0 | N/A |
| Mutant | 0 | N/A | N/A | 0 | 0 | ||
| Wild‐type | 9 | 12 | 56 | 0 | 0 | ||
| Ganglioglioma (Gr. 1) | 6 | 0% | 22 | 67 | 0 | 0 | N/A |
| Mutant | 0 | N/A | N/A | 0 | 0 | ||
| Wild‐type | 6 | 22 | 67 | 0 | 0 |
*P = 0.0037; **P = 0.0001; ***P = 0.0018. The association with IDH mutation (Fisher’s exact test). GBM, glioblastoma; GBMO, glioblastoma with oligodendroglioma component; N/A, not analyzed; n.s., not significant.
Association between IDH mutation and 1p/19q co‐deletion, TP53 mutation, or MGMT promoter methylation. The frequencies of 1p/19q co‐deletion and TP53 mutation and their relationship with IDH mutations are shown in Table 1. As expected, 1p/19q co‐deletion was common in oligodendroglial tumors, especially those without an astrocytic component (OG 76%, AOG 67%), whereas TP53 mutations were common in lower‐grade astrocytomas (DA 45%); these genetic aberrations were never coincident. In OG, 1p/19q co‐deletion was significantly correlated with IDH mutation (P < 0.001), and almost all oligodendroglial tumors with 1p/19q co‐deletion had an IDH mutation (28/30, 93%).
The TP53 mutation was more prevalent in DA (45%) than in AA (34%) or primary GBM (22%). However, when IDH mutation was present, TP53 mutation was more frequent, and TP53 mutations were found in 12/17 (71%) DA, 5/8 (63%) AA, and 3/6 (50%) primary GBMs that also had an IDH mutation. The rates of IDH mutation in astrocytic tumors with TP53 mutation were higher than those with wild‐type TP53 (92% vs 31% in DA, 50% vs 16% in AA, and 13% vs 4% in primary GBM), and the association between TP53 and IDH mutation was significant in DA (P = 0.0018), but not in AA or GBM. The majority of DA tumors with TP53 mutation had IDH mutation (12/13, 92%); in contrast, only a few primary GBM tumors with TP53 mutation also had an IDH mutation (3/23, 13%).
Of a total 250 gliomas, MGMT promoter methylation status was analyzed for 132 gliomas (grade 2, 3, and 4) resected at the University of Tokyo hospital. Methylation was evident in 37/69 GBM (54%), 5/18 AA (28%), 10/17 DA (59%), 8/10 AOG/AOA (80%), and 13/18 OG/OA (72%) (Table 1). The association between IDH mutation and MGMT methylation was significant in grade 2 (P < 0.001) and grade 3 gliomas (P = 0.02), but not in grade 4 gliomas (P = 0.11).
Prognostic value of IDH mutation and other genetic alterations. We evaluated the potential prognostic value of IDH mutation and other genetic alterations in WHO grade 2, 3, and 4 gliomas. For patients with grade 2 gliomas, univariate analysis showed that IDH mutation was not associated with OS (P = 0.07) or PFS (P = 0.29). Codeleted 1p/19q and wild‐type TP53 each slightly correlated with increased PFS (P = 0.014 and P = 0.029, respectively), but they were not correlated with OS, and neither of these genetic alterations showed significant association with prognosis in multivariate analysis (Table 2). MGMT promoter methylation was also not associated with prognosis. Similarly, we did not observe a significant association of IDH mutation with better prognosis for DA (OS, P = 0.10; PFS, P = 0.58).
Table 2.
Prognostic value of common genetic alterations for overall survival (OS) and progression‐free survival (PFS) in gliomas (multivariate analysis)
| PFS | OS | |||||
|---|---|---|---|---|---|---|
| P‐value | Hazard ratio | 95% CI | P‐value | Hazard ratio | 95% CI | |
| Grade 2 glioma | ||||||
| IDH mutation | 0.4408 | 0.602 | 0.1678–2.1535 | 0.1573 | 0.329 | 0.0728–1.5270 |
| 1p/19q co‐deletion | 0.3591 | 0.495 | 0.1083–2.2020 | 0.7988 | 1.237 | 0.2353–6.0194 |
| TP53 mutation | 0.2904 | 2.036 | 0.5526–7.7157 | 0.4693 | 0.537 | 0.0685–2.6350 |
| Grade 3 glioma | ||||||
| IDH mutation | <0.0001† | 0.059 | 0.0086–0.2395 | 0.0403† | 0.319 | 0.0985–0.9519 |
| 1p/19q co‐deletion | 0.0016† | 0.055 | 0.0025–0.3904 | 0.0170† | 0.184 | 0.0271–0.7567 |
| TP53 mutation | 0.4144 | 0.646 | 0.2045–1.7994 | 0.0300† | 0.294 | 0.0786–0.8937 |
| Primary GBM | ||||||
| IDH mutation | 0.8456 | 0.898 | 0.2575–2.4255 | 0.8560 | 0.905 | 0.2609–2.4203 |
| TP53 mutation | 0.1533 | 0.605 | 0.2792–1.1944 | 0.3089 | 0.705 | 0.3354–1.3613 |
| Methylated MGMT promoter | 0.0031† | 0.407 | 0.2216–0.7375 | 0.0058† | 0.429 | 0.2324–0.7820 |
†Significant value. Cox proportional hazard modeling for OS or PFS was applied for the major variable for prognostic factors. CI, confidence interval; GBM, glioblastoma.
In grade 3 gliomas, univariate analysis showed that the association between IDH mutation and prolonged survival (OS, P = 0.0004; PFS, P < 0.0001) was significant and that 1p/19q co‐deletion was associated with prolonged survival (OS, P = 0.028; PFS, P = 0.0025), but that neither TP53 mutation nor MGMT promoter status was associated with prognosis. Although IDH mutation and 1p/19q co‐deletion were tightly associated with one another, the multivariate analysis further indicated that these alterations were independent indicators of a favorable prognosis (Table 2). IDH mutation was present in almost all tumors with the 1p/19q co‐deletion.( 26 ) Therefore, grade 3 gliomas were divided into three genetic subgroups: (i) 1p/19q codeleted tumors, most of which carry IDH mutation and show oligodendroglial phenotype; (ii) tumors without 1p/19q co‐deletion and with mutant IDH; and (iii) tumors without 1p/19q co‐deletion and with wild‐type IDH. Grade 3 gliomas were assessed with regard to the association between the genetic alterations and disease course (Fig. 1). In these genetic subgroups, grade 3 gliomas without 1p/19q co‐deletion and with wild‐type IDH were revealed to have markedly worse OS (P < 0.0001) (Fig. 1C) and PFS (P < 0.0001) (Fig. 1D), but these lower survival rates were not observed for patients with grade 2 gliomas lacking 1p/19q co‐deletion and IDH mutation (Fig. 1A,B). Grade 3 gliomas without 1p/19q co‐deletion were predominantly AA (28 AAs, 5 AOAs, and 5 AOGs), and only 1 AA had the 1p/19q co‐deletion (Table 1). The IDH mutation was significantly associated with increased OS (P = 0.0064) and PFS (P = 0.0001) for patients with AA based on the univariate analysis. TP53 mutation was also correlated with increased PFS (P = 0.013), but MGMT promoter methylation showed no significant association with PFS or OS.
Figure 1.
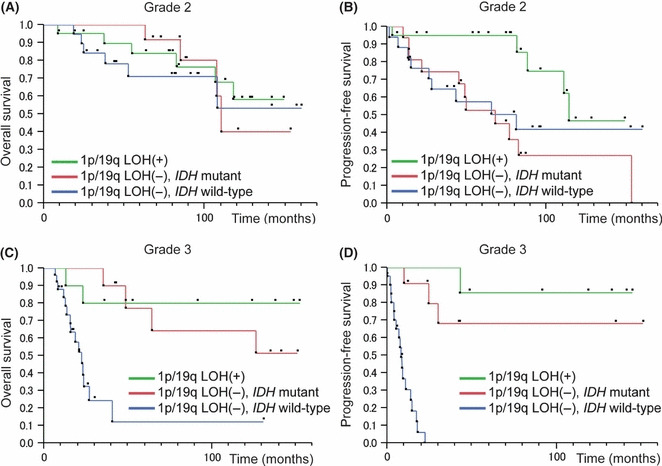
Overall survival (OS) and progression‐free survival (PFS) curves for patients with grade 2 and 3 gliomas with or without 1p/19q loss of heterozygosity (LOH) and/or isocitrate dehydrogenase (IDH) mutation. Overall survival (A) and PFS (B) in grade 2 gliomas; OS (C) and PFS (D) in grade 3 gliomas.
In primary GBM, our univariate analysis showed that neither IDH mutation, 1p/19q co‐deletion, nor TP53 mutation was associated with PFS or OS, but MGMT promoter methylation was significantly associated with increased OS (P = 0.0043) and PFS (P = 0.0038).
Discussion
Here we report that IDH mutation, which was tightly associated with 1p/19q co‐deletion and MGMT promoter methylation, was common in grade 2 gliomas and also, but to a lesser extent, in grade 3 gliomas. Moreover, we found that IDH mutation would be an especially useful genetic marker for evaluating the malignancy of grade 3 gliomas that do not have a 1p/19q co‐deletion and that these gliomas were predominantly AA.
The frequencies and patterns of IDH mutation in our glioma samples from Japanese patients were largely comparable to those in previous reports.( 2 , 3 , 4 , 5 , 6 , 7 ) IDH mutation was found predominantly in grade 2 glioma, such as DA, OA, and OG. IDH mutation frequencies were lower in higher‐grade gliomas, and less than 10% of GBM had an IDH mutation; however, nearly half of secondary GBM, which developed from malignant transformation of lower‐grade glioma, had an IDH mutation. These observations supported the notion that the IDH mutation has a crucial role in the development of the majority of grade 2 gliomas. In grade 3 gliomas, the oligodendroglial tumors had higher frequency of IDH mutation than astrocytic tumors (OG 76% > DA 59%, P = 0.18; AOG 67% > AA 28%, P < 0.05; Pearson’s chi‐square‐test). No IDH mutation was detected in any grade 1 glioma, PA or GGL; this observation indicated that these tumors had a different genetic etiology from that of grade 2 and 3 infiltrative astrocytic and oligodendroglial tumors. This observation also supported the usefulness of IDH mutations along with BRAF alterations for differential diagnosis of PA.( 27 ) However, our results differed from two previous reports that detected IDH1 mutation (8–38%) in GGL.( 5 , 28 ) Further studies are needed to clarify the biological and clinical significance of IDH1 mutation in GGL.
As reported previously,( 23 ) TP53 mutation and co‐deletion of chromosomes 1p and 19q were frequent alterations in grade 2 and grade 3 gliomas. The 1p/19q co‐deletions were mostly found in oligodendroglial lineage gliomas, whereas TP53 mutations were more frequent in gliomas derived from the astrocytic lineage. IDH mutation is currently believed to precede 1p/19q LOH and TP53 mutation during the early stage of gliomagenesis,( 4 , 6 ) and consistent with this hypothesis, most of our grade 2 gliomas that had 1p/19q co‐deletion or TP53 mutations also harbored an IDH mutation. In one study, all the gliomas with deletions of the entire 1p and 19q arms carried an IDH1 or IDH2 mutation;( 26 ) however, we found a few exceptions in which there was 1p/19q LOH, but no IDH mutation. These apparent exceptions might have been artifacts due to our imperfect methods for detecting the extent of 1p/19q LOH, specifically microsatellite analysis or MLPA; these methods do not effectively differentiate partial chromosomal loss from typical entire 1p/19q hemizygous deletion, which is generally found in OG harboring IDH mutation. It would be better to carefully evaluate the extent of 1p/19q LOH in such exceptional cases. TP53 mutation was also associated with IDH mutation in DA. However, there was no association between IDH and TP53 mutation in AA or primary GBM. This observation suggested that TP53 mutation promoted tumor growth independently of IDH mutation, especially in higher grade gliomas. Most gliomas with an IDH mutation had either 1p/19q LOH or TP53 mutation, further supporting the hypothesis that combinations of IDH mutation and subsequent genetic alteration are common pathways leading to low‐grade glioma. However, there were also a few other IDH‐mutated gliomas that had neither 1p/19q co‐deletion nor TP53 mutation. In these gliomas, the kind of alterations subsequent to IDH mutation that caused progenitor cells to give rise to low‐grade glioma remains to be elucidated.
Methylation at the MGMT promoter was associated with IDH mutation, especially for low‐grade gliomas. Some IDH enzymes with a mutation in the catalytic domain acquire a novel enzymatic activity( 9 , 11 ) that causes accumulation of 2‐HG, and 2‐HG is known to inhibit enzymes such as 5‐methylcytosine hydroxylases and histone demethylases. As a result, IDH mutations bring about genome‐wide hypermethylation, which might lead to tumor initiation.( 12 , 29 ) Reportedly, the majority of low‐grade gliomas have hypermethylated CpG islands throughout the genome; this phenomenon is called the glioma CpG island methylator phenotype, and these tumors frequently harbor IDH mutation.( 30 , 31 , 32 ) Therefore, frequent MGMT promoter methylation in IDH‐mutated low‐grade glioma was possibly simply a reflection of hypermethylation of a plethora of genes resulting from the methylator phenotype. In contrast, the majority of GBM had MGMT promoter hypermethylation without also having IDH mutation, indicating that MGMT promoter methylation occurs independent of IDH‐related hypermethylation in most GBM. Probably because of such a background, the prognostic values of MGMT promoter methylation for IDH‐mutated and IDH‐wild‐type gliomas are not equal. In GBM, MGMT promoter methylation is a predictive factor for the efficacy of temozolomide, which is a common alkylating agent used in the chemotherapeutic treatment of malignant glioma.( 33 , 34 ) However, the predictive value of MGMT promoter methylation for chemosensitivity in grade 2 and grade 3 glioma is controversial.( 19 , 35 )
The prognostic significance of IDH mutation differed among WHO tumor grades. Unlike previous reports, IDH mutation was not associated with the PFS or OS of our GBM patients; however, this finding might result from insufficient numbers of GBM patients with IDH mutation. A methylated MGMT promoter, which reflects the sensitivity of a tumor to temozolomide, was associated with favorable PFS and OS for patients with GBM, further emphasizing the importance of detecting MGMT promoter methylation status in GBM.
Among patients with grade 2 gliomas, IDH mutation was also not associated with prognosis. Wild‐type TP53 and 1p/19q co‐deletion were each associated with prolonged PFS, probably because these two genetic alterations were mutually exclusive and tumors with wild‐type TP53 likely have a 1p/19q co‐deletion, which is a recognized favorable prognostic factor. The prognostic value and predictability of temozolomide efficacy associated with IDH mutation in low‐grade gliomas has been controversial. Consistent with our results, Kim et al. ( 20 ) showed that IDH1 and IDH2 mutations are not prognostic in low‐grade gliomas, but that TP53 mutation is a significant prognostic indicator of shorter survival and 1p/19q loss is prognostic of longer survival. However, Sanson et al. ( 15 ) reported a different result, specifically that IDH1 mutation is associated with a better outcome in grade 2 gliomas. Dubbink et al. ( 18 ) showed that IDH mutation is associated with better outcomes for relapsed astrocytomas previously treated with radiotherapy, but there was no relationship between IDH mutation and temozolomide responsiveness. Houillier et al. ( 19 ) showed that IDH1 or IDH2 mutations predict better prognosis of glioma treated with temozolomide, but they did not appear to influence the course of untreated low‐grade glioma. Thus, the prognostic value of IDH mutation is different from that of 1p/19q co‐deletion, which is prognostic as well as predictive for responsiveness to temozolomide in low‐grade gliomas. These inconsistent results on the association between IDH mutation and survival in cases of low‐grade gliomas might be caused by the variable numbers of OG and DA included in these studies. Almost all oligodendroglial tumors with 1p/19q co‐deletion also have an IDH mutation;( 26 ) therefore, many cases of OG with favorable prognoses may affect and confound measurements of survival rate in the whole group of low‐grade gliomas with IDH mutations. To avoid the confounding influence of OG, we also focused on DA with wild‐type IDH; these tumors generally have neither 1p/19q LOH nor TP53. However, they had outcomes comparable to those of DA with IDH mutation. This finding indicated that DA with wild‐type IDH was not more malignant than DA with an IDH mutation. This observation differed from the observation that AA with wild‐type IDH had markedly worse outcomes than AA with an IDH mutation (OS, P = 0.0064; PFS, P = 0.0001).
In contrast with grade 2 and 4 gliomas, the prognostic significance of IDH mutation was evident for grade 3 gliomas, and this finding was consistent with previous reports.( 15 , 17 , 36 ) Almost all gliomas with 1p/19q co‐deletion have an IDH mutation,( 26 ) and anaplastic oligodendroglial tumors often harbor 1p/19q co‐deletion; therefore, monitoring of IDH mutation might have more clinical significance for patients with grade 3 gliomas with intact 1p/19q, and these tumors are predominantly AA. In fact, as the histopathological differential diagnosis of AA from GBM or DA is often subjective and diagnoses frequently differ between pathologists,( 23 ) a pathological diagnosis of AA may not always indicate sameness between gliomas and similar prognosis. However, accurate determination of the pathological group of a tumor is clinically critical for planning adjuvant therapy, such as radiation and chemotherapy. Therefore, genetic analyses, which may reflect causative origins of tumors, are expected to reveal biological traits with less inter‐observer variation, as is the case of 1p/19q co‐deletion in oligodendroglial tumors. Because IDH mutations have a defined role in gliomagenesis and indicate, to some extent, the nature of the original tumor cell, monitoring IDH mutational status may allow for accurate assignment of diagnosed AA to low‐grade gliomas that frequently harbor IDH mutation or to primary GBM that usually have intact IDH. Therefore, we believe that monitoring IDH mutation in combination with 1p/19q co‐deletion, which genetically differentiates oligodendroglial and astrocytic tumors, could be a useful genetic marker of prognostic value, especially for grade 3 glioma patients.
Disclosure Statement
The authors have no conflicts of interest.
Abbreviations
- 2‐HG
2‐hydroxyglutarate
- AA
anaplastic astrocytoma
- AOA
anaplastic oligoastrocytoma
- AOG
anaplastic oligodendroglioma
- DA
diffuse astrocytoma
- GBM
glioblastoma
- GBMO
glioblastoma with oligodendroglioma component
- GGL
ganglioglioma
- IDH
isocitrate dehydrogenase
- LOH
loss of heterozygosity
- MLPA
multiplex ligation‐dependent probe amplification
- OA
oligoastrocytoma
- OG
oligodendroglioma
- OS
overall survival
- PA
pilocytic astrocytoma
- PFS
progression‐free survival
Acknowledgments
We appreciate the technical assistance of Reiko Matsuura (Department of Neurosurgery, University of Tokyo, Tokyo, Japan). This work was supported in part by a Grant‐in‐Aid for Scientific Research (C) (No. 20591706) to A.M. and a Grant‐in‐Aid for Young Scientists (B) (No. 22791334) to K.S. from the Japan Society for the Promotion of Science. A.M. was also supported by the Takeda Science Foundation.
References
- 1. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, eds. WHO Classification of Tumours of the Central Nervous System, 4th edn Lyon: International Agency for Research on Cancer, 2007. [Google Scholar]
- 2. Parsons DW, Jones S, Zhang X et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008; 321: 1807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol (Berl) 2008; 116: 597–602. [DOI] [PubMed] [Google Scholar]
- 4. Ichimura K, Pearson DM, Kocialkowski S et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol 2009; 11: 341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sonoda Y, Kumabe T, Nakamura T et al. Analysis of IDH1 and IDH2 mutations in Japanese glioma patients. Cancer Sci 2009; 100: 1996–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 Mutations Are Early Events in the Development of Astrocytomas and Oligodendrogliomas. Am J Pathol 2009; 174: 1149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yan H, Parsons DW, Jin G et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009; 360: 765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mardis ER, Ding L, Dooling DJ et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 2009; 361: 1058–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ward PS, Patel J, Wise DR et al. The Common Feature of Leukemia‐Associated IDH1 and IDH2 Mutations Is a Neomorphic Enzyme Activity Converting α‐Ketoglutarate to 2‐Hydroxyglutarate. Cancer Cell 2010; 17: 225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bleeker FE, Lamba S, Leenstra S et al. IDH1 mutations at residue p.R132 (IDH1 R132) occur frequently in high‐grade gliomas but not in other solid tumors. Hum Mutat 2009; 30: 7–11. [DOI] [PubMed] [Google Scholar]
- 11. Dang L, White DW, Gross S et al. Cancer‐associated IDH1 mutations produce 2‐hydroxyglutarate. Nature 2009; 462: 739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu W, Yang H, Liu Y et al. Oncometabolite 2‐Hydroxyglutarate Is a Competitive Inhibitor of α‐Ketoglutarate‐Dependent Dioxygenases. Cancer Cell 2011; 19: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lesniak M, Jin G, Reitman ZJ et al. 2‐Hydroxyglutarate Production, but Not Dominant Negative Function, Is Conferred by Glioma‐Derived NADP+‐Dependent Isocitrate Dehydrogenase Mutations. PLoS ONE 2011; 6: e16812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 Mutations as Molecular Signature and Predictive Factor of Secondary Glioblastomas. Clin Cancer Res 2009; 15: 6002–7. [DOI] [PubMed] [Google Scholar]
- 15. Sanson M, Marie Y, Paris S et al. Isocitrate Dehydrogenase 1 Codon 132 Mutation Is an Important Prognostic Biomarker in Gliomas. J Clin Oncol 2009; 27: 4150–4. [DOI] [PubMed] [Google Scholar]
- 16. Van den Bent MJ, Dubbink HJ, Marie Y et al. IDH1 and IDH2 Mutations Are Prognostic but not Predictive for Outcome in Anaplastic Oligodendroglial Tumors: a Report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res 2010; 16: 1597–604. [DOI] [PubMed] [Google Scholar]
- 17. Shibahara I, Sonoda Y, Kanamori M et al. IDH1/2 gene status defines the prognosis and molecular profiles in patients with grade III gliomas. Int J Clin Oncol 2011; DOI: 10.1007/s10147-011-0323-2 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 18. Dubbink HJ, Taal W, Van Marion R et al. IDH1 mutations in low‐grade astrocytomas predict survival but not response to temozolomide. Neurology 2009; 73: 1792–5. [DOI] [PubMed] [Google Scholar]
- 19. Houillier C, Wang X, Kaloshi G et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low‐grade gliomas. Neurology 2010; 75: 1560–6. [DOI] [PubMed] [Google Scholar]
- 20. Kim Y‐H, Nobusawa S, Mittelbronn M et al. Molecular Classification of Low‐Grade Diffuse Gliomas. Am J Pathol 2010; 177: 2708–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jha P, Suri V, Sharma V et al. IDH1 mutations in gliomas: first series from a tertiary care centre in India with comprehensive review of literature. Exp Mol Pathol 2011; 91: 385–93. [DOI] [PubMed] [Google Scholar]
- 22. Qi SA, Yu L, Lu YT et al. IDH mutations occur frequently in Chinese glioma patients and predict longer survival but not response to concomitant chemoradiotherapy in anaplastic gliomas. Oncol Rep 2011; 26: 1479–85. [DOI] [PubMed] [Google Scholar]
- 23. Ueki K, Nishikawa R, Nakazato Y et al. Correlation of histology and molecular genetic analysis of 1p, 19q, 10q, TP53, EGFR, CDK4, and CDKN2A in 91 astrocytic and oligodendroglial tumors. Clin Cancer Res 2002; 8: 196–201. [PubMed] [Google Scholar]
- 24. Mukasa A, Ueki K, Matsumoto S et al. Distinction in gene expression profiles of oligodendrogliomas with and without allelic loss of 1p. Oncogene 2002; 21: 3961–8. [DOI] [PubMed] [Google Scholar]
- 25. Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6‐methylguanine‐DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 1999; 59: 793–7. [PubMed] [Google Scholar]
- 26. Labussiere M, Idbaih A, Wang XW et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2 . Neurology 2010; 74: 1886–90. [DOI] [PubMed] [Google Scholar]
- 27. Korshunov A, Meyer J, Capper D et al. Combined molecular analysis of BRAF and IDH1 distinguishes pilocytic astrocytoma from diffuse astrocytoma. Acta Neuropathol 2009; 118: 401–5. [DOI] [PubMed] [Google Scholar]
- 28. Horbinski C, Kofler J, Yeaney G et al. Isocitrate dehydrogenase 1 analysis differentiates gangliogliomas from infiltrative gliomas. Brain Pathol 2011; 21: 564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Figueroa ME, Abdel‐Wahab O, Lu C et al. Leukemic IDH1 and IDH2 Mutations Result in a Hypermethylation Phenotype, Disrupt TET2 Function, and Impair Hematopoietic Differentiation. Cancer Cell 2010; 18: 553–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Christensen BC, Smith AA, Zheng S et al. DNA Methylation, Isocitrate Dehydrogenase Mutation, and Survival in Glioma. J Natl Cancer Inst 2010; 103: 143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laffaire J, Everhard S, Idbaih A et al. Methylation profiling identifies 2 groups of gliomas according to their tumorigenesis. Neuro Oncol 2010; 13: 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Noushmehr H, Weisenberger DJ, Diefes K et al. Identification of a CpG Island Methylator Phenotype that Defines a Distinct Subgroup of Glioma. Cancer Cell 2010; 17: 510–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hegi ME, Diserens AC, Gorlia T et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005; 352: 997–1003. [DOI] [PubMed] [Google Scholar]
- 34. Rivera AL, Pelloski CE, Gilbert MR et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol 2010; 12: 116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van den Bent MJ, Dubbink HJ, Sanson M et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J Clin Oncol 2009; 27: 5881–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hartmann C, Hentschel B, Wick W et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1‐mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol (Berl) 2010; 120: 707–18. [DOI] [PubMed] [Google Scholar]


