Abstract
Objective
To investigate cross-sectional associations of CSF levels of neurofilament light chain (NfL) and of the newly emerging marker chitinase 3–like protein 1 (CHI3L1) with brain and spinal cord atrophy, which are established MRI markers of disease activity in MS, to study CHI3L1 and NfL in relapsing (RMS) and progressive MS (PMS), and to assess the expression of CHI3L1 in different cell types.
Methods
In a single-center study, 131 patients with MS (42 RMS and 89 PMS) were assessed for NfL and CHI3L1 concentrations in CSF, MRI-based spinal cord and brain volumetry, MS subtype, age, disease duration, and disability. We included 42 matched healthy controls receiving MRI. CHI3L1 expression of human brain cell types was examined in 2 published single-cell RNA sequencing data sets.
Results
CHI3L1 was associated with spinal cord volume (B = −1.07, 95% CI −2.04 to −0.11, p = 0.029) but not with brain volumes. NfL was associated with brain gray matter (B = −7.3, 95% CI −12.0 to −2.7, p = 0.003) but not with spinal cord volume. CHI3L1 was suitable to differentiate between progressive or relapsing MS (p = 0.015, OR 1.0103, CI for OR 1.002–1.0187), and its gene expression was found in MS-associated microglia and macrophages and in astrocytes of MS brains.
Conclusions
NfL and CHI3L1 in CSF were differentially related to brain and spinal cord atrophy. CSF CHI3L1 was associated with spinal cord volume loss and was less affected than NfL by disease duration and age, whereas CSF NfL was associated with brain gray matter atrophy. CSF NfL and CHI3L1 measurement provides complementary information regarding brain and spinal cord volumes.
Classification of evidence
This study provides Class II evidence that CSF CHI3L1 is associated with spinal cord volume loss and that CSF NfL is associated with gray matter atrophy.
In recent years, MS research has progressed, especially in the field of immune therapies of MS, raising the need for reliable markers of individual disease activity and treatment response.1
Neurofilament (NF) proteins, as part of the neuronal cytoskeleton,2 have been proposed as markers of neuronal damage in MS and various other neurologic disorders. Because neuroaxonal degeneration causes their release into the adjacent body fluid compartment, NF levels can be detected in the CSF and in the blood serum. In MS, associations have been reported between NF light chain (NfL) levels and disease activity, progression, and long-term clinical outcome.3,4 Increased CSF NfL levels also predicted the risk of conversion from a clinically isolated syndrome (CIS) to relapsing-remitting MS (RRMS).5 Recently, the relevance of serum NfL for discerning CIS from newly diagnosed MS has been demonstrated6; however, CSF NfL displayed a higher sensitivity for disease activity than serum NfL.7,8 Moreover, NfL levels were related with MRI-derived markers, such as brain MS lesion load, atrophy of the brain and spinal cord, and with previous radiologic disease activity.4,7,9,10
Besides NfL, other CSF markers of tissue damage have been proposed in MS, e.g., chitinase 3–like protein 1 (CHI3L1), a glycoprotein which is secreted by activated glia cells in the CNS. Increased CHI3L1 levels have been associated with disease progression11,12 and with conversion from CIS to MS.13 Recently, the benefit of combining NFL and CHI3L1 measures in CSF to discriminate MS phenotypes and to predict disease progression in RRMS was demonstrated.14
In the present study on patients with MS, we studied the associations of CHI3L1 and NfL levels in CSF with atrophy of the brain and spinal cord. Brain and spinal cord atrophy served as established paraclinical markers of disease progression that are known to be associated with axonal degeneration and demyelination in early and chronic MS subtypes.15,16 We hypothesized that the CSF CHI3L1 and NfL levels were related to both brain and spinal cord atrophy. A second aim was to test whether CSF levels of NfL and CHI3L1 acquired in the mixed cohort of patients with relapsing MS or progressive MS can distinguish between these 2 subtypes. Finally, the expression of CHI3L1 in different cell types from healthy and MS brains was examined to further investigate its relevance with respect to MS pathogenesis.
Methods
Participants
A total of 131 patients (table 1) with clinically definite MS or CIS with subsequent conversion to MS,17 availability of CSF samples for assessment of CHI3L1 and NfL, and an associated MRI examination were assessed for the analysis. Clinical data, i.e., Expanded Disability Severity Scale (EDSS) and disease duration (DD) at the times of CSF assessment and MRI, were extracted from the patient files.
Table 1.
Demography and MRI results of patients and HCs, clinical status, and CSF results for patients and patient subgroups
For the evaluation of CNS atrophy, MRI data of 42 healthy controls (HCs), which had been acquired with the same scanner and identical MRI protocol as the patients, were included from a preexisting in-house MRI database. Patient and control recruitment was performed over a 4-year period from 2011 to 2014.
Standard protocol approvals, registrations, and patient consents
The study protocol was approved by the ethics committee of the Medical Faculty of the Ruhr University Bochum (Approval No. 15-5268) and the research ethics boards of all participating centers. All individuals gave their written informed consent according to the Declaration of Helsinki.
CSF analysis and NfL/CHI3L1 measurements
The concentrations of CHI3L1 (also called YKL-40) and NfL in CSF were analyzed using the assays MicroVueTM YKL-40 EIA (Quidel, San Diego, CA) and NF-light ELISA (IBL International, Hamburg, Germany), respectively. The limit of detection was 5.4 ng/mL for CHI3L1 and 32 pg/mL for NfL. The intra- and interassay coefficients of variation for MicroVueTM YKL-40 EIA were <5.6%–6.6% and <6.0%–7.0%. The intra- and interassay coefficients of variation for NF-light ELISA were <5% and <9%.
MRI and quantitative image analysis
All participants underwent MRI scans at a single 1.5T scanner, which included 3D T1-weighted (T1w) and T2-weighted fluid-attenuated inversion recovery imaging. The MRI protocol is provided in the electronic supplement (links.lww.com/NXI/A329).
The MRI analysis aimed at quantification of the mean upper cervical cord area (MUCCA), the volumes of brain gray and white matter (BGM and BWM), and the brain MS lesion load. MUCCA was calculated on the basis of the T1w brain image series using a previously described semi-automated method and normalized relative to the distance between the C1 and C3 vertebrae to compensate for physiologic differences related to body size.15
To calculate volumes of BGM and BWM, we used the VBM8 preprocessing and segmentation tools with default parameters.18 All 3D T1w series were corrected for WM lesions before volumetric analysis using the lesion growths algorithm as implemented in the LST toolbox (version 1.2.2).19 Intracranial cavity volumes were determined by summation of the volumes of GM, WM, and CSF.
We normalized BGM and BWM with regard to head size differences by normalization according to the intracranial cavity volume and corrected for physiologic aging effects compared with HC.
Expression of CHI3L1 in different cell types from healthy and MS brains
CHI3L1 expression of the human brain was examined in 2 published single-cell RNA sequencing (scRNA-seq) data sets (Jäkel et al. and Masuda et al. data sets).20,21 Both data sets were analyzed using the RaceID algorithm. The Jäkel et al.20 data set was prepared from white matter areas of postmortem human brain from patients with MS and from unaffected controls using a nuclear sequencing protocol. It provided an overview of different cell types from healthy and MS brains and was acquired from different MS lesions.20 The findings of the Jäkel et al. data set were validated in an scRNA-seq data set that was acquired from stereotactic biopsies and fluorescence-activated cell sorting sorted for CD45+ cells of patients with MS and HCs (Masuda et al. data set).21 This data set contained cells from chronically active lesions. The results were grouped into 10 clusters (Hu-C1 to Hu-C10), representing the transcriptome signatures of different cell types.
Statistical analysis
Association study between MRI and CHI3L1 and NfL concentrations in CSF
We used IBM SPSS for Windows version 24 (IBM Corp., Armonk, NY) for statistical analyses. Results were considered statistically significant at p < 0.05.
All parameters were tested for normality with a Kolmogorov-Smirnov test. Intergroup comparisons of CNS volumes, age, and DD were assessed using 1-way analysis of variance (normally distributed data), with post hoc pairwise tests adjusted for multiple comparisons with Games-Howell correction when more than 2 groups were compared. Intergroup differences of relapsing MS (RMS) and progressive MS (PMS) regarding EDSS, brain lesion load, and CHI3L1 and NfL levels were studied by using Mann-Whitney U tests (ordinal or not normally distributed data). We used χ2 tests to investigate differences in the male-female distribution within the subgroups.
Because we detected no differences related to sex in the variables (CHI3L1, NfL, age, DD, EDSS, MUCCA, BGM, or BWM by using Mann-Whitney U tests or Student t tests), the results of female and male patients were pooled for further analyses.
We evaluated potential associations between CHI3L1, NfL, and clinical variables (age, DD, and EDSS) and between CHI3L1 and NfL using Spearman rank correlation analyses. Estimates of standard errors and CIs for the correlation coefficients were derived using the bootstrapping method implemented in SPSS (1,000 samples).
Univariate simple linear regression and multiple linear regression analyses with backward selection of variables (exclusion criteria: probability of F ≥ 0.1) for CHI3L1 or NfL as dependent variables and MUCCA, BGM, and BWM as independent variables served to investigate whether the CSF biomarker levels were related brain and/or spinal cord atrophy.
Discrimination between the RMS (CIS/early RRMS and RRMS >2 years) and PMS (primary progressive and secondary progressive) by CHI3L1 and NfL levels was tested through multiple binary logistic regression analyses with MS type as dependent variable (RMS coded: 0, PMS: coded 1) and CHI3L1 and NfL as independent variables using backward conditional exclusion (pin = 0.05, pout = 0.10).
Expression of CHI3L1 in different cell types from healthy and MS brains
Published scRNA-seq data sets were examined for the expression of CHI3L1. Both data sets were analyzed using the RaceID algorithm using standard settings.22 CHI3L1 expression was presented using mean and standard error of the mean. Data analysis and visualization of the genetic data was performed using the R programming suite and the tidyverse package.23
Data availability
Anonymized data used for this study are available from the corresponding author on reasonable request.
Results
Patient characteristics
Of the 131 patients with MS, 42 (32%) had relapsing MS (CIS and early RRMS with DD ≤2 years [n = 17] or RRMS with DD >2 years [n = 25]), and 89 (68%) patients had progressive MS (secondary progressive [n = 69] or primary progressive subtype [n = 20]). Mean/median values of age, DD, and EDSS were higher in the PMS group than in the RMS group. Clinical and demographic details of the entire patient group, RMS, PMS, and HC are shown in table 1.
CSF ELISA analyses of CHI3L1 and NfL concentrations
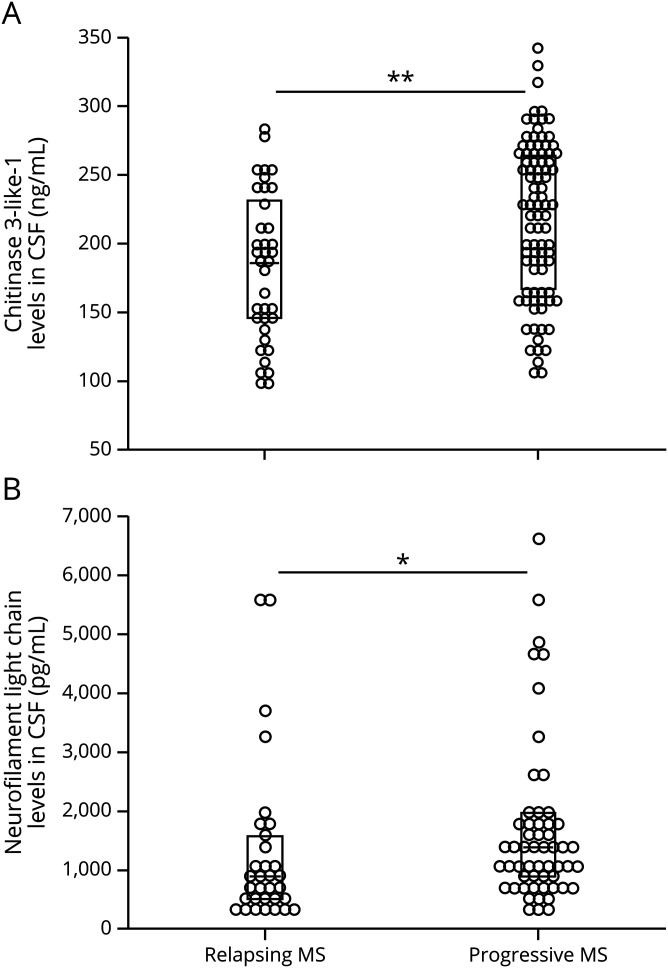
The median of CHI3L1 concentration was higher in PMS than in the RMS subgroup, and maximal values of CHI3L1 concentrations were observed progressive MS (table 1, figure 1, left panel). Similarly, the median of NfL concentration was higher in the PMS group than in RMS (figure 1, right panel). Compared with CHI3L1, the NfL concentrations of the entire patient group spanned a considerably wider relative range (factor 0.24–6.4 of the whole group median in NfL compared with 0.5–1.6 of the median of the entire group in CHI3L1).
Figure 1. CHI3L1 (A) and NfL (B) concentrations in CSF of PMS and RMS.
The 2D plots depict the distribution of the results; the boxes mark the median and interquartile ranges. The 2D plots depict the distribution of the results; the boxes mark the median and interquartile ranges. *p < 0.050, **p < 0.001 in Mann-Whitney U tests. CHI3L1 = chitinase 3–like protein 1; NfL = neurofilament light chain; PMS = progressive MS; RMS = relapsing MS.
Although correlations between CHI3L1 and age, DD, and EDSS were moderate (Spearman ρ [95% CI], p: 0.241 [0.064–0.419], p = 0.006; 0.198 [0.011–0.368], p = 0.198; 0.217 [0.010–0.407], p = 0.015), NfL showed stronger associations with these clinical parameters (ρ [95%CI], p: 0.556 [0.352–0.715], p < 0.001; 0.452 [0.248–0.624], p < 0.001; 0.279 [0.060–0.469], p = 0.009), especially with age (reflected by higher correlation coefficients and lower p values).
The CSF concentrations of CHI3L1 and NfL were correlated (Spearman ρ [95% CI], p: 0.373 [0.177–0.552], p < 0.001), A scatter plot of the association between CHI3 and NfL is provided in the supplement (figure e-1, links.lww.com/NXI/A329).
MRI-derived atrophy of the cervical cord and the brain in patients with MS
We observed MUCCA atrophy in the entire MS group as compared to HC (table 1 and figure e-2A, links.lww.com/NXI/A329), which was driven by MUCCA reduction in the PMS subgroup Furthermore, the mean of MUCCA in the PMS group was smaller than in the RMS subtype. In the latter group, no significant group differences compared with HC were observed. Atrophy of BGM was observed in the entire patient group compared with HC and in PMS compared with RMS (figure e-2B). Brain lesion load was higher in chronic MS compared with RMS. We observed no BWM atrophy in the patient groups.
Associations between CSF biomarkers and cervical cord area or brain volumes addressed using backward linear regression analyses
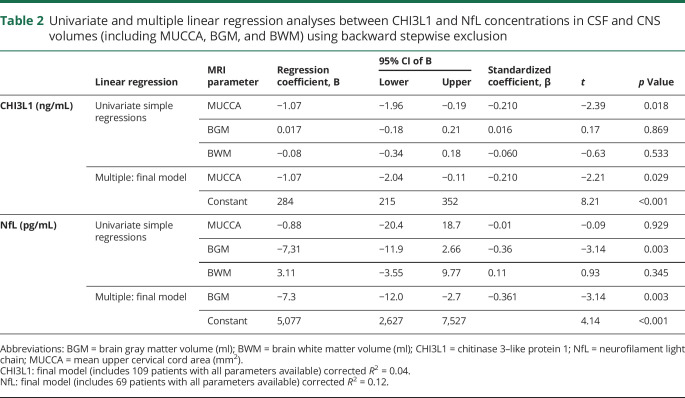
To study associations between the CSF biomarkers and the MRI-derived CNS volumetry measures, we used univariate and multiple linear regression analyses separately for CHI3L1 or NfL as dependent variables including MUCCA, brain GM, and WM as independent variables. The multiple regressions used backward stepwise exclusion (table 2). As a prerequisite for the applicability of the multiple linear regressions, we checked conditions, such as independence and normal distribution of the residuals, no collinearity of the independent variables, and occurrence of potential outliers. As a consequence, we excluded 5 samples with NfL >3,500 ng/mL from the analysis.
Table 2.
Univariate and multiple linear regression analyses between CHI3L1 and NfL concentrations in CSF and CNS volumes (including MUCCA, BGM, and BWM) using backward stepwise exclusion
CHI3L1: In the univariate regression analyses, MUCCA showed the strongest associations with CHI3L1 (reflected by the t statistic and lowest p value). In the final multiple regression model, MUCCA was the only predictive MRI volumetry variable for CHI3L1. Still, the corrected R2 of the final model reflected merely a small effect size.24 According to the negative regression coefficient, a decrease of MUCCA of 10 mm2 predicted an average increase in CHI3L1 of 11 ng/mL in this regression model.
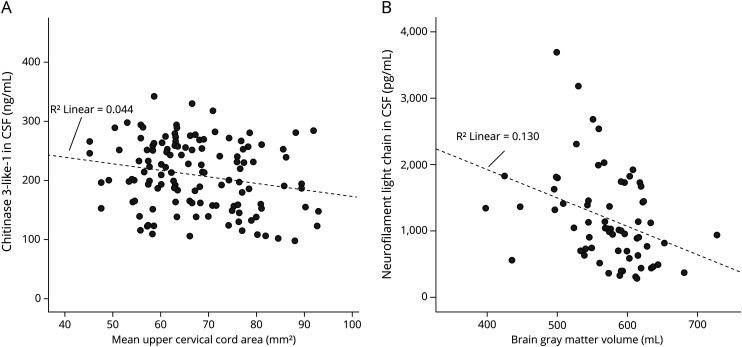
NfL: For NfL, the univariate regressions showed the strongest associations between BGM and NfL. BGM was found to be the only predictive MRI volumetry variable in the final multiple regression model. Within this model, a decrease of 10 mL in BGM predicted an average increase of 73 pg/mL NfL. The final model explained 12% of the variance of NfL, corresponding to a small to medium effect size24; figure 2, A and B further illustrates the associations between CHI3L1 and MUCCA and between NfL and BGM.
Figure 2. Association between (A) CHI3L1 and MUCCA and (B) NfL and BGM.
The line represents simple linear regressions between the variable pairs with R2: coefficient of determination. BGM = brain gray matter; CHI3L1 = chitinase 3–like protein 1; NfL = neurofilament light chain; MUCCA = mean upper cervical cord area.
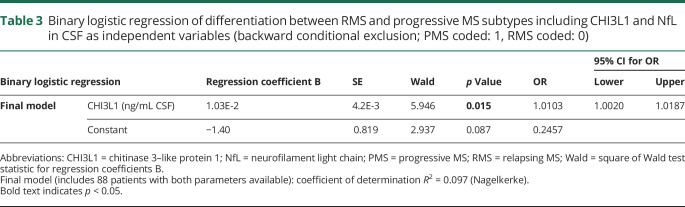
Discrimination between the RMS and chronic MS types by CHI3L1 and NfL CSF levels
To address the impact of CHI3L1 and NfL levels in CSF to discriminate between the RMS and progressive MS types, we used multiple binary logistic regression analyses (backward conditional exclusion) with type (RMS coded: 0, PMS: coded 1) as dependent variable and CHI3L1 and NfL as independent variables (table 3). CHI3L1 turned out to be most suitable for differentiating between PMS or RMS types. Thus, in the final model (χ2(1) = 6.445, p = 0.011, N = 88), the odds of having PMS are 1.11 times larger for a patient with a 10 ng/mL higher CHI3L1 level, Nagelkerke R2 = 0.097 of the final model corresponded to a medium effect size, Cohen f = 0.33.24
Table 3.
Binary logistic regression of differentiation between RMS and progressive MS subtypes including CHI3L1 and NfL in CSF as independent variables (backward conditional exclusion; PMS coded: 1, RMS coded: 0)
Expression of CH3L1 in different cell types from healthy and MS brains
In the Jäkel et al. data set, reflecting CH3L1 in MS and healthy BWM, CHI3L1 was mainly expressed in 2 astrocyte subsets and in microglia and macrophages (figure e-2A, links.lww.com/NXI/A329). Of interest, in MS samples, CHI3L1 expression was mainly found in MS-associated microglia and macrophages, but not in other immune cells, such as monocytes and lymphocytes (figure e-2B).
These findings were validated using the Masuda et al. scRNA-Seq data set. Herein, MS-associated microglia and macrophages (clusters Hu-C2, Hu-C3, and Hu-C8) expressed elevated levels of CHI3L1,21 while CH3L1 was almost undetectable in the homeostatic clusters (figure e-2C, links.lww.com/NXI/A329). Moreover, like in the Jäkel et al. data set, hematogenic immune cells (monocytes and lymphocytes) showed no detectable and barely detectable levels of CHI3L1. Overall, specific CHI3L1 gene expression was found in MS-associated microglia and macrophages, but not in homeostatic microglia or other immune cells. In contrast, astrocytes expressed CHI3L1 in healthy control samples and moreover in active and chronically active MS lesions.
Discussion
Contrary to our hypothesis, CHI3L1 and NfL were not associated equally with both cervical cord area and brain volumes in our study. Although CHI3L1 was associated with cervical cord volume loss (MUCCA reduction) but not with brain volumes, NfL was related to BGM decrease and not to MUCCA.
To date, relations between CSF biomarkers and spinal cord atrophy in MS have rarely been investigated. CSF NF heavy chain levels were associated with late spinal cord atrophy progression, but not with baseline spinal cord measures in a longitudinal study including a small cohort of patients with MS.25 Similarly, NfL in blood serum was associated with long-term progression of spinal cord volume loss but not with baseline measures in a large longitudinal study including a mixed MS cohort7 This association between NfL and spinal cord volume might seem contradictory to our findings, but differences in the study design (longitudinal spinal cord volume loss was assessed, while the patient group did not include patients with later-stage secondary PMS) and assessment of serum NfL instead of CSF NfL may be reasons for differing results.8
Our findings of associations between NfL levels and baseline reduction of BGM are corroborated by recent findings based on serum NfL.26 Other studies reported relations between NfL in CSF or serum with longitudinal brain volume changes.7,8
These differing patterns of associations, especially referring to the association of CHI3L1 with MUCCA, may be based on different pathophysiologic sources of CHI3L1 and NfL CSF concentrations in the patient subgroups.
In line with previous studies, average levels of CHI3L1 and NfL were higher in PMS compared with RMS.11,27–29 Still, CHI3L1 was better suited to differentiate between both subtypes than NfL. This might be due to broader ranges of values in NfL and cases of high NfL levels both in the RMS and in the PMS group.
The relevance of CSF CHI3L1 to neurologic disability in PMS was recently reported.30 In PMS compared with RMS, higher levels of spinal cord atrophy are common findings.31
Spinal cord atrophy in MS, correlating with clinical disability, has been shown to be at least in part independent of local inflammation. Thus remote effects of Wallerian degeneration secondary to neuroaxonal tissue damage in the brain have been suggested as drivers of spinal cord atrophy in MS.15 In PMS, neurodegenerative rather than inflammatory processes are the main drivers of disease progression. Neurodegeneration generates higher relative volume losses in the spinal cord than in the brain due to a lack of compensatory capabilities in this relatively small structure. We hypothesize that diffuse neurodegeneration along with ongoing astrocytic and microglial activation results in higher levels of CSF CHI3L1 in progressive MS, which correlates with cervical spinal cord volume reduction. Thus, CHI3L1 may have a closer association to spinal cord atrophy than NfL.
Brain GM atrophy, on the other hand, is driven by inflammatory activity reflected by new or gadolinium-enhancing brain lesions in relapsing-remitting disease courses and by diffuse neurodegeneration and chronical inflammation in progressive MS.16,25,32 NfL has widely been described as a marker for axonal damage secondary to acute inflammatory processes in MS,2,4,28 which might explain the strong association between CSF NfL and BGM atrophy.
We also could verify a differing pattern of CHI3L1 and NfL related to age and DD. In contrast to CHI3L1, NfL showed stronger associations with age and DD. In line with our CSF-based results, increasing levels of serum NfL with age in HC and in patients with MS have been previously reported,7 suggesting a limitation for the use of NfL as disease marker in older patients with longer DD.33,34 The weaker impact of age and DD on CHI3L1 may underline its suitability for use as biomarker in PMS.
Although elevated CHI3L1 and NfL levels are not MS specific,35,36 both markers have been associated with inflammatory processes in MS, whereby CHI3L1 has a regulatory function on inflammation, whereas NfL is a product of axonal degeneration.28 Furthermore, the expression of CHI3L1 and NfL can be considerably increased by various inflammatory and immunologic conditions.37
The occurrence of CHI3L1 in inflammatory environments as described38,39 was further substantiated by the analysis of cell-specific CHI3L1 expression patterns: CHI3L1 expression was mainly found in active and chronically active MS lesions in activated macrophages/microglia of MS and in activated astrocytes.
Our study is not without limitations. A drawback is its retrospective design and a lack of CSF probes of HC, which excluded the discrimination between physiologic and disease-related age dependencies. Another limitation is the cross-sectional design. In particular, to clarify whether CHI3L1 baseline levels might be sensitive markers for future spinal cord atrophy progression, longitudinal studies including CHI3L1 and spinal cord atrophy are warranted.
Conclusions
NfL and CHI3L1 in CSF were differentially related to brain and spinal cord atrophy. We demonstrated associations between CHI3L1 and spinal cord volume. In contrast, NfL was not correlated with MUCCA, but we confirmed recently reported correlations of NfL with BGM reduction. CHI3L1 was suited to differentiate between relapsing and progressive MS and was less affected by DD and age than NfL. Consistent with a recent study showing that the pattern of combined measurement of CSF-based NfL and CHI3L1 was useful to discriminate MS phenotypes,14 the combined measurement of CSF NfL and CHI3L1 provides complementary information with regard to brain and spinal cord volumes.
Glossary
- BGM
brain gray matter
- BWM
brain white matter
- CHI3L1
chitinase 3–like protein 1
- CIS
clinically isolated syndrome
- DD
disease duration
- EDSS
Expanded Disability Severity Scale
- HC
healthy control
- MUCCA
mean upper cervical cord area
- NfL
neurofilament light chain
- PMS
progressive MS
- RMS
relapsing MS
- RRMS
relapsing-remitting MS
- scRNA-seq
single-cell RNA sequencing
- T1w
T1 weighted
Appendix. Authors
Study funding
This work was funded by FoRUM (Ruhr University Bochum) [grant number F 839-14].
Disclosure
The authors report no relevant disclosures. Go to Neurology.org/NN for full disclosures.
References
- 1.Haghikia A, Hohlfeld R, Gold R, Fugger L. Therapies for multiple sclerosis: translational achievements and outstanding needs. Trends Mol Med 2013;19:309–319. [DOI] [PubMed] [Google Scholar]
- 2.Teunissen CE, Khalil M. Neurofilaments as biomarkers in multiple sclerosis. Mult Scler 2012;18:552–556. [DOI] [PubMed] [Google Scholar]
- 3.Kuhle J, Plavina T, Barro C, et al. . Neurofilament light levels are associated with long-term outcomes in multiple sclerosis. Mult Scler Epub 2019 Nov 4. [DOI] [PMC free article] [PubMed]
- 4.Håkansson I, Tisell A, Cassel P, et al. . Neurofilament light chain in cerebrospinal fluid and prediction of disease activity in clinically isolated syndrome and relapsing-remitting multiple sclerosis. Eur J Neurol 2017;24:703–712. [DOI] [PubMed] [Google Scholar]
- 5.Arrambide G, Espejo C, Eixarch H, et al. . Neurofilament light chain level is a weak risk factor for the development of MS. Neurology 2016;87:1076–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bittner S, Steffen F, Uphaus T, et al. . Clinical implications of serum neurofilament in newly diagnosed MS patients: a longitudinal multicentre cohort study. EBioMedicine 2020;56:102807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barro C, Benkert P, Disanto G, et al. . Serum neurofilament as a predictor of disease worsening and brain and spinal cord atrophy in multiple sclerosis. Brain 2018;141:2382–2391. [DOI] [PubMed] [Google Scholar]
- 8.Håkansson I, Tisell A, Cassel P, et al. . Neurofilament levels, disease activity and brain volume during follow-up in multiple sclerosis. J Neuroinflammation 2018;15:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novakova L, Axelsson M, Khademi M, et al. . Cerebrospinal fluid biomarkers as a measure of disease activity and treatment efficacy in relapsing-remitting multiple sclerosis. J Neurochem 2017;141:296–304. [DOI] [PubMed] [Google Scholar]
- 10.Uher T, Schaedelin S, Srpova B, et al. . Monitoring of radiologic disease activity by serum neurofilaments in MS. Neurol Neuroimmunol Neuroinflamm 2020;7:e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantó E, Tintoré M, Villar LM, et al. . Chitinase 3-like 1: prognostic biomarker in clinically isolated syndromes. Brain 2015;138(pt 4):918–931. [DOI] [PubMed] [Google Scholar]
- 12.Modvig S, Degn M, Roed H, et al. . Cerebrospinal fluid levels of chitinase 3-like 1 and neurofilament light chain predict multiple sclerosis development and disability after optic neuritis. Mult Scler 2015;21:1761–1770. [DOI] [PubMed] [Google Scholar]
- 13.Comabella M, Fernández M, Martin R, et al. . Cerebrospinal fluid chitinase 3-like 1 levels are associated with conversion to multiple sclerosis. Brain 2010;133(pt 4):1082–1093. [DOI] [PubMed] [Google Scholar]
- 14.Gil-Perotin S, Castillo-Villalba J, Cubas-Nuñez L, et al. . Combined cerebrospinal fluid neurofilament light chain protein and chitinase-3 like-1 levels in defining disease course and prognosis in multiple sclerosis. Front Neurol 2019;10:1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukas C, Sombekke MH, Bellenberg B, et al. . Relevance of spinal cord abnormalities to clinical disability in multiple sclerosis: MR imaging findings in a large cohort of patients. Radiology 2013;269:542–552. [DOI] [PubMed] [Google Scholar]
- 16.Sastre-Garriga J, Pareto D, Battaglini M, et al. . MAGNIMS consensus recommendations on the use of brain and spinal cord atrophy measures in clinical practice. Nat Rev Neurol 2020;16:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polman CH, Reingold SC, Edan G, et al. . Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann Neurol 2005;58:840–846. [DOI] [PubMed] [Google Scholar]
- 18.Gaser C. VBM by Structural Brain Mapping Group. 2008. Available at: dbm.neuro.uni-jena.de/wordpress/vbm/. Accessed November 1, 2020.
- 19.Schmidt P, Gaser C, Arsic M, et al. . An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage 2012;59:3774–3783. [DOI] [PubMed] [Google Scholar]
- 20.Jäkel S, Agirre E, Mendanha Falcão A, et al. . Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 2019;566:543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda T, Sankowski R, Staszewski O, et al. . Author correction: spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 2019;568:E4. [DOI] [PubMed] [Google Scholar]
- 22.Herman JS, Sagar, Grün D. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat Methods 2018;15:379–386. [DOI] [PubMed] [Google Scholar]
- 23.R-Foundation, Tidyverse. The R Project for Statistical Computing, and the Tidyverse Collection of R packages for data science. Available at: R-project.org/ and tidyverse.org. 2018.
- 24.Cohen J. A power primer. Psychol Bull 1992;112:155–159. [DOI] [PubMed] [Google Scholar]
- 25.Petzold A, Steenwijk MD, Eikelenboom JM, Wattjes MP, Uitdehaag BM. Elevated CSF neurofilament proteins predict brain atrophy: a 15-year follow-up study. Mult Scler 2016;22:1154–1162. [DOI] [PubMed] [Google Scholar]
- 26.Jakimovski D, Kuhle J, Ramanathan M, et al. . Serum neurofilament light chain levels associations with gray matter pathology: a 5-year longitudinal study. Ann Clin Transl Neurol 2019;6:1757–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinsinger G, Galéotti N, Nabholz N, et al. . Chitinase 3-like proteins as diagnostic and prognostic biomarkers of multiple sclerosis. Mult Scler 2015;21:1251–1261. [DOI] [PubMed] [Google Scholar]
- 28.Mane-Martinez MA, Olsson B, Bau L, et al. . Glial and neuronal markers in cerebrospinal fluid in different types of multiple sclerosis. J Neuroimmunol 2016;299:112–117. [DOI] [PubMed] [Google Scholar]
- 29.Cantó E, Reverter F, Morcillo-Suárez C, et al. . Chitinase 3-like 1 plasma levels are increased in patients with progressive forms of multiple sclerosis. Mult Scler 2012;18:983–990. [DOI] [PubMed] [Google Scholar]
- 30.Pérez-Miralles F, Prefasi D, García-Merino A, et al. . CSF chitinase 3-like-1 association with disability of primary progressive MS. Neurol Neuroimmunol Neuroinflamm 2020;7:e815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casserly C, Seyman EE, Alcaide-Leon P, et al. . Spinal cord atrophy in multiple sclerosis: a systematic review and meta-analysis. J Neuroimaging 2018;28:556–586. [DOI] [PubMed] [Google Scholar]
- 32.Romme Christensen J, Börnsen L, Khademi M, et al. . CSF inflammation and axonal damage are increased and correlate in progressive multiple sclerosis. Mult Scler 2013;19:877–884. [DOI] [PubMed] [Google Scholar]
- 33.Lu CH, Macdonald-Wallis C, Gray E, et al. . Neurofilament light chain: a prognostic biomarker in amyotrophic lateral sclerosis. Neurology 2015;84:2247–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutphen CL, Jasielec MS, Shah AR, et al. . Longitudinal cerebrospinal fluid biomarker changes in preclinical Alzheimer disease during middle age. JAMA Neurol 2015;72:1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giovannoni G. Peripheral blood neurofilament light chain levels: the neurologist's C-reactive protein? Brain 2018;141:2235–2237. [DOI] [PubMed] [Google Scholar]
- 36.Kušnierová P, Zeman D, Hradílek P, Zapletalová O, Stejskal D. Determination of chitinase 3-like 1 in cerebrospinal fluid in multiple sclerosis and other neurological diseases. PLoS One 2020;15:e0233519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeo IJ, Lee CK, Han SB, Yun J, Hong JT. Roles of chitinase 3-like 1 in the development of cancer, neurodegenerative diseases, and inflammatory diseases. Pharmacol Ther 2019;203:107394. [DOI] [PubMed] [Google Scholar]
- 38.Recklies AD, Ling H, White C, Bernier SM. Inflammatory cytokines induce production of CHI3L1 by articular chondrocytes. J Biol Chem 2005;280:41213–41221. [DOI] [PubMed] [Google Scholar]
- 39.Bonneh-Barkay D, Bissel SJ, Kofler J, Starkey A, Wang G, Wiley CA. Astrocyte and macrophage regulation of YKL-40 expression and cellular response in neuroinflammation. Brain Pathol 2012;22:530–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Anonymized data used for this study are available from the corresponding author on reasonable request.