Abstract
Objective
To develop and evaluate a model for staging cortical amyloid deposition using PET with high generalizability.
Methods
Three thousand twenty-seven individuals (1,763 cognitively unimpaired [CU], 658 impaired, 467 with Alzheimer disease [AD] dementia, 111 with non-AD dementia, and 28 with missing diagnosis) from 6 cohorts (European Medical Information Framework for AD, Alzheimer's and Family, Alzheimer's Biomarkers in Daily Practice, Amsterdam Dementia Cohort, Open Access Series of Imaging Studies [OASIS]-3, Alzheimer’s Disease Neuroimaging Initiative [ADNI]) who underwent amyloid PET were retrospectively included; 1,049 individuals had follow-up scans. With application of dataset-specific cutoffs to global standard uptake value ratio (SUVr) values from 27 regions, single-tracer and pooled multitracer regional rankings were constructed from the frequency of abnormality across 400 CU individuals (100 per tracer). The pooled multitracer ranking was used to create a staging model consisting of 4 clusters of regions because it displayed a high and consistent correlation with each single-tracer ranking. Relationships between amyloid stage, clinical variables, and longitudinal cognitive decline were investigated.
Results
SUVr abnormality was most frequently observed in cingulate, followed by orbitofrontal, precuneal, and insular cortices and then the associative, temporal, and occipital regions. Abnormal amyloid levels based on binary global SUVr classification were observed in 1.0%, 5.5%, 17.9%, 90.0%, and 100.0% of individuals in stage 0 to 4, respectively. Baseline stage predicted decline in Mini-Mental State Examination (MMSE) score (ADNI: n = 867, F = 67.37, p < 0.001; OASIS: n = 475, F = 9.12, p < 0.001) and faster progression toward an MMSE score ≤25 (ADNI: n = 787, hazard ratio [HR]stage1 2.00, HRstage2 3.53, HRstage3 4.55, HRstage4 9.91, p < 0.001; OASIS: n = 469, HRstage4 4.80, p < 0.001).
Conclusion
The pooled multitracer staging model successfully classified the level of amyloid burden in >3,000 individuals across cohorts and radiotracers and detects preglobal amyloid burden and distinct risk profiles of cognitive decline within globally amyloid-positive individuals.
PET can identify amyloid-β (Aβ) plaques in vivo with high sensitivity and specificity in clinical populations.1 The clinical standard for determining amyloid abnormality is a dichotomous visual assessment, while classification based on thresholds of the global standardized uptake value ratio (SUVr) is generally used for research purposes. However, both procedures might miss emerging amyloid pathology, and the extent of the pathologic burden is generally disregarded.2,3
Recently, staging of amyloid burden with PET imaging has shown promising results to address the limitations of signal dichotomization.4–6 For example, by separately classifying the cortical and striatal PET signal, a system of 3 stages of amyloid deposition has been proposed, which corresponded to distinct risk of cognitive decline.4 A more fine-grained 4-stage model was also constructed by a separate group based on [18F]florebetapir PET data from cognitively unimpaired (CU) individuals, succesfully staging 410 (98%) individuals from the same cohort.5 More recently, a longitudinally valid PET staging model was developed using cerebrospinal fluid (CSF) as reference, and validated in an independent cohort using a different radiotracer.6 However, the model construction could not be replicated, and contrasting CSF/PET groups could have hampered the ability to identify more fine-grained stages.6 Therefore, further evidence is required regarding the generalizability of single-tracer models to multitracer studies.
The present work aimed to construct and apply a data-driven and generalizable PET-based model for staging cortical amyloid burden. Cross-sectional and longitudinal relationships were assessed between amyloid stages and cognitive status, age, genetic risk, CSF, and risk of future cognitive decline.
Methods
Standard protocol approvals, registrations, and patient consents
The protocol, patient information, consent form, and other relevant study documentation were approved by the Ethics Committee or Institutional Review Board of each site before study initiation. The studies were performed in accordance with the Declaration of Helsinki and were consistent with Good Clinical Practice. Before enrollment, all patients provided written informed consent.
Cohorts
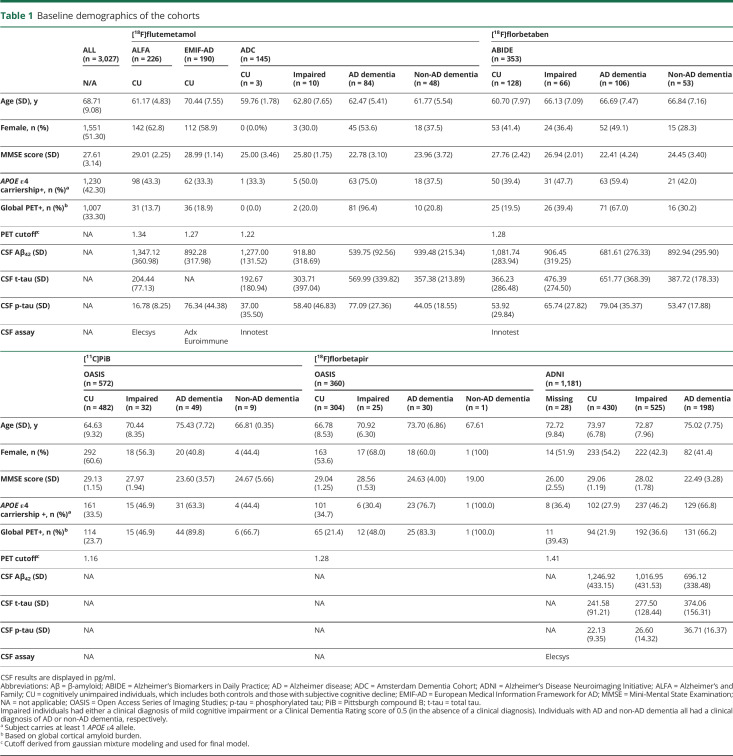
From 6 cohorts, all participants with available amyloid PET scans of sufficient quality for quantification were retrospectively included in this study (table 1). In detail, [18F]flutemetamol scans of 226 CU individuals from the Alzheimer's and Family (ALFA) cohort of the Barcelonaβeta Brain Research center,7 145 memory clinic patients from the Amsterdam Dementia Cohort (ADC) of the Amsterdam University Medical Center,8 and 190 CU individuals from the Innovative Medicine Initiative European Medical Information Framework for AD (EMIF-AD)9 project (emif.eu/emif-ad-2/) were included. [18F]florbetaben scans of 353 memory clinic patients were obtained from the Alzheimer's Biomarkers in Daily Practice (ABIDE) study.10 Regional SUVr values were obtained from [18F]florbetapir scans of 360 individuals and [11C]Pittsburgh compound B (PiB) scans of 572 individuals from the Open Access Series of Imaging Studies (OASIS)-3 dataset (oasis-brains.org/).11 Regional SUVr values from [18F]florbetapir scans of 1,179 individuals were included from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu/). The ADNI study was launched in 2003 as a public-private partnership led by principal investigator Michael W. Weiner, MD. The primary goal of ADNI is to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment and early Alzheimer disease (AD).
Table 1.
Baseline demographics of the cohorts
Across cohorts, 3,027 individuals (1,763 CU, 658 impaired, 467 with AD dementia, 111 with non-AD dementia, and 28 with missing diagnosis at time of baseline PET) were included. In total, 4,783 PET scans were included; 1,049 individuals had at least 2 amyloid PET scan sessions (OASIS and ADNI), with an average of 2.7 PET scans per subject (range 2–5). For each study, missing data were assumed to be missing at random.
Image acquisition and processing
[18F]flutemetamol scans from the ALFA cohort consisted of 4 frames (4 × 5 minutes) acquired 90 to 110 minutes after injection. Images were checked for motion, and PET and accompanying structural T1-weighted MRIs were warped into Montreal Neurological Institute space using SPM12. [18F]flutemetamol scans from the ADC studies and [18F]florbetaben scans from ABIDE were processed as described previously,12 with static scans consisting of 4 frames (4 × 5 minutes) acquired 90 to 110 minutes after injection. In addition, [18F]flutemetamol EMIF-AD scans were acquired with a dual-time-window protocol (0–30 minutes after injection, 60-minute break, 90–110 minutes after injection).13 Images were checked for motion, and accompanying structural T1-weighted MRIs were coregistered to PET with Vinci software (Max Planck Institute for Neurologic Research, Cologne, Germany) and then warped into Montreal Neurological Institute space using SPM12. [18F]florbetapir (50–70 minutes after injection) and [11C]PiB (30–60 minutes after injection) data from the OASIS platform were processed with FreeSurfer and the PET Unified Pipeline (github.com/ysu001/PUP).14
Acquisition and standardized preprocessing steps of MR and PET data in ADNI have been reported previously and are described in detail on the ADNI website (adni.loni.usc.edu/methods/pet-analysis-method/pet-analysis). Briefly, [18F]florbetapir PET scans consisted of 4 frames (4 × 5 minutes) acquired 50 to 70 minutes after injection and were processed with FreeSurfer.
The Desikan-Killiany atlas was used to extract regional SUVr values normalized to cerebellar gray matter.15 Small regions enclosed within high-signal areas were excluded due to risk for spill-in and misregistration errors (pericalcarine and banks superior temporal), and small/intermediate regions were merged (volume-weighted average) to reduce noise-related variability. The resulting 27 regions were anterior, isthmus, and posterior cingulate cortices, inferior frontal gyrus (pars opercularis, pars triangularis, and pars orbitalis), lateral and medial orbitofrontal cortex, middle frontal gyrus (caudal and rostral middle frontal), superior frontal gyrus, frontal pole, inferior temporal gyrus, middle temporal gyrus, superior temporal gyrus (superior temporal and transverse temporal), fusiform gyrus, entorhinal cortex, parahippocampal gyrus, lingual gyrus, lateral occipital gyrus temporal pole, insula, inferior parietal gyrus, supramarginal gyrus, precuneus, superior parietal gyrus, precentral gyrus, postcentral gyrus, paracentral gyrus, and cuneus. Global SUVr was determined as a volume-weighted average of these 27 regions.
Model development
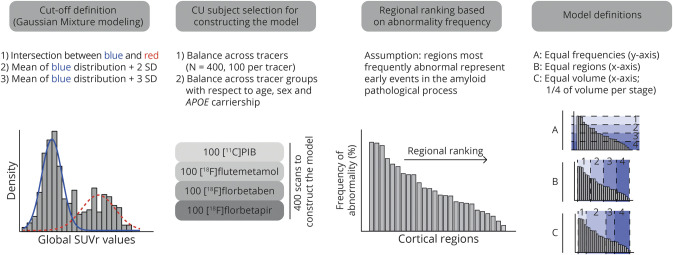
To construct a PET-based cortical amyloid staging model, regions were ranked as suggested in Grothe et al.,5 assuming that those most frequently abnormal in a CU population reflect early events in the pathologic process, similar to how pathology studies are performed.16,17
Gaussian mixture modeling (mixtools and AdaptGauss packages from R statistical software program) was used to identify 2 distributions from global cortical SUVr values, and on the basis of each complete cohort, tracer and cohort specific cutoffs were determined from the mean plus 2 SDs from the gaussian (i.e., normal) distribution corresponding to the amyloid-negative group (data available from Dryad, figure S1, doi.org/10.5061/dryad.7wm37pvp9).
Using an in-house algorithm for random optimal allocation subsampling, we randomly selected 400 baseline scans balanced across tracers (i.e., 100 per tracer) from the CU group for model construction to minimize demographic differences (i.e., APOE ε4 carriership, age, and sex) (table S1 available from Dryad, doi:10.5061/dryad.7wm37pvp9).
Single-tracer regional ranking
First, 4 different regional rankings were computed for each radiotracer (n = 100 per tracer) to assess comparability. The cohort- and tracer-specific cutoffs were applied to each region, and these were ranked according to the frequency of abnormality (percent of individuals for whom the SUVr from their region was larger than the cutoff) across each radiotracer group. Correlations between the 4 rankings ([11C]PiB, [18F]flutemetamol, [18F]florbetaben, and [18F]florbetapir) were assessed by Spearman rank correlation analysis.
Multitracer regional ranking
Next, the complete selection of 400 baseline scans from CU individuals was pooled to create a multitracer regional ranking as per above. Agreement between the multitracer and each of the single-tracer rankings was also assessed.
Staging model
To overcome differences between single-tracer rankings identified in the previous steps, the model construction was performed on the basis of the pooled multitracer data from 400 CU individuals.
In the construction step, we assessed the impact of strategies for cutoff and stage definition on the applicability of the model. Therefore, 2 additional data-driven cutoffs were defined as (1) the Bayes optimal classifier (i.e., intersection between the 2 distributions) and (2) mean plus 3 SD from the gaussian distribution corresponding to the amyloid-negative group (figure S1 available from Dryad, doi:10.5061/dryad.7wm37pvp9). Subsequently, these additional cutoffs were also used to rank regions according to the frequency of abnormality across the 400 CU individuals. Finally, 4 stages of amyloid burden were defined on the basis of 3 independent strategies: (1) equal frequencies: regions were grouped in 4 categories, each with equal frequencies5; (2) equal number of regions: regions were grouped so that each stage had an equal number of regions; and (3) equal volume: regions were grouped such that each stage contained one-fourth of the total cortical brain volume.
The steps described above (figure 1) resulted in 9 candidate staging models (data available from Dryad, figure S2, doi.org/10.5061/dryad.7wm37pvp9). Furthermore, to assess the impact of known confounding factors (e.g., spill-in, partial volume effect and cerebral blood flow effect) on the regional ranking, the regional ranking was repeated for available partial volume corrected SUVr (EMIF-AD) and distribution volume ratio (DVR) values (EMIF-AD and OASIS) and reported in figures S3 and S4 in the supplementary results in Dryad.18
Figure 1. Construction of cortical amyloid staging models.
Schematic representation of the work flow for constructing 9 staging models. First, 3 different data-driven cutoffs per cohort were determined with gaussian mixture modeling and each entire cohort. Then, 400 of all cognitively unimpaired (CU) individuals were selected for model construction using a iterative algorithm, followed by regional ranking of the cortical regions in their frequency of abnormality across these people. Finally, 3 definitions of stages were applied to the ranking. SUVr = standardized uptake value ratio.
Model application and assessment
All scans (n = 4,783) were classified into 5 stages (0–4) according to all 9 models. A stage was attributed when >50% of the encompassed regions displayed SUVr above the cutoff.5 In addition, the model was hierarchical by design; i.e., higher stages are achieved only once the staging conditions are also met for previous stages; otherwise, the scan was considered unclassifiable.
The optimal model used for all subsequent analyses was defined as the one minimizing the number of unclassifiable individuals across cohorts. This performance indicator was chosen to assess the applicability of the model to a heterogeneous dataset.
Statistical analysis
SPSS version 22 (IBM, Armonk, NY) was used for all statistical analyses.
Cross-sectional relationships
We used χ2 tests to assess the distribution of stages across dichotomous amyloid PET status (based on global SUVr), syndromic diagnosis, and APOE ε4 alleles. One-way analysis of covariance was performed to assess the relationship between stages and age, Mini-Mental State Examination (MMSE) score, and CSF measures, with cohort as a covariate and excluding individuals with a clinical diagnosis of non-AD dementia. A Bonferroni post hoc test was used to assess between-stage differences, and effect size was reported according to the Cohen guidelines (η2 = 0.01 small, η2 = 0.06 medium, and η2 = 0.14 large), with significance set at p < 0.05.
CSF measures of Aβ40, Aβ42, phosphorylated tau, and total tau were available in a subset of individuals (table 1). These were standardized to z scores (tau CSF after log transformation due to skewedness) within each dataset based on the mean and SD of CU individuals to correct for cross-cohort differences in assays.
Longitudinal PET
Based on first and last PET scans, individuals were considered stable (no changes in stage or global PET status), progressors (increase in stage or global PET status conversion from normal to abnormal), or reversed (decrease in stage or global PET status conversion from abnormal to normal). From all PET scans, the monotonicity of the trajectory was determined. Differences in follow-up time between stage stable and progressors were tested with one-way analysis of variance.
Longitudinal cognition
Longitudinal cognitive data were available only for subsets of ADNI and OASIS cohorts, and all individuals with non-AD dementia were excluded from these analyses. The effects of (1) baseline stage and (2) global SUVr classification on cognitive decline were investigated with the use of a linear mixed model analysis with MMSE score as outcome measure (corrected for age, sex, clinical diagnosis, and time between repeated measures) and Kaplan-Meier survival analysis with an MMSE score ≤25 as event (excluding individuals with MMSE score ≤25 at baseline). In parallel, a Cox regression analysis was performed to obtain stage- and global status–related hazard ratios. For both the linear mixed model and Kaplan-Meier survival analyses, pairwise comparison with Bonferroni correction was performed to assess between-stage differences.
Data availability
The data that support the findings of this study can be made available on request to the study-specific principal investigator (i.e., ABIDE, EMIF-AD, ADC, ALFA) or are openly available (i.e., ADNI and OASIS open-source databases).
Results
Demographics
Single- and cross-cohort baseline demographics are shown in table 1.
Model development
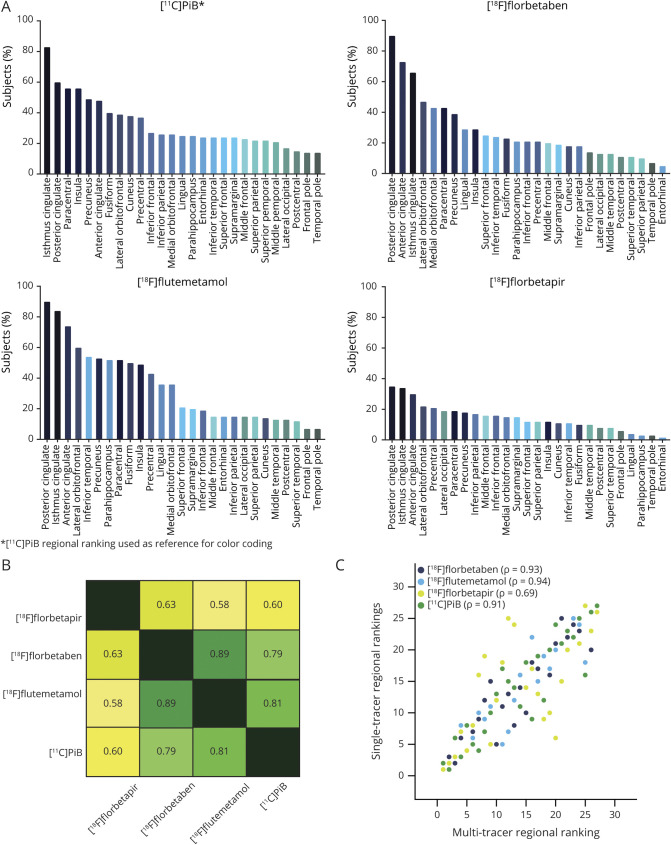
Single-tracer regional rankings can be found in figure 2A. Correlation of the regional rankings was highest between [18F]florbetaben and [18F]flutemetamol (ρ = 0.89), followed by [11C]PiB and [18F]flutemetamol (ρ = 0.81) and then by [11C]PiB and [18F]florbetaben (ρ = 0.79). Lower correlations (ρ = 0.58–0.63) were observed between [18F]florbetapir and all other tracers (figure 2B). The multitracer regional ranking showed improved correlations (ρ = 0.69–0.94) with each single-tracer ranking compared to the correlations of the single-tracer rankings between themselves (figure 2C) and was used to construct the 9 potential staging models (data available from Dryad, figure 2, doi.org/10.5061/dryad.7wm37pvp9).
Figure 2. Regional ranking per radiotracer.
(A) Frequency of regional abnormality across 100 cognitively unimpaired (CU) individuals per radiotracer. (B) Heat map showing Spearman rank correlation (ρ) between the tracer-specific regional rankings. (C) Correlation matrix displaying the correlation between each single-tracer regional ranking and the multitracer regional ranking based on the pooled data of 400 CU individuals. PiB = Pittsburgh compound B.
The model defined on the basis of equal frequencies using the mean plus 2 SD cutoffs of the gaussian distribution corresponding to the amyloid-negative group (table 1) resulted in the smallest amount of unclassifiable individuals (n = 23, 0.8%) and was therefore selected for all subsequent analyses. Of note, 2 of 23 unclassifiable individuals were represented in model construction. The results for the application of all 9 models can be found in data available from Dryad (table S2, doi.org/10.5061/dryad.7wm37pvp9).
Model application and assessment
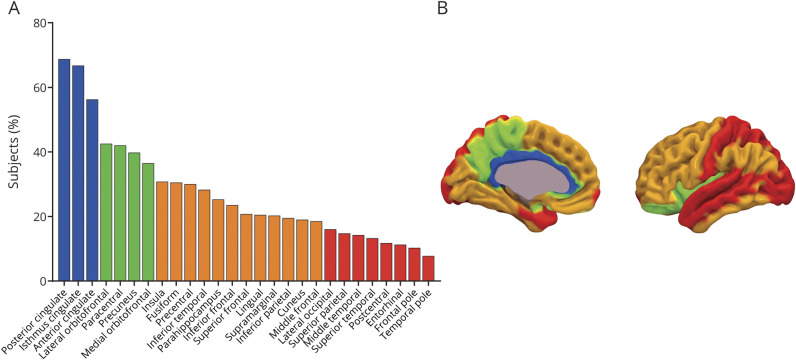
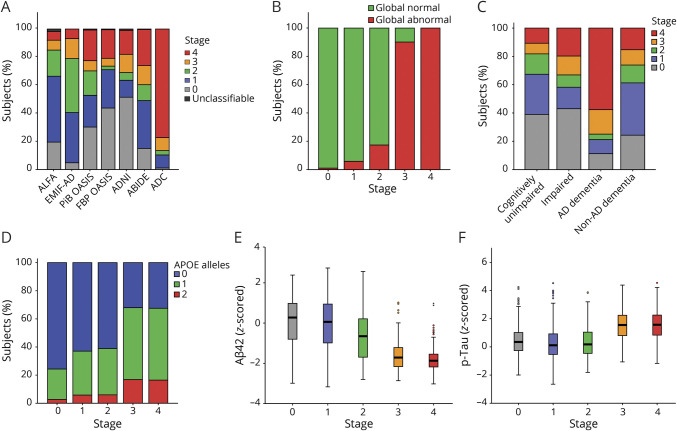
According to the final 5-stage model (figure 3), the ranking of regional frequency of abnormality starts with the cingulate regions (≈60% of individuals), followed by precuneus, paracentral gyrus, lateral orbital cortex, and insula (≈40% of individuals), as well as basal temporal, frontal, and more associative cortices (≈20%–30% of individuals), and ends with other temporal and occipital regions (≈10% of individuals).
Figure 3. Five-stage (mean + 2 SD equal frequencies) cortical amyloid staging model.
(A) Frequency of regional abnormality across 400 cognitively unimpaired individuals used to construct the model. Colors represent the 4 different stages as defined by the equal frequency approach. (B) Anatomic image displaying the brain regions involved in each stage. A stage was attributed when >50% of the encompassed regions displayed a standardized uptake value ratio greater than the cutoff (mean + 2 SD cohort- and tracer-specific cutoff). Higher stages were achieved only once the staging conditions were also met for previous stages.
The model successfully classified virtually all 4,783 scans, with only 48 (1.0%) unclassifiable scans. These scans were not from any particular cohort, tracer, diagnostic group, or age group (data not shown). Most unclassifiable scans (n = 38, 79.2%) met the requirements for stages 1 and 3 but failed to fulfill stage 2. The remaining (n = 10, 21.8%) displayed sufficient abnormal stage 2 regions without achieving stage 1.
Cross-sectional relationships
At baseline, 1,058 (35.0%) individuals were classified as stage 0, 694 (22.9%) as stage 1, 336 (11.1%) as stage 2, 319 (10.5%) as stage 3, and 597 (19.7%) as stage 4. The distribution of stages per cohort is depicted in figure 4A. The association between stages and age was small (n = 2,866, F = 30.55, η2 = 0.04). Global SUVr classification identified positivity in 1.0%, 5.5%, 17.9%, 90.0%, and 100.0% in individuals in stage 0 to 4, respectively (n = 3,004, χ2 = 2,439.67) (figure 4B). There was a significant association between stage and syndromic diagnosis (n = 2,977, χ2 = 679.00), with most (≈60%) CU individuals and those with non-AD dementia classified as stage 0 to 1, ∼40% the impaired individuals were classified as stage 0, while the remaining individuals were distributed across stages, and individuals with AD dementia were mostly classified as stages 3 and 4 (figure 4C). Baseline stage was related to number of APOE ε4 alleles (n = 2,887, χ2 = 400.16) (figure 4D). Higher stages were associated with lower baseline MMSE scores across diagnostic groups (n = 2,852, F = 129.64, η2 = 0.15), within CU individuals (n = 1,743, F = 3.27, η2 = 0.01), and within impaired individuals (n = 654, F = 14.12, η2 = 0.08) with distinct effect sizes. Increasing stages were associated with decreases in Aβ42 levels (n = 1,462, F = 235.35, η2 = 0.39). More specifically, individuals in stages 3 and 4 had lower Aβ42 levels compared to all other stages; stage 2 individuals had lower levels compared to those in stage 0/1; and individuals in stage 1 had lower levels compared to those in stage 0 (figure 4E). Finally, cortical amyloid stage was related to phosphorylated tau levels, with stages 3 and 4 associated with higher z scores compared to the other stages but not different from each other (n = 1,461, F = 108.58, η2 = 0.22) (figure 4F).
Figure 4. Cross-sectional analyses.
(A) Baseline distribution of staging classification per cohort. Classification based on the amyloid staging model vs (B) global amyloid PET classification, (C) syndromic diagnosis, (D) genetic risk, (E) z-scored CSF β-amyloid42 (Aβ42) levels, and (F) log-transformed z-scored phosphorylated tau (p-tau) values. AD = Alzheimer disease.
Longitudinal PET
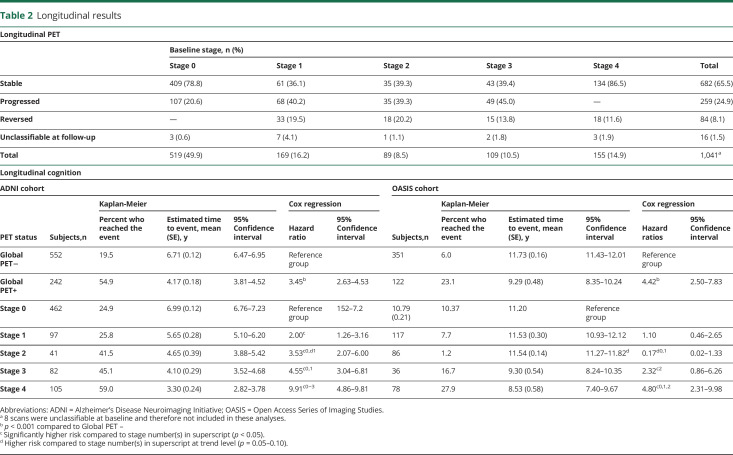
In total, 1,049 individuals (ADNI: n = 741; OASIS: n = 308) had longitudinal PET available, with a mean follow-up period of 3.97 years (SD 1.84, range 0.86–9.61 years). On the basis of stage at the first and last PET scans, 65.5% of individuals were stable, 24.9% individuals were progressors, 8.1% reversed, and 1.5% had an unclassifiable scan at follow-up. In comparison, based on global amyloid PET status, 89.0% of individuals were stable, 6.7% were progressors, and 3.9% reversed. Progression and reversal rates were relatively consistent across stages of amyloid burden (table 2). Considering all scans, 14.8% of individuals displayed a nonmonotonic behavior based on the staging model compared to 6.8% based on global PET.
Table 2.
Longitudinal results
Furthermore, stage progressors (meanyears = 4.61, SDyears = 1.92, 95% CI 4.38–4.85) had a significantly longer follow-up period compared to stage stable (Myears = 3.77, SDyears = 1.75, 95% CI 3.64–3.91) individuals (n = 941, F = 40.68, η2 = 0.04). Global progressors (Myears = 4.45, SDyears = 1.98, 95% CI 3.98–4.92) also had a significantly longer follow-up period compared to global stable (Myears = 3.94, SDyears = 1.82, 95% CI 3.82–4.06) individuals (n = 1,008, F = 5.00, η2 = 0.01).
Longitudinal cognition
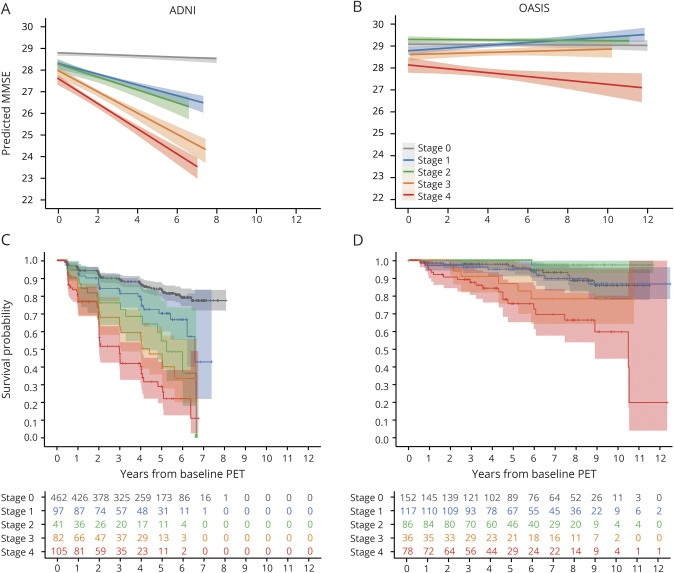
Linear mixed model analysis showed that baseline stage, corrected for age, sex, clinical diagnosis, and time between follow-up visits (repeated measures), predicts longitudinal MMSE score decline (ADNI: n = 867, F = 67.37; OASIS: n = 475, F = 9.12). More specifically, in ADNI, post hoc analyses showed that individuals in stage 1 to 4 declined faster compared to those in stage 0 (95% CI −1.13 to 0.42, −1.49 to −0.44, −2.25 to 1.47, and −2.74 to −2.01 respectively), those in stage 4 declined faster than those in stage 1/2 (95% CI −2.05 to −1.14 and −2.00 to −0.81, respectively), and those in stage 3 declined faster than individuals in stage 1/2 (95% CI −1.56 to −0.60 and −1.50 to −0.28, respectively). In OASIS, individuals in stage 4 declined faster than individuals in stage 0/1/2 (95% CI −1.13 to −0.45, −1.09 to −0.39, and 1.35 to −0.60 respectively). No other significant differences between stages were observed (figure 5, A and B).
Figure 5. Longitudinal analyses.
Results from the linear mixed model analyses displaying the effect of baseline amyloid stage on subsequent Mini-Mental State Examination (MMSE) scores with colored bands representing the 95% confidence interval for (A) Alzheimer’s Disease Neuroimaging Initiative (ADNI) and (B) Open Access Series of Imaging Studies (OASIS) separately. Kaplan-Meier survival plot displaying risk of progression to MMSE score ≤25 per baseline amyloid stage for (C) ADNI and (D) OASIS separately.
Furthermore, in ADNI, higher baseline stage predicted faster decline toward an MMSE score ≤25 (n = 787) (figure 5C). Post hoc analyses showed a number of stage-wise differences in time to reach the event (table 2). This effect was driven mainly by the impaired subgroup (figure S7 available from Dryad, doi:10.5061/dryad.7wm37pvp9). In comparison, a global SUVr dichotomization showed that amyloid-positive individuals (n = 242) reached the event faster than amyloid-negative individuals (n = 552) (table 2).
In OASIS, a faster decline toward an MMSE score ≤25 was observed only for individuals at baseline stages 3 and 4 (n = 469) (figure 5D). In comparison, global SUVr dichotomization showed that amyloid-positive individuals (n = 122) reached the event faster compared to amyloid-negative ones (n = 351). Mean estimated time to event and hazard ratios for all survival analyses can be found in table 2.
Discussion
Our pooled analysis of >3,000 individuals demonstrates that a PET-based multitracer model can reliably assign cortical amyloid stage across 6 cohorts. More specifically, the 5-stage model classified >99% of 4,783 scans from a large heterogeneous sample across diagnostic groups scanned with 4 different amyloid PET radiotracers. The model provides information on amyloid burden beyond global dichotomized classification and shows a clear relationship with genetic risk, CSF values, and clinical variables, thereby supporting its value.
The excellent cross-cohort performance of the presented model can be attributed to its multitracer aspect. While previous groups achieved high applicability either within a single cohort4,5 or between 2 studies,6 neither fully replicated the model in an independent sample. In this work, we were able to perform a head-to-head comparison of the same methodology applied to a balanced sample of individuals across 4 different tracers, which showed marked differences in regional rankings (figure 2A), especially for [18F]florbetapir. This particular tracer effect could be due to the choice of target and reference region in this work, which has previously been deemed suboptimal for ADNI scans.19 However, a consistent ROI definition across cohorts was prioritized in this work to allow the pooling of the datasets. This finding suggests that the direct translation of a single-tracer model to multitracer studies could bring challenges. As the field moves toward large-scale integrative studies, the need for harmonizing results across radiotracers and cohorts greatly increases, as can already be appreciated by initiatives such as the Centiloid Project.20
The observed regional ordering of the 5-stage model is somewhat in disagreement with established neuropathologic literature.16,17 The main difference would be the early appearance of the cingulate in our model, while it is reported only after neocortical involvement by Thal et al.17 and after basal portions of the frontal, temporal and occipital lobe by Braak and Braak.16 In addition, Braak and Braak report an early involvement of basal temporal regions, a pattern that is not consistently observed in PET literature. Except for the work of Grothe et al.,5 several PET studies fail to identify early temporal involvement despite distinct methodologies and instead point to the cingulate and precuneus as early regions, including the most recent and extensive staging work to date.6,21,22 This consistent discrepancy between neuropathology and PET-only studies has been described previously23 and is likely to be influenced by signal distortion present in PET imaging. Due to the proximity of medial regions to white matter and the additional gray matter signal spill-in from the contralateral hemisphere, medial regions such as those in stage 1 and 2 in this work are more frequently classified as abnormal in PET imaging compared to lateral counterparts, even when levels of pathology are comparable.24
While the discrepancy with pathologic studies might be due to sample size, resolution limitations and differences in technique such as the intrinsic signal distortion of PET imaging, remaining differences across PET studies, could also be explained by methodologic choices, including the use of partial volume correction. However, our results from partial volume effect–corrected SUVr values did not substantially differ from the main model (data available from Dryad, figure S3, doi.org/10.5061/dryad.7wm37pvp9), suggesting that spill-in from white matter (of special concern for 18F-labeled radiotracers25) has only a limited effect on the regional ordering. In addition, the similar ordering observed with DVR indicates that the overall impact of SUVr confounders on this method is limited (data available from Dryad, figure S4, doi.org/10.5061/dryad.7wm37pvp9).26,27
The observed relationships between stages and several demographic, clinical, and CSF measures support the validity of the staging model. In particular, the lower CSF Aβ42 values observed in individuals in stage 2 compared to those in stage 0/1 suggest that regional amyloid PET may be more sensitive for early pathologic detection compared to standard dichotomization of global amyloid burden. In addition, the comparable levels of CSF Aβ42 between individuals in stage 3 and those in stage 4 indicate that an amyloid-PET staging system can differentiate pathologic burden even after a plateau is reached in CSF levels. Furthermore, the cortical amyloid staging model seems valuable for prognosis because baseline amyloid stage relates to distinct risk profiles of subsequent cognitive decline as measured with the MMSE (figure 5). In particular, distinct patterns can be observed per cohort in which a step-wise effect of stage on cognitive decline is apparent mostly in the impaired individuals of ADNI, while only later stages related to faster decline in CU individuals. Furthermore, a staging system captures differences in cognitive decline between individuals in stages 3 and 4 whereby the latter group declines faster than a globally positive group, indicating that the extent of the global burden provides further prognostic information. These results demonstrate the risk related to amyloid pathology alone, and to fully assess an individual's risk profile, it is necessary to incorporate additional biomarkers.2
In view of these results, the main added value of this cortical amyloid staging model compared to current approaches in PET quantification is 2-fold. First, it is able to detect regional abnormality before global positivity is assigned; >90% of individuals in stage 1, ≈80% of those in stage 2, and 10% of people in stage 3 are classified as negative on the basis of global SUVr. This points to the limitations of dichotomizing amyloid PET and provides further evidence for the possibility for early pathologic identification with amyloid PET imaging. Second, even after global positivity is assigned, the model allows a detailed assessment of the extent of amyloid burden and corresponding risk of cognitive decline (figure 5, individuals in stage 3 vs 4). Therefore, independently of global PET status, the stage classifications can help identify individuals with Alzheimer pathologic changes who have greater risk of amyloid-related long-term cognitive decline, a relevant population for secondary prevention trials.28 In addition, the intrinsic grouping of individuals with similar levels of amyloid pathology with this staging ensures that treatment arms are appropriately balanced29 and may be relevant for modeling expected treatment effects on cognition.30
The staging model better captures the accumulation process compared to a simple conversion from negative to positive. In this study, the model identified 24.7% of individuals with longitudinal PET as progressors compared to 6.7% of converters according to global SUVr. On the other hand, the staging model relies on classifying regional SUVr, which might be more sensitive than composite or global signal to local blood flow changes, segmentation, and coregistration errors, all common challenges in longitudinal PET analyses.19 This can be appreciated by the identification of the model of 14.8% nonmonotonic trajectories, of which approximately half are also observed with dichotomous global SUVr. This suggests that creating a composite ROI per stage, as previously proposed,6 could reduce the variability observed in our results, although likely not below the effect already present with global SUVr. Because the choice of reference region could also have affected these results, future work could assess the performance of different regions to improve longitudinal plausibility.
One potential solution to minimize the impact of regional sensitivity in a staging system while maintaining a more fine-grained description of the accumulation process would be to create stages based on global SUVr alone. For such a model to be applicable across tracers, a harmonization tool such as the Centiloid scale would be required, which would avoid single-tracer cutoffs.20 Conversion to Centiloid was out of the scope of this work; in fact, the staging model can be considered an alternative and simple method for cross-tracer analysis of amyloid PET images. Nonetheless, a supplementary analysis was performed in which ADNI data were divided into quintiles to create 5 stages based on global SUVr. Although such a global system showed a low agreement (κ = 0.32) with the stages defined in this work (data available from Dryad, table S3 and figure S8, doi.org/10.5061/dryad.7wm37pvp9), in general, a similar pattern of increased risk per level of amyloid burden was observed. However, hazard ratios are consistently higher for the 5-stage model compared to a quintiles-based approach, and only the 5-stage model associates the first stage with increased risk compared to amyloid-negative individuals. These results suggest that while global analyses are able to provide risk stratification, the use of regional information is particularly relevant for the earliest changes in amyloid burden and better distinguishes risk profiles across the spectrum.
In this work, a total of 9 model definitions were assessed without an available standard of truth to determine the underlying regional levels of amyloid burden. While the results add to the evidence of a temporal evolution of the topography of amyloid pathology, this methodology is not sufficient to shed light on the potential underlying mechanism. In fact, our results reflect the ordering of regions as detected by PET, which are likely to be affected by the distortions in the PET signal between medial and lateral regions.24 Therefore, this staging model should be seen as a tool, applicable across tracers and cohorts, with which one is able to assess a subject's level of brain amyloidosis and its associated risks in terms of cognitive decline. Thus, the model is optimized to classify as many individuals as possible, and while defining additional stages could provide a more detailed picture of the pathologic process, it would likely increase the number of unclassifiable scans due to between-subject variability. In addition, when this model is used within a particular application, image processing and cutoff definitions can be optimized accordingly, e.g., using more lenient cutoffs to detect risk of subsequent amyloid accumulation or more conservative and neuropathology-grounded cutoffs for trial inclusion.
The main limitations of this study are the lack of pathologic confirmation, the use of a global cutoff for regional SUVr values, the omission of subcortical structures, and the atlas-based definition of regions of interest. First, no pathologic evaluation has been made at the level of detail available from PET scans used in this analysis, thereby limiting the availability of a standard of truth to this work. However, current studies find similar relationships between global and regional SUVr and postmortem data.31 Second, the use of a global SUVr cutoff to define regional abnormality has not been validated, and such a validation would require a reference method (regional visual assessment or regional histopathologic data), with its application across datasets potentially posing major challenges. In addition, recent work suggests that while regional cutoffs would differ from global ones, the final regional ordering would be highly consistent with results from global cutoff.32–34 Third, including subcortical structures in the model would likely require region-specific cutoffs, as indicated by previous work.4 However, our model could be combined with previous work by Hanseeuw et al.4 for a complete cortical and subcortical staging of amyloid deposition. Finally, atlas-based approaches could be limiting the power to detect signal increases that do not follow anatomic boundaries, and a voxel-wise model could be considered to extend the current work.35
A multitracer 5-stage cortical amyloid staging model based solely on amyloid PET scans from 400 CU individuals is able to classify the level of amyloid burden in >3,000 individuals across cohorts and radiotracers. Amyloid stage is strongly related to demographic and clinical measures, can detect preglobal amyloid burden, and can distinguish cognitive decline risk profiles within amyloid-positive individuals. The model is easily applicable to any cohort after a cohort-specific global SUVr cutoff is defined and the Desikan-Killiany atlas is applied, and it has the potential to improve targeted subject trial inclusion and to support routine clinical work. In relation to identification of preglobal amyloid pathology, further comparison of this model with other established methods is warranted to minimize false positives.
Acknowledgment
The authors acknowledge all data donors and collaborators (ADNI, EMIF-AD, OASIS-3, ABIDE, ALFA, and ADC). This research would not have been possible without the time and dedication of every participant included in these studies. The authors thank GE Healthcare for kindly providing 18F-flutemetamol doses of ALFA participants and Roche Diagnostics International Ltd for kindly providing the kits for the CSF analysis of the ALFA+ participants. Additional funding has been obtained by Project RTI2018-102261-B-I00, funded by European Regional Development Fund/Ministry of Science and Innovation–State Research Agency (Spain). Data were provided by OASIS-3: principal investigators: T. Benzinger, D. Marcus, J. Morris; NIH P50AG00561, P30NS09857781, P01AG026276, P01AG003991, R01AG043434, UL1TR000448, R01EB009352. AV-45 doses were provided by Avid Radiopharmaceuticals, a wholly owned subsidiary of Eli Lilly.
Glossary
- Aβ
β-amyloid
- ABIDE
Alzheimer's Biomarkers in Daily Practice
- AD
Alzheimer disease
- ADC
Amsterdam Dementia Cohort
- ADNI
Alzheimer's Disease Neuroimaging Initiative
- ALFA
Alzheimer's and Family
- CU
cognitively unimpaired
- EMIF-AD
European Medical Information Framework for AD
- MMSE
Mini-Mental State Examination
- OASIS
Open Access Series of Imaging Studies
- PiB
Pittsburgh compound B
- SUVr
standardized uptake value ratio
Appendix 1. Authors

Appendix 2. ALFA Study group coinvestigators

Appendix 3. AMYPAD coinvestigators

Contributor Information
for the ALFA Study, for the Alzheimer's Disease Neuroimaging Initiative, on behalf of the AMYPAD Consortium:
Eider M Arenaza-Urquijo, Annabella Beteta, Anna Brugulat-Serrat, Raffaele Cacciaglia, Alba Cañas Boccagni, Yelena G. Bodien, Marta Crous-Bou, Carme Deulofeu, Ruth Dominguez, Karine Fauria, Carles Falcon, Marta Félez-Sánchez, Jose María González de Echavarri, Oriol Grau-Rivera, Laura Hernández, Gema Huesa, Jordi Huguet, MarÍa LeÓn, Paula Marne, Tania Menchón, Marta Milà-Alomà, Grégory Operto, Carolina Minguillon, Maria Pascual, Albina Polo, Sandra Pradas, Aleix Sala-Vila, Gonzalo Sánchez-Benavides, Mahnaz Shekari, Anna Soteras, Marc Suárez-Calvet, Laia Tenas, Marc Vilanova, and Natalia Vilor-Tejedor
Study funding
The project leading to this article has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement 115952. This Joint Undertaking receives the support from the European Union's (EU’s) Horizon 2020 research and innovation program and European Federation of Pharmaceutical Industries and Associations (EFPIA). F. Barkhof is supported by the National Institute for Health Research University College London Hospital (NIHR UCLH) biomedical research center. Data collection and sharing for this project were funded by the EU/EFPIA Innovative Medicines Initiative Joint Undertaking EMIF grant agreement (grant 115372). F. Barkhof, V. Wottschel, and A.M. Wink are supported by the European Union's Horizon 2020 research and innovation program under grant agreement 666992. Research of the Alzheimer Center Amsterdam is part of the neurodegeneration research program of Amsterdam Neuroscience. The clinical database structure was developed with funding from Stichting Dioraphte. Alzheimer Center Amsterdam is supported by Alzheimer Nederland and Stichting VUmc fonds. The ABIDE project was supported by a ZonMW-Memorabel grant (ABIDE; project 733050201), in the context of the Dutch Deltaplan Dementie, and through a grant of Piramal Imaging (PET scan costs) to the Stichting Alzheimer & Neuropsychiatrie, Amsterdam. Data collection and sharing for this project were funded by the ADNI (NIH Grant U01 AG024904) and DOD ADNI (Department of Defense award W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, by the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc; Biogen; Bristol-Myers Squibb Co; CereSpir, Inc; Cogstate; Eisai Inc; Elan Pharmaceuticals, Inc; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc; Fujirebio; GE Healthcare; IXICO Ltd; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Lumosity; Lundbeck; Merck & Co, Inc; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corp; Pfizer Inc; Piramal Imaging; Servier; Takeda Pharmaceutical Co; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the NIH (fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. The ALFA Study is funded “la Caixa” Foundation (LCF/PR/GN17/10300004), the Alzheimer's Association, and an international anonymous charity foundation through the TriBEKa Imaging Platform project.
Disclosure
L.E. Collij, F. Heeman, G. Salvadó, S. Ingala, D. Altomare, A. de Wilde, E. Konijnenberg, M. van Buchem, M. Yaqub, P. Markiewicz, S.S.V. Golla, V. Wottschel, and A.M. Wink report no disclosures relevant to the manuscript. P.J. Visser has served as member of the advisory board of Roche Diagnostics. P.J. Visser received nonfinancial support from GE Healthcare, research support from Biogen, and grants from Bristol-Myers Squibb, EU/EFPIA Innovative Medicines Initiative Joint Undertaking, EU Joint Programme–Neurodegenerative Disease Research (JPND and ZonMw). C. Teunissen has functioned in advisory boards of Roche, received nonfinancial support in the form of research consumables from ADxNeurosciences and Euroimmun, and performed contract research or received grants from Probiodrug, Biogen, Esai, Toyama, Janssen Prevention Center, Boehringer, AxonNeurosciences, EIP farma, PeopleBio, and Roche. C.E. Teunissen received grants from the European Commission, the Dutch Research Council (ZonMW), Association of Frontotemporal Dementia/Alzheimer's Drug Discovery Foundation, The Weston Brain Institute, and Alzheimer Netherlands. A.A. Lammertsma reports no disclosures relevant to the manuscript. P. Scheltens received grants from GE Healthcare, Piramal, and Merck paid to his institution; he has received speaker's fees paid to the institution from the Alzheimer Center, VU University Medical Center, Lilly, GE Healthcare, and Roche. W.M. van der Flier, R. Boellaard, and B.N.M. van Berckel report no disclosures relevant to the manuscript. J.L. Molinuevo received advisory honoraria from Alergan, Roche Diagnostics, Genentech, Novartis, Lundbeck, Oryzon, Biogen, Lilly, Janssen, Green Valley, MSD, Eisai, Alector, and Raman Health. He also received research support from the EU/EFPIA Innovative Medicines Initiative Joint Undertaking AMYPAD grant agreement 115952; the EU/EFPIA Innovative Medicines Initiative Joint Undertaking EPAD grant agreement 115736; the EU/EFPIA Innovative Medicines Initiative Joint Undertaking AETIONOMY grant 115568; and “la Caixa” Foundation. J.D. Gispert has received speaker's fees from Biogen and Philips. In addition he holds a “Ramón y Cajal” fellowship (RYC-2013-13054) and has received research support from the EU/EFPIA Innovative Medicines Initiative Joint Undertaking AMYPAD grant agreement 115952 and from Ministerio de Ciencia y Universidades (grant agreement RTI2018-102261). M.E. Schmidt received funding from Janssen Pharmaceutica NV. F. Barkhof received payment and honoraria from Bayer Genzyme, Biogen-Idec, TEVA, Merck, Novartis, Roche, IXICO Ltd, GeNeuro, and Apitope Ltd for consulting; payment from IXICOLtd and MedScape for educational presentations; research support via grants from EU/EFPIA Innovative Medicines Initiative Joint Undertaking (AMYPAD consortium), EuroPOND (H2020), UK MS Society, Dutch MS Society, PICTURE (IMDI-NWO), NIHR UCLH Biomedical Research Centre, and ECTRIMS-MAGNIMS. I. Lopes Alves reports no disclosures relevant to the manuscript. Go to Neurology.org/N for full disclosures.
References
- 1.Morris E, Chalkidou A, Hammers A, et al. Diagnostic accuracy of (18)F amyloid PET tracers for the diagnosis of Alzheimer's disease: a systematic review and meta-analysis. Eur J Nucl Med Mol Imaging 2016;43:374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jack CR Jr, Bennett DA, Blennow K, et al. NIA-AA research framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement 2018;14:535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thal DR, Beach TG, Zanette M, et al. [(18)F]flutemetamol amyloid positron emission tomography in preclinical and symptomatic Alzheimer's disease: specific detection of advanced phases of amyloid-beta pathology. Alzheimers Dement 2015;11:975–985. [DOI] [PubMed] [Google Scholar]
- 4.Hanseeuw BJ, Betensky RA, Mormino EC, et al. PET staging of amyloidosis using striatum. Alzheimers Dement 2018;14:1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grothe MJ, Barthel H, Sepulcre J, et al. In vivo staging of regional amyloid deposition. Neurology 2017;89:2031–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattsson N, Palmqvist S, Stomrud E, et al. Staging beta-amyloid pathology with amyloid positron emission tomography. JAMA Neurol Epub 2019 Jul 18. [DOI] [PMC free article] [PubMed]
- 7.Molinuevo JL, Gramunt N, Gispert JD, et al. The ALFA project: a research platform to identify early pathophysiological features of Alzheimer's disease. Alzheimers Dement 2016;2:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Flier WM, Scheltens P. Amsterdam dementia cohort: performing research to optimize care. J Alzheimers Dis 2018;62:1091–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konijnenberg E, Carter SF, Ten Kate M, et al. The EMIF-AD PreclinAD study: study design and baseline cohort overview. Alzheimers Res Ther 2018;10:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Wilde A, van Maurik IS, Kunneman M, et al. Alzheimer's Biomarkers in Daily Practice (ABIDE) project: rationale and design. Alzheimers Dement (Amst) 2017;6:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcus DS, Fotenos AF, Csernansky JG, et al. Open access series of imaging studies: longitudinal MRI data in nondemented and demented older adults. J Cogn Neurosci 2010;22:2677–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collij L, Konijnenberg E, Reimand J, et al. Assessing amyloid pathology in cognitively normal subjects using [(18)F]flutemetamol PET: comparing visual reads and quantitative methods. J Nucl Med 2019;60:541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heeman F, Yaqub M, Lopes Alves I, et al. Optimized dual-time-window protocols for quantitative [(18)F]flutemetamol and [(18)F]florbetaben PET studies. EJNMMI Res 2019;9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su Y, D'Angelo GM, Vlassenko AG, et al. Quantitative analysis of PiB-PET with FreeSurfer ROIs. PLoS One 2013;8:e73377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006;31:968–980. [DOI] [PubMed] [Google Scholar]
- 16.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 1991;82:239–259. [DOI] [PubMed] [Google Scholar]
- 17.Thal DR, Rub U, Orantes M, et al. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology 2002;58:1791–1800. [DOI] [PubMed] [Google Scholar]
- 18.Alves IL. Supplementary material for “Multi-tracer model for staging cortical amyloid deposition using PET imaging,” v3, Dryad, Dataset. 2020. doi: 10.5061/dryad.7wm37pvp9. [DOI] [Google Scholar]
- 19.Landau SM, Fero A, Baker SL, et al. Measurement of longitudinal beta-amyloid change with 18F-florbetapir PET and standardized uptake value ratios. J Nucl Med 2015;56:567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klunk WE, Koeppe RA, Price JC, et al. The Centiloid Project: standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement 2015;11:1–15 e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmqvist S, Scholl M, Strandberg O, et al. Earliest accumulation of beta-amyloid occurs within the default-mode network and concurrently affects brain connectivity. Nat Commun 2017;8:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrell ME, Chen X, Rundle MM, et al. Regional amyloid accumulation and cognitive decline in initially amyloid-negative adults. Neurology 2018;91:e1809–e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fantoni E, Collij L, Alves IL, et al. The spatial-temporal ordering of amyloid pathology and opportunities for PET imaging. J Nucl Med 2020;61:166–171. [DOI] [PubMed] [Google Scholar]
- 24.Smith A, Buckley C. [18F]flutemetamol PET image representation of Ab pathology; differences between lateral and medial image intensity for equivalent levels of pathology. Presented at the 10th Human Amyloid Imaging; January 13–15, 2016; Miami: PP77.
- 25.Catafau AM, Bullich S. Amyloid PET imaging: applications beyond Alzheimer's disease. Clin Transl Imaging 2015;3:39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Berckel BN, Ossenkoppele R, Tolboom N, et al. Longitudinal amyloid imaging using 11C-PiB: methodologic considerations. J Nucl Med 2013;54:1570–1576. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt ME, Chiao P, Klein G, et al. The influence of biological and technical factors on quantitative analysis of amyloid PET: points to consider and recommendations for controlling variability in longitudinal data. Alzheimers Dement 2015;11:1050–1068. [DOI] [PubMed] [Google Scholar]
- 28.Ritchie CW, Molinuevo JL, Truyen L, et al. Development of interventions for the secondary prevention of Alzheimer's dementia: the European Prevention of Alzheimer's Dementia (EPAD) project. Lancet Psychiatry 2016;3:179–186. [DOI] [PubMed] [Google Scholar]
- 29.Guo T, Dukart J, Brendel M, et al. Rate of β-amyloid accumulation varies with baseline amyloid burden: implications for anti-amyloid drug trials. Alzheimers Demen 2018;14:1387–1396. [DOI] [PubMed] [Google Scholar]
- 30.Liu E, Schmidt ME, Margolin R, et al. Amyloid-beta 11C-PiB-PET imaging results from 2 randomized bapineuzumab phase 3 AD trials. Neurology 2015;85:692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo SW, Ayakta N, Grinberg LT, et al. Regional correlations between [(11)C]PIB PET and post-mortem burden of amyloid-beta pathology in a diverse neuropathological cohort. Neuroimage Clin 2017;13:130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopes-Alves I, Collij L, Heeman F, et al. Event-based modeling of the temporal ordering of regional β-amyloid deposition in the brain. Alzheimers Demen 2018;14:P887–P88. [Google Scholar]
- 33.Salvadó G, Collij L, Niñerola-Baizán A, et al. Voxel-based amyloid pet staging for the whole Alzheimer's disease continuum. Alzheimers Demen 2019;15:P425–P26. [Google Scholar]
- 34.Jelistratova I, Bugla K, Teipel S, et al. Reconsidering In-Vivo Models of Regional Amyloid Pathology Progression: Region-Specific Thresholds and Longitudinal Data. Presented at the 13th Human Amyloid Imaging; January 16-18, 2019; Miami, FL; P24. [Google Scholar]
- 35.Bilgel M, Prince JL, Wong DF, et al. A multivariate nonlinear mixed effects model for longitudinal image analysis: application to amyloid imaging. Neuroimage 2016;134:658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study can be made available on request to the study-specific principal investigator (i.e., ABIDE, EMIF-AD, ADC, ALFA) or are openly available (i.e., ADNI and OASIS open-source databases).