Abstract
Cell reprogramming reverts cells to multipotent, preprogrammed states by re‐establishing epigenetic markers. It can also induce considerable malignant phenotype modification. Because key events in cancer relapse and metastasis, including epithelial–mesenchymal transition phenotypes, are regulated primarily by reversible and transient epigenetic modifications rather than the accumulation of irreversible and stable genetic abnormalities, studying dynamic mechanisms regulating these biological processes is important. Transcription factors for induced pluripotent stem cells and non‐coding microRNAs allow pluripotent phenotype induction. We present the current knowledge of the possible applications of cell reprogramming in reducing aggressive phenotype expression, which can induce tumor cell hibernation and maintain appropriate phenotypes, thereby minimizing relapse and metastasis after surgical resection of gastrointestinal cancer. (Cancer Sci 2012; 103: 393–399)
During cell reprogramming, mature cells revert to an immature, preprogrammed (undifferentiated) state, and usually acquire multidifferentiated characteristics following the loss and re‐establishment of important epigenetic markers including DNA methylation.( 1 ) This deregulation of important genomic and epigenomic factors is commonly associated with the abnormal cell differentiation characteristics of different cancers.( 2 , 3 , 4 , 5 , 6 , 7 ) Emerging data suggest that epigenetic modifications and cell reprogramming‐like processes are important for cellular transformation and the development of malignant cancer phenotypes.( 8 , 9 , 10 , 11 , 12 ) Understanding the underlying process of epigenome reprogramming facilitates the use of regenerative medicine and cancer therapy. Here we discuss whether the reprogramming‐like phenomenon observed in normal cells can be adapted for developing novel therapies.
Programming and Reprogramming of Cells
Mammalian tissues develop from a totipotent zygote. During cell differentiation, a less specialized cell (i.e. stem or progenitor cell) continuously produces more specialized cell types through cell division, and thus, a complex tissue system containing increasingly differentiated and specialized cells is established. Subsequently, pluripotent primitive ectodermal cells in the inner cell mass of blastocysts develop from the totipotent zygote.( 13 , 14 ) Following blastocyst implantation, pluripotent epiblast cells differentiate into somatic cells. Repression of the somatic program and re‐expression of pluripotency‐specific genes through epigenetic modifications are necessary for germ cell development,( 13 , 15 ) indicating that differentiated cells retain all the genetic information necessary to generate an entire organism. This was initially tested by cloning animals from differentiated cells by nuclear transfer. Mouse( 16 , 17 ) and human ES cells( 18 ) are derived from their respective blastocysts. The recently developed iPSCs( 19 , 20 ) can produce derivatives of each germ layer. Differentiation occurs both during the developmental stages and in adults;( 21 ) multipotent tissue stem cells produce completely differentiated daughter cells during normal cell turnover in adult tissues and during tissue repair.( 22 ) Differentiation is associated with dynamic alterations in cell morphology, cell metabolism, and responsiveness to cell signaling,( 21 , 23 ) which occur largely because of highly regulated gene expression through mRNA regulation( 23 ) and non‐coding miRNA expression.( 24 , 25 , 26 )
Defined factor‐mediated reprogramming. Considering the ethical issues regarding the use of fertilized oocytes for establishing and producing ES cells, and the immunological compatibility that occurs in case of unrelated donors, a great breakthrough was reported by Takahashi and Yamanaka( 19 ), who discovered that complete reprogramming can be achieved by introducing defined biological factors, such as Oct4 (also known as Pou5f1), Sox2, Klf4, and c‐Myc, in mouse and human fibroblasts.( 20 ) The initial experimental injection of Fbx15‐selected iPSCs into mouse blastocysts revealed that iPSCs alone could not efficiently produce chimeric mice, presumably because of substantial methylation of immature gene (including Nanog and Oct4) promoters.( 19 ) Subsequent studies indicated that modified selection methods of completely reprogrammed cells through expression of endogenous Nanog( 27 , 28 ) and Oct4( 28 ) allowed the successful generation of viable chimeras and detectable transmission into the germline.
Complete pluripotency. During stable Oct4 and Nanog selection, although the overall appearance of the colonies was similar,( 27 , 28 ) quantitative differences existed between the two selection strategies. Oct4‐selected ES‐like colonies provided more stable and homogenous iPSC lines than Nanog‐selected ES‐like colonies.( 28 ) Eventually, the fraction of ES‐like colonies from Oct4‐selected MEF cultures was two or threefold higher than that from Nanog‐selected cultures, although initially fewer colonies existed with Oct4‐selected MEF‐derived iPSCs. This suggests that although the Nanog locus was more easily activated, a higher fraction of colonies from Oct4‐selected MEF cells was reprogrammed to pluripotency.( 28 ) These studies established that selection for Oct4 and Nanog expression results in germline‐competent iPSCs with increased ES cell‐like gene expression and DNA methylation patterns compared with Fbx15‐iPSCs. Whereas one clone from seven Nanog‐iPSC clones was transmitted through the germline to the next generation,( 27 ) Oct4‐iPSCs injected into tetraploid blastocysts can generate live late‐term embryos.( 28 ) The biological potency and epigenetic state of iPSCs and ES cells are the same. The overall estimated efficiency (0.05–0.10%) to establish iPSC lines from MEFs was similar between Oct4 and Nanog selection, despite the larger number of total Nanog‐iPSC colonies( 28 ) (Table 1).
Table 1.
Summary of current studies of normal somatic cell reprogramming
| Method for factor delivery | Factor | Starting material | Efficiency | Ref. |
|---|---|---|---|---|
| Retroviral vector | OSKM | Mouse embryonic and adult fibroblast | n.d. | (19) |
| Retroviral vector | OSKM | Human fibroblast | 10 colonies/5 × 104 | (20) |
| Lentiviral | OSNL | Human fetal fibroblast | 198 colonies/0.9 × 106 | (71) |
| Plasmid transfection | OSKM | MEF | Lower than the viral delivery method | (41) |
| Adenovirus (non‐integrating vector) | OSKM | MEF and hepatocyte | n.d. | (72) |
| Retrovirus | OSKM | Adult mouse liver and stomach cells | n.d. | (73) |
| Retrovirus | OSKM OK OM | Mouse neural stem cell | 3.6% 0.11% Slower than OK | (74) |
| Retrovirus | OSKM+5’Aza OSK+VPA/5’Aza | MEF | 0.5% 11.8% 100 × higher than the OSKM method | (75) |
| Doxycycline‐inducible lentiviruses | OSKM | Secondary somatic cells containing Dox‐inducible OSKM expression (MEF, intestinal epithelium) | 20–50 × higher than the direct infection method | (76) |
| Retroviral | OSKM | Adult human adipose stem cells | 0.2% | (77) |
| Repeated protein transduction | OSKM + VPA OSK + VPA | MEF | Slower kinetic than the viral delivery method | (49) |
| Retrovirus | OK + BIX/BayK compound | Neural progenitor cells | 12 colonies/3.5 × 104 | (78) |
| Doxycycline‐inducible transcription factors delivered by PiggyBac transposition | OSKM | Murine and human embryonic fibroblasts | n.d. | (43) |
| Retrovirus for OSK Transfection of miRNA mimics | OSK+miR‐291‐3p, miR‐294, miR‐295 | MEF | 0.1–0.3% | (79) |
| 2A‐peptide linked reprogramming cassette introduced by nucleofection | OSKM | MEF | 2.5% | (42) |
| Sendai virus | OSKM | Human terminally differentiated circulating T cells | 0.1% | (45) |
| Lentivirus | Oct4 + small compound (A‐83‐01, PD0325901, PS48, NaB) | Neonatal human epidermal keratinocytes, HUVECs, and amniotic fluid‐derived cells | 4–6 colonies/1 × 106 slower kinetic | (50) |
| Repeated transfection of synthetic modified messenger RNAs | OSKM | Primary human neonatal epidermal keratinocytes, BJ human neonatal foreskin fibroblasts, human fetal lung fibroblasts, and human fetal skin fibroblasts | 1.4%, 36‐fold higher than retrovirus | (80) |
| Lentivirus | miR‐302a,b,c,d, miR‐367 + VPA miR‐302a,b,c,d, miR‐367 | MEF Human fibroblast | Faster kinetics efficiency 2 × higher compared with OSKM Efficiency 10000 × higher | (39) |
| Repeated transfection | miR‐302s, miR‐369‐3p, miR‐369‐5p, miR‐200c | Human and mouse adipose stromal cells, dermal fibroblast | 5 colonies/5 × 104 | (51) |
| Retroviral Repeated transfection of miRNA mimics | OSKM miR‐106b, miR‐93, miR‐106a, miR‐17 | MEF | Addition of miRNA enhanced efficiency by 4–6‐fold (miR‐106b, 93), 3–4‐fold (miR‐106a,17) | (81) |
| Retrovirus Repeated transfection of miRNA mimics | OSKM OSK miR‐302b, miR‐372, miR‐294 | Human fibroblast | Addition of miRNA enhanced efficiency by promoting MET | (82) |
BIX, the small molecule BIX‐01294, an inhibitor of the G9a histone methyltransferase; K, Klf4; L, Lin28; M, c‐Myc; MEF, mouse embryonic fibroblast; MET, mesenchymal–epithelial transition; miR, microRNA; N, Nanog; n.d.: not determined; O, Oct3/4; Ref., reference; S, Sox2; VPA, valproic acid, a HDAC inhibitor.
Reprogramming barriers by tumor suppressors. Several factors can enhance the efficiency of iPSC generation, such as cell cycle checkpoints mediated by the cyclin‐dependent kinase inhibitor family. The CDKN2b–CDKN2a locus on human chromosome 9p21 (mice chromosome 4) is frequently lost in cancer. The locus encodes three cyclin‐dependent kinase inhibitors of the cell cycle: p15INK4b, p16INK4a, and p14ARF (p19Arf in mice) encoded by CDKN2b, CDKN2a, and an alternative reading frame of CDKN2a, respectively.( 29 ) These inhibitors are endogenously expressed in differentiated cells and downregulated by aberrant mitogenic signaling. The study of double KO (Ink4ab −/−) and triple KO of all three ORFs (Cdkn2ab −/−) showed that p15Ink4b can act as a critical backup for p16Ink 4 a, suggesting a rationale for frequent loss of the complete CDKN2b–CDKN2a locus in human tumors.( 30 ) Endogenous p19Arf,( 31 ) p16Ink4a,( 32 ) and Trp53 (also known as p53), all inactivated in several tumors,( 33 , 34 , 35 ) can limit reprogramming and inhibit pathways leading to an increased level of iPSC generation. In mice, Arf, rather than Ink4a, blocks important reprogramming pathways through p53 and p21 (encoded by Cdkn1a) activation. However, in humans, INK4a is more important than ARF.( 32 ) Loss of replicative potential may prevent cell reprogramming. The acquisition of cell immortality is a rate‐limiting step for establishing pluripotency in somatic cells.( 31 , 32 ) The transient inhibition of these proteins may significantly improve iPSC generation,( 31 , 32 , 33 , 34 , 35 ) although the ability of the resultant iPSCs to become tumorigenic is not completely understood. During reprogramming, cells increase their intolerance to different types of DNA damage. A p53‐mediated DNA damage response limits reprogramming to ensure iPSC genomic integrity and prevent genomic instability.( 35 ) This phenomenon emphasizes the similarities between induced pluripotency and tumorigenesis. Eventually, approximately 20% of the offspring developed tumors attributable to c‐Myc transgene reactivation. Retroviral c‐Myc introduction should be avoided for clinical application.( 27 ) Studies of the other barriers indicated that increased iPSC generation efficiency is observed after treating cells with butyrate( 36 ) or vitamin C( 37 ) or after exposing them to hypoxia.( 38 )
Reprogramming using miRNA. Considering the future application of reprogramming technology, two major non‐mutually exclusive issues that should be solved are safety and efficiency. The introduction and addition of specific non‐coding miRNA( 25 ) can, for example, improve reprogramming efficiency.( 9 , 39 , 40 ) Regarding safety, genomic modification, which is critical to carcinogenesis, is an important concern. The introduction of genes involved in reprogramming events is often facilitated by viral vector‐mediated transduction, which can involve random insertions of exogenous sequences into the genome.( 19 , 20 ) iPSCs can be obtained using virus‐free, removable PiggyBac transposons or episomal systems,( 41 , 42 , 43 , 44 ) but these approaches still use DNA constructs; thus, the possibility of genomic integration of introduced sequences is still a potential problem. Alternatively, the Sendai virus has been used; iPSCs were generated from human terminally differentiated circulating T cells( 45 ) using Sendai RNA virus vectors.( 46 , 47 ) Reprogramming events using just protein or mRNA has also been reported, but the protocols involved are technically challenging.( 48 , 49 , 50 )
Recently, two independent studies from the Morrisey group and our group have demonstrated that human and mouse somatic cells can be reprogrammed to iPSCs through forced miRNA expression, completely eliminating the need for ectopic protein expression.( 39 , 51 ) Morrisey group( 39 ) revealed that lentiviral‐mediated transfection of immature miR‐302/367 sequences generated reprogrammed cells (miR‐302/367 iPSCs) displaying characteristics similar to those of Oct4/Sox2/Klf4/Myc‐iPSCs, including pluripotency marker expression, teratoma formation, and chimera contribution and germline contribution for mouse cells. miR‐367 expression is required for miR‐302/367‐mediated reprogramming, activation of Oct4 expression, and Hdac2 suppression.( 39 ) Conversely, direct transfection of direct mature double‐stranded miRNAs (miR‐200c + miR‐302s + miR‐369s) resulted in PSC generation from differentiated adipose‐derived stem cells in humans and mice.( 51 ) This reprogramming method does not require vector‐based gene transfer, and thus holds significant potential in biomedical research and regenerative medicine.
Other reports have indicated that electroporation of the polycistronic cassette of hsa‐miR‐302a/b/c/d resulted in the reprogramming of human hair follicle cells.( 40 ) This reprogramming mechanism functioned through miR‐302‐targeted cosuppression of four epigenetic regulators: AOF2 (also known as KDM1 or LSD1), AOF1, MECP1‐p66, and MECP2.( 40 ) Furthermore, retroviral expression of the polycistronic cassette of hsa‐miR‐302a/b/c/d allowed the development of iPSC‐like phenotypes from human skin cancer cells.( 9 ) Because these methods were carried out without transcription factors, the introduction of miRNAs may play critical roles in differentiated cell reprogramming in humans and mice.
The underlying mechanism of miRNA reprogramming is not completely understood. Generally, miRNAs are involved in translation inhibition, mRNA destabilization, and coding mRNA function suppression.( 52 , 53 ) We hypothesize that miRNA expression fine‐tunes cell reprogramming mainly by inhibiting mRNA signaling, although evidence also suggests that miRNAs may have other functions including translation stimulation through an unknown mechanism. For example, miR‐369‐3p, which was used for reprogramming,( 51 ) acts as a unique switch for regulating translation repression and activation.( 54 ) miR‐302,( 39 , 40 , 51 , 55 ) which targets TGFβ receptor 2 and antagonizes EMT,( 55 ) was also reported to suppress AOF2, AOF1, MECP1‐p66, and MECP2,( 40 ) indicating that the miR‐302 pathway is fundamental for reprogramming. Inhibition or reversion of EMT could be stimulated by miR‐302,( 39 , 40 , 55 ) miR‐367,( 39 , 55 ) and miR‐200c.( 51 ) TGFβ modulates reprogramming by EMT signaling, whereas Klf4 stimulated E‐cadherin expression, a hallmark of MET, which is involved in the stimulation of important reprogramming events.( 56 ) When mammary epithelial cells, which express endogenous Klf4 (MET expression is unnecessary), were used as the starting material, iPSCs were successfully developed only by introducing Sox2 and Oct4 without adding Klf4.( 56 ) This suggests that the requirements needed for EMT inhibition may be dependent on cellular context.
Effect of reprogramming on cancer cells. Retrovirus‐mediated gene transfer in gastrointestinal cancer cells resulted in the induction of ES‐like gene and protein expression (patterns induced from the endoderm of the gastrointestinal tract to the mesoderm and ectoderm).( 10 ) Interestingly, retrovirus‐mediated exogenous expression of Oct4/Sox2/Klf4/Myc or Oct4/Sox2/Klf4 sensitized gastrointestinal cancer cells to vitamins and other chemotherapeutic agents.( 10 ) In vivo experiments involving short‐term cultured reprogrammed cells showed an inhibition of tumorigenicity in DLD‐1 colorectal cancer cells.( 10 ) The study also revealed changes in DNA methylation and histone modification and revealed that the epigenome of DLD‐1 cells resembled that of ES cells. The promoter region of p16Ink4a was demethylated similar to the heavily demethylated state.( 10 ) Long‐term cultured reprogrammed cells with gain‐of‐function mutations, including TP53R175H and KRASG 12 D, elicit a malignant transformation with c‐Myc activation in KRAS and TP53‐mutated HuCC‐T1 cholangiocellular carcinoma cells, suggesting a role of such oncogenic mutations in malignant phenotype reactivation.( 57 ) Recent studies have indicated that decreasing the p53 expression level enables the development of murine fibroblasts into iPSCs capable of generating germline‐transmitting chimeric mice, indicating that p53 may not be necessary for reprogramming. Silencing p53 will significantly increase the reprogramming efficiency of human somatic cells.( 33 , 34 , 58 ) Gain‐of‐function TP53 oncogenic mutations enhance defined factor‐mediated cell reprogramming,( 59 ) suggesting that the TP53 mutation context is influenced by the quality and quantity of reprogramming events. Reprogramming efficiency was increased in hypoxia,( 38 ) an effect observed in cancer cells (Masaki Mori, unpublished data, 2011).
Transfection of miR‐302 induces ES‐like phenotypes of skin cancer.( 9 ) MiR‐302 also inhibits tumorigenecity by coordinating suppression of the Cdk2 and Cdk4/6 cell cycle pathways.( 60 ) The study indicated that concurrent silencing of BMI‐1, an miR‐302‐targeted CSC marker, further promoted tumor suppressor functions of p16Ink4a and p14/p19Arf directed against Cdk4/6‐mediated cell proliferation. Also, miR‐302 inhibits human pluripotent stem cell tumorigenicity by enhancing the multiple G1 phase arrest pathways.( 60 ) Another study of glioma indicated that the miR‐302–367 cluster drastically affects the self‐renewal and infiltration properties of glioma‐initiating cells through Cxcr4 repression and consequent disruption of the Shh‐Gli‐Nanog network.( 61 ) This indicates that the miR‐302–367 cluster can efficiently trigger a cascade of inhibitory events leading to the disruption of CSC‐like and tumorigenic properties.( 61 ) Taken together, further study of novel reprogramming‐based therapeutic approaches that could prove beneficial for treatment of tumors with p53 inactivation( 33 , 34 , 58 ) and/or of CSCs, which can survive in a region of hypoxia,( 38 ) is warranted (Table 2; Fig. 1).
Table 2.
Summary of current studies of cancer cell reprogramming
| Method | Type of cancer | Malignant‐related phenotype | Characterization | Ref. |
|---|---|---|---|---|
| Nuclear transfer | Medulloblastoma (primary culture, mouse, Ptc1 heterozygous) | Suppressed proliferation, restore normal differentiation, normal proliferation in cultured blastocyst | Cloned blastocyst can support postimplantation development, as the embryo appeared normal and showed extensive differentiation, although not viable after E8.5 | (83) |
| Nuclear transfer (two‐step cloning) | Melanoma RAS+/Ink4a/Arf−/− | NT ES‐cell chimeric mice developed various types of tumors with shorter latency and higher penetrance compared with the donor mouse model | NT ES cells could form teratoma and generate chimera. Injection into tetraploid blastocyst resulted in a normal embryo viable until E9.5 | (84) |
| Nuclear transfer | EC | Dependent on donor ECs, one NT ES cell chimera suffered from head and neck EC and was inviable, and the other resulting NT ES cells showed a broad differentiation potential into teratomas and broad contribution to normal‐appearing mid‐gestation embryos | Nuclei from EC can direct preimplantation development, resulting in normal appearing blastocyst, higher efficiency of producing an ES cell line compared with the differentiated cells, although the degree of differentiation depends on the cell line character | (85) |
| Embryonic microenvironment | Metastatic melanoma, breast cancer | Reduced invasion, tumor growth, increased apoptosis Downregulated Nodal signaling through Lefty activation | n.d. | (86) |
| microRNA (miR‐302a, b, c, d) | Melanoma (Colo), prostate cancer cell line (PC3) | Reduced migration ability, reduced expression of cell cycle‐related genes (CCND1, CCND2, CDK2), and DNA methylation facilitator, MeCP2 MECP1‐p66, and some melanoma oncogenes | Expression of pluripotency markers Nanog, Oct4, Sox2, SSEA3, SSEA4 Demethylation of Oct4 Teratoma (+) | (9) |
| Defined transcription factor (OKM) | Melanoma (R545) | Chimeras were tumor‐free at 5 months of age | Teratoma (+), chimera (+), ES cell marker expression, demethylation of Nanog and Oct4 | (87) |
| Defined transcription factor (OSKM) | Gastrointestinal cancer (colon, liver, pancreatic cancer) DLD‐1 completely characterized | Differentiated iPC (post‐iPC) showed sensitivity to chemotherapy, reduced invasion, and reduced tumorigenicity, showed higher expression of p16 and p53 as compared to the parental cell | Expression of pluripotency marker, demethylation of Nanog, in vitro differentiation into adipocyte, epithelial, mesenchymal, and neural lineage, teratoma (−) | (10) |
| Defined transcription factor (OSKM) | KBM7 cells derived from blast crisis stage chronic myeloid leukemia (CML) | Completely resistant to imatinib, loss of BCR‐ABL‐dependent signaling | ES cell marker expression (+), demethylation of Oct4 and Nanog, teratoma (+) | (88) |
| Defined transcription factor (OSLN) | A549 lung cancer | Increased tumorigenic properties when transplanted into a NOD/SCID mouse, more aggressive and invasive, teratoma (−) | Demethylation of Oct4 promoter expressed endogenous Nanog and Oct4 although lower than HES cell ALP(+), teratoma (−) Reprogramming efficiency was higher compared with normal primary lung fibroblast | (89) |
| Oocyte extract | Breast cancer (MCF7 and HCC1945 cell lines) | Re‐expression of tumor suppressor genes RARB, CST6, CCND2, CDKN2A through demethylation and remodeling of histone marks to a more euchromatic state No changes in DNA methylation at pluripotency gene promoters Oct4/Nanog Reduced colony formation | n.d. | (90) |
ALP, alkaline phosphatase (staining); EC, embryonal carcinoma; HES, human embryonic stem; iPC, induced pluripotent stem (iPS)‐like cancer cells; K, Klf4; L, Lin28; M, c‐Myc; N, Nanog; n.d., not determined; NT ES, nuclear transfer‐generated embryonic stem cells; O, Oct3/4; Ref., reference; S, Sox2.
Figure 1.
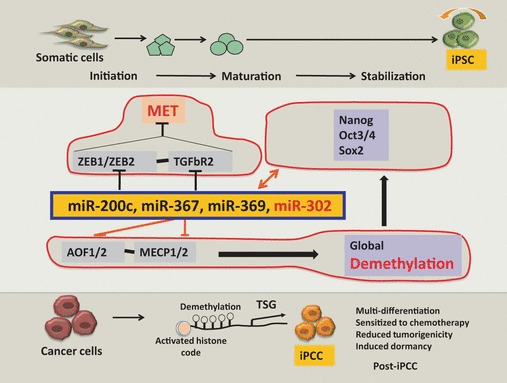
Cellular reprogramming in normal and cancer cells. Cellular reprogramming in normal and cancer cells can be viewed globally as a mechanism of phenotype reversal of parental cells through the modulation of epigenetic status into a more undifferentiated state. Defined transcription factors (Yamanaka cocktail)‐induced reprogramming is involved in the regulation of mesenchymal–epithelial transition (MET), which is controlled by a group of microRNAs (miR) through ZEB1/ZEB2 and TGFβR2. Those miRNAs play a role in global demethylation through AOF1/2 and MECP1/2. In contrast to normal cell reprogramming (upper panel), cancer cell reprogramming (lower panel) remains obscure. The reverse of MET, epithelial–mesenchymal transition (EMT), results in a chemotherapy‐resistant phenotype. Thus, reprogramming is supposed to open the silent chromatin through DNA demethylation and activate histone codes, which would elicit re‐expression of tumor suppressor genes, pushing cancer cells into a more benign phenotype. Further investigation would provide insight into how much of the tumor phenotype could be reversed through the contribution of reversible epigenetic and irreversible genetic changes in cancer. Reprogramming cancer cells might become a promising method for reversing or attenuating malignancy for therapeutic purposes. iPCC, induced pluripotent stem cell‐like cancer cell; iPSC, induced pluripotent stem cell; TGF, transforming growth factor; TSG, tumor suppressor gene.
Defined Factor‐Induced Reprogramming and CSCs
The differential mechanisms between cancer cells, which undergo a mutated form of reprogramming, and naturally occurring CSCs remain unclear.
Gastrointestinal cancer cells. Recently, it has been proposed that two types of stem cells coexist in normal and cancer cells and that these stem cells are transiently regulated by epigenetic controls.( 62 , 63 , 64 , 65 ) Emerging evidence indicates that quiescent and active stem cell subpopulations that are in lower metabolic and proliferative states, respectively, may coexist in several tissues.( 62 ) It has been proposed that these stem cell populations have separate but cooperative functional roles, and these adult stem cells are crucial for physiological tissue renewal and regeneration after injury.( 62 ) Generally, a stem cell divides asymmetrically into a new stem cell (self‐renewal) and a committed progenitor (differentiation). Whereas the asymmetric architecture of the stem cell niche in Drosophila and Caenorhabditis elegans is apparent, mammalian adult stem cells are generally detected in a predominantly quiescent state.( 63 , 64 ) Quiescent stem cells have been proposed to produce transit‐amplifying cells in rapidly regenerating tissue, which differentiate into mature cells and provide tissue architecture. Considering that transit‐amplifying cells have a short lifespan and limited self‐renewal capabilities, recent studies suggest that stem cell populations that are long‐lived yet constantly cycling are involved in the maintenance of tissue homeostasis.( 62 ) A new model describes the coexistence of quiescent and active adult stem cell subpopulations in bone marrow, intestinal epithelium, and hair follicles.( 62 ) In contrast to physiological tissues, serial transplantation experiments indicated that liver CSCs are composed of quiescent and active CSCs. This system plays a role in the exertion of resistance against chemoradiotherapy. During the study of CSCs, we identified CD13+ CSCs as a subpopulation of quiescent stem cells of the liver.( 65 ) Our study indicated that TGFβ induced the development of a CD13+ CSC population (Masaki Mori, submitted). CD13+ CSCs express immature genes often connected with a lower differentiation state, an observation that might explain why CD13+ CSCs exhibit aggressive behaviors (Masaki Mori, unpublished data, 2011). Considering TGFβ signaling counteracts the induction of cell reprogramming from normal differentiated cells, the outcome of reprogramming‐like stimulation should be investigated.
Reprogramming effect on CSCs. Endogenous expression levels of ES‐like genes could be relevant to tumor cell malignancy.( 66 ) The concept that a small population is contained in adult tissues may be relevant to CSCs in a tumor.( 67 ) The involvement of a very small embryonic/epiblast‐like stem cell population in carcinogenesis could support century‐old concepts involving embryonic rest‐ or germline‐origin hypotheses of cancer development;( 67 ) however, this working hypothesis requires further direct experimental confirmation.( 67 ) Further evidence indicates that tissues contain a unique population of mesenchymal stem cells or Muse cells,( 68 ) and that Muse cells are a primary source of iPSCs in human fibroblasts.( 69 ) By using immunocytochemistry to express Nanog, Oct3/4, and Sox2 and TRA‐1–81 to assess reprogramming efficiency, the authors showed that iPSC lines were generated with an efficiency of 0.001% from naive human skin fibroblasts, whereas Muse‐iPSCs were formed with an efficiency of 0.03%, indicating that Muse cells generate iPSCs 30‐fold more efficiently than naive fibroblasts.( 69 ) This type of subpopulation study elicits a challenging notion that a subset of pre‐existing adult stem cells in adult human tissues (or fibroblasts), which are somewhat similar to iPSCs, selectively become iPSCs, whereas the remaining cells make no contribution to iPSC generation.( 69 ) Nevertheless, at least two issues should be considered. First, the efficiency of iPSC generation in this study is much lower than that reported in other studies (≥0.02%; Table 1).( 20 , 70 ) Although the susceptibility to each cell reprogramming may be presumably based on pre‐existing conditions of epigenetic and transcription factor networks, underestimation cannot be excluded without adjusting the complete reprogramming technology. Second, given that higher efficiencies of reprogramming have been reported (up to approximately 10%, see Table 1) than the pre‐existing frequency of Muse cells in tissues, (1.1–1.3% of human fibroblasts or bone marrow stromal cells formed Muse cell‐derived cell clusters in naive populations without long‐term trypsin incubation), cells other than Muse cells may generate iPSCs. Taken together, it may be too early to conclude whether the defined factor‐induced reprogramming fits the elite model,( 69 ) rather than the stochastic model of iPSC generation.( 70 ) To reconcile these issues, further investigation is necessary to improve the reprogramming efficiency and understand the mechanism by which cellular reprogramming functions, especially in subpopulations of susceptible clones subjected to defined factor‐induced reprogramming. Considering that ES‐like genes expressing CSCs and unique populations including very small embryonic/epiblast‐like stem cells and Muse cells could be essential in cancer development, further research is necessary to determine the presence of these cell subpopulations in tumor tissues, relevancy to epithelial cancerous cells, and susceptibility of reprogramming events in these cell populations.
Perspective
Tissue homeostasis is a carefully balanced process controlled by epigenome regulation and efficient interplay between stem cells, their progeny, and the microenvironment (e.g. recently reviewed in intestinal stem cells( 23 )). Epigenome deregulation and malignant stem cell formation lead to tumor cell development. Reprogramming technology or epigenome modification through transfection of iPSC factors can lead to ES‐like gene expression patterns and considerable malignant phenotype modification,( 10 , 60 ) indicating that this technology could be used to create novel therapeutic targets against CSCs by combining small non‐coding RNAs with efficient drug delivery systems.
Disclosure Statement
This work was supported in part by: a grant‐in‐aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (H.I., N.H., H.N., Y.D., M.M.); the Ministry of Health, Labour, and Welfare; the Princess Takamatsu Foundation, Japan (H.I., M.M.); the Takeda Foundation, Japan (H.I.); the Senshin Medical Research Foundation (H.I.); Chugai Pharmaceutical Corporation, Japan (H.I., N.H. D.S., T.S.); and Yakult Corporation, Japan (H.I., N.H. D.S., T.S.). Patents pending on PCT/JP2011/053457, PCT/JP2011/054287, and PCT/JP2010/068406.
Abbreviations
- CSC
cancer stem cell
- EMT
epithelial–mesenchymal transition
- iPSC
induced pluripotent stem cell
- MEF
mouse embryonic fibroblast
- MET
mesenchymal–epithelial transition
- miRNA
microRNA
- Muse (cells)
multilineage‐differentiating stress‐enduring
- TGF
transforming growth factor
References
- 1. Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science 2001; 293: 1089–93. [DOI] [PubMed] [Google Scholar]
- 2. Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet 1993; 9: 138–41. [DOI] [PubMed] [Google Scholar]
- 3. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100: 57–70. [DOI] [PubMed] [Google Scholar]
- 4. Barker N, Ridgway RA, van Es JH et al. Crypt stem cells as the cells‐of‐origin of intestinal cancer. Nature 2009; 457: 608–11. [DOI] [PubMed] [Google Scholar]
- 5. Irons RD, Stillman WS. The process of leukemogenesis. Environ Health Perspect 1996; 104 (Suppl 6): 1239–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonnet D, Dick JE. Human acute leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997; 3: 730–37. [DOI] [PubMed] [Google Scholar]
- 7. Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature 2001; 414: 105–11. [DOI] [PubMed] [Google Scholar]
- 8. Kasemeier‐Kulesa JC, Teddy JM, Postovit LM et al. Reprogramming multipotent tumor cells with the embryonic neural crest microenvironment. Dev Dyn 2008; 237: 2657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin SL, Chang DC, Chang‐Lin S et al. Mir‐302 reprograms human skin cancer cells into a pluripotent ES‐cell‐like state. RNA 2008; 14: 2115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miyoshi N, Ishii H, Nagai K et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci USA 2010; 107: 40–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J, Emadali A, Le Bescont A, Callanan M, Rousseaux S, Khochbin S. Induced malignant genome reprogramming in somatic cells by testis‐specific factors. Biochim Biophys Acta 2011; 1809: 221–5. [DOI] [PubMed] [Google Scholar]
- 12. Ben‐David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer 2011; 11: 268–77. [DOI] [PubMed] [Google Scholar]
- 13. Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell 2007; 128: 747–62. [DOI] [PubMed] [Google Scholar]
- 14. Rasmussen TP, Corry GN. Epigenetic pre‐patterning and dynamics during initial stages of mammalian preimplantation development. J Cell Physiol 2010; 225: 333–6. [DOI] [PubMed] [Google Scholar]
- 15. Surani MA, Durcova‐Hills G, Hajkova P, Hayashi K, Tee WW. Germ line, stem cells, and epigenetic reprogramming. Cold Spring Harb Symp Quant Biol 2008; 73: 9–15. [DOI] [PubMed] [Google Scholar]
- 16. Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981; 292: 154–6. [DOI] [PubMed] [Google Scholar]
- 17. Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 1981; 78: 7634–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thomson JA, Itskovitz‐Eldor J, Shapiro SS et al. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282: 1145–7. [DOI] [PubMed] [Google Scholar]
- 19. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–76. [DOI] [PubMed] [Google Scholar]
- 20. Takahashi K, Tanabe K, Ohnuki M et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–72. [DOI] [PubMed] [Google Scholar]
- 21. Beers MF, Morrisey EE. The three R’s of lung health and disease: repair, remodeling, and regeneration. J Clin Invest 2011; 121: 2065–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herdrich BJ, Lind RC, Liechty KW. Multipotent adult progenitor cells: their role in wound healing and the treatment of dermal wounds. Cytotherapy 2008; 10: 543–50. [DOI] [PubMed] [Google Scholar]
- 23. Medema JP, Vermeulen L. Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nature 2011; 474: 318–26. [DOI] [PubMed] [Google Scholar]
- 24. Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell‐specific MicroRNAs. Dev Cell 2003; 5: 351–8. [DOI] [PubMed] [Google Scholar]
- 25. Mattick JS, Makunin IV. Non‐coding RNA. Hum Mol Genet 2006; 1: R17–29. [DOI] [PubMed] [Google Scholar]
- 26. Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell 2010; 40: 205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Okita K, Ichisaka T, Yamanaka S. Generation of germline‐competent induced pluripotent stem cells. Nature 2007; 448: 313–7. [DOI] [PubMed] [Google Scholar]
- 28. Wernig M, Meissner A, Foreman R et al. In vitro reprogramming of fibroblasts into a pluripotent ES‐cell‐like state. Nature 2007; 448: 318–24. [DOI] [PubMed] [Google Scholar]
- 29. Gil J, Peters G. Regulation of the INK4b–ARF–INK4a tumour suppressor locus: all for one or one for all. Nature Rev Mol Cell Biol 2006; 7: 667–77. [DOI] [PubMed] [Google Scholar]
- 30. Krimpenfort P, IJpenberg A, Song JY, van der Valk M, Nawijn M, Zevenhoven J. Berns A. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature 2007; 448: 943–6. [DOI] [PubMed] [Google Scholar]
- 31. Utikal J, Polo JM, Stadtfeld M et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 2009; 460: 1145–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H, Collado M, Villasante A et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 2009; 460: 1136–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hong H, Takahashi K, Ichisaka T et al. Suppression of induced pluripotent stem cell generation by the p53‐p21 pathway. Nature 2009; 460: 1132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawamura T, Suzuki J, Wang YV et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2009; 460: 1140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marión RM, Strati K, Li H et al. A p53‐mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 2009; 460: 1149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mali P, Chou B.‐K, Yen J et al. Butyrate Greatly Enhances Derivation of Human Induced Pluripotent Stem Cells by Promoting Epigenetic Remodeling and the Expression of Pluripotency‐Associated Genes. Stem Cell 2010; 28: 713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Esteban MA, Wang T, Qin B et al. Vitamin C Enhances the Generation of Mouse and Human Induced Pluripotent Stem Cells. Cell Stem Cell 2010; 6: 71–9. [DOI] [PubMed] [Google Scholar]
- 38. Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell 2009; 5: 237–41. [DOI] [PubMed] [Google Scholar]
- 39. Anokye‐Danso F, Trivedi CM, Juhr D et al. Highly efficient miRNA‐mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011; 8: 376–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DT. Regulation of somatic cell reprogramming through inducible mir‐302 expression. Nucleic Acids Res 2011; 39: 1054–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 2008; 322: 949–53. [DOI] [PubMed] [Google Scholar]
- 42. Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus‐free induction of pluripotency and subsequent excision of reprogramming factors. Nature 2009; 458: 771–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Woltjen K, Michael IP, Mohseni P et al. piggybac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 2009; 458: 766–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jia F, Wilson KD, Sun N et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods 2010; 7: 197–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Seki T, Yuasa S, Oda M et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell 2010; 7: 11–4. [DOI] [PubMed] [Google Scholar]
- 46. Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene‐free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci 2009; 85: 348–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ban H, Nishishita N, Fusaki N et al. Efficient generation of transgene‐free human induced pluripotent stem cells (iPSCs) by temperature‐sensitive Sendai virus vectors. Proc Natl Acad Sci USA 2011; 108: 14234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim D, Kim CH, Moon JI et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 2009; 4: 472–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou H, Wu S, Joo JY et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 2009; 4: 381–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Warren L, Manos PD, Ahfeldt T et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010; 7: 618–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miyoshi N, Ishii H, Nagano H et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 2011; 8: 633–8. [DOI] [PubMed] [Google Scholar]
- 52. Ambros V. The functions of animal microRNAs. Nature 2004; 431: 350–5. [DOI] [PubMed] [Google Scholar]
- 53. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol 2004; 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vasudevan S, Tong Y, Steitz JA. Switching from Repression to Activation: microRNAs Can Up‐Regulate Translation. Science 2007; 318: 1931–4. [DOI] [PubMed] [Google Scholar]
- 55. Liao B, Bao X, Liu L et al. MicroRNA cluster 302‐367 enhances somatic cell reprogramming by accelerating a mesenchymal‐to‐epithelial transition. J Biol Chem 2011; 286: 17359–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li R, Liang J, Ni S et al. A mesenchymal‐to‐epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010; 7: 51–63. [DOI] [PubMed] [Google Scholar]
- 57. Nagai K‐i, Ishii H, Miyoshi N et al. Long‐term culture following ES‐like gene‐induced reprogramming elicits an aggressive phenotype in mutated cholangiocellular carcinoma cells. BiochemBiophysic Res Commun 2010; 395: 258–63. [DOI] [PubMed] [Google Scholar]
- 58. Zhao Y, Yin X, Qin H et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell 2008; 3: 475–9. [DOI] [PubMed] [Google Scholar]
- 59. Moon JH, Ishii H, Dewi DL et al. Gain‐of‐function oncogenic mutations in TP53 enhance defined factor‐mediated cellular reprogramming. Nature Precedings, 2011.
- 60. Lin SL, Chang DC, Ying SY, Leu D, Wu DT. MicroRNA miR‐302 inhibits the tumorigenecity of human pluripotent stem cells by coordinate suppression of the CDK2 and CDK4/6 cell cycle pathways. Cancer Res 2010; 70: 9473–82. [DOI] [PubMed] [Google Scholar]
- 61. Fareh M, Turchi L, Virolle V et al. The miR 302‐367 cluster drastically affects self‐renewal and infiltration properties of glioma‐initiating cells through CXCR4 repression and consequent disruption of the SHH‐GLI‐NANOG network. Cell Death Differ 2011; DOI: 10.1038/cdd.2011.89. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science 2010; 327: 542–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Arai F, Hirao A, Ohmura M et al. Tie2/angiopoietin‐1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 2004; 118: 149–61. [DOI] [PubMed] [Google Scholar]
- 64. Fuchs E, Segre JA. Stem cells: a new lease on life. Cell 2000; 100: 143–55. [DOI] [PubMed] [Google Scholar]
- 65. Haraguchi N, Ishii H, Mimori K et al. CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest 2010; 120: 3326–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ben‐Porath I, Thomson MW, Carey VJ et al. An embryonic stem cell‐like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ratajczak MZ, Shin DM, Liu R et al. Epiblast/germ line hypothesis of cancer development revisited: lesson from the presence of Oct‐4+ cells in adult tissues. Stem Cell Rev 2010; 6: 307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kuroda Y, Kitada M, Wakao S et al. Unique multipotent cells in adult human mesenchymal cell populations. Proc Natl Acad Sci USA 2010; 107: 8639–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wakao S, Kitada M, Kuroda Y et al. Multilineage‐differentiating stress‐enduring (Muse) cells are a primary source of induced pluripotent stem cells in human fibroblasts. Proc Natl Acad Sci USA 2011; 108: 9875–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature 2009; 460: 49–52. [DOI] [PubMed] [Google Scholar]
- 71. Yu J, Vodyanik MA, Smuga‐Otto K et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 2007; 318: 1917–20. [DOI] [PubMed] [Google Scholar]
- 72. Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science 2008; 322: 945–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Aoi T, Yae K, Nakagawa M et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 2008; 321: 699–702. [DOI] [PubMed] [Google Scholar]
- 74. Kim JB, Zaehres H, Wu G et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 2008; 454: 646–50. [DOI] [PubMed] [Google Scholar]
- 75. Huangfu D, Maehr R, Guo W et al. Induction of pluripotent stem cells by defined factors is greatly improved by small‐molecule compounds. Nat Biotechnol 2008; 26: 795–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wernig M, Lengner CJ, Hanna J et al. A drug‐inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol 2008; 26: 916–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sun N, Panetta NJ, Gupta DM et al. Feeder‐free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci USA 2009; 106: 15720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shi Y, Do JT, Desponts C, Hahm HS, Schöler HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2008; 2: 525–8. [DOI] [PubMed] [Google Scholar]
- 79. Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell‐specific microRNAs promote induced pluripotency. Nat Biotechnol 2009; 27: 459–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhu S, Li W, Zhou H et al. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 2010; 7: 651–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li Z, Yang CS, Nakashima K, Rana TM. Small RNA‐mediated regulation of iPS cell generation. EMBO J 2011; 30: 823–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Subramanyam D, Lamouille S, Judson RL et al. Multiple targets of miR‐302 and miR‐372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 2011; 29: 443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li L, Connelly MC, Wetmore C, Curran T, Morgan JI. Mouse embryos cloned from brain tumors. Cancer Res 2003; 63: 2733–36. [PubMed] [Google Scholar]
- 84. Hochedlinger K, Blelloch R, Brennan C et al. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev 2004; 18: 1875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Blelloch RH, Hochedlinger K, Yamada Y et al. Nuclear cloning of embryonal carcinoma cells. Proc Natl Acad Sci USA 2004; 101: 13985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Postovit LM, Margaryan NV, Seftor EA et al. Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc Natl Acad Sci USA 2008; 105: 4329–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci 2009; 122: 3502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S. Generation of iPSCs from cultured human malignant cells. Blood 2010; 115: 4039–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res 2011; 71: 4640–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Allegrucci C, Rushton MD, Dixon JE et al. Epigenetic reprogramming of breast cancer cells with oocyte extracts. Mol Cancer 2011; 10: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]


