Abstract
Toll‐like receptors (TLRs) function as pattern‐reorganization receptors in mammals and play an essential role in innate immunity. After recognizing certain ligands, TLRs activate a cascade of signal pathways to establish a guard environment. For the first time, we report that in vitro treatment with recombinant calcineurin subunit B (CNB) upregulated several TLR‐related genes. Calcineurin subunit B interacted with the ectodomain of TLR4 in vitro. On further investigation, phosphorylation of interferon regulatory factor 3 and degradation of IκB‐α were observed by CNB stimulation. In addition to pro‐inflammatory cytokines, transcription and production of β‐interferon were also enhanced after CNB stimulation. Thus, CNB could be further explored as a cancer and virus immunotherapy drug. (Cancer Sci 2012; 103: 515–521)
Innate immunity provides immediate and efficient defense against microbial infection. Toll‐like receptors play an important role in innate immunity. They detect microbes, viruses, and endogenous ligands to mediate the induction of genes encoding inflammatory cytokines, chemokines, interferons, and interferon‐related molecules.( 1 ) Toll‐like receptor 4 is the only member of the TLR family that functions through both the MyD88 and TRIF signal pathways. After receiving certain kinds of stimulation, TLR4 promotes IRF3 and NF‐κB translocation to the nucleus to activate IFN‐β and pro‐inflammatory cytokines transcription. Long‐term IFN‐β therapy decreases the recurrence rate of hepatocellular carcinoma after curative therapy.( 2 ) Interferons do not possess the ability to eliminate viruses or tumors; instead, they activate a cascade of signaling pathways to inhibit virus replication or tumor proliferation.( 3 )
Calcineurin is regulated by Ca2+, which in turn dephosphorylates and induces the nuclear localization of cytoplasmic components of the nuclear factor of activated T cells transcription complexes.( 4 ) Subunit A of calcineurin is a catalytic subunit, whereas CNB is a regulatory subunit that regulates CN’s activity by binding to the CNB binding helix.( 5 ) Subunit B of calcineurin is highly hydrophobic and conserved. CNB−/− mice showed a higher risk for squamous cell carcinoma.( 6 ) The CNB potentiates the activation of procaspase‐3 by accelerating its proteolytic maturation.( 7 ) In plants, CNB‐like proteins have been associated with many fundamental processes, such as salt tolerance or K+ uptake.( 8 , 9 ) These results reveal that CNB might possess some biological functions independent of CNA. To further test the function of CNB alone, CNB has been successfully characterized and purified in our laboratory.( 10 ) Suppression of certain types of carcinoma cells was observed by i.p. injection of CNB into mice pre‐inoculated with carcinoma.( 11 ) Moreover, CNB activates human PBMC‐derived and mice marrow‐derived DCs to secrete pro‐inflammatory cytokines,( 12 ) but the underlying mechanisms have not been well described.
Here, for the first time, we report that CNB interacted with TLR4 ectodomain in vitro, which in turn led to phosphorylaiton of IRF3 and degradation of IκB‐α. Secretion of IFN‐β and pro‐inflammatory cytokines were induced after CNB stimulation. This provides a molecular mechanism for CNB’s immune booster and anticancer mechanism.
Materials and Methods
Mice and cell line. Male BALB/c mice, 5–6 weeks old, were purchased from Vital River Laboratory Animal Technology (Beijing, China) and kept under specific pathogen‐free and humane conditions. All experiments involving the use of mice were carried out in accordance with protocols approved by the Animal Care and Use Committee of Beijing Normal University (Beijing, China). Raw264.7 cells were cultured in DMEM (Gibco, Grand Island, NY, USA) supplemented with 10% (v/v) FBS (Invitrogen, Carlsbad, CA, USA) in a humidified atmosphere containing 5% CO2 at 37°C. U937 cells were cultured in 1640 (Gibco) supplemented with 10% FBS (Invitrogen) in a humidified atmosphere containing 5% CO2 at 37°C. Sf9 insect cells were maintained in TNM‐FH insect medium (Sigma, St. Louis, MO, USA) with 10% (v/v) FBS in a humidified atmosphere at 27°C.
Recombinant CNB and reagents. Construction of vectors, expression, and purification of CNB were accomplished in our previous work.( 10 ) Endotoxin contamination was <8 U/mg as determined by the Limulus amebocyte lysate assay (Beijing Institute for Drug Control, Beijing, China). Lipopolysaccharide derived from Escherichia coli 0111:B4 was obtained from Sigma. Antibodies against TLR4, β‐actin, TRAF3, IRF3, and CNB were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against p‐IRF3 and IκB‐α were bought from Cell Signaling Technology (Beverly, MA, USA). Antibodies against HA and Myc tag were obtained from Genetex (Irvine, CA, USA). Horseradish peroxidase‐coupled secondary antibodies were from Golden Bridge (Beijing, China). The ELISA kit for mouse IFN‐β was from PBL Biomedical Laboratories (Piscataway, NJ, USA). FuGENE HD transfection reagent was purchased from Roche (Mannheim, Germany). The pGL3‐Enhancer and pGL3‐IFNb‐reporter plasmids were gifts from Xuetao Cao (Second Military Medical University, Shanghai, China),( 13 ) the pGL6‐6xκB plasmid was from Beyotime (Jiangsu, China), and the pRL‐TK‐Renilla‐luciferase plasmid was a gift from Rui Huang (Beijing Normal University). The dual reporter assay kit was purchased from Promega (Madison, WI, USA). XFI‐Myc‐IRF3 and pcDNA3‐HA‐TRAF3 were generous gifts from Rik Derynck (University of Massachusetts, Worcester, MA, USA)( 14 ) and Micheal Karin (University of California San Diego, La Jolla, CA, USA),( 1 ) respectively. BaculoGold Linearized virus genome DNA was purchased from BD Pharmingen (Franklin Lakes, NJ, USA). pAcGP67a‐hTLR4‐ectodomain (amino acids 27–631) plasmid was a kind gift from J.O. Lee (KAIST, Daejeon, Korea).( 15 ) The plasmid was modified by standard molecular methods to add 6× His tag at the C‐terminus of hTLR4, and was named pAcGP67a‐His‐hTLR4‐ectodomain.
Enzyme‐linked immunosorbent assay. The ELISA for mouse IFN‐β was carried out according to the manufacturer’s instructions. Mice were i.p. injected with 20 mg/kg CNB or 0.5 mg/kg LPS (body weight) dissolved in PBS, or PBS alone (control group) every day for 1 week, and blood was collected from the cut tail on the day 7. Data were based on serum samples from the cut tail blood. Raw264.7 cells were seeded in 96‐well plates at 1 × 105 cells per well. After culture for 24 h, cells were incubated with 1 μM or 3 μM TAK‐242 (Invitrogen) for 4 h, subsequently stimulated with 8 μM CNB for 12 h. The concentrations of IFN‐β in culture medium were determined with mouse IFN‐β ELISA kits.
GeneChip assay. The method for GeneChip assay has been reported previously.( 16 ) In brief, total RNA of U937 cells treated or untreated with 1 μM CNB for 3 h was prepared by Trizol (Invitrogen, Gaithersburg, MD, USA) according to the recommended protocol. The NucleoSpin RNA clean‐up kit (Macherey‐Nagel, Düren, Germany) was used to further purify the total RNA. The quality of RNA was confirmed by use of a formaldehyde agarose gel and the concentration of RNA was determined by reading the absorbance at 260/280 nm. The procedure for labeling, hybridization, and washing was carried out as previously described.( 16 ) After drying, the slides were scanned for the fluorescence intensity of both the Cy5 and Cy3 Fluors using LuxScan 10KA confocal laser scanner and analyzed by LuxScan 3.0 (CapitalBio Company, Beijing, China). The biological interpretation of the gene clusters was further completed by KEGG pathways annotation.
Assay of luciferase reporter gene. For the IFN‐β reporter assay, Raw264.7 cells were cotransfected with a mixture of 100 ng IFN‐β luciferase reporter plasmid, 10 ng pRL‐TK‐Renilla‐luciferase plasmid, and various amounts of IRF3 or TRAF3 construct. For the κB reporter assay, Raw264.7 cells were cotransfected with a mixture of 100 ng κB luciferase reporter plasmid and 10 ng pRL‐TK‐Renilla‐luciferase plasmid. Either CNB or LPS (1 μg/mL) was added to the medium 6 h post transfection. The luciferase activity in the cells was measured with a Dual‐Luciferase Reporter Assay system according to the manufacturer’s instructions (Promega) 24 h post transfection. Empty vector was added to each sample to obtain a final concentration of 0.02 μg/μL. Data were normalized for transfection efficiency by division of firefly luciferase activities with that of Renilla luciferase.
Quantitative PCR and immunoblot analysis. Raw264.7 cells were stimulated with LPS (1 μg/mL) or CNB. Total RNA was extracted with an RNA extraction kit according to the manufacturer’s instructions (Bioteke, Beijing, China). The quality of RNA was confirmed by use of a formaldehyde agarose gel and the concentration of RNA was determined by reading the absorbance at OD 260/280 nm. The 7500 Realtime PCR cycler (ABI, Foster City, CA, USA), RevertAid First Strand synthesis kit (Fermentas, Vilnius, Lithuania) and SYBR Green Mix (Toyobo, Osaka, Japan) were used for quantitative PCR analysis. Data were normalized to GAPDH expression in each sample. The primers used for IFN‐β, RANTES, TAK‐1, TBK‐1, and GAPDH have been described.( 17 , 18 , 19 )
His‐tag pull‐down assay and IRF3 dimerization assay. In brief, 2 μg pAcGP67a‐His‐hTLR4‐ectodomain (amino acids 27–631) was cotransfected with 0.5 μg BD BaculoGold Linearized Baculovirus DNA into Sf9 cells using FuGENE HD transfection reagent (Roche). Cotransfection efficiencies were assayed by end‐point dilution assay according to the manufacturer’s instructions. Virus titer higher than 2 × 108 was used for further protein production. Cells were pelleted by centrifugation. Supernatant (20 mL) was incubated with 40 μL Ni Sepharose High Performance resin (GE Healthcare, San Diego, CA, USA) at 4°C overnight in a sterile tube. The resin was then centrifuged at 2000g and washed five times in ice‐cold buffer A (20 mM Tris, 20 mM imidazole, 120 mM NaCl, and protease inhibitor cocktail [pH 7.4]). The resin was then boiled for 5 min for detection of hTLR4. The infected supernatant was incubated with CNB for 12 h in buffer A, then incubated with Ni resin for 2 h, and washed five times with buffer A. The resin was boiled for 5 min in 2× SDS loading buffer for SDS gel separation and blotting analysis. The IRF3 dimerization assay has been described previously.( 20 ) In brief, 5 × 106 Raw264.7 cells were stimulated with 4 μM CNB or 1 μg/mL LPS for 2 h or 1 h, respectively. The cells were lysed in Western and IP Buffer (Beyotime), and the lysates were electrophoresed on 7% polyacrylamide native gels containing 1% deoxycholate. Both IRF3 monomer and dimer were detected by Western blotting assay.
Statistical analyses. Data are expressed as the means ± SE of three independent experiments. Statistical significance was determined with the two‐tailed Student’s t‐test, with a P‐value of less than 0.05 considered statistically significant.
Results
Subunit B of calcineurin upregulates several TLRs and apoptosis‐related genes. To address the possible pathways stimulated by CNB, we generated two RNA samples from U937 cells treated or untreated with CNB, and analyzed the data by gene chip (Fig. 1A,B). Among the 897 assayed genes, 30 genes were upregulated and two genes were downregulated. Analysis using the KEGG database showed that 43% of the upregulated genes were TLR pathway‐related genes and 34% were apoptosis‐related genes. Genes with a twofold change or higher are listed in Table 1. These results accorded with our previous findings that DCs stimulated with CNB enhanced the secretion of cytokines tumor necrosis factor‐α, IL‐12p70, IL‐1β, and IL‐6 and chemokines IL‐8, IP‐10, MIP‐1α, and MIP‐1β by ELISA or qPCR.( 12 )
Figure 1.
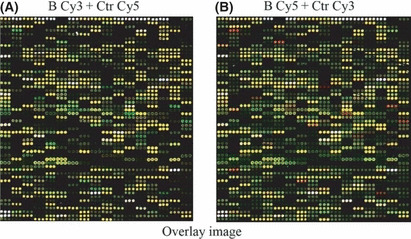
Expression of various Toll‐like receptor‐related genes was upregulated by subunit B of calcineurin (CNB) stimulation in U937 cells. (A) Microarray‐based expression profiling experiments using a two‐color design: total RNA extracted from CNB‐induced U937 cells labeled with Cy3 (green); and that of medium control with Cy5 (red). Green dot, gene is upregulated; red dot, gene is downregulated; yellow, gene is not sensitive to CNB stimulation. B, CNB‐treated group; Ctr, control group. (B) Fluorophore exchange in microarray analysis. Green, control group treated with Cy3; red, CNB‐treated group labeled with Cy5.
Table 1.
Genes upregulated by in vitro treatment with recombinant calcineurin subunit B
| Gene symbol | GenBank accession no. | Ratio | Signaling pathway in KEGG |
|---|---|---|---|
| NF‐κB | NM_003998 | 3.500 | hsa04010, hsa04210, hsa04620, hsa04660 |
| MIP‐1α | NM_002983 | 2.228 | hsa04060, hsa04620 |
| MIP‐1β | NM_002984 | 2.769 | hsa04060, hsa04620 |
| IκB‐α | NM_020529 | 3.200 | hsa04210, hsa04620, hsa04660, hsa04662, hsa04920, hsa05120, hsa05215, hsa05220, hsa05225 |
| TNF‐α | NM_000594 | 4.108 | hsa04010, hsa04060, hsa04210, hsa04350, hsa04620, hsa04640, hsa04650, hsa04660, hsa04664, hsa04920, hsa04930, hsa04940, hsa05010, hsa05060 |
| TRAF3 | AF110908 | 2.700 | hsa04620, hsa05222 |
| IKK‐ε | NM_014112 | 2.200 | hsa04620 |
| IL‐8 | NM_000584 | 11.68 | hsa04060, hsa04620, hsa05120, hsa05219 |
| NF‐κB‐2 | S76638 | 3.767 | hsa04662, hsa04920, hsa05120, hsa05212, hsa05215, hsa05220, hsa05221, hsa05222 |
| TRAF1 | NM_005658 | 2.996 | hsa05222 |
| ADRB2 | M15169 | 3.398 | hsa04020, hsa04080 |
| GADD45 | L24498 | 2.199 | hsa04010, hsa04110, hsa04115 |
| NFκBIE | NM_004556 | 2.711 | hsa04660, hsa04662, hsa04920 |
| Collagenase‐1 | NM_002421 | 2.714 | hsa03320, hsa05219 |
| Fas/Apo‐1 | NM_000043 | 2.133 | hsa04010, hsa04060, hsa04115, hsa04210, hsa04650, hsa04940 |
| ICAM1 | NM_000201 | 4.035 | hsa04514, hsa04650, hsa04670 |
| IFNGR‐2 | NM_005534 | 2.801 | hsa04060, hsa04630, hsa04650 |
| MKP‐3 | NM_001946 | 2.098 | hsa04010 |
| IAP‐1 | AF070674 | 2.058 | hsa04210, hsa04510, hsa05222 |
| IRS2 | AF073310 | 2.155 | hsa04910, hsa04920, hsa04930 |
| IRAK2 | NM_001570 | 2.315 | hsa04210 |
| Mcp‐1 | NM_002982 | 5.991 | hsa04060 |
Ratio, describes fold change. hsa04620, Toll‐like receptor‐related signaling pathway in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
Activation of IRF3 by CNB stimulation in Raw264.7 and U937 cells. IRF3 is an essential transcription factor downstream of TLR. To test whether CNB promotes activation of IRF3, we added purified CNB to the medium of U937 or Raw264.7 cells. IRF3 phosphorylation was observed in both cell lines (Fig. 2A,B). The time to reach the peak for IRF3 phosphorylation differed between Raw264.7 cells (approximately 30 min) and U937 cells (maintained 1–2 h). We detected the constitutive formation of the IRF3 dimer and its further augmentation by CNB stimulation in Raw264.7 cells. Consistent with IRF3 phosphorylation blotting, the IRF3 dimer was detected 1 h post CNB stimulation (LPS was used as a positive control)( 21 ) (Fig. 2C). The protein level of TRAF3 was detected by blotting. It was previously shown that CNB cannot promote TRAF3 degradation, which is different from LPS (Fig. 2D).( 1 ) These data indicated that CNB activated a signaling pathway leading to activation of IRF3.
Figure 2.
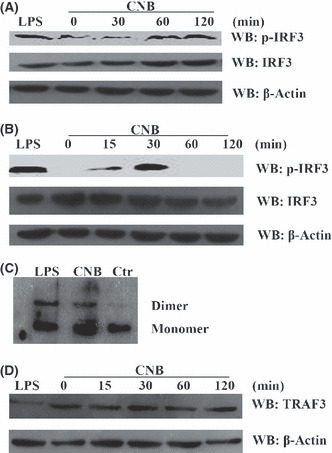
Phosphorylation and dimerization of interferon regulatory factor 3 (IRF3) were induced by subunit B of calcineurin (CNB) stimulation. (A) Detection of p‐IRF3 in U937 cells. Cells were treated with either 1 μg/mL lipopolysaccharide (LPS) for 30 min as a positive control or 1 μM CNB for 0 min, 30 min, 1 h, or 2 h. WB, Western blotting. (B) Detection of p‐IRF3 in Raw264.7 cells. Cells were treated with either 1 μg/mL LPS for 30 min as a positive control or 2 μM CNB for 0 min, 15 min, 30 min, 1 h, or 2 h. IRF3 and β‐actin in the whole cell lysates were used as endogenous controls. (C) Dimerization of IRF3 in Raw264.7 cells. Cells were treated with 1 μg/mL LPS for 1 h, 2 μM CNB for 2 h, or left untreated (Ctrl, control). Cell lysates were separated by native PAGE and detected by WB. (D) Detection of tumor necrosis factor receptor‐associated factor 3 (TRAF3) degradation. The samples were the same as in (B).
Degradation of IκB‐α by CNB stimulation. Our previous research also implied that CNB undertook the NF‐κB pathway.( 12 ) Therefore, we examined IκB‐α expression levels after CNB stimulation. The degradation of IκB‐α was observed in a time‐dependent manner (Fig. 3A). To confirm this result, we transfected κB reporter plasmid into Raw264.7 cells. The CNB enhanced expression of the κB reporter gene in a dose‐dependent and time‐dependent manner in Raw264.7 cells (Fig. 3B,C).
Figure 3.
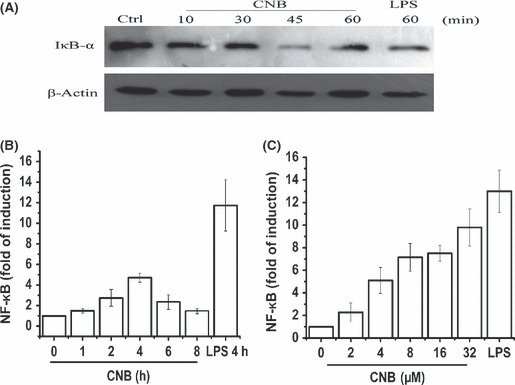
Nuclear factor‐κB (NF‐κB) activated by subunit B of calcineurin (CNB) stimulation. (A) Detection of IκB‐α degradation. RAW264.7 cells were treated with 1 μg/mL lipopolysaccharide (LPS) for 60 min as a positive control (Ctrl) or 4 μM CNB for 0 min, 10 min, 30 min, 45 min, or 1 h. (B) Luciferase assay of the induction of NF‐κB‐related genes in lysates of Raw264.7 cells transfected with 100 ng pGL6‐6xκB reporter plasmid and 10 ng pTK‐Renilla‐luciferase. Six hours post transfection, 4 μM CNB was added to the medium. Activity of κB was detected at different time points. Empty control vector was added to each sample to ensure transfection of the same amount of DNA in each. Luciferase activity was normalized to Renilla luciferase activity. (C) Transfection procedure was the same as (B). Different amounts of CNB (0, 2, 4, 8, 16, or 32 μM) were added after transfection. Activity of κB was detected at 4 h post stimulation. LPS was used as a positive control.
Effects of CNB on IFN‐β transcription. Given the results that CNB promoted both phosphorylation of IRF3 and degradation of IκB‐α, we tested whether CNB enhanced IFN‐β transcription. The indicated amounts of recombinant CNB were added to the medium of Raw264.7 cells, and the mRNA level for IFN‐β was detected using qPCR. The IFN‐β mRNA level was upregulated by CNB stimulation in a time‐dependent way (Fig. 4A). The time point to reach the peak was approximately 1 h post CNB stimulation, which was consistent with previous result (30 min to reach the peak for p‐IRF3) (Fig. 2B). As expected, the mRNA level of another IRF3‐related gene, RANTES, was also upregulated by CNB stimulation (Fig. 4B). The IFN‐αn mRNA (IFN‐αn was designed to detect the mRNAs for IFN‐a1, a2, a7, a11, and a12 subtypes) was not detected as stimulated with either LPS or CNB (data not shown) by qPCR, consistent with a previous report.( 22 )
Figure 4.
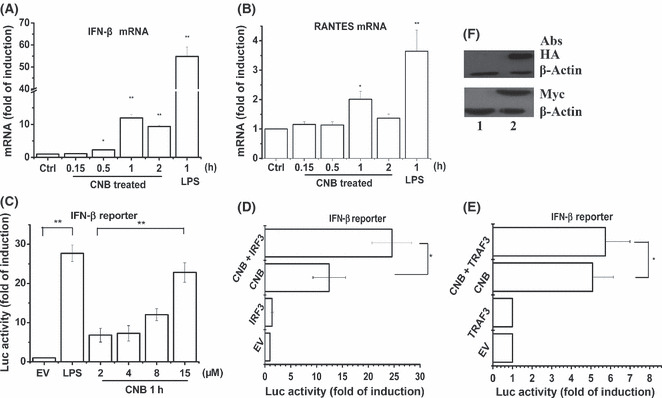
Subunit B of calcineurin (CNB) enhanced transcription of genes encoding β‐interferon (IFN‐β) in Raw264.7 cells in a tumor necrosis factor receptor‐associated factor 3/interferon regulatory factor 3 (TRAF3/IRF3)‐dependent and dose‐dependent way. (A,B) Real‐time PCR of IFN‐β and RANTES mRNA in RAW264.7 cells stimulated with CNB or lipopolysaccharide (LPS; 1 μg/mL) for the indicated time (horizontal axis), presented as “fold of induction” relative to GAPDH. (C,D,E) Luciferase assay of the induction of genes encoding IFN‐β in lysates of Raw264.7 cells transfected with 100 ng pGL3‐IFNb reporter plasmid and 10 ng pTK‐Renilla‐luciferase, and 100 ng IRF3 expression plasmid (D), or 100 ng TRAF3 expression plamid (E). Empty control vector (EV) was added to each sample to ensure transfection of the same amount of DNA in each. Luciferase activity was normalized to Renilla luciferase activity. (F) Detection of IRF3 (lower, lane 2) or TRAF3 (upper, lane 2) expression in the lysates of (D) or (E). Lane 1, EV control. *P < 0.05; **P < 0.01.
Expression of the IFNb reporter gene was enhanced by CNB in a dose‐dependent way in Raw264.7 cells (Fig. 4C). To further test the function of IRF3 and TRAF3 in CNB stimulated pathways, we transfected Raw264.7 cells with different amounts of IRF3 or TRAF3 constructs. Cells were stimulated with indicated amounts of CNB 6 h post transfection. Activation of the IFNb reporter gene was also further upregulated by IRF3 or TRAF3 (Fig. 4D,E). Protein expression of IRF3 (lower) or TRAF3 (upper) was detected in whole cell lysates by Western blotting (Fig. 4F). These results indicated that CNB increased IRF3/TRAF3‐mediated transcription of genes encoding IFN‐β.
Involvement of TAK‐1 and TBK‐1 in CNB signal pathway. Nuclear translocation of IRF3 and degradation of IκB‐α both require signal‐dependent phosphorylation. Previous reports indicated that TBK‐1 and TAK‐1 were essential for IRF3 phosphorylation and NF‐κB activation, respectively.( 23 , 24 ) To seek kinases involved in the CNB signal pathway, TAK‐1 and TBK‐1 mRNA were analyzed after CNB stimulation. After CNB stimulation, TAK‐1 and TBK‐1 were upregulated 1.7‐fold and 2.2‐fold, respectively (Fig. 5A,B). Thus, TBK‐1 and TAK‐1 might be responsible for IRF3 phosphorylation and NF‐κB activation in the CNB stimulated pathway.
Figure 5.
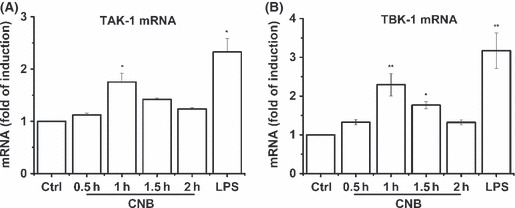
Analysis of transforming growth factor β‐activated kinase 1 (TAK‐1) and Tank binding kinase 1 (TBK‐1) mRNA level by subunit B of calcineurin (CNB) stimulation. (A,B) Real‐time PCR of TAK‐1 and TBK‐1 mRNA in RAW264.7 cells stimulated with CNB or lipopolysaccharide (LPS; 1 μg/mL) for the indicated time (horizontal axis), presented as “fold of induction” relative to GAPDH.
Production of IFN‐β upregulated by CNB. To further investigate the function of CNB in vivo, we i.p. injected mice with CNB, LPS, or PBS for a period of 7 days. The level of IFN‐β in serum was measured by ELISA (Fig. 6A). Subunit B of calcineurin showed a mild degree of IFN‐β induction (35.75 ± 6.78 pg/mL) compared to LPS treatment (215.37 ± 20.76 pg/mL). A TLR4‐specific inhibitor, TAK‐242, was used to test whether CNB activates IFN‐β expression through TLR4.( 25 ) As expected, inhibition of TLR4 by TAK‐242 attenuated the production of IFN‐β in vitro (Fig. 6B). This result indicated that CNB probably undertook the TLR4 signal pathway.
Figure 6.
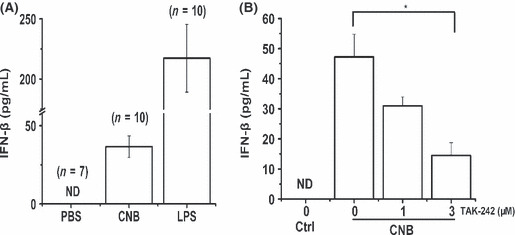
Enzyme‐linked immunosorbent assay of β‐interferon (IFN‐β). (A) Effect of subunit B of calcineurin (CNB) in vivo. Mice were i.p. injected with PBS (control), lipopolysaccharide (LPS), or CNB. β‐interferon in serum samples from cut tail blood was measured by ELISA. (B) Effect of transforming growth factor β‐activated kinase 242 (TAK‐242) on CNB‐induced IFN‐β production. RAW264.7 cells were incubated with 1 or 3 μM TAK‐242 for 4 h then stimulated with CNB for 12 h. The concentrations of IFN‐β in the culture medium were determined by ELISA. Ctrl, control group; ND, not detected. * P < 0.05.
Interaction between CNB with TLR4 in vitro. Activation of IRF3 is found to confer a TLR3/TLR4 specificity, which mediates a specific gene program for innate immunity response.( 26 ) Moreover, from previous results (Fig. 6B), we speculated that CNB might interact with TLR4. To further address this hypothesis, we expressed the His‐tagged ectodomain of TLR4 (amino acids 27–631) in Sf9 insect cells. The TLR4 ectodomain expression was monitored by SDS‐PAGE (Fig. 7A) or Western blotting using anti‐TLR4 or anti‐His antibody (Fig. 7B). As expected, CNB interacted with the TLR4 ectodomain, as shown by the His‐tag pull‐down assay in vitro (Fig. 7C).
Figure 7.
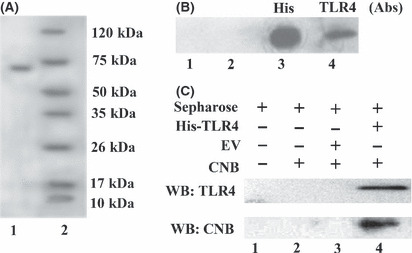
Subunit B of calcineurin (CNB) interacts with Toll‐like receptor 4 (TLR4) in vitro. (A) SDS‐PAGE detection of TLR4 ectodomain. Lane 1, 20 mL supernatant of hTLR4‐infected Sf9 cells was incubated with 40 μL Ni Sepharose and washed five times with buffer (20 mM Tris, 20 mM imidazole, 120 mM NaCl [pH 7.4]); resin was boiled with 2× SDS loading buffer for TLR4 ectodomain detection. Lane 2, protein marker. (B) Western blotting detection of TLR4 ectodomain. Lane 1, protein marker; lane 2, supernatant of Sf9 without infection; lanes 3,4, supernatant of Sf9 infected with His‐tag hTLR4 ectodomain baculo‐virus expression vectors (BEVs), detection of His‐tag hTLR4 ectodomain with either anti‐His mAb (lane 3) or TLR4 (H180) pAbs (lane 4). (C) His‐tag pull‐down assay. Lane 1, mock, resin only. Ten micrograms of CNB was incubated with resin only (lane 2), uninfected Sf9 supernatant (lane 3), or pAcGP67a‐hTLR4/BEVs‐infected Sf9 supernatant (lane 4) overnight at 4°C. Then Ni resin was added to each sample, incubated for 2 h at 4°C, then washed five times. Resin was boiled for 5 min with 2× SDS loading buffer. The interaction was detected by Western blotting. EV, empty control vector.
Discussion
Our previous results have shown that the phagocytic activities of peritoneal macrophages and the natural killer activities of murine spleen lymphocytes increased after CNB injection.( 27 ) Subunit B of calcineurin also inhibits the growth of H22 liver ascites cancer cells and S180 solid cancer cells in vivo.( 11 ) Moreover, CNB activates human PBMC‐derived and mice marrow‐derived DCs to secrete pro‐inflammatory cytokines.( 12 ) To elucidate the molecular mechanism, we designed a gene chip assay. From the assay, we found that genes for IKKε, TRAF3, and NF‐κB were upregulated by CNB stimulation in U937 cells (Fig. 1). The TRAF3–TBK1–IKKε–IRF3 complex is of great importance to both TLR‐dependent and TLR‐independent production of type I IFN production.( 28 , 29 ) IKKε can form a heterodimer with TBK1, and may act as a kinase to phophorylate IRF3.( 23 , 28 ) Both TRAF3 and IRF3 are uniquely required for the TRIF‐dependent interferon response.( 29 ) The regulatory elements for IFN‐β included κB sites, IRES sites, and AP‐1 sites, which bind NF‐κB, IRFs, or c‐Jun/ATF2 heterodimer, respectively.( 30 ) Previous reports have also implied that CNB undertakes NF‐κB signal pathways to activate DCs to produce several pro‐inflammatory cytokines.( 12 ) To further investigate the immune booster mechanism, we carried out experiments to detect NF‐κB and IRF3 activation. As expected, CNB promoted degradation of IκB‐α in both Raw264.7 cells (Fig. 3) and U937 cells (Wu Wu, Beijing Normal University, Beijing, China, unpublished data). Phosphorylation and dimerization of IRF3 was also observed after CNB stimulation (Fig. 2). Transcription of IFN‐β in Raw264.7 cells was enhanced in a TRAF3‐ and IRF3‐dependent way by CNB stimulation (Fig. 4). All of these results indicate that CNB stimulation may lead to IFN‐β production as well as pro‐inflammatory cytokines.
Stimulation of LPS caused degradation of TRAF3 in Raw264.7 cells and upregulation of RANTES in A549 cells, which has been reported previously.( 1 , 31 ) Subunit B of calcineurin does not promote degradation of TRAF3, which is different from LPS (Fig. 2). TRAF3 is a key regulator for balancing type I interferon and pro‐inflammatory cytokines production. K48 ubiquitination of TRAF3 induced by LPS is designed for proteasomal degradation, whereas K63 ubiquitination of TRAF3 is essential for IFN production.( 1 ) TRAF3−/− cells are defective in type I IFN responses activated by several different TLRs.( 28 ) TRAF3 associates with the TLR adaptors TRIF and IRAK1, as well as downstream IRF3/7 kinases TBK1 and IKKε, suggesting that TRAF3 serves as a critical link between TLR adaptors and downstream regulatory kinases important for IRF activation. We speculated that cells receiving CNB stimulation might “preferentially” promote K63 ubiquitination of TRAF3 for type I IFN production, which needs further research.
The MKK4/7‐JNK or TBK‐1/IKKε complex functions as an upstream kinase for IRF3.( 23 , 32 ) TAK1 forms a complex with TAB 1, TAB 2, and TAB 3. The activated TAK1 complex phosphorylates IκB kinases, which activates NF‐κB.( 33 ) Thus, we detected TAK‐1 and TBK‐1 mRNA levels after CNB stimulation by qPCR (Fig. 5). The mRNA levels for TAK‐1 and TBK‐1 were both upregulated after addition of CNB. These results implied that the kinases for IRF3 and IκB‐α phosphorylation in our CNB stimulation pathways were TBK‐1 and TAK‐1, respectively.
β‐interferon itself does not kill the tumor or virus. Instead, it initiates a cascade of signal pathways to inhibit virus replication or tumor cell proliferation.( 3 ) β‐interferon eliminates activated DCs by caspase‐3/11 activation, which plays an essential role in re‐establishing homeostasis.( 34 ) Antitumor effects mediated by IFN‐β have also been reported.( 34 , 35 , 36 ) β‐interferon KO mice showed a higher risk for sustained inflammation, cytokine production, and tissue damage with consequent chronic neurological deficits.( 37 ) Injection of CNB upregulated IFN‐β production to a mild degree compared to LPS‐stimulated mice (Fig. 6). This result explained the efficient antitumor and immune booster effects of CNB.( 11 , 12 )
Activation of IRF3 confers a TLR3/4, rather than TLR2/9, specificity.( 26 ) Moreover, from our gene chip assay results, approximately half of the genes changed by twofold or more were TLR‐related genes (Fig. 1). To seek the membrane receptor for CNB, we focused mainly on TLRs. An atypical member of the leucine‐rich repeats family, TLR4 is composed of N‐terminal, central, and C‐terminal domains. The primary function of these motifs provides a versatile structural framework for the formation of protein–protein interactions. The leucine‐rich repeat domain of TLR4 interacts with a large range of hydrophobic ligands such as HSP70/60, MD‐2, fusion protein, envelope protein, and flavolipin.( 38 , 39 ) The structural basic of TLR4 also provides a possibility for our highly hydrophobic CNB to bind. We validated our speculation by pull‐down assay (Fig. 7). Moreover, CNB binds to endogenous TLR4 by IP experiment in U937 cell lines and TAK‐242 (TLR4‐specific inhibitor) pre‐incubation attenuates CNB stimulated signals (Wu Wu, Beijing Normal University, Beijing, China, unpublished data; Fig. 6B). Indeed, CNB interacted with TLR4 and activated a signal cascade.
In conclusion, CNB upregulates IFN‐β and several pro‐inflammatory cytokines by activation of the IRF3 and NF‐κB signal pathways through TLR4 signal pathways. Our findings imply that CNB could be useful for enhancing immune response and preventing cancer and virus infections.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- CNA
subunit A of calcineurin
- CNB
subunit B of calcineurin
- DC
dendritic cell
- IFN‐β
β‐interferon
- IL
interleukin
- IP
immunoprecipitation
- IRF3
interferon regulatory factor 3
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LPS
lipopolysaccharide
- MIP
macrophage inflammatory protein
- NF‐κB
nuclear factor κB
- qPCR
quantitative PCR
- TAB
TAK1‐binding protein
- TAK1
transforming growth factor β‐activated kinase 1
- TBK1
Tank‐binding kinase 1
- TLR
Toll‐like receptor
- TRAF3
tumor necrosis factor receptor‐associated factor 3
- TRIF
TIR‐domain‐containing adapter‐inducing interferon‐β
Acknowledgments
This work was partly supported by the National Natural Science Foundation of China (Grant No. 30970636), the International Cooperation Project (2008DFR30500), the National Important Novel Medicine Research Project (2009ZX09103‐643), the Fundamental Research Funds for Central Universities and the Excellent Doctor’s Funds of Beijing Normal University.
References
- 1. Tseng P‐H, Matsuzawa A, Zhang W, Mino T, Vignali DAA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol 2010; 11: 70–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ikeda K, Arase Y, Saitoh S et al. Interferon beta prevents recurrence of hepatocellular carcinoma after complete resection or ablation of the primary tumor‐A prospective randomized study of hepatitis C virus‐related liver cancer. Hepatology 2000; 32: 228–32. [DOI] [PubMed] [Google Scholar]
- 3. Decker T, Muller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol 2005; 5: 675–87. [DOI] [PubMed] [Google Scholar]
- 4. Klee CB, Ren H, Wang X. Regulation of the calmodulin‐stimulated protein phosphatase, calcineurin. J Biol Chem 1988; 273: 13367–70. [DOI] [PubMed] [Google Scholar]
- 5. Kissinger CR, Parge HE, Knighton DR et al. Crystal structures of human calcineurin and the human FKBP12‐FK506‐calcineurin complex. Nature 1995; 387: 641–4. [DOI] [PubMed] [Google Scholar]
- 6. Wu X, Nguyen BC, Dziunycz P et al. Opposing roles for calcineurin and ATF3 in squamous skin cancer. Nature 2010; 465: 368–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saeki M, Irie Y, Ni L et al. Calcineurin potentiates the activation of procaspase‐3 by accelerating its proteolytic maturation. J Biol Chem 2007; 282: 11786–94. [DOI] [PubMed] [Google Scholar]
- 8. Liu J, Zhu JK. A calcium sensor homolog required for plant salt tolerance. Science 1998; 280: 1943–5. [DOI] [PubMed] [Google Scholar]
- 9. Xu J, Li HD, Chen LQ et al. A protein kinase, interacting with two calcineurin B‐like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 2006; 125: 1347–60. [DOI] [PubMed] [Google Scholar]
- 10. Wei Q, Lee EY. Expression and reconstitution of calcineurin A and B subunits. Biochem Mol Biol Int 1997; 41: 169–77. [DOI] [PubMed] [Google Scholar]
- 11. Wei Q, Lian ML, Jing FZ et al. Studies of calcineurin B subunit from genetic engineering for use in medicine. Drug Develop Res 2002; 56: 40–3. [Google Scholar]
- 12. Li J, Guo J, Su Z, Hu M, Liu W, Wei Q. Calcineurin subunit B activates dendritic cells and acts as a cancer vaccine adjuvant. Int Immunol 2011; 23: 327–34. [DOI] [PubMed] [Google Scholar]
- 13. An HZ, Hou J, Zhou J et al. Phosphatase SHP‐1 promotes TLR‐ and RIG‐I‐activated production of type I interferon by inhibiting the kinase IRAK1. Nat Immunol 2008; 9: 542–50. [DOI] [PubMed] [Google Scholar]
- 14. Qin BY, Liu C, Lam SS et al. Crystal structure of IRF‐3 reveals mechanism of autoinhibition and virus‐induced phosphoactivation. Nat Struct Mol Biol 2003; 10: 913–21. [DOI] [PubMed] [Google Scholar]
- 15. Kim HM, Park BS, Kim JI et al. Crystal structure of the TLR4‐MD‐2 complex with bound endotoxin antagonist eritoran. Cell 2007; 130: 906–17. [DOI] [PubMed] [Google Scholar]
- 16. Lin R, Lu G, Wang J et al. Time course of gene expression profiling in the liver of experimental mice infected with Echinococcus multilocularis. PLoS ONE 2011; 6: e14557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ikeda K, Arase Y, Saitoh S et al. Interferon beta prevents recurrence of hepatocellular carcinoma after complete resection or ablation of the primary tumor—a prospective randomized study of hepatitis C virus–related liver cancer. Hepatology 2000; 32: 228–33. [DOI] [PubMed] [Google Scholar]
- 18. Ricour Cl, Borghese F, Sorgeloos Fdr, Hato SV, Kuppeveld FJMv, Michiels T. Random Mutagenesis Defines a Domain of Theiler’s Virus Leader Protein That Is Essential for Antagonism of Nucleocytoplasmic Trafficking and Cytokine Gene Expression. J Virol 2009; 83: 11223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chiang S‐H, Bazuine M, Lumeng CN et al. The protein kinase IKKε regulates energy expenditure, insulin sensitivity and chronic inflammation in obese mice. Cell 2009; 138: 961–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barro M, Patton JT. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc Natl Acad Sci USA 2005; 102: 4114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Poltorak A, He X, Smirnova I et al. Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr Mice: mutations in Tlr4 Gene. Science 1998; 282: 2085–8. [DOI] [PubMed] [Google Scholar]
- 22. Sakaguchi S, Negishi H, Asagiri M et al. Essential role of IRF‐3 in lipopolysaccharide‐induced interferon‐beta gene expression and endotoxin shock. Biochem Biophys Res Commun 2003; 306: 860–6. [DOI] [PubMed] [Google Scholar]
- 23. Fitzgerald KA, McWhirter SM, Faia KL et al. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 2003; 4: 491–7. [DOI] [PubMed] [Google Scholar]
- 24. Gaynor RB, Takaesu G, Surabhi RM, Park KJ, Ninomiya‐Tsuji J, Matsumoto K. TAK1 is critical for I kappa B kinase‐mediated activation of the NF‐kappa B pathway. J Mol Biol 2003; 326: 105–15. [DOI] [PubMed] [Google Scholar]
- 25. Matsunaga N, Tsuchimori N, Matsumoto T, Ii M. TAK‐242 (resatorvid), a small‐molecule inhibitor of Toll‐like receptor (TLR) 4 signaling, binds selectively to TLR4 and interferes with interactions between TLR4 and its adaptor molecules. Mol Pharmacol 2011; 79: 34–41. [DOI] [PubMed] [Google Scholar]
- 26. Doyle SE, Vaidya SA, O’Connell R et al. IRF3 Mediates a TLR3/TLR4‐Specific Antiviral Gene Program. Immunity 2002; 17: 251–63. [DOI] [PubMed] [Google Scholar]
- 27. Jin FZ, Lian ML, Wang X, Wei Q. Studies of the anticancer effect of calcineurin B. Immunopharmacol Immunotoxicol 2005; 27: 199–210. [DOI] [PubMed] [Google Scholar]
- 28. Oganesyan G, Saha SK, Guo BC et al. Critical role of TRAF3 in the Toll‐like receptor‐dependent and ‐independent antiviral response. Nature 2006; 439: 208–11. [DOI] [PubMed] [Google Scholar]
- 29. Hacker H, Redecke V, Blagoev B et al. Specificity in Toll‐like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 2006; 439: 204–7. [DOI] [PubMed] [Google Scholar]
- 30. Sato M, Suemori H, Hata N et al. Distinct and essential roles of transcription factors IRF‐3 and IRF‐7 in response to viruses for IFN‐alpha/beta gene induction. Immunity 2000; 13: 539–48. [DOI] [PubMed] [Google Scholar]
- 31. Berube J, Bourdon C, Yao Y, Rousseau S. Distinct intracellular signaling pathways control the synthesis of IL‐8 and RANTES in TLR1/TLR2, TLR3 or NOD1 activated human airway epithelial cells. Cell Signal 2009; 21: 448–56. [DOI] [PubMed] [Google Scholar]
- 32. Yoshizawa T, Hammaker D, Sweeney SE, Boyle DL, Firestein GS. Synoviocyte Innate Immune Responses: I. Differential Regulation of Interferon Responses and the JNK Pathway by MAPK Kinases. J Immunol 2008; 181: 3252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sayama K, Hanakawa Y, Nagai H et al. Transforming growth factor‐beta‐activated kinase 1 is essential for differentiation and the prevention of apoptosis in epidermis. J Biol Chem 2006; 281: 22013–20. [DOI] [PubMed] [Google Scholar]
- 34. Yen JH, Ganea D. Interferon beta induces mature dendritic cell apoptosis through caspase‐11/caspase‐3 activation. Blood 2009; 114: 1344–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shibata S, Okano S, Yonemitsu Y et al. Induction of Efficient Antitumor Immunity Using Dendritic Cells Activated by Recombinant Sendai Virus and Its Modulation by Exogenous IFN‐beta Gene. J Immunol 2006; 177: 3564–76. [DOI] [PubMed] [Google Scholar]
- 36. Qin XQ, Tao N, Dergay A et al. Interferon‐beta gene therapy inhibits tumor formation and causes regression of established tumors in immune‐deficient mice. Proc Natl Acad Sci USA 1998; 95: 14411–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Teige I, Treschow A, Teige A et al. IFN‐beta gene deletion leads to augmented and chronic demyelinating experimental autoimmune encephalomyelitis. J Immunol 2003; 170: 4776–84. [DOI] [PubMed] [Google Scholar]
- 38. Gay NJ, Gangloff M. Structure and function of toll receptors and their Ligands. Annu Rev Biochem 2007; 76: 141–65. [DOI] [PubMed] [Google Scholar]
- 39. Kobe B, Kajava AV. The leucine‐rich repeat as a protein recognition motif. Protein 2001; 11: 725–32. [DOI] [PubMed] [Google Scholar]


