Abstract
Objective
To determine whether atrophy relates to phenotypical variants of posterior cortical atrophy (PCA) recently proposed in clinical criteria (i.e., dorsal, ventral, dominant-parietal, and caudal) we assessed associations between latent atrophy factors and cognition.
Methods
We employed a data-driven Bayesian modeling framework based on latent Dirichlet allocation to identify latent atrophy factors in a multicenter cohort of 119 individuals with PCA (age 64 ± 7 years, 38% male, Mini-Mental State Examination 21 ± 5, 71% β-amyloid positive, 29% β-amyloid status unknown). The model uses standardized gray matter density images as input (adjusted for age, sex, intracranial volume, MRI scanner field strength, and whole-brain gray matter volume) and provides voxelwise probabilistic maps for a predetermined number of atrophy factors, allowing every individual to express each factor to a degree without a priori classification. Individual factor expressions were correlated to 4 PCA-specific cognitive domains (object perception, space perception, nonvisual/parietal functions, and primary visual processing) using general linear models.
Results
The model revealed 4 distinct yet partially overlapping atrophy factors: right-dorsal, right-ventral, left-ventral, and limbic. We found that object perception and primary visual processing were associated with atrophy that predominantly reflects the right-ventral factor. Furthermore, space perception was associated with atrophy that predominantly represents the right-dorsal and right-ventral factors. However, individual participant profiles revealed that the large majority expressed multiple atrophy factors and had mixed clinical profiles with impairments across multiple domains, rather than displaying a discrete clinical–radiologic phenotype.
Conclusion
Our results indicate that specific brain behavior networks are vulnerable in PCA, but most individuals display a constellation of affected brain regions and symptoms, indicating that classification into 4 mutually exclusive variants is unlikely to be clinically useful.
Posterior cortical atrophy (PCA) is a clinical–radiologic syndrome defined by progressive loss of higher-order visual functions and atrophy that markedly affects posterior brain regions.1–4 Whereas multiple pathologies may underlie the PCA syndrome, the most common biological substrate is Alzheimer disease (AD).5,6 The dominant features of PCA are visuoperceptual and visuospatial symptoms but there exists considerable phenotypical heterogeneity between individuals, which has motivated efforts to categorize PCA into phenotypical variants.7 The 2 best characterized variants are the occipitotemporal (ventral) and temporoparietal (dorsal) variants, which reflect the functional organization of the visual system (i.e., ventral and dorsal streams), and are characterized by the presence of prominent visuoperceptual and visuospatial deficits, respectively.8–10 Recent consensus criteria7 describe 2 additional variants: a primary visual (caudal) variant, characterized by primary visual processing deficits,9,11,12 and a dominant parietal variant, which presents with prominent nonvisual parietal function deficits like dyscalculia, dyslexia, and apraxia.13–15 These PCA variants are mainly based on single case studies or studies of limited sample sizes, and previous attempts to identify consistent clinical and neuroimaging correlates to these variants have failed.16–18 Consequently, in the consensus criteria it is emphasized that current literature provides insufficient cognitive or neuroimaging evidence to support the existence of discrete PCA subtypes and that more research is needed.7 With this in mind, we employed a data-driven Bayesian modeling approach to detect endophenotypes on MRI among a relatively large set of extensively phenotyped PCA participants, and assessed associations between these phenotypes and PCA-specific clinical symptoms.
Methods
Standard protocol approvals, registrations, and patient consents
Written informed consent was obtained from all participants, and the local medical ethics review committees of the Amsterdam University Medical Center (UMC) and University of California, San Francisco (UCSF) approved the study.
Participants
We selected participants with PCA from 2 independent expert centers: the Amsterdam Dementia Cohort of the Amsterdam UMC, the Netherlands,19 and the UCSF Alzheimer's Disease Research Center. All participants underwent dementia screening between June 2000 and July 2017, and inclusion into the present study was based on the following criteria: (1) a syndrome diagnosis of PCA as defined by published diagnostic criteria6,7,20 and established by consensus in a multidisciplinary meeting and (2) availability of an MRI scan including a structural T1-weighted sequence. We excluded participants who had negative biomarkers for β-amyloid (Aβ) pathology (either CSF molecular profile21 or amyloid PET visual rating22). These criteria yielded 69 participants from Amsterdam UMC and 50 from UCSF. Participants from the 2 cohorts were merged into one combined cohort. Of the 119 participants in this combined cohort, 91 (76%) were Aβ-positive (40 [34%] on CSF, 28 [24%] on PET, and 23 [19%] on both PET and CSF) and for 28 (24%) the Aβ status was unknown. We also selected 121 Aβ-negative cognitively normal individuals (age 57.4 ± 8.9, 41% male, Mini-Mental State Examination [MMSE] 29.0 ± 0.8) from the Amsterdam UMC cohort, who served as a reference group for voxelwise contrasts and were also used to standardize gray matter density images (see Imaging analyses).
Cognition
Neuropsychological test scores covered 2 higher-order visual processing domains in both cohorts: object perception (fragmented letters) and space perception (number location and dot counting).23 The visual test battery administered in the UCSF sample included more tests than the Amsterdam UMC sample and 2 additional domains could be assessed within the UCSF cohort only: nonvisual dominant parietal functions (calculations, spelling, and reading) and primary visual processing (point location, figure discrimination, shape discrimination, hue discrimination, visual acuity, size discrimination, letter cancellation, static circle detection, and motion coherence). Additional neuropsychological test scores covered the following nonvisual cognitive domains: memory (Amsterdam UMC: Rey auditory verbal learning test–immediate and delayed recall [15 items/5 trials]; UCSF: California Verbal Learning Test–immediate and delayed recall [9 items/4 trials]), executive function (Amsterdam UMC and UCSF: digit-span forwards and backwards; letter fluency [D]), and language (Amsterdam UMC and UCSF: verbal fluency [animal naming]).24,25 MMSE scores were used as a measure of global cognition.
Before combining neuropsychological data from the 2 cohorts, all test scores were converted into z scores using the mean and SD of each separate cohort and then combined. This was done to account for center-specific effects on cognition. Furthermore, educational attainment levels were measured using a qualitative scale in the Amsterdam UMC cohort and these were converted to years of education before combining the samples. Cognitive data obtained closest to the MRI date (maximally 6 months, mean follow-up: 0.2 ± 1.1 months) were used for the analyses. Availability of cognitive data across neuropsychological tests is presented in table 1.
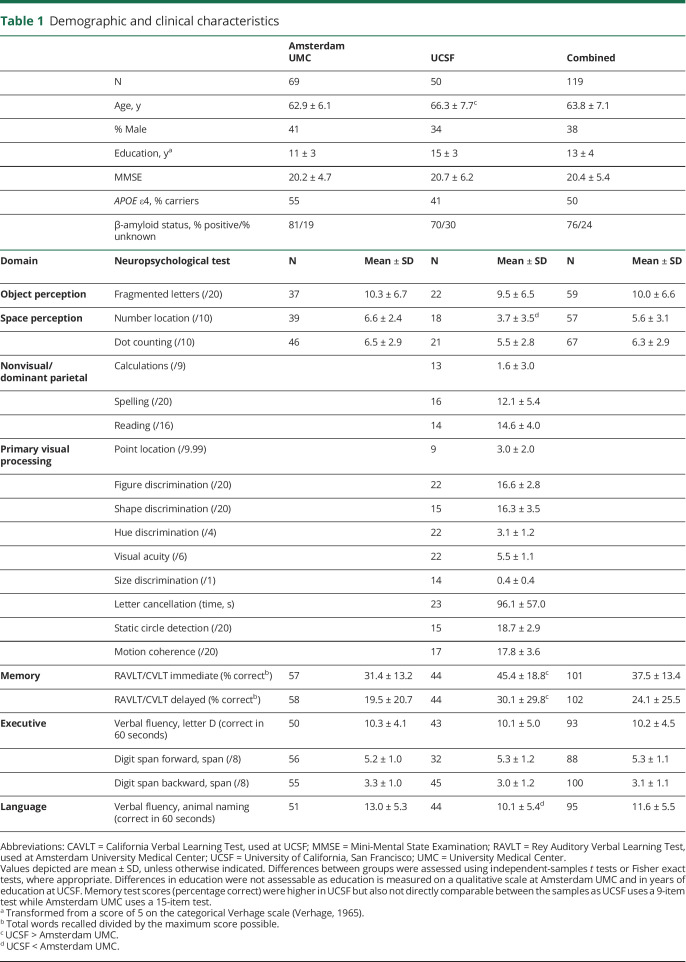
Table 1.
Demographic and clinical characteristics
Neuroimaging
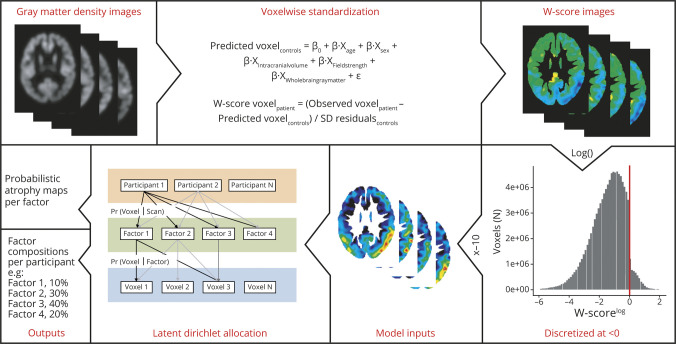
MRIs from Amsterdam UMC were acquired on 8 different scanners using previously described standardized acquisition protocols (data available from Dryad, table 1, doi.org/10.5061/dryad.jdfn2z37p) and with a scanner field strength of 1.5T or 3T. MRIs from UCSF were acquired on a 1.5T Magnetom Avanto, a 3T Siemens Tim Trio, or a 3T Siemens Prisma Fit scanner. Proportions of participants scanned on a 1.5T scanner were balanced between the 2 samples, 22% in Amsterdam UMC and 26% in UCSF, and scanner field strength was used as a covariate in all imaging analyses. T1-weighted images were segmented, smoothed, weighted, modulated, and spatially normalized to a common space using a standard SPM12 (Wellcome Trust Centre for Neuroimaging, UK) preprocessing pipeline described elsewhere.25 The resulting normalized gray matter density images were used to assess the whole-brain spatial distribution of atrophy by performing voxelwise contrasts between participants with PCA and controls. Next, the gray matter density images were converted into W-score maps (i.e., control-normalized z scores adjusted for covariates)4,26 by performing voxelwise standardization to the control group, regressing out the effects of age, sex, intracranial volume, scanner field strength, and whole-brain gray matter volume (operationalized as gray matter to intracranial volume ratios). The resulting W-maps represent voxelwise atrophy adjusted for covariates and are used as input in the Bayesian modeling framework (figure 1).
Figure 1. A Bayesian model to compute latent atrophy factors based on structural MRI.
The estimated parameters are the probability that a participant expresses a particular factor (i.e., Pr[factor|scan]) and the probability that a factor is associated with atrophy at an MRI voxel (i.e., Pr[voxel|factor]). To achieve these estimations, the Bayesian modeling framework uses continues W-score maps as inputs. W-scores are obtained by performing regression analyses in the control group to determine predicted voxel values (voxelcontrols) based on age, sex, intracranial volume, scanner field strength, and whole-brain gray matter volume. W-scores are then calculated by subtracting voxelcontrols from the observed voxel values (voxelpatient) and dividing by the residuals from the regression analysis in controls (SDresidual). These W scores were log-transformed and then discretized so that a W score of <0 at a given voxel of a particular scan would imply above-average atrophy at the voxel relative to the controls (adjusted for the effects of age, sex, intracranial volume, scanner field strength, and whole-brain gray matter volume). W-scores >0 were set to zero (values above 0 reflect gray matter density greater than the control group). Then, the W scores were multiplied by −10 and rounded to the nearest integer, so that larger positive values indicated more severe atrophy. Given the discretized voxelwise atrophy of the PCA participants and the number of latent atrophy factors K, the variational expectation maximization algorithm (cs.princeton.edu/∼blei/lda-c/) was applied to estimate Pr(factor|scan) and Pr(voxel|factor). Each participant expresses one or more factors to a certain degree and each factor is associated with distinct but possibly overlapping patterns of brain atrophy. The algorithm was rerun with 20 different random initializations, and the solution with the best model fit (based on log-likelihood) was selected. Sixty iterations were run for each random initialization, although each run plateaued after around 30–50 iterations. Adjusted with permission from Zhang et al.27
Bayesian modeling
We employed a Bayesian modeling approach based on latent Dirichlet allocation (LDA) to discover atrophy patterns that covary across participants in order to identify latent atrophy factors present within the PCA sample. This method has been adapted for structural MRI data in a previous study including patients with AD27 and estimates atrophy factors in a voxelwise, spatially unconstrained manner. LDA has previously outperformed supervised methods like canonical correlation analyses.28 The LDA model (figure 1) considers each scan as an unordered collection of voxels associated with a predefined number of latent atrophy factors (K), and allows each individual's scan to be associated with multiple factors and each factor to be associated with multiple voxels. More specifically, given a dataset of scans (W-score images), the algorithm estimates the probability of atrophy at a particular voxel given a latent atrophy factor (Pr[voxel|factor]) and the probability that a factor is associated with a particular scan (Pr[factor|scan]). A latent factor (Pr[voxel|factor]) can be visualized as a probabilistic atrophy map. Pr(factor|scan) is a probability distribution over latent atrophy factors, representing the factor composition of the participant (scan). For example, in a 4-factor model (K = 4), Pr(factor|scan) might be 10% factor 1, 30% factor 2, 40% factor 3, and 20% factor 4 (figure 1). These factor compositions add up to 100%, and the individual components will henceforth be referred to as (atrophy) factor expressions, while the combination of the factor expressions constitutes an individual's factor composition. Because the factor expressions add up to 100%, an individual's expression of a particular factor could be regarded as the proportion of atrophy falling into a specific (but not necessarily localized) anatomical region rather than in the anatomical regions encompassed by the other factors. Therefore, factor expressions and factor compositions are reflective of an individual's spatial distribution of atrophy rather than its severity. An important model parameter is the number of latent factors (K). We ran models allowing for 4 factors (K = 4) in accordance with the number of PCA variants proposed in the clinical criteria (i.e., dorsal, ventral, caudal, and dominant parietal variant).7 Models, as well as all preprocessing steps and voxel-based morphometry analyses, were also performed in the 2 separate cohorts and visual inspection of the spatial distribution of the atrophy factors revealed that these were highly similar (data available from Dryad, figures 1–2, doi.org/10.5061/dryad.jdfn2z37p). Therefore, we will present results on the factors obtained in the combined sample in the main text, while results from the separate samples are presented in the supplement (data available from Dryad, figures 1–4, doi.org/10.5061/dryad.jdfn2z37p). In addition, we assessed LDA models allowing for K = 2 through 6 in the combined cohort, which are also presented in the supplement (data available from Dryad, figures 5–7, doi.org/10.5061/dryad.jdfn2z37p).
Statistical analyses
Statistical analyses were performed using R version 3.5.2. To assess cross-sectional associations between atrophy factor expressions and cognition, we used multiple linear regression analyses, adjusted for education and the temporal delay between neuropsychological assessment and MRI, using the lme4 package. Note that factor expressions were already adjusted for age, sex, whole-brain gray matter volume, intracranial volume, and scanner field strength effects in the LDA model. We included 3 of the 4 factors in the predictor set and the fourth was implicitly modeled because factor expressions of the 4 factors add up to 100%. The relative effects of the 3 directly modeled factors were calculated using the implicitly modeled fourth factor as a reference and all models were repeated using a different atrophy factor implicitly modeled to obtain pairwise differences between all factors (K1 vs K2, K1 vs K3, K1 vs K4, K2 vs K3, K2 vs K4 and K3 vs K4). As factor expressions represent the proportion of atrophy falling into a specific region rather than in the others, a negative association of factor X with cognition Z would state that individuals with a greater proportion of atrophy in regions associated with factor X, rather than factor Y, have worse scores on domain Z. Statistical significance for all models was set at α = 0.05 and we performed post hoc adjustment for multiple comparisons using the false discovery rate (FDR) method. Both uncorrected and FDR-corrected results are presented.
Data availability
The code for the Bayesian modeling approach is publicly available at github.com/ThomasYeoLab/CBIG/tree/master/stable_projects/disorder_subtypes/Zhang2016_ADFactors. Anonymized data used in the present study may be available upon request to the corresponding author.
Results
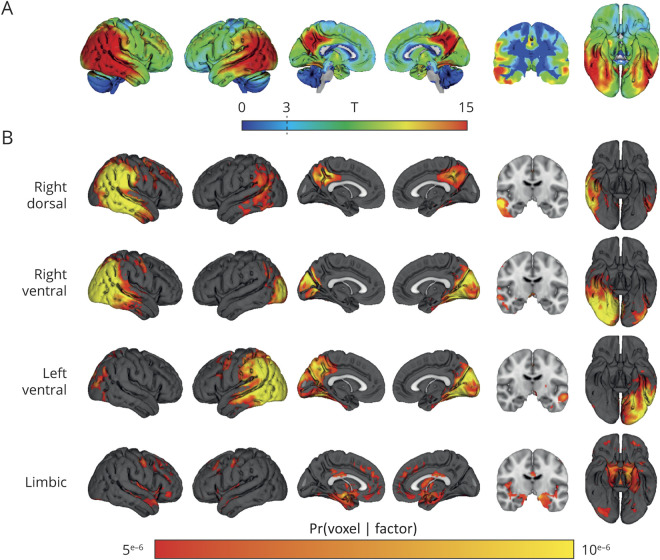
Demographic and clinical characteristics are presented in table 1. Mean age of the total sample was 63.8 ± 7.1, 38% were men, and MMSE was 20.4 ± 5.4. Voxelwise contrasts compared to controls revealed a classical PCA pattern, covering the middle and inferior temporal gyrus, inferior and medial parietal areas, and the occipital cortex (figure 2A). The atrophy pattern was slightly lateralized to the right hemisphere.
Figure 2. Voxel-wise contrasts between posterior cortical atrophy participants and controls and atrophy factors revealed by latent Dirichlet allocation (K = 4).
(A) Voxelwise T maps, adjusted for the effects of age, sex, intracranial volume, whole-brain gray matter volume, and scanner field strength. Significant voxels at T > 3. (B) Atrophy factors revealed by the latent Dirichlet allocation model (K = 4). Intensity of voxels signify the probability Pr(voxel|factor) of a voxel belonging to 1 of the 4 factors. Scale is truncated at Pr(voxel|factor) = 5e−6, and the cerebellum was removed from the template for visualization purposes.
Latent atrophy factors
The Bayesian model (K = 4) revealed 4 distinct yet partially overlapping latent atrophy factors (figure 2B). The first factor (right-dorsal) included the right lateral temporoparietal cortex as well as bilateral medial parietal regions. The second factor (right-ventral) included the right medial and lateral occipital cortex, extended inferiorly into the temporal cortex, and also covered part of the inferior parietal cortex. The third factor (left-ventral) included the left medial and lateral occipital cortex, inferior temporal cortex, and inferior parietal cortex. The fourth factor (limbic) mainly included bilateral medial-temporal areas as well as medial frontal regions (figure 2B).
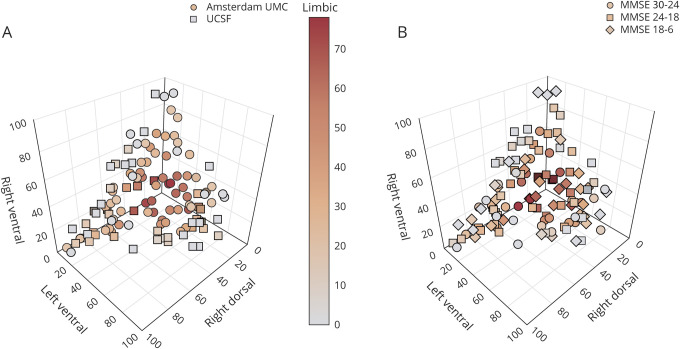
Individual factor compositions
Factor compositions of the combined sample reveal that the majority of PCA participants expressed a combination of multiple atrophy factors rather than predominantly expressing only one of the factors (figure 3A; data available from Dryad, figure-e3A, doi.org/10.5061/dryad.jdfn2z37p). A similar distribution was observed when we stratified individuals according to clinical disease severity (MMSE: 30–24 vs 23–18 vs 17–6; figure 3B; data available from Dryad, figure-e3B, doi.org/10.5061/dryad.jdfn2z37p). To assess whether factor expressions were partly driven by global atrophy, we examined the relationship between factor expressions and whole brain gray matter to intracranial volume ratios. We observed a significant correlation only between the limbic factor and whole-brain gray matter to intracranial volumes ratios (lower values indicate more atrophy; r = −0.43, p < 0.001), while the other factors did not show a correlation (range r = 0.11–0.19, all p > 0.05). Furthermore, we observed a significant correlation only between the right-dorsal factor and age (r = −0.26, p = 0.005) but there were no associations between factor expressions and sex (range t = −1.51 to 1.75, all p > 0.05), APOE ε4 (positive/negative; range: t = −1.28 to 1.00, all p > 0.05), or handedness (right/non-right-handed; range: t = −0.36 to 1.31, all p > 0.05).
Figure 3. Atrophy factors compositions for the combined sample.
(A) The 4D plot displays the factors right dorsal, left ventral, and right ventral on the x, y, and z axes and the limbic factor is displayed by the color gradient of the markers. Displayed factor compositions are for the combined sample and each marker represents one participant. Expressions of the 4 factors adds up to 100%. (B) Factor compositions of the combined sample and the markers represent 3 clinical disease severity groups. See also these figures in the provided interactive .html format (data available from Dryad, figure-e3A and e3B, doi.org/10.5061/dryad.jdfn2z37p). MMSE = Mini-Mental State Examination; UCSF = University of California, San Francisco; UMC = University Medical Center.
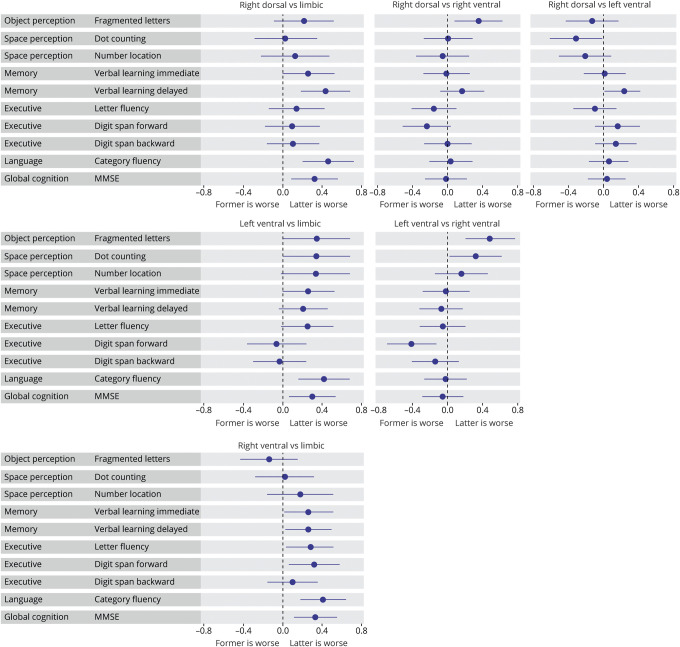
Associations between factor expression and higher-order visual processing
Fragmented letter scores (object perception) were negatively associated with right-ventral factor expression compared to right-dorsal and left-ventral factor expressions (β [confidence interval (CI)] = −0.35 [−0.63 to −0.09], p = 0.008 uncorrected; β [CI] = −0.48 [−0.76 to −0.020], p = 0.001 FDR-corrected; figure 4). Furthermore, dot counting scores (space perception) were negatively associated with right-ventral, limbic, and right-dorsal factor expression compared to the left-ventral factor (β [CI] = −0.32 [−0.62 to −0.03], p = 0.030 uncorrected, β [CI] = −0.31 [−0.61 to −0.00], p = 0.044 uncorrected, β [CI] = −0.32 [−0.61 to −0.02], p = 0.031 uncorrected). This same pattern was observed for number location scores (space perception), although none of the effects reached statistical significance (figure 4; data available from Dryad, table 2, doi.org/10.5061/dryad.jdfn2z37p).
Figure 4. Associations between factor expressions and neuropsychological tests assessing visual functions and memory, executive, and language functions.
The forest plots contain relative cross-sectional effects from linear regression models. As in each model, one of the factors is implicitly modeled, one of the factors serves as a reference to assess effects of all the others, and each subplot displays results from the different factor comparisons. Lines indicate the 95% confidence intervals and a significant effect (uncorrected for multiple comparisons) is denoted by confidence intervals not including x = 0. MMSE = Mini-Mental State Examination.
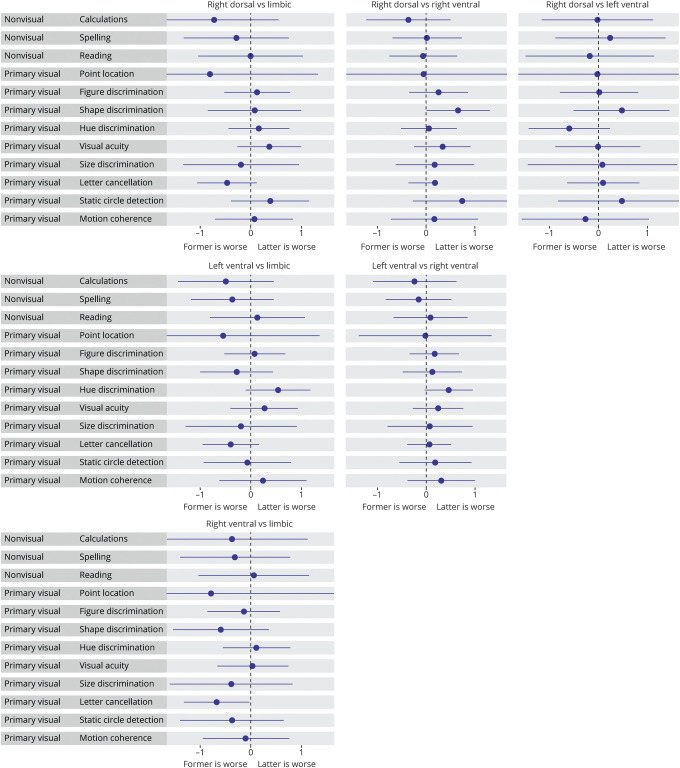
Associations between factor expression and nonvisual dominant parietal and primary visual processing functions
Primary visual processing was negatively associated with right-ventral compared to left-ventral (hue discrimination: β [CI] = −0.59 [−1.23 to 0.04], p = 0.048 uncorrected), limbic (letter cancellation: β [CI] = −0.59 [−1.16 to −0.03], p = 0.028 uncorrected), and right-dorsal factor expression (shape discrimination: β [CI] = −0.59 [−1.18 to 0.00], p = 0.027 uncorrected). With regard to nonvisual parietal functions, we observed a trend towards worse calculations and spelling scores in the right-dorsal, left-ventral, and right-ventral factors, compared to limbic (figure 5; data available from Dryad, table 2, doi.org/10.5061/dryad.jdfn2z37p).
Figure 5. Associations between factor expressions and neuropsychological tests assessing dominant parietal and primary visual functions.
The forest plots contain relative cross-sectional effects from linear regression models. As in each model, one of the factors is implicitly modeled, one of the factors serves as a reference to assess effects of all the others, and each subplot displays results from the different factor comparisons. Lines indicate the 95% confidence intervals and a significant effect (uncorrected for multiple comparisons) is denoted by confidence intervals not including x = 0. This plot only includes participants from the University of California, San Francisco (UCSF) cohort as tests assessing nonvisual/dominant parietal functions were only available in the UCSF cohort.
Associations between atrophy and MMSE, memory, and executive and language functioning
Beyond the visual processing domains, we examined associations between factor expressions and memory and executive and language functions, as well as global cognition measured by MMSE. Across verbal learning, letter fluency, digit span, and category fluency tests, we found negative associations with the limbic factor compared to the other factors. For MMSE, we also found more associations with limbic factor expression compared to the other factors, while associations between the extralimbic factors were sparse (figure 4).
Case series of participants corresponding to posterior cortical atrophy variants
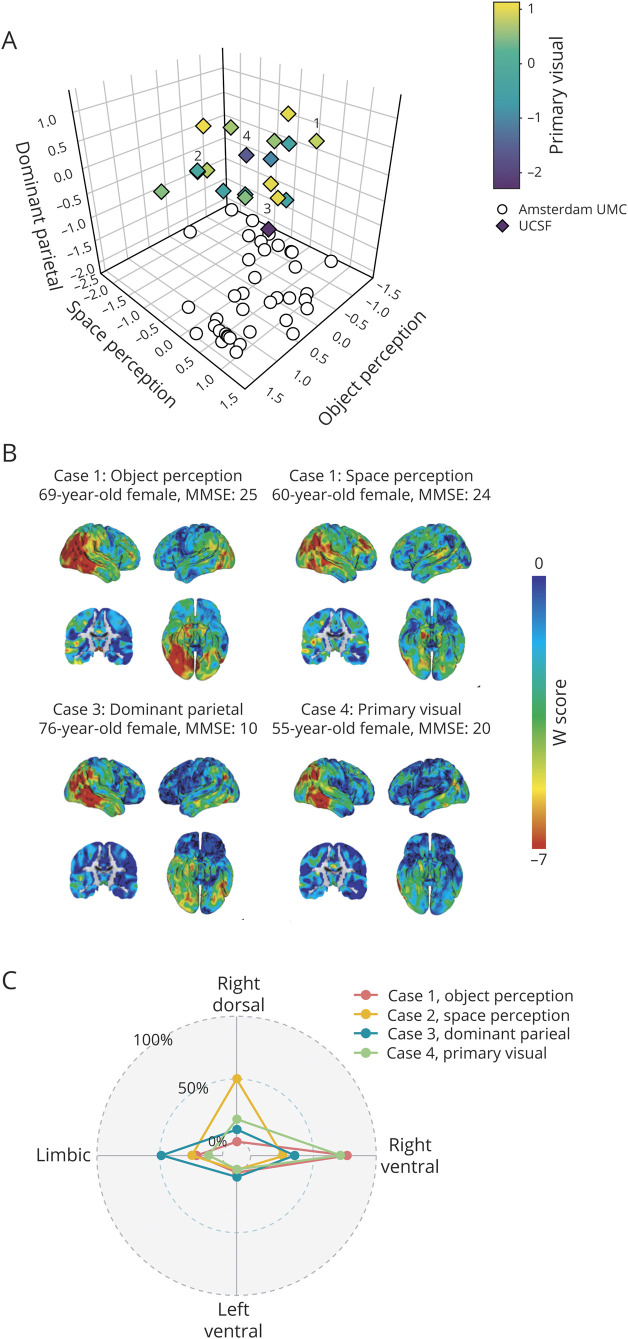
Individual factor compositions indicated that the majority of participants express atrophy across multiple factors rather than in one primarily and our results therefore do not support the notion that discrete phenotypical variants of PCA are common. To provide an explanation for the description of these variants in earlier studies, we include a case description of 4 participants who were selected based on an isolated relative impairment in one of the cognitive domains most relevant to PCA: object perception, space perception, nonvisual/dominant parietal functions, or primary visual processing (figure 6A). These scores were obtained by averaging scores across neuropsychological tests within each domain. From figure 6A it is evident that, similar to what we observed for the factor compositions, most participants have impairments across multiple cognitive domains, and only a few had a clinical phenotype that was characterized by isolated impairments (see annotated markers in figure 6A). We outlined the clinical and radiologic characteristics of these 4 cases in figure 6, B and C. Case 1 was selected based on pronounced object perception impairment and showed an atrophy pattern compatible with the occipitotemporal (ventral) variant of PCA described in literature, and this participant mainly expressed the right-ventral factor (80% loading). Case 2 was selected based on pronounced space perception deficits and showed atrophy that matches with the temporoparietal (dorsal) variant, and this participant had a relatively high right-dorsal factor expression (50% loading). Case 3 was selected based on low scores on nonvisual, dominant parietal functions (but also showed low scores on the other 3 domains) yet did not display prominent parietal atrophy, and factor expression was mainly in the limbic factor (50% loading). Finally, case 4 was selected based on low primary visual processing scores and had high right-ventral factor expression (70% loading) but the atrophy pattern was not markedly caudal (figure 6, B and C).
Figure 6. Case series of extreme clinical phenotypes.
(A) 4D plot with scores on the object perception, space perception, and nonvisual/dominant parietal domains on the x, y, and z axes. Primary visual processing scores are displayed by the color gradient of the markers. As the Amsterdam University Medical Center (UMC) cohort did not include any nonvisual/dominant parietal or primary visual processing tests, these scores are projected onto the x and y axes, and colorless. We selected cases with isolated relative impairments, one for each domain. Selected cases within this distribution are annotated by numbers: 1 = object perception, 2 = space perception, 3 = dominant parietal, and 4 = primary visual. Only participants with scores on all 4 domains (from the University of California, San Francisco [UCSF] sample) were eligible for selection. The plot in this panel is better viewed in the provided interactive .html format (data available from Dryad, figure-e6A, doi.org/10.5061/dryad.jdfn2z37p). (B) Clinical characteristics of the 4 selected cases as well as the regional spread of atrophy indicated by voxelwise W-scores. Lower W-scores represent more atrophy. (C) Radar plot displays individual factor compositions of the 4 selected cases. MMSE = Mini-Mental State Examination.
Discussion
In the present study, we employed a data-driven approach to identify phenotypical variants of PCA by detecting latent atrophy factors and assessing associations between these factors and cognitive domains known to be affected in PCA (i.e., object perception, space perception, nonvisual dominant parietal and primary visual processing). A Bayesian modeling framework was used to detect atrophy patterns that covary across participants and identified 4 distinct but partially overlapping atrophy factors: right-dorsal, right-ventral, left-ventral, and limbic. When we evaluated individual expressions of these atrophy factors, we observed that the vast majority of participants expressed multiple factors rather than primarily expressing only a single factor. This indicates that most participants have atrophy that extends across multiple regions rather than focal atrophy confined to a single region. Furthermore, we found that object perception was associated with atrophy that predominantly affects the right-ventral region and that space perception was associated with atrophy that predominantly affects right-dorsal and right-ventral regions (compared to left-ventral). Primary visual functions were also associated with atrophy that predominantly affects the right-ventral factor but we found no associations for dominant-parietal functions. These findings indicate that atrophy patterns within participants were associated with particular cognitive functions, mostly in line with known brain–behavior relationships. However, similar to expressions across atrophy factors, scores across cognitive domains revealed that most participants had impairments on multiple visual processing and nonvisual parietal functions, rather than being primarily impaired in one. Four participants selected based on a relative impairment on a single domain revealed individual atrophy patterns that were largely in accordance with the hypothesized variants of PCA, but these cases constituted the exception rather than the norm and even then were not mutually exclusive. Taken together, our Bayesian modeling approach captures atrophy factors that are in accordance with the most well-described phenotypical variants of PCA (i.e., dorsal and ventral variants) and these brain regions are individually associated with specific clinical features. However, the majority of participants display a constellation of affected brain regions and symptoms, and classification into 4 overarching phenotypical variants is, therefore, unlikely to be clinically useful.
Our results are in line with previous studies with more limited sample sizes that have tried to identify PCA variants using neuroimaging techniques.16,17 A diffusion tensor imaging investigation of PCA found that all investigated participants had ventral white matter tract abnormalities (e.g., inferior longitudinal fasciculus), while some had additional dorsal stream abnormalities.17 Another study assessed regional cortical thickness in object vs space perception subgroups and found trends towards thinner cortex in focal dorsal and ventral areas, respectively. However, these differences were subtle and the substantial anatomical overlap between subgroups indicates that there was insufficient evidence for the existence of distinct PCA variants.16 Here, we found that right-ventral atrophy was negatively associated with object perception compared to right-dorsal atrophy but dorsal vs ventral associations were not detected with regard to space perception. We did observe that both object and space perception, as well as primary visual functions, were associated with the right-ventral and right-dorsal factors compared to the left-ventral factor. Whereas higher-order visual processing is not clearly lateralized,29 it has consistently been found that PCA presents with a tendency towards right-lateralized atrophy.3,30 Because visual processing impairments are the hallmark feature of PCA, a link to vulnerability of the right hemisphere is conceivable.
Deficits in visual processing functions are, by definition, the most prominent features of PCA, but memory, executive, and language function impairment are also often observed, although these impairments—especially language18,31,32—are often only present in the later stages of PCA.7,33 We found that individuals for whom the majority of their atrophy was specific to limbic regions had worse memory, executive, and language scores compared to those for whom the majority of the atrophy was localized to the extralimbic factors. It has been reported in a previous study that performance on verbal learning is associated with volume of the inferior parietal lobules in PCA,34 rather than the medial temporal lobe, which contrasts with our findings. However, individuals with atrophy that predominantly affected the limbic regions also showed worse global cognition, indicating that disproportionate limbic atrophy indicates worse cognition overall. As this factor was also the only one associated with global atrophy, it seems that high limbic factor expression might be a feature of late-stage PCA, which is in accordance with findings reported in previous studies.35,36
Classical neuroscientific literature describes the ventral and dorsal pathways of visual processing, which together constitute the “2-streams hypothesis,” sometimes called the “what” and “where” pathways.29 These processing streams respectively encompass occipitotemporal and temporoparietal areas, and one may assume that atrophy in one of these regions leads to specific clinical phenotypes in PCA. In the present study, we found 3 distinct (although partly overlapping) atrophy factors that roughly corresponded to the ventral and dorsal visual processing pathways, namely the right- and left-ventral factors, and the right-dorsal factor. We found that these factors were, in accordance with the 2-streams hypothesis, associated with object and space perception, although space perception was not discretely associated with the (right-)dorsal factor. It might be that our method was not able to delineate this association accurately, but a previous study implementing the same approach to structural MRI data in a mild cognitive impairment and AD dementia population did find distinct brain–behavior associations in a biologically plausible manner.27 Moreover, previous examinations that have focused on dorsal vs ventral neuroimaging features and clinical symptoms16,17 have been unable to provide definitive results. The explanation for this may lie in the fact that individuals do not exclusively express atrophy in either the dorsal or the ventral regions, as illustrated by the factor compositions in the present study. Likewise, clinical impairments are also not limited to a single domain but spread across multiple domains. This combination indicates that the dorsal and ventral stream variants are either too rare or too much overlapping to be discernible.
Evidence for the existence of the other 2, admittedly less well-defined, variants of PCA described in literature (i.e., the caudal and dominant parietal variants) is even more limited. The dominant parietal variant has been proposed to be characterized by prominent impairments in nonvisual parietal functions (i.e., agraphia, alexia, and apraxia), symptoms that are often present in PCA.7,18,20 In the present study, we were unable to discern clear associations between any of the atrophy factors and dominant-parietal functions. Furthermore, outlining the impairments across cognitive domains revealed that none of the participants had a clearly isolated nonvisual dominant parietal impairment, and the case we selected also had severe impairments on other domains and a low MMSE. The discrepancy between our findings and earlier studies, which formed the basis for the hypothesized dominant parietal variant, may again be that these were based on small studies or single case studies selected based on this particular phenotype. Also, these previous studies primarily focused on apraxia (not assessed in the present study).13–15 Another possible explanation for why we did not observe patients with isolated impairments in nonvisual dominant parietal functions is that these individuals might have been less likely to be included because the clinical criteria for PCA rely primarily on prominent visual features.7,20,37
We did detect atrophy factors that might be related to the caudal variant of PCA, characterized by primary visual processing function deficits, namely the left- and right-ventral factors. These factors encompassed occipital regions proposed to be associated with the caudal variant. However, these factors also included inferior temporal and inferior parietal regions, so we were unable to discern a clearly caudal factor associated with primary visual processing. An explanation for the lack of clear caudal factors may be found in earlier observations that PCA starts in the most posteriorly located brain regions and then spreads to lateral parieto-temporal cortices,38,39 following a posterior-to-anterior pathway in line with the network-based degeneration hypothesis.40 This suggests that descriptions of isolated impairments on primary visual processing and caudally located atrophy might be based on individuals in an early stage of the disease, which would be in line with the observation that all participants with PCA show impairments in at least one primary visual domain.16 The caudal variant may, therefore, represent an early disease stage–related phenomenon, rather than a distinct variant of PCA.
The main strengths of the present study include the relatively large, multicenter sample of extensively phenotyped participants with PCA. Furthermore, our data-driven approach allowed atrophy factors to be partly overlapping instead of completely distinct and allowed participants to express each atrophy factor to a certain degree. These characteristics make this approach more biologically plausible than a priori categorization of participants into mutually exclusive subgroups or selection of regions of interest to investigate. Whereas this results in partly overlapping atrophy patterns, complicating the interpretation of the associations between factor expressions and cognition, functionally related pathways are likely also structurally interconnected and would be expected to degenerate together when pathology occurs. Another strength is that we excluded participants with negative Aβ biomarkers in order to increase the likelihood that individuals had PCA due to AD,5,6,41 thereby minimizing possible confounding effects of differences in underlying pathology. However, this also constitutes a possible limitation as individuals with PCA due to non-AD pathology may show a different pattern of neurodegeneration41 and we were not able to assess whether non-AD pathology could have formed the basis for some of the hypothesized phenotypical variants of PCA. Another possible limitation is the retrospective inclusion of participants assessed from 2000 to 2017, which resulted in participants being selected based on different clinical criteria.6,7,20 However, there were no associations between date of inclusion and atrophy factor expression (range: r = −0.14 to 0.18, all p > 0.05; data available from Dryad, figure 8, doi.org/10.5061/dryad.jdfn2z37p). Furthermore, our sample (n = 119), while large for a PCA cohort, was relatively small, and we performed many comparisons. After correction for multiple comparisons, several of the associations between factor expressions and neuropsychological tests lost statistical significance. However, we have included effect sizes in the results to allow the reader to draw their own conclusions. Finally, performance on some of our tests is interrelated (data available from Dryad, table 3, doi.org/10.5061/dryad.jdfn2z37p) and individual tests might assess multiple aspects of visual functioning. For example, the visuo-perceptive fragmented letters test also includes a visuospatial component, thus future studies with more specific tests are warranted.
Akin to classifying patients with AD into atypical variants (e.g., logopenic variant primary progressive aphasia or the dysexecutive/behavioral variant42,43), subtyping PCA into variants constrains a wealth of clinical and radiologic variability into categorical entities. These classifications are mostly useful in a clinical setting, to aid in an early and accurate diagnosis and to direct patient care as well as aiding in selection for clinical trials.44 However, when only the extremes of an already relatively rare syndrome are captured by this classification, clinical utility becomes limited and, for clinical purposes, categorizing PCA as a single entity might be sufficiently specific. Elucidating the link between clinical heterogeneity and neurobiological differences may, however, be useful in a research setting to assess mechanisms leading to selective vulnerability in neurodegenerative diseases.45 Hypothetical models of AD suggest that tau aggregation and hypometabolism precede neurodegeneration.46 This indicates that successfully identifying phenotypical variants of PCA may rely on early detection using, e.g., tau-PET or FDG-PET, which have already been shown to distinguish PCA from typical AD.2,30,47,48 Another possible avenue to detect phenotypical variants of PCA may be the assessment of functional connectivity,49 as emerging evidence points to the spread of neurodegeneration along intrinsic functional brain networks. Aside from neuroimaging factors related to regional vulnerability, it has also been shown that genetic risk factors convey a specific risk to PCA.33 In addition, a recent study has found that mathematical and visuospatial learning difficulties are related to visuospatial predominant clinical syndromes, which indicates that neurodevelopment might also be related to vulnerability of specific brain networks that predisposes an individual to show network failure in these systems when neurodegenerative diseases emerge in later life.50
These emerging findings help to elucidate the intricate pathways that eventually result in discrete clinical syndromes and indicate that regional susceptibility to pathology is most likely multifactorial. Considering the interplay between different susceptibility factors, future examinations assessing regional vulnerability will therefore require multimodal assessment with large sample sizes. Owing to the relatively low prevalence of PCA, obtaining sufficient cohorts exclusively containing individuals with PCA will remain challenging. For now, it might be prudent to focus on the entire AD spectrum and examine factors related to particular vulnerability for developing PCA rather than specific variations of this already relatively rare syndrome.
Acknowledgment
The authors thank Sarah Remmers, MSc (Amsterdam UMC) for comments on the neuropsychological data, J.Q. Alida Chen, BSc, for helping to design the figures, and Paul K. Crane, MD, for proofreading the manuscript. Research of the Alzheimer Center Amsterdam is part of the neurodegeneration research program of the Neuroscience Campus Amsterdam. The Alzheimer Center Amsterdam is supported by Alzheimer Nederland and Stichting Amsterdam UMC Fonds. The clinical database structure was developed with funding from Stichting Dioraphte. Wiesje M. van der Flier holds the Pasman chair.
Glossary
- Aβ
β-amyloid
- AD
Alzheimer disease
- CI
confidence interval
- FDR
false discovery rate
- LDA
latent Dirichlet allocation
- MMSE
Mini-Mental State Examination
- PCA
posterior cortical atrophy
- UCSF
University of California, San Francisco
- UMC
University Medical Center
Appendix. Authors
Study funding
Study funded by the National Institute on Aging of the NIH within the US Department of Health and Human Services (R01 AG029672 to P.K.C., PI, R.O. Co-PI), National Institute on Aging grants (R01-AG045611 to G.D.R., P50-AG023501 to B.L.M., H.J.R., and G.D.R.), and John Douglas French Alzheimer's Foundation. B.T.T.Y. is funded by the Singapore National Research Foundation (NRF) Fellowship (Class of 2017). F.B. is supported by the NIHR Biomedical Research Centre at UCLH.
Disclosure
C. Groot, B.T.T. Yeo, J.W. Vogel, X. Zhang, N. Sun, E.C. Mormino, Y.A.L. Pijnenburg, B.L. Miller, H.J. Rosen, and R. La Joie report no disclosures relevant to the manuscript. F. Barkhof reports research support from GE Healthcare, Biogen, Novartis, and TEVA and scientific advisory boards for Roche, Biogen, Merck, Roche, Lundbeck, and IXICO. P. Scheltens and W.M. van der Flier report no disclosures relevant to the manuscript. G.D. Rabinovici reports research support from Avid Radiopharmaceuticals, GE Healthcare, Eli Lilly, and Life Molecular Imaging; scientific advisory boards for Axon Neurosciences, Eisai, Merck, and Roche; and is Associate Editor for JAMA Neurology. R. Ossenkoppele reports no disclosures relevant to the manuscript. Go to Neurology.org/N for full disclosures.
Publication history
This manuscript was previously published as a pre-print in bioRxiv: doi: https://doi.org/10.1101/679225. Received by Neurology October 25, 2019. Accepted in final form April 6, 2020.
References
- 1.Benson DF, Davis RJ, Snyder BD. Posterior cortical atrophy. Arch Neurol 1988;45:789–793. [DOI] [PubMed] [Google Scholar]
- 2.Whitwell JL, Jack CR, Kantarci K, et al. Imaging correlates of posterior cortical atrophy. Neurobiol Aging 2007;28:1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crutch SJ, Lehmann M, Schott JM, Rabinovici GD, Rossor MN, Fox NC. Posterior cortical atrophy. Lancet Neurol 2012;11:170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ossenkoppele R, Cohn-Sheehy BI, La Joie R, et al. Atrophy patterns in early clinical stages across distinct phenotypes of Alzheimer's disease. Hum Brain Mapp 2015;36:4421–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renner JA, Burns JM, Hou CE, McKeel DW, Storandt M, Morris JC. Progressive posterior cortical dysfunction: a clinicopathologic series. Neurology 2004;63:1175–1180. [DOI] [PubMed] [Google Scholar]
- 6.Tang-Wai DF, Graff-Radford NR, Boeve BF, et al. Clinical, genetic, and neuropathologic characteristics of posterior cortical atrophy. Neurology 2004;63:1168–1174. [DOI] [PubMed] [Google Scholar]
- 7.Crutch SJ, Schott JM, Rabinovici GD, et al. Consensus classification of posterior cortical atrophy. Alzheimers Dement 2017;13:870–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross SJ, Graham N, Stuart-Green L, et al. Progressive biparietal atrophy: an atypical presentation of Alzheimer's disease. J Neurol Neurosurg Psychiatry 1996;61:388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galton CJ, Patterson K, Xuereb JH, Hodges JR. Atypical and typical presentations of Alzheimer's disease: a clinical, neuropsychological, neuroimaging and pathological study of 13 cases. Brain 2000;123:484–498. [DOI] [PubMed] [Google Scholar]
- 10.Grossi D, Soricelli A, Ponari M, et al. Structural connectivity in a single case of progressive prosopagnosia: the role of the right inferior longitudinal fasciculus. Cortex 2014;56:111–120. [DOI] [PubMed] [Google Scholar]
- 11.Chan D, Crutch SJ, Warrington EK. A disorder of colour perception associated with abnormal colour after-images: a defect of the primary visual cortex. J Neurol Neurosurg Psychiatry 2001;71:515–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine DN, Lee JM, Fisher CM. The visual variant of Alzheimer's disease: a clinicopathologic case study. Neurology 1993;43:305. [DOI] [PubMed] [Google Scholar]
- 13.Aharon-Peretz J, Israel O, Goldsher D, Peretz A. Posterior cortical atrophy variants of Alzheimer's disease. Dement Geriatr Cogn Disord 1999;10:483–487. [DOI] [PubMed] [Google Scholar]
- 14.Green RC, Goldstein FC, Mirra SS, Alazraki NP, Baxt JL, Bakay RA. Slowly progressive apraxia in Alzheimer's disease. J Neurol Neurosurg Psychiatry 1995;59:312–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Renzi E. Slowly progressive visual agnosia or apraxia without dementia. Cortex 1986;22:171–180. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann M, Barnes J, Ridgway GR, et al. Basic visual function and cortical thickness patterns in posterior cortical atrophy. Cereb Cortex 2011;21:2122–2132. [DOI] [PubMed] [Google Scholar]
- 17.Migliaccio R, Agosta F, Scola E, et al. Ventral and dorsal visual streams in posterior cortical atrophy: a DT MRI study. Neurobiol Aging 2012;33:2572–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMonagle P, Deering F, Berliner Y, Kertesz A. The cognitive profile of posterior cortical atrophy. Neurology 2006;66:331–338. [DOI] [PubMed] [Google Scholar]
- 19.van der Flier WM, Scheltens P. Amsterdam dementia cohort: performing research to optimize care. J Alzheimers Dis 2018;62:1091–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendez MF, Ghajarania M, Perryman KM. Posterior cortical atrophy: clinical characteristics and differences compared to Alzheimer's disease. Dement Geriatr Cogn Disord 2002;14:33–40. [DOI] [PubMed] [Google Scholar]
- 21.Tijms BM, Willemse EAJ, Zwan MD, et al. Unbiased approach to counteract upward drift in cerebrospinal fluid amyloid-β 1–42 analysis results. Clin Chem 2018;64:576–585. [DOI] [PubMed] [Google Scholar]
- 22.Rabinovici GD, Furst AJ, Alkalay A, et al. Increased metabolic vulnerability in early-onset Alzheimer's disease is not related to amyloid burden. Brain 2010;133:512–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd CD, Tierney M, Wassermann EM, et al. Visuoperception test predicts pathologic diagnosis of Alzheimer disease in corticobasal syndrome. Neurology 2014;83:510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer JH, Jurik J, Sha SJ, et al. Distinctive neuropsychological patterns in frontotemporal dementia, semantic dementia, and Alzheimer disease. Cogn Behav Neurol 2003;16:211–218. [DOI] [PubMed] [Google Scholar]
- 25.Groot C, Van Loenhoud ACAC, Barkhof F, et al. Differential effects of cognitive reserve and brain reserve on cognition in Alzheimer disease. Neurology 2018;90:e149–e156. [DOI] [PubMed] [Google Scholar]
- 26.La Joie R, Perrotin A, Barré L, et al. Region-specific hierarchy between atrophy, hypometabolism, and β-amyloid (Aβ) load in Alzheimer's disease dementia. J Neurosci 2012;32:16265–16273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Mormino EC, Sun N, Sperling RA, Sabuncu MR, Yeo BTT. Bayesian model reveals latent atrophy factors with dissociable cognitive trajectories in Alzheimer's disease. Proc Natl Acad Sci USA 2016;113:E6535–E6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun N, Mormino EC, Chen J, Sabuncu MR, Yeo BTT. Multi-modal latent factor exploration of atrophy, cognitive and tau heterogeneity in Alzheimer's disease. Neuroimage 2019;201:116043. [DOI] [PubMed] [Google Scholar]
- 29.Ungerleider LG, Haxby JV. “What” and “where” in the human brain. Curr Opin Neurol 1994;4:157–165. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann M, Ghosh PM, Madison C, et al. Diverging patterns of amyloid deposition and hypometabolism in clinical variants of probable Alzheimer's disease. Brain 2013;136:844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schott JM, Crutch SJ. Posterior cortical atrophy. Contin Lifelong Learn Neurol 2019;25:52–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crutch SJ, Lehmann M, Warren JD, Rohrer JD. The language profile of posterior cortical atrophy. J Neurol Neurosurg Psychiatry 2013;84:460–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schott JM, Crutch SJ, Carrasquillo MM, et al. Genetic risk factors for the posterior cortical atrophy variant of Alzheimer's disease. Alzheimers Dement 2016;12:862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed S, Loane C, Bartels S, et al. Lateral parietal contributions to memory impairment in posterior cortical atrophy. Neuroimage Clin 2018;20:252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips JS, Da Re F, Irwin DJ, et al. Longitudinal progression of grey matter atrophy in non-amnestic Alzheimer's disease. Brain 2019;142:1701–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann M, Barnes J, Ridgway GR, et al. Global gray matter changes in posterior cortical atrophy: a serial imaging study. Alzheimers Dement 2012;8:502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai PH, Teng E, Liu C, Mendez MF. Posterior cortical atrophy. Am J Alzheimers Dis Other Demen 2011;26:413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennedy J, Lehmann M, Sokolska MJ, et al. Visualizing the emergence of posterior cortical atrophy. Neurocase 2012;18:248–257. [DOI] [PubMed] [Google Scholar]
- 39.Firth NC, Primativo S, Marinescu RV, et al. Longitudinal neuroanatomical and cognitive progression of posterior cortical atrophy. Brain 2019;142:2082–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron 2009;62:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montembeault M, Brambati SM, Lamari F, et al. Atrophy, metabolism and cognition in the posterior cortical atrophy spectrum based on Alzheimer's disease cerebrospinal fluid biomarkers. Neuroimage Clin 2018;20:1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorno-Tempini ML, Hillis AE, Weintraub S, et al. Classification of primary progressive aphasia and its variants. Neurology 2011;76:1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ossenkoppele R, Pijnenburg YAL, Perry DC, et al. The behavioural/dysexecutive variant of Alzheimer's disease: clinical, neuroimaging and pathological features. Brain 2015;138:2732–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheltens NME, Tijms BM, Koene T, et al. Cognitive subtypes of probable Alzheimer's disease robustly identified in four cohorts. Alzheimers Dement 2017;13:1226–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattsson N, Schott JM, Hardy J, Turner MR, Zetterberg H. Selective vulnerability in neurodegeneration: insights from clinical variants of Alzheimer's disease. J Neurol Neurosurg Psychiatry 2016;87:1000–1004. [DOI] [PubMed] [Google Scholar]
- 46.Jack CR, Holtzman DM. Biomarker modeling of Alzheimer's disease. Neuron 2013;80:1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ossenkoppele R, Schonhaut DR, Baker SL, et al. Tau, amyloid, and hypometabolism in a patient with posterior cortical atrophy. Ann Neurol 2015;77:338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Day GS, Gordon BA, Jackson K, et al. Tau-PET binding distinguishes patients with early-stage posterior cortical atrophy from amnestic Alzheimer disease dementia. Alzheimer Dis Assoc Disord 2017;31:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehmann M, Madison C, Ghosh PM, et al. Loss of functional connectivity is greater outside the default mode network in nonfamilial early-onset Alzheimer's disease variants. Neurobiol Aging 2015;36:2678–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller ZA, Rosenberg L, Santos-Santos MA, et al. Prevalence of mathematical and visuospatial learning disabilities in patients with posterior cortical atrophy. JAMA Neurol 2018;75:728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The code for the Bayesian modeling approach is publicly available at github.com/ThomasYeoLab/CBIG/tree/master/stable_projects/disorder_subtypes/Zhang2016_ADFactors. Anonymized data used in the present study may be available upon request to the corresponding author.