Abstract
Background
Alterations in gut microbiota (GMB) and host metabolites have been noted in individuals with HIV. However, it remains unclear whether alterations in GMB and related functional groups contribute to disrupted host metabolite profiles in these individuals.
Methods
This study included 185 women (128 with longstanding HIV infection, 88% under antiretroviral therapy; and 57 women without HIV from the same geographic location with comparable characteristics). Stool samples were analyzed by 16S rRNA V4 region sequencing, and GMB function was inferred by PICRUSt. Plasma metabolomic profiling was performed using liquid chromatography–tandem mass spectrometry, and 133 metabolites (amino acids, biogenic amines, acylcarnitines, and lipids) were analyzed.
Results
Four predominant bacterial genera were identified as associated with HIV infection, with higher abundances of Ruminococcus and Oscillospira and lower abundances of Bifidobacterium and Collinsella in women with HIV than in those without. Women with HIV showed a distinct plasma metabolite profile, which featured elevated glycerophospholipid levels compared with those without HIV. Functional analyses also indicated that GMB lipid metabolism was enriched in women with HIV. Ruminococcus and Oscillospira were among the top bacterial genera contributing to the GMB glycerophospholipid metabolism pathway and showed positive correlations with host plasma glycerophospholipid levels. One bacterial functional capacity in the acetate and propionate biosynthesis pathway was identified to be mainly contributed by Bifidobacterium; this functional capacity was lower in women with HIV than in women without HIV.
Conclusions
Our integrative analyses identified altered GMB with related functional capacities that might be associated with disrupted plasma metabolite profiles in women with HIV.
Keywords: HIV infection, gut microbiota, metabolomics, integrative analysis
We identified altered gut microbiota associated with disrupted plasma metabolite profiles in women with HIV. Two genera, Ruminococcus and Oscillospira, with functional capacities in the glycerophospholipid metabolism pathway were enriched, while Bifidobacterium, a probiotic genera, was depleted in these women.
The human gut microbiome has been suggested to play an important role in the host immune system [1]. Previous studies suggested that human immunodeficiency virus (HIV) infection may alter the gut microbiota (GMB)—for example, by decreasing gut microbial α-diversity [2]. The effects of HIV infection on the gut microbiome, and especially the relationships of HIV infection with individual taxa, are still unclear. Earlier studies found a shift from Bacteroides predominance to Prevotella predominance in individuals with HIV [3, 4], while subsequent studies reported that the shift was associated with sexual preference (eg, men who have sex with men [MSM]) rather than HIV infection [5, 6]. Most previous gut microbiome studies in HIV infection were limited by small sample sizes, and women with HIV were understudied. In a recent meta-analysis of HIV infection and gut microbial α-diversity, only 24% of the 1032 participants from 11 different studies were women (74 HIV negative, 167 HIV positive); yet, that analysis showed that gender had a significant impact on gut microbial α-diversity in the context of HIV infection [2]. Many previous studies focused on individuals newly infected with HIV or those not receiving antiretroviral therapy (ART) [7–9]. With the spectacular success of ART, HIV infection is now considered to be a chronic condition in many settings.
It has been suggested that altered GMB may contribute to host immune and metabolic activities in HIV infection through their molecular agents (eg, metabolites) [10, 11]. For example, alterations in host tryptophan and kynurenine metabolites have been associated with HIV infection and comorbidities [12, 13], which might be related to HIV-induced GMB dysbiosis [13]. Lipid metabolism disruption is a feature of HIV infection and recent studies have suggested that lipid profiles differ between individuals with and without HIV [14, 15]. It remains unknown whether alterations in GMB and related pathways may contribute to disrupted host lipid profiles in individuals with HIV.
In the present study, through integrative analyses of gut microbiome features, imputed microbiome functional contents, and host plasma metabolomics, we aimed to identify HIV-associated bacteria and examine their relationships with host metabolite profiles among women from the Women’s Interagency HIV Study (WIHS).
METHODS
Study Population
The WIHS is an ongoing prospective cohort study in women with or at risk of HIV infection [16]. Every 6 months, WIHS participants undergo a core visit with a comprehensive physical examination, providing biological specimens and completing interviewer-administered questionnaires. Demographic, clinical, and laboratory variables were collected using standardized protocols at semiannual core study visits [17]. In this study, we included 185 women (128 HIV positive, 57 HIV negative) whose fecal samples were collected using a home-based self-collection kit [18, 19] during 2015–2017 in the WIHS Bronx site. The study was reviewed and approved by the Institutional Review Board at the Albert Einstein College of Medicine. All participants provided written informed consent.
Microbiome Measurement
The 16S ribosomal RNA (rRNA) V4 region sequencing was performed on DNA extracted from fecal samples using the MiSeq platform (Illumina) [18]. After quality control, the average coverage was approximately 35 000 reads per sample (Supplementary Figure 1). Microbiome bioinformatics analysis was performed using the Quantitative Insights Into Microbial Ecology (QIIME) software package, version 1.9.1 [20]. The functional potential of the GMB community was imputed by Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) [21]. Detailed methods are described in the Supplementary Methods.
Metabolomic Profiling
Metabolomic profiling was performed on plasma samples that were collected at the semiannual core study visits closest in time to fecal sample collection. Using the AbsoluteIDQ p180 kit (Biocrates Life Sciences AG), metabolomics was performed by liquid chromatography and flow-injection analysis mass spectrometry [22]. A total of 133 metabolites were included in the current analysis (21 amino acids, 6 biogenic amines, 18 acylcarnitines, 74 glycerophospholipids, and 14 sphingolipids), and all metabolites had coefficients of variation less than 30% and missing rates of less than 25%. Metabolites with missing data (under detectable levels) were imputed with half of the minimum values for a given metabolite.
Statistical Analysis
Characteristics between women with and without HIV were compared using the Mann–Whitney U test (continuous variables) and χ 2 test (categorical variables). Mann–Whitney U tests were applied to compare the differences in the microbial α-diversity indices (Shannon index, Simpson index, and Chao-1 index) by HIV status. Permutational multivariate ANOVA (PERMANOVA) and principal-coordinate analysis (PCoA) were carried out for the microbial β-diversity analyses. Cumulative sum scaling normalization was conducted for the genus-level abundance of taxonomic units [23] before analyses. Linear discriminant analysis (LDA) effect size (LefSe) was used to identify bacterial genera associated with HIV infection, with LDA score of 2.0 as a cutoff [24]. Linear regression models were further used to examine the associations between HIV infection and identified bacterial genera, adjusting for age, race, income, education, body mass index (BMI), marijuana use, probiotics use, and antibiotics use.
Spearman correlation was employed to estimate correlation coefficients among plasma metabolites and their correlations with identified bacterial genera. Partial least-squares discriminant analysis (PLSDA) [25], with a variable importance in projection (VIP) score of 1.5 as a cutoff, was used to identify plasma metabolite signatures of HIV infection. We further used linear regression models to examine the associations between HIV infection and identified metabolites, adjusting for age, race, income, education, and BMI. The false-discovery rate–corrected P value was calculated using Benjamini and Hochberg’s method [26]. For gut microbiome functional analyses, Mann–Whitney U test was used to compare the relative abundances of KEGG ortholog groups, and Welch’s t test was used for KEGG pathways by HIV status. R packages vegan, DESeq2, and phyloseq were used for the statistical analyses [27–29].
RESULTS
Participant Characteristics
Supplementary Table 1 shows characteristics of study participants. The participants living with and without HIV infection were generally similar in terms of demographic variables (age, race/ethnicity, education, and annual income), antibiotic use, and commercial probiotic use (all P > .05). Body mass index was similar among participants with and without HIV infection. Thirty-five percent of the participants undiagnosed HIV infected and 20% of the participants diagnosed HIV infected reported marijuana use. The majority of individuals with HIV had normal CD4+ T-cell counts (73%; ≥500 cells/mm3) and undetectable HIV-1 viral load (73%; <20 copies/mL), and reported current ART use (88%) and long-term ART use (under ART ≥2 years; 83%). Current ART users were more like to have normal CD4+ T-cell counts and undetectable HIV-1 viral load compared with nonusers, but other characteristics were comparable.
Human Immunodeficiency Virus Infection and Gut Microbiome
Bacterial community α-diversity indices (Shannon index, Chao-1 index, and Simpson’s index) were not significantly different between groups with and without HIV (all P > .05) (Supplementary Figure 2A). No clear separation was observed by HIV status in the PCoA plot of Bray-Curtis dissimilarity (Supplementary Figure 2B). No significant associations of β-diversity with HIV status and related factors (CD4+ T-cell count, viral load, and ART use) were observed in the PERMANOVA analysis (all R2 < .1, P > .05).
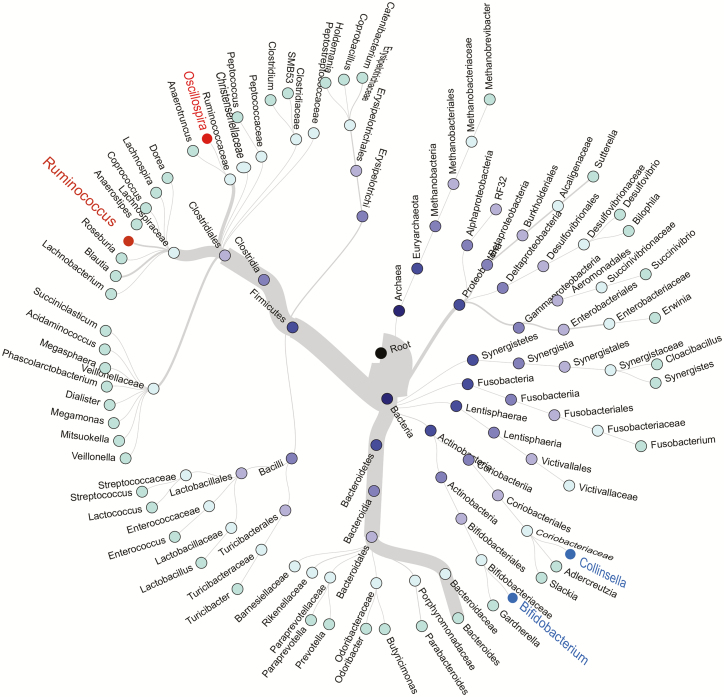
A total of 1015 unique operational taxonomic units were identified from sequenced specimens. The most abundant phylum was Firmicutes, followed by Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria (Figure 1). The LEfSe analysis indicated that HIV infection was associated with higher abundances of Ruminococcus, Oscillospira, and Anaerotruncus and lower abundances of Bifidobacterium, Collinsella, and Pyramidobacter (all LDA scores >2.5) (Supplementary Figure 3A). Since the relative abundance of Anaerotruncus (median, 0.02%) and Pyramidobacter (0.01%) were very low, we focused on 4 major HIV-associated genera—Ruminococcus (3.56%), Oscillospira (0.62%), Bifidobacterium (1.16%), and Collinsella (1.32%)—in the subsequent analyses. Ruminococcus and Oscillospira belong to the same order, Clostridiales, within the most abundant phylum Firmicutes (Figure 1), while Bifidobacterium and Collinsella belong to the phylum Actinobacteria.
Figure 1.
Hierarchical structure tree of microbial communities. Taxa from the inner to outer circle represent the super kingdom to genus level. The branch widths reflect the relative abundance of each detected taxon. Red and blue labels depict bacterial genera with higher (red) or lower (blue) relative abundance in individuals with HIV compared with those without HIV, respectively. Genera whose average relative abundance if >0.01% are included. Abbreviation: HIV, human immunodeficiency virus.
After multivariate adjustment, the associations of HIV infection with the 4 bacterial genera did not change materially (Supplementary Table 2). We also observed some suggestive associations between covariates and these 4 bacterial genera. For example, BMI was inversely associated with Oscillospira (P = .057) and positively associated with Collinsella (P = .001). Marijuana use was positively associated with Collinsella (P = .073). The magnitude of the HIV infection and Collinsella association (β = −.624, P = .035) was attenuated after adjusting for marijuana use (β = −.54, P = .069). The association between HIV infection and Ruminococcus was stronger in individuals without marijuana use compared with those with marijuana use, with a marginally significant interaction (P for interaction = .09) (Supplementary Table 3). Associations of HIV infection with these 4 bacterial genera did not change after excluding antibiotics users or women with HIV without ART use (data not shown).
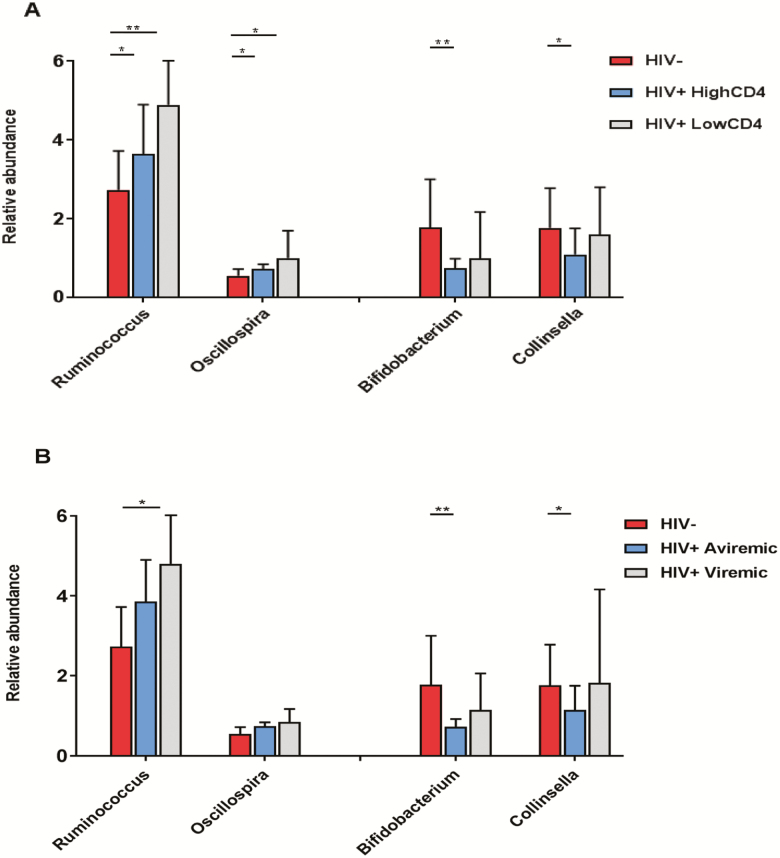
There was an increasing trend for Ruminococcus and Oscillospira, across the group without HIV, the group with HIV with a high CD4+ T-cell count (≥500 cells/mm3), and the group with HIV with a low CD4+ T-cell count (<500 cells/mm3) (Figure 2A). Bifidobacterium and Collinsella showed the highest relative abundance in the group without HIV, but there was no significant difference in relative abundance between high and low CD4+ T-cell count groups of individuals with HIV (Figure 2A). Similar patterns of bacterial abundance for these 4 genera were observed across the group without HIV, the group with aviremic HIV, and the group with viremic HIV (Figure 2B). No significant differences in these genera were observed between individuals with HIV with and without ART use (Supplementary Table 4). We further examined the associations of these genera with specific classes of ART drugs including protease inhibitors, nonnucleoside reverse transcriptase inhibitors, nucleoside reverse transcriptase inhibitors, and integrase inhibitors, but did not find significant associations.
Figure 2.
Differentially abundant genera according to HIV infection and related factors. Data are means (SD) of relative abundance of 4 genera (Ruminococcus, Oscillospira, Bifidobacterium, and Collinsella) among (A) 57 individuals without HIV, 93 individuals with HIV with high CD4 counts (≥500 cells/mm3), and 35 individuals with HIV with low CD4 counts (<500 cells/mm3) and (B) 57 individuals without HIV, 94 individuals with aviremic HIV (undetectable viral load ≤20 copies/mL), and 34 individuals with viremic HIV (viral load >20 copies/mL). *P < .05; **P < .01. Abbreviations: HIV, human immunodeficiency virus; SD, standard deviation; –, negative; +, positive.
Human Immunodeficiency Virus Infection and Host Plasma Metabolomic Profiles
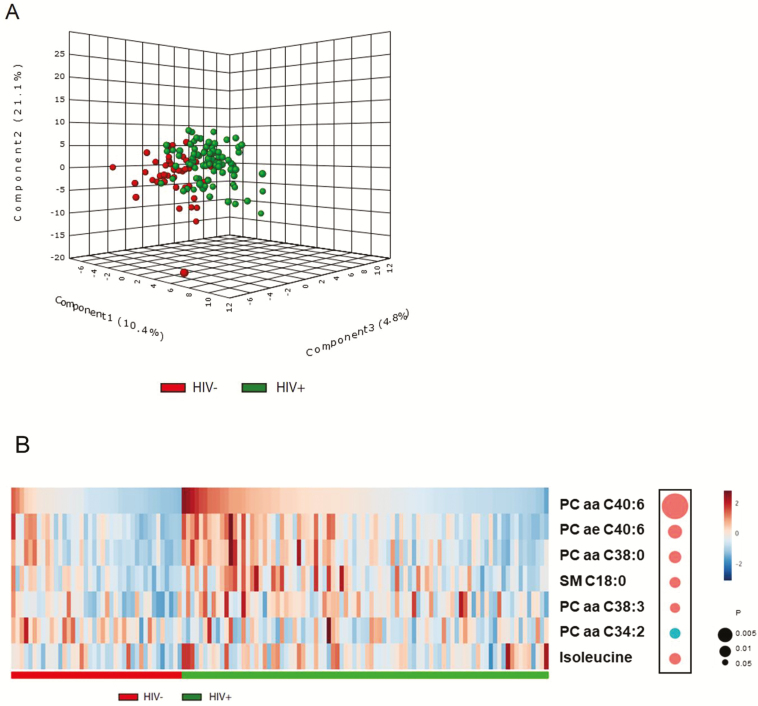
Most metabolites within the same categories showed moderate-to-high correlations with each other, especially for lipids (Supplementary Figure 4). Partial least-squares discriminant analysis of all metabolites revealed a distinction between groups with and without HIV, although these 2 groups were not fully separated (Figure 3A). A total of 15 metabolites that had major contributions to the distinction (VIP scores >1.5) were selected by PLSDA (Supplementary Table 5), including 10 glycerophospholipids, 1 sphingolipid (SM C18:0), 2 amino acids (isoleucine and lysine), and 2 biogenic amines (ɑ-AAA and kynurenine); and 7 of these (5 glycerophospholipids, sphingolipid C18:0, and isoleucine) showed significant differences between individuals with and without HIV after multivariate adjustment (false-discovery rate–adjusted P < .05). As shown in Figure 3B, the top metabolites altered in HIV infection were glycerophospholipids. In addition, as previously reported [13], a higher kynurenine-to-tryptophan ratio was associated with HIV infection (P = .021). The associations of HIV infection with these metabolites did not change after excluding women with HIV without ART use. No significant difference in metabolite levels were observed between participants with HIV with and without ART use, but it should be noted that the number of participants without ART use was relatively small in this analysis (n = 15).
Figure 3.
Plasma metabolomic profiles and HIV infection. A, Three-dimensional PLSDA score plot by HIV status, based on 133 plasma metabolites. B, Heatmap of the top metabolites altered in individuals with HIV: 5 glycerophospholipids (phosphatidyl-choline diacyl PC aa C40:6, PC aa C38:0, PC aa C38:3, PC aa C34:2; phosphatidyl-choline acyl-alkyl PC ae C40:6), 1 sphingolipid (SM C18:0), and 1 amino acid (isoleucine). The concentration was inverse normal transformed. These metabolites were selected first by PLSDA (VIP scores ≥1.5) and then by linear regression model, adjusted for age, race, income, education, and BMI (FDR P < .05). The diameters of the bubbles represent the P values in the linear regression model. Red and blue bubbles depict metabolites that showed increased (red) and decreased (blue) concentration in individuals with HIV, respectively. Abbreviations: BMI, body mass index; FDR, false-discovery rate; HIV, human immunodeficiency virus; PLSDA, partial least-squares discriminant analysis; VIP, variable importance in projection; –, negative; +, positive.
Gut Microbiome and Host Plasma Metabolites
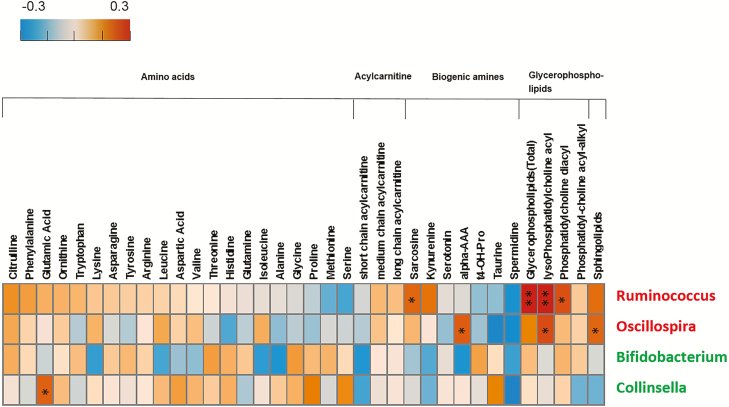
As shown in Figure 4, 2 HIV-increased bacterial genera, notably Ruminococcus, showed positive correlations with plasma glycerophospholipids but did not have correlations with amino acids or biogenic amines. Two HIV-decreased microbial genera, Bifidobacterium and Collinsella, were not correlated with plasma metabolites, except for a positive correlation between Collinsella and glutamic acid.
Figure 4.
Correlations between plasma metabolites and 4 HIV-associated bacterial genera. The positive Spearman rank correlation coefficients are denoted by orange squares and negative correlation coefficients by blue squares. Red and green labels depict bacterial genera that showed increased (red) and decreased (green) relative abundance in individuals with HIV. The glycerophospholipid column represents total plasma glycerophospholipid concentration (sum of all glycerophospholipid species); the lysoPhosphatidyl-choline acyl, phosphatidyl-choline diacyl, and phosphatidyl-choline acyl-alkyl columns represent the total concentration for each glycerophospholipid subcategory, respectively. *P < .05; **P < .01. Abbreviation: HIV, human immunodeficiency virus.
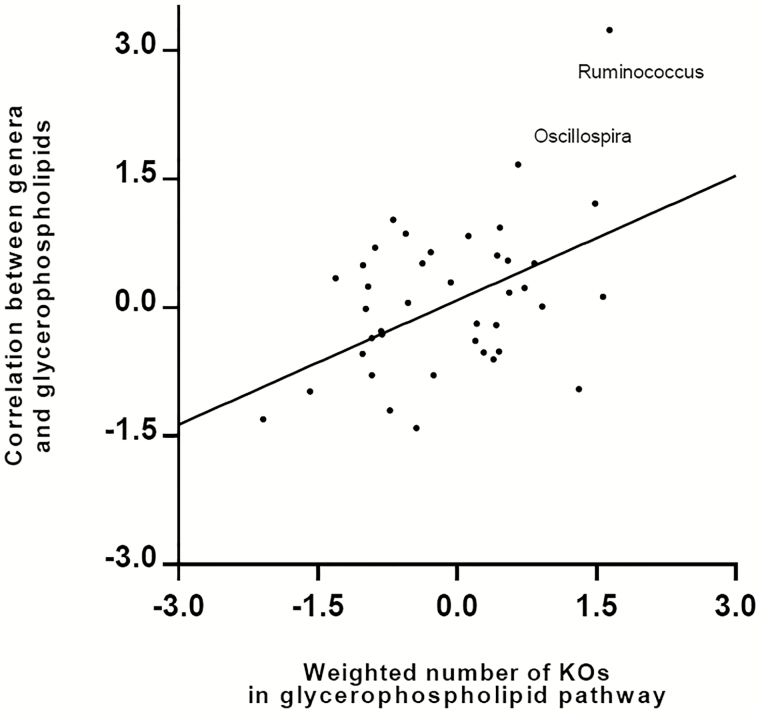
The PICRUSt analysis revealed that the microbial lipid metabolism pathway was significantly enriched in the participants with HIV (P = .01). We thus focused on the microbial lipid metabolism pathway to explore the relationships of HIV-increased bacterial genera with host lipid metabolites. In the glycerophospholipid metabolism pathway, a total of 31 homologs to glycerophospholipid metabolism pathway enzymes (measured by KEGG ortholog groups [KOs]) were encoded by the predominant genera in this study (Supplementary Figure 5). These homologs were genetically encoded in numerous bacterial genera while each genus could also encode multiple enzyme homologs. As shown in Figure 5, bacterial genera with greater numbers of glycerophospholipid metabolism pathway enzyme homologs (weighted by relative abundance of respective genera) exhibited stronger correlations with host plasma glycerophospholipid levels compared with those with fewer numbers of homologs. For example, Ruminococcus had the greatest weighted number of enzyme homologs in the glycerophospholipid metabolism pathway and showed the strongest correlation with host plasma glycerophospholipid levels.
Figure 5.
Relationships among predominant bacterial genera, host plasma glycerophospholipids, and bacteria’s contribution to the glycerophospholipids metabolism pathway. Each node represents 1 genus, and all the predominant genera with average relative abundance >0.05% were included in this analysis. The x axis describes the weighted bacteria’s enzyme contribution to the glycerophospholipid metabolism pathway (inverse normal transformed). The y axis depicts the Spearman correlation coefficients (inverse normal transformed) between the relative abundance of bacterial genera and plasma glycerophospholipid metabolite concentration. Each genus’s contribution to the glycerophospholipid metabolism pathway was first measured by the number of KOs, which were genetically encoded by the bacteria, according to PICRUSt-imputed functional content analysis. Then this genetic contribution was weighted by the relative abundance of each genus. Abbreviation: KO, KEGG ortholog group; PICRUSt, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States.
Although Bifidobacterium was not correlated with any plasma metabolites measured, 1 bacterial functional capacity (K11263) in the acetate and propionate biosynthesis pathway was identified to be mainly contributed by Bifidobacterium, and this functional capacity was lower in women with HIV than that in women without HIV (P = .007) (Supplementary Figure 6A and B).
DISCUSSION
The extent to which HIV infection and the resultant changes in the immune system alter the gut microbiome remains controversial [30]. In the meta-analysis of 167 women with HIV and 74 women without HIV [2], HIV infection was found to be associated with decreased α-diversity, while our study did not find such results. This inconsistency might be due to the heterogeneous nature of the studies in this literature [2]. Women with HIV included in our study had relatively long-term duration of infection and most of them were on stable and effective drug therapy, while participants included in that meta-analysis were mixed with newly infected women and more than half did not have ART use [2]. It is also important to note that our study included a comparable group without HIV with very similar demographic characteristics (eg, location, race/ethnicity) and behaviors (eg, sexual behaviors) [16], which may help eliminate the potential confounding effects. Indeed, we did not replicate a previously reported association between HIV infection and Prevotella. This supports that Prevotella might be associated with other confounders rather than HIV infection itself [5].
Our study identified 2 microbial genera, Ruminococcus and Oscillospira, which were enriched in women with HIV. Ruminococcus is a gram-positive anaerobic genus commonly found in the human intestine. In support of our findings, the relative abundance of Ruminococcus gnavus was found to be higher in stool samples of 33 patients with HIV infection (14 with ART for 3 months, 19 without treatment) compared with 35 healthy controls in a study in South China, and no significant effects of ART on this species was observed [31]. In contrast, another study reported a depletion of Ruminococcus associated with HIV infection. However, rectal mucosal samples from men with HIV without ART were examined [32], which is different from our study. In addition, a study of HIV-positive MSM reported that marijuana use was associated with an increased abundance of Ruminococcus [7]. We did not find such an association, but interestingly, the association between HIV infection and Ruminococcus was stronger in individuals without marijuana use compared with those with marijuana use, suggesting a potential effect modification, which warrants further investigation.
Oscillospira is a nonculturable bacteria that belongs to the same order (Clostridiales) as Ruminococcus. Consistent with previous findings that Oscillospira was enriched in lean individuals or individuals with a low BMI [33], we also observed a similar trend. Our multivariate-adjusted models and stratified analyses suggested that the observed association between HIV infection and Oscillospira might be independent of obesity. Nevertheless, given the close relationships of HIV infection and ART use with adiposity and fat distribution, future studies are warranted to investigate the potential involvement of Oscillospira and other bacteria in the link between HIV infection and obesity.
Consistent with previous metabolomic/lipidomic studies [34, 35], we also found a distinct plasma metabolite profile in women with HIV compared with those without HIV infection featuring elevated glycerophospholipid levels. Interestingly, we found that the HIV-increased bacterial genera, Ruminococcus and Oscillospira, were correlated with host plasma lipid profiles. In our prior work in the WIHS, markers of T-cell activation and senescence [36] and serum markers of inflammation and immune activation [39] were elevated in individuals with HIV, and alterations in plasma lipidome were observed in individuals with HIV, especially among those using ART [37]. A recent study also linked lipid abnormalities to markers of inflammation and microbial translocation in individuals with HIV receiving protease inhibitors [14]. Our current gut microbiome data expand previous findings and provide further evidence supporting a potential contribution of GMB to host lipid metabolite profiles. Our PICRUSt analysis indicated that Ruminococcus and Oscillospira were among the top taxa that have the most genes matching the glycerophospholipid metabolism pathway. In line with our study, the glycerophospholipid pathway has been correlated with the lung microbiota in individuals with HIV [38], and the potential implication of gut Ruminococcus and the glycerophospholipid metabolism pathway was also reported in cardiovascular disease [39]. However, our PICRUSt results should be interpreted with caution, and the direct biological links of Ruminococcus and other bacteria with host glycerophospholipid profiles need to be elucidated. Another interesting finding of our study is that Bifidobacterium, a common probiotic bacterial genus in the human gut, was decreased in women with HIV, and this finding is partially supported by a previous study in children [40]. Our functional analysis indicated that 1 inferred bacterial function in the acetate and propionate biosynthesis pathway was lower in women with HIV compared with those without HIV infection, and this function was mainly contributed by Bifidobacterium in our samples. This is consistent with the fact that Bifidobacterium can produce short-chain fatty acids (SCFAs) [41, 42]. A few small intervention studies have suggested that probiotics containing Lactobacillus could help improve immune function in adults with HIV infection [43, 44]. However, the implication of our current findings in clinical practice remains unclear, and future studies are needed to examine potential benefits of probiotics containing Bifidobacterium on individuals with HIV.
The genus Collinsella is a dominant taxon of the family Coriobacteriaceae, and recent studies have linked this genus with obesity, insulin resistance, and diabetes in populations without HIV [45]. We confirmed the positive association between Collinsella and BMI in this study, but our further analyses indicated that the observed association between this genus and HIV infection might not be influenced by obesity status. It is potentially interesting that we found a marginal association between increased Collinsella abundance and marijuana use. In a previous study of MSM with HIV infection, there was no association between marijuana use and Collinsella, but methamphetamine use was associated with decreased Collinsella abundance [7]. Unfortunately, we were unable to test this association because only 2 participants reported methamphetamine use in our study. A large number of pharmacological agents have been found to influence the GMB [46], and this may also be the case for recreational drugs commonly used in the population with HIV.
Our study has several limitations. Due to the nature of observational study design, our study in human participants is unable to demonstrate causal relationships between HIV infection, GMB, and plasma metabolite profiles. Our metabolomics did not capture SCFAs in plasma, and we were unable to perform metabolomics on stool samples collected by RNAlater [18]. Dietary intake, a major factor influencing the gut microbiome, was not examined in this analysis. Although the inferred functional contents by PICRUSt have been shown to be robust, particularly for the human gut microbiome [21], they should be interpreted with caution and need to be verified by shotgun metagenomic sequencing. Finally, this study only included women, and our findings need to be validated in men and other HIV studies.
In summary, this study identified altered GMB, especially the genus Ruminococcus, with potential functional contents in the glycerophospholipid metabolism pathway, correlating with elevated plasma glycerophospholipid levels in women with chronic HIV infection. In addition, one of the common probiotic bacterial genera, Bifidobacterium, was found to be depleted in women with HIV. These findings suggest alterations in GMB and their potential influences on host metabolite profiles in chronic HIV infection. Our study supports the concept of a potential therapeutic role of modulating the GMB in people living with HIV infection.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. Q. Q. and Z. W. led the statistical modeling/bioinformatics analysis and drafted the manuscript. Q. Q., R. C. K., and R. D. B. designed the study and supervised the process of the project. Z. W., S. H., K. A., and Q. Q. collected data from WIHS participants; M. U., C. C. S., A. G., and R. D. B. processed fecal samples and performed sequencing experiments. Y. Q. and I. J. K. performed metabolomics experiments. Z. W., T. W., M. U., and Y. Q. contributed to data analyses. J. W., K. L., A. L. L., R. K., and all coauthors provided critical comments and revised the manuscript.
Disclaimer. Data in this article were collected by the Women’s Interagency HIV Study (WIHS). The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health.
Financial support. This work was supported by the National Heart, Lung, and Blood Institute (NHLBI) (grant number R01HL140976). Other related funding sources included NHLBI grant number K01HL129892 and a Feldstein Medical Foundation Research grant to Q. Q. R. C. K was supported by NHLBI grants 5R01HL126543 and R01 HL132794 and the National Institute on Mental Health (NIMH) grant number 5R01MD011389-03. Y. Q. and I. J. K. were supported by the Metabolomics Core Facility of the Einstein-Mount Sinai Diabetes Research and Training Center of the Albert Einstein College of Medicine (National Institutes of Health/National Cancer Institute [NIH/NCI] grant number P60DK020541). Other funding sources include NHLBI grants R01HL083760 and R01HL095140, the National Institute of Allergy and Infectious Diseases (NIAID) (grant number U01 AI035004), and the Einstein Cancer Research Center (grant number P30CA013330), the Einstein Liver Research Center (grant number P30DK041296), the Einstein-Rockefeller-CUNY Center for AIDS Research funded by the NIAID (grant number P30AI124414), and the Stable Isotope and Metabolomics Core Facility of the Einstein–Mount Sinai Diabetes Research Center of the Albert Einstein College of Medicine funded by the NCI (grant number P60DK020541). Women’s Interagency HIV Study (WIHS; Principal Investigators): University of Alabama at Birmingham and University of Mississippi WIHS (Mirjam-Colette Kempf and Deborah Konkle-Parker), U01-AI-103401; Atlanta WIHS (Ighovwerha Ofotokun and Gina Wingood), U01-AI-103408; Bronx WIHS (Kathryn Anastos and Anjali Sharma), U01-AI-035004; Brooklyn WIHS (Howard Minkoff and Deborah Gustafson), U01-AI-031834; Chicago WIHS (Mardge Cohen and Audrey French), U01-AI-034993; Metropolitan Washington WIHS (Seble Kassaye), U01-AI-034994; Miami WIHS (Margaret Fischl and Lisa Metsch), U01-AI-103397; University of North Carolina (UNC) WIHS (Adaora Adimora), U01-AI-103390; Connie Wofsy Women’s HIV Study, Northern California (Ruth Greenblatt, Bradley Aouizerat, and Phyllis Tien), U01-AI-034989; WIHS Data Management and Analysis Center (Stephen Gange and Elizabeth Golub), U01-AI-042590; Southern California WIHS (Joel Milam), U01-HD-032632 (WIHS I–WIHS IV). The WIHS is funded primarily by the NIAID, with additional co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the NCI, the National Institute on Drug Abuse, and the NIMH. Targeted supplemental funding for specific projects is also provided by the National Institute of Dental and Craniofacial Research, the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Deafness and other Communication Disorders, and the NIH Office of Research on Women’s Health. WIHS data collection is also supported by grants UL1-TR000004 (University of California–San Francisco Clinical and Translational Science Awards [CTSA]), UL1-TR000454 (Atlanta CTSA), and P30-AI-050410 (UNC Center for AIDS Research).
Potential conflicts of interest. R. K. reports personal fees from Biota, Commence, CoreBiome, DayTwo, GenCirq, and Prometheus, outside the submitted work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity 2017; 46:562–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tuddenham SA, Koay WLA, Zhao N, et al. . The impact of HIV infection on gut microbiota alpha-diversity: an individual level meta-analysis. Clin Infect Dis 2019. doi:10.1093/cid/ciz258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mutlu EA, Keshavarzian A, Losurdo J, et al. . A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 2014; 10:e1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lozupone CA, Li M, Campbell TB, et al. . Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013; 14:329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noguera-Julian M, Rocafort M, Guillén Y, et al. . Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 2016; 5:135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Armstrong AJS, Shaffer M, Nusbacher NM, et al. . An exploration of Prevotella-rich microbiomes in HIV and men who have sex with men. Microbiome 2018; 6:198,018-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fulcher JA, Hussain SK, Cook R, et al. . Effects of substance use and sex practices on the intestinal microbiome during HIV-1 infection. J Infect Dis 2018; 218:1560–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dillon SM, Lee EJ, Kotter CV, et al. . An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 2014; 7:983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vesterbacka J, Rivera J, Noyan K, et al. . Richer gut microbiota with distinct metabolic profile in HIV infected elite contr ollers. Sci Rep 2017; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Serrano-Villar S, Rojo D, Martínez-Martínez M, et al. . Gut bacteria metabolism impacts immune recovery in HIV-infected individuals. EBioMedicine 2016; 8:203–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vázquez-Castellanos JF, Serrano-Villar S, Jiménez-Hernández N, et al. . Interplay between gut microbiota metabolism and inflammation in HIV infection. ISME J 2018; 12:1964–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vujkovic-Cvijin I, Dunham RM, Iwai S, et al. . Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Trans Med 2013; 5:193ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qi Q, Hua S, Clish CB, et al. . Plasma tryptophan-kynurenine metabolites are altered in human immunodeficiency virus infection and associated with progression of carotid artery atherosclerosis. Clin Infect Dis 2018; 67:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cassol E, Misra V, Holman A, Kamat A, Morgello S, Gabuzda D. Plasma metabolomics identifies lipid abnormalities linked to markers of inflammation, microbial translocation, and hepatic function in HIV patients receiving protease inhibitors. BMC Infect Dis 2013; 13:203,2334-13-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scarpellini B, Zanoni M, Sucupira MC, et al. . Plasma metabolomics biosignature according to HIV stage of infection, pace of disease progression, viremia level and immunological response to treatment. PLoS One 2016; 11:e0161920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bacon MC, von Wyl V, Alden C, et al. . The Women’s Interagency HIV Study: an observational cohort brings clinical sciences to the bench. Clin Diagn Lab Immunol 2005; 12:1013–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanna DB, Post WS, Deal JA, et al. . HIV infection is associated with progression of subclinical carotid atherosclerosis. Clin Infect Dis 2015; 61:640–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Z, Zolnik CP, Qiu Y, et al. . Comparison of fecal collection methods for microbiome and metabolomics studies. Front Cell Infect Microbiol 2018; 8:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moon JY, Zolnik CP, Wang Z, et al. . Gut microbiota and plasma metabolites associated with diabetes in women with, or at high risk for, HIV infection. EBioMedicine 2018; 37:392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caporaso JG, Kuczynski J, Stombaugh J, et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Langille MG, Zaneveld J, Caporaso JG, et al. . Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Romisch-Margl W, Prehn C, Bogumil R, Rohring C, Suhre K, Adamski J. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics 2012; 8:133–42. [Google Scholar]
- 23. Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods 2013; 10:1200–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Segata N, Izard J, Waldron L, et al. . Metagenomic biomarker discovery and explanation. Genome Biol 2011; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Westerhuis JA, Hoefsloot HCJ, Smit S, et al. . Assessment of PLSDA cross validation. Metabolomics 2008; 4:81–9. [Google Scholar]
- 26. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B 1995; 57:289–300. [Google Scholar]
- 27. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oksanen J. Multivariate analysis of ecological communities in R: Vegan tutorial. version 2.3-0. 2015. Available at: http://cc.oulu.fi/~jarioksa/opetus/metodi/vegantutor.pdf. Accessed 10 June 2015. [Google Scholar]
- 29. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu J, Williams B, Frank D, Dillon SM, Wilson CC, Landay AL. Inside out: HIV, the gut microbiome, and the mucosal immune system. J Immunol 2017; 198:605–14. [DOI] [PubMed] [Google Scholar]
- 31. Zhou Y, Ou Z, Tang X, et al. . Alterations in the gut microbiota of patients with acquired immune deficiency syndrome. J Cell Mol Med 2018; 22:2263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McHardy IH, Li X, Tong M, et al. . HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 2013; 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goodrich JK, Waters JL, Poole AC, et al. . Human genetics shape the gut microbiome. Cell 2014; 159:789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scarpellini B, Zanoni M, Sucupira MC, et al. . Plasma metabolomics biosignature according to HIV stage of infection, pace of disease progression, viremia level and immunological response to treatment. PLoS One 2016; 11:e0161920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Belury MA, Bowman E, Gabriel J, et al. . Prospective analysis of lipid composition changes with antiretroviral therapy and immune activation in persons living with HIV. Pathog Immun 2017; 2:376–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaplan RC, Sinclair E, Landay AL, et al. . T cell activation and senescence predict subclinical carotid artery disease in HIV-infected women. J Infect Dis 2011; 203:452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chai JC, Deik AA, Hua S, et al. . Association of lipidomic profiles with progression of carotid artery atherosclerosis in HIV infection. JAMA Cardiol 2019. doi:10.1001/jamacardio.2019.4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cribbs SK, Uppal K, Li S, et al. . Correlation of the lung microbiota with metabolic profiles in bronchoalveolar lavage fluid in HIV infection. Microbiome 2016; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jie Z, Xia H, Zhong SL, et al. . The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 2017; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Claassen-Weitz S, Gardner-Lubbe S, Nicol P, et al. . HIV-exposure, early life feeding practices and delivery mode impacts on faecal bacterial profiles in a south african birth cohort. Sci Rep 2018; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Z, Qi Q. Gut microbial metabolites associated with HIV infection. Future Virol 2019; 14:335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. LeBlanc JG, Chain F, Martin R, Bermudez-Humaran LG, Courau S, Langella P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact 2017; 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Falasca K, Vecchiet J, Ucciferri C, Di Nicola M, D’Angelo C, Reale M. Effect of probiotic supplement on cytokine levels in HIV-infected individuals: a preliminary study. Nutrients 2015; 7:8335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim CJ, Walmsley SL, Raboud JM, et al. . Can probiotics reduce inflammation and enhance gut immune health in people living with HIV: study designs for the Probiotic Visbiome for Inflammation and Translocation (PROOV IT) pilot trials. HIV Clin Trials 2016; 17:147–57. [DOI] [PubMed] [Google Scholar]
- 45. Hou YP, He QQ, Ouyang HM, et al. . Human gut microbiota associated with obesity in Chinese children and adolescen ts. Biomed Res Int 2017; 2017:7585989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maier L, Pruteanu M, Kuhn M, et al. . Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018; 555:623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.