Abstract
Objectives:
To determine if identifiable hepatic textural features are present at abdominal CT in patients with colorectal cancer (CRC) prior to the development of CT-detectable hepatic metastases.
Methods:
Four filtration-histogram texture features (standard deviation, skewness, entropy, and kurtosis) were extracted from the liver parenchyma on portal venous phase CT images at staging and post-treatment surveillance. Surveillance scans corresponded to the last scan prior to the development of CT-detectable CRC liver metastases in 29 patients (median time interval, 6 months), and these were compared with interval-matched surveillance scans in 60 CRC patients who did not develop liver metastases. Predictive models of liver metastasis-free survival and overall survival were built using regularized Cox proportional hazards regression.
Results:
Texture features did not significantly differ between cases and controls. For Cox models using all features as predictors, all coefficients were shrunk to zero, suggesting no association between any CT texture features and outcomes. Prognostic indices derived from entropy features at surveillance CT incorrectly classified patients into risk groups for future liver metastases (p < 0.001).
Conclusions:
On surveillance CT scans immediately prior to the development of CRC liver metastases, we found no evidence suggesting that changes in identifiable hepatic texture features were predictive of their development.
Keywords: CT texture analysis, hepatic metastases, colorectal cancer, CT, colorectal metastases
Introduction:
Colorectal cancer remains the 2nd leading cause of cancer worldwide despite expansion of screening programs and advances in treatment [1, 2]. Significant mortality is attributable to disease recurrence in colorectal cancer survivors, with an estimated occurrence of 29 – 63% of patients diagnosed with stage II-III disease [3]. The American Society of Clinical Oncology has highlighted the importance of identifying new prognostic factors associated with disease recurrence in order to improve surveillance guidelines [4]. Identifying risk factors may allow clinicians to tailor surveillance strategies for patients at higher risk for recurrence, particularly among those with stage II-III disease. Advances in computing power have given radiomics, the process of using quantitative image features as clinical data, the potential for earlier identification of metastatic recurrence that could positively impact outcomes and help guide post-treatment surveillance strategies [5].
Radiomic features of the liver, the most common site of distant metastatic spread in patients with colorectal cancer, may provide information about underlying physiology [6]. It has been shown that hepatic textural features at staging portal-venous enhanced computed tomography (CT) appear correlated with hepatic perfusion values in colorectal cancer patients [7, 8]. For example, entropy, a textural feature that increases with overall image ‘disorder,’ [9] appears to be inversely correlated with hepatic perfusion indices [8]. Such physiologic measurements may be altered in the setting of early occult liver metastases, where reduced portal venous blood flow and increased hepatic arterial blood flow have been observed in mouse and rat models [10, 11].
Given that CT texture features may reflect changes in hepatic perfusion, monitoring changes in these features could potentially alert radiologists to the imminent development of hepatic metastases. In other words, micro-metastatic hepatic disease may be present, detectable by CT texture features, which may be later seen at imaging as discrete lesions. However, to our knowledge, no studies regarding the change in hepatic texture features at CT during post-treatment surveillance of colorectal cancer patients to portend subsequent development of hepatic metastases have been reported. The purpose of this study was twofold. First, a major aim was to determine if CT texture changes of the hepatic parenchyma are present in patients shortly before the development of hepatic metastases compared with control patients with colorectal cancer who did not develop hepatic metastases, and if these changes can predict their development. And secondly, whether such hepatic textures predictive changes are present at the initial staging CT.
Materials and Methods:
This study was HIPAA-compliant and IRB-approved; the need for informed consent was waived.
Patient population
A flowchart of patient participation and sample selection is given in figure 1. A database of 923 patients treated with FOLFOX, FOLFOX + bevacizumab, FOLFIRI, or FOLFIRI + bevacizumab at our institution from 2003 to 2016 was searched to identify potential patients with colorectal cancer. Colorectal cancer cases were identified through manual review of pathology reports in the electronic health record. Date of diagnosis was taken as the date of colonoscopy biopsy or surgical pathology report of the resected tumor. TNM staging information was collected from the surgical pathology report of the primary tumor resection. Imaging follow-up was obtained through the picture archiving and communication system (PACS). Staging CT images were reviewed by three board-certified abdominal radiologists (DK, MGL, PJP) to exclude the presence of liver metastases at the time of initial cancer diagnosis. Then, the earliest CT images demonstrating visible hepatic metastases were identified. We then searched for the most recent surveillance CT study prior to the development of CT-detectable liver metastases (no earlier than one year prior to demonstrable metastatic disease). 60 control patients were selected such that the distribution of TNM stages at diagnosis and timing of surveillance scans were similar to the case cohort, yielding a final sample size of n = 89. Chart review was performed by a single reader (SJL).
Figure 1:
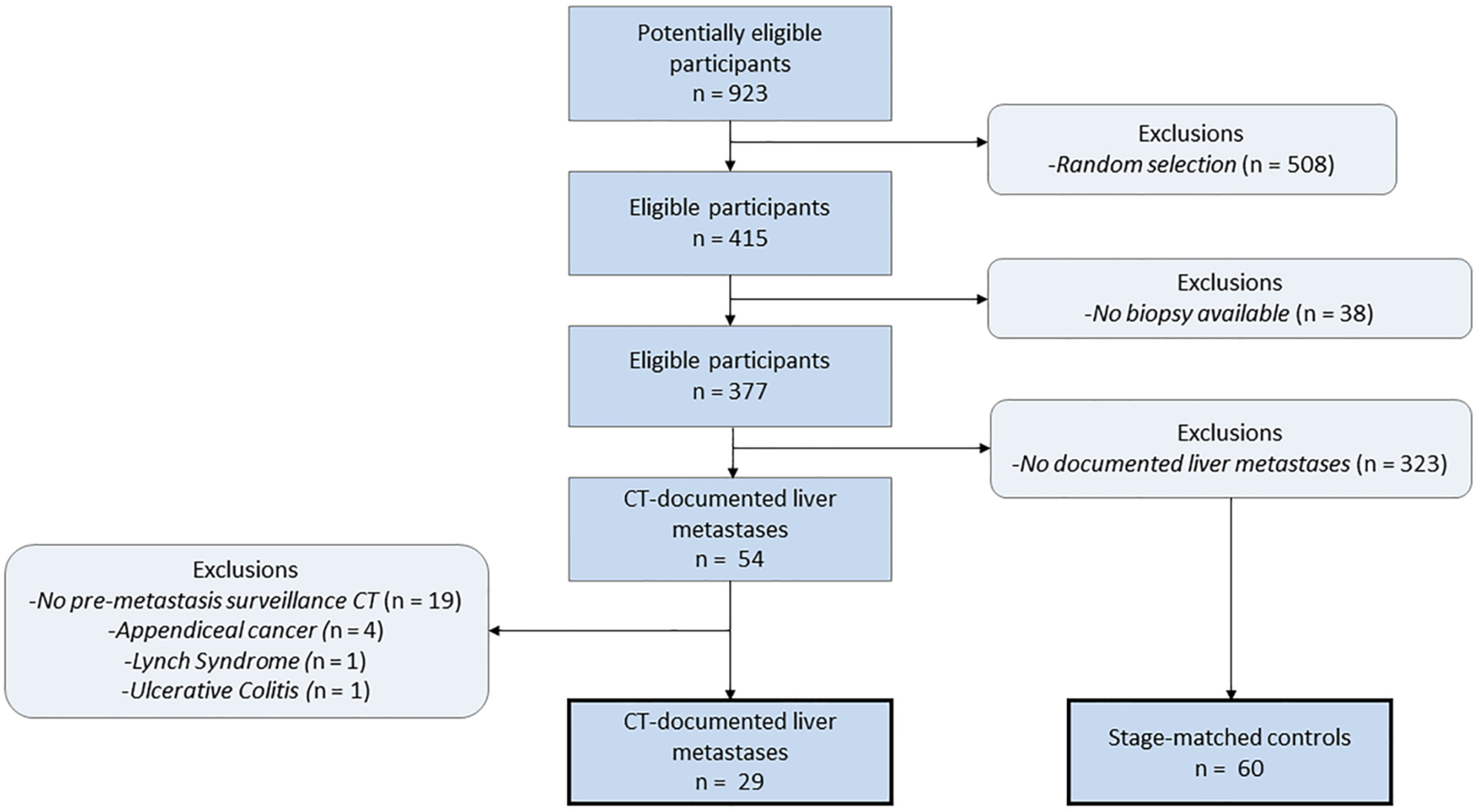
Flow diagram for case and control patient selection
CT imaging
All texture measurements were performed on anonymized contrast-enhanced portal venous-phase exams. CT imaging studies were performed on a variety of scanners (predominately GE, but also Siemens and Toshiba in a small minority of cases). Seven scans were performed at 140 kV, two were performed at 130 kV, and one was performed at 100 kV; all others were performed at 120 kV. Our standard IV contrast protocol consists of weight-based contrast dosing (range, 80–150 ml) with iohexol (300 mgI/ml), followed by 40 ml saline chaser, all at 3 ml/second. Portal venous phase imaging is initiated by a liver enhancement threshold of 50 HU, typically 60–70 seconds after initiation of contrast injection. The CT series were reconstructed at a slice thickness of 5 mm at 3-mm intervals.
Texture analysis
A single image slice at the level of the porta hepatis was selected for texture analysis of the liver parenchyma, similar to prior studies (Figure 2) [12]. All images were reviewed and appropriate slices were selected by an abdominal radiologist. These anonymized images were then uploaded to a commercially-available texture analysis program (TexRAD Ltd, Somerset, UK). A region of interest was manually drawn around the margin of the liver parenchyma by a single reader (SJL), excluding the large proximal branches of the portal vein (Figure 2). To perform texture analysis, the software uses a filtration– histogram method in which an initial filtration step is performed that highlights image features of a specified size, followed by histogram analysis of the filtered image. The initial filtration step uses a Laplacian of Gaussian (LoG) spatial band-pass filter to selectively extract features of different sizes. Informed in part by prior work, the following histogram-based texture features were calculated to characterize intrahepatic heterogeneity and complexity:
Standard deviation of pixel attenuation histogram (SD)
Entropy of pixel attenuation histogram
Skewness of pixel attenuation histogram
Kurtosis of pixel attenuation histogram
Figure 2:
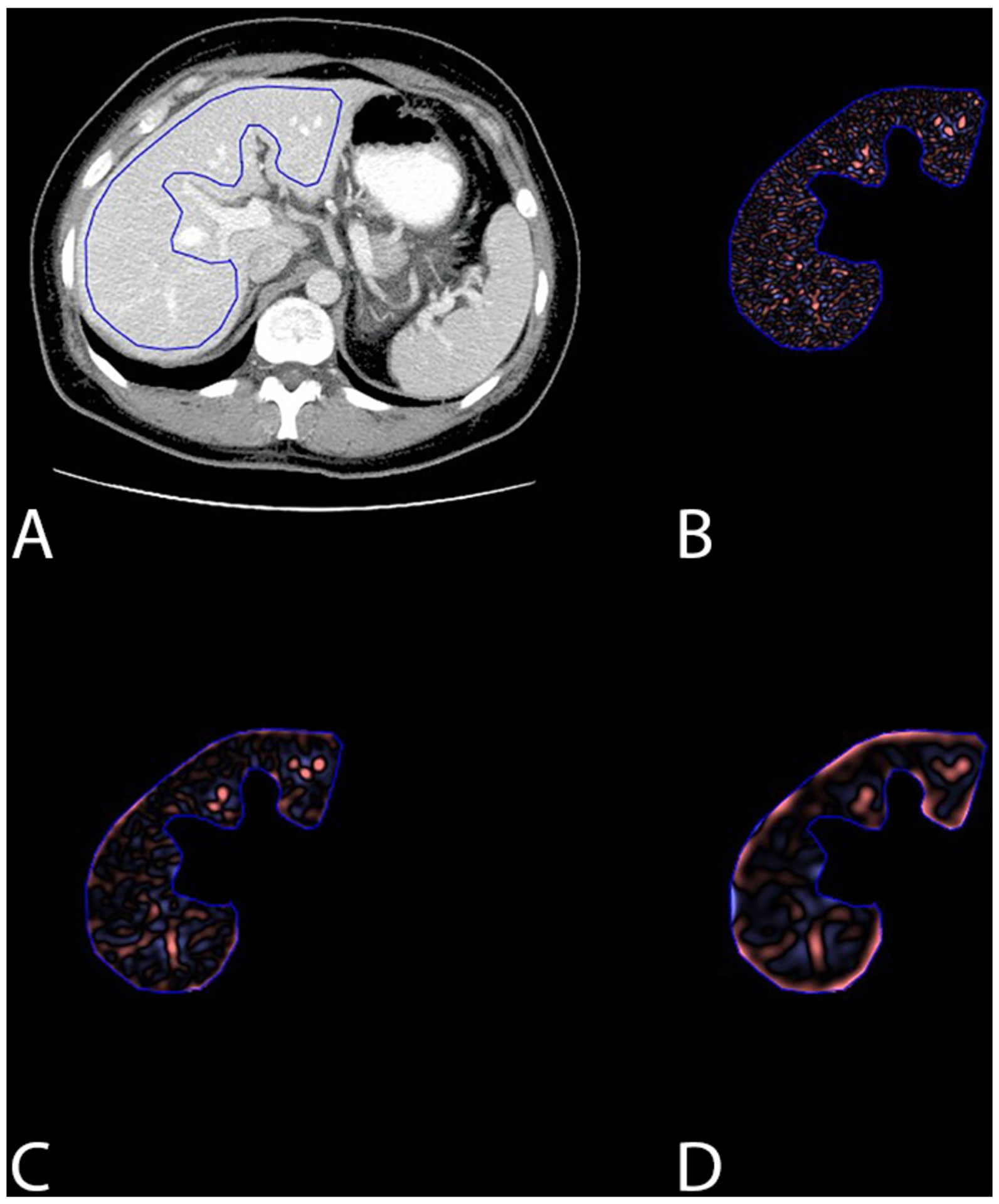
Images showing (A) the region-of-interest utilized for CT texture analysis, and (B-D) the subsequent outputrom (B) fine, (C) medium, and (D) coarse filtering of the image. The anatomic level shown was seleceted for segementation in all patients, performed on the last negative CT prior to the development of hepatic metastases for cases, and matched for controls.
The number of variables was deliberately limited to minimize type I error (multiple hypothesis testing). Each of these features were calculated at six filter sizes, ranging between fine (spatial scaling factor (ssf=0,2), medium (ssf=3,4), and coarse (ssf=5–6), yielding a total of 24 texture features.
Statistical Analysis
Texture features for cases and controls were separately compared at staging and surveillance CT with Mann-Whitney U-tests. Bonferroni correction for type 1 error was applied. Hypothesis tests for 24 texture features were performed at both staging and surveillance CT; the corrected threshold for statistical significance was p = 0.05/48.
For Cox models predicting freedom from metachronous metastatic liver disease, time-to-event was defined as the number of months between the date of tissue diagnosis of colorectal cancer and the date of CT-detectable liver metastases. Control patients were right-censored at their last follow-up date at our institution, or at date of death from any cause (if recorded). For models predicting overall survival, time-to-event was defined as the number of months between date of tissue diagnosis and date of death from any cause; control patients in overall survival models were right-censored at their last follow-up date at our institution. We also noted the use of oxaliplatin in each patient’s chemotherapy regimen. Colorectal cancer patients treated with oxaliplatin frequently develop sinusoidal obstruction syndrome, which affects hepatic hemodynamics and thus may affect texture measurements. We tested for a difference in proportion of patients receiving oxaliplatin between the liver metastases group and control group with a Pearson’s chi-squared test.
To test our specific hypotheses that hepatic entropy values are associated with the development of metachronous liver metastases or overall survival, we built separate Cox proportional hazards models using staging and surveillance CT entropy values as the predictor variables. Linear predictor values for each patient were obtained by leave-one-out cross-validation (LOOCV) and were used to create a prognostic index based on their median value. For a Cox model, the linear predictor is the sum of covariate values weighted by the regression coefficients, and it represents the log(relative hazard) compared to a hypothetical observation whose linear predictor value is 0. When the outcome of interest is an adverse event, higher linear predictor values indicate a greater risk of occurrence [13]. Using this paradigm, we assigned patients into “low risk” or “high risk” groups based on the median value of the linear predictors. Kaplan-Meier estimators were then fit to the data using the linear predictor categorization, and log-rank statistics were calculated to determine if the survival functions were significantly different between the two groups [14].
For models using all 24 texture features as predictors, we utilized least absolute shrinkage and selection operator (LASSO) regression and LOOCV to perform model selection [15]. The number of features allowed in the final model is subjected to a penalty determined by λ, a tuning parameter. By varying λ, any number of coefficients in the model may be shrunk to zero, effectively removing them. The optimal value for λ and thus the number of included features in the final model is determined by calculating the partial likelihood deviance at each λ value through LOOCV, and selecting the model with the minimal value [16]. This process reduces type 1 error rates by reducing the probability of overfitting the model to the ‘noise’ in the dataset, which becomes a significant concern when there is a large number of predictor variables relative to the number of observations [17]. LASSO regression and cross-validation were implemented using the software package, glmnet, in R [18].
Results:
Patient characteristics are listed in Table 1. The median interval between staging and surveillance CT scans from which texture features was calculated were 14 months for patients who developed liver metastases (on the subsequent CT scan) and 22 months for patients who did not develop metastases. For cases, the median interval between surveillance CT and the CT showing hepatic metastases was 6 months (IQR 4.5 – 11.25 mo). The distribution of stage at diagnosis was well-matched between cases and controls, with the majority being stage III (69% for cases, 72% for controls). There was no evidence that the proportion of patients receiving oxaliplatin in their chemotherapy regimen differed between cases and controls (p = 0.81 by Chi-Squared test). The distributions of entropy values for cases and controls at staging and surveillance CT are shown in Figure 3.
Table 1:
Patient Demographics
| Cases (n = 29) | Controls (n = 60) | |
|---|---|---|
| 58.1 ± 12.4 | 61.8 ± 13.3 | |
| Sex | ||
| Male | 18 | 24 |
| Female | 11 | 36 |
| Median total follow-up (months) | 24 (range 3 – 66) | 47.5 (range 11 – 165) |
| Median CT interval (months)* | 14 (range 2 – 38) | 22 (range 1 – 42) |
| Alive at last follow-up | 10/29 (34.5%) | 47/60 (78.3%) |
| Stage at Diagnosis | ||
| I | 0 | 3 (5%) |
| II | 6 (21%) | 12 (20%) |
| III | 20 (69%) | 43 (72%) |
| IV | 3 (10%) | 2 (3%) |
CT interval is time from initial staging CT to “pre-metastatic” surveillance CT in cases (and approximately matched in controls)
Figure 3:
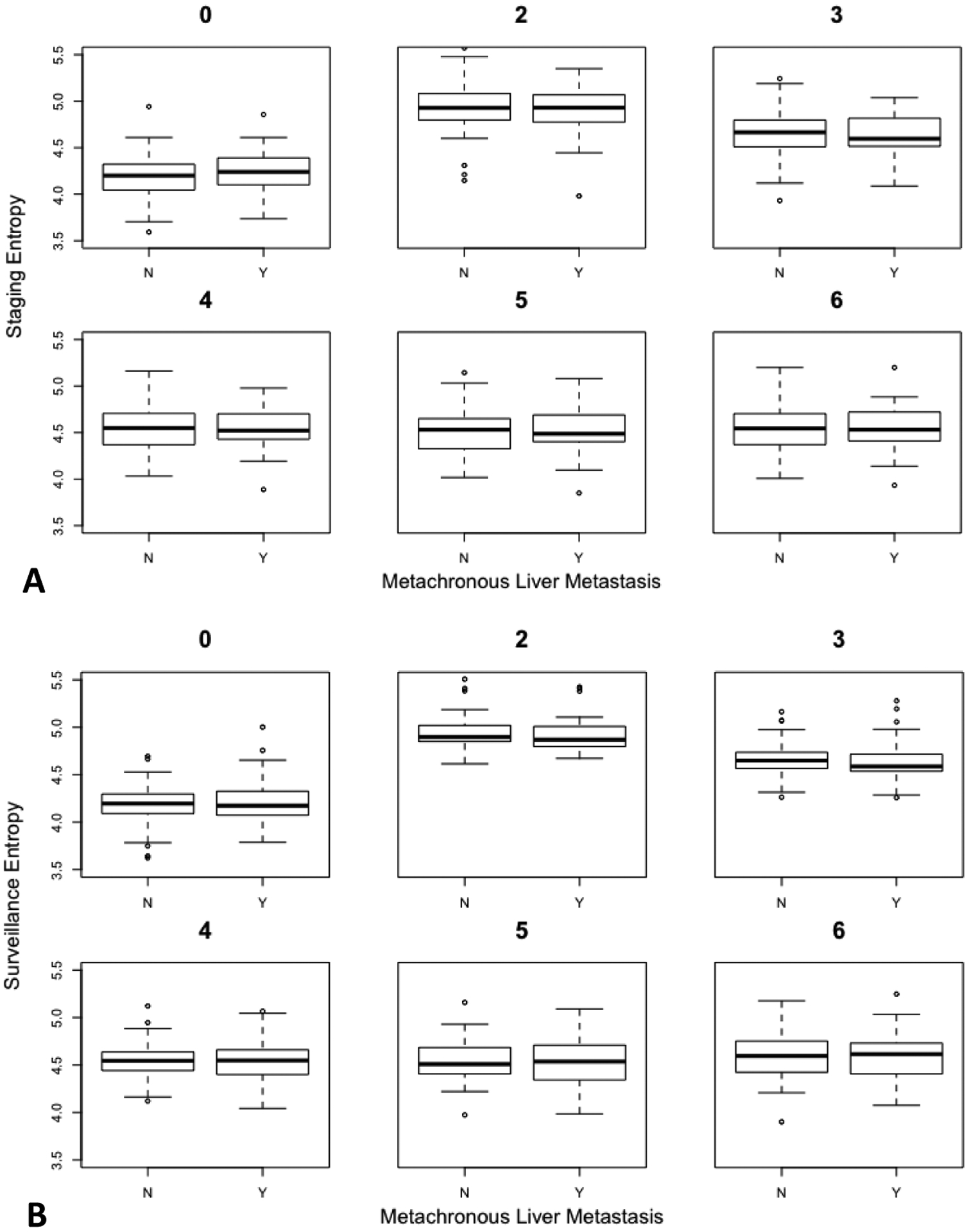
Box-and-whisker plots show the change in liver parenchyma entropy values at both the initial staging and the surveillance CT scans before imminent development of metastasis amongst cases (Y) - and matched for controls (N). Each plot represents entropy values at different spatial scaling factors (fine (ssf=0,2), medium (ssf=3,4), and coarse (ssf=5–6). The whiskers extend to the furthest measurement within (1.5 × interquartile range). Panels (A), (B) correspond to staging and surveillance entropy, respectively. Note the lack of separation for entropy values between cases and controls for any filter at either time point. This demonstrates the lack of predictive ability for identifying those patients who subsequently went on to develop identifiable metastases at the next CT scan after the surveillance scan depicted.
Table 2 displays the results of Mann-Whitney U-tests comparing texture features between cases and controls at staging and surveillance CT. Two of the tests suggested that kurtosis values (ssf = 3,4) differed between cases and controls (p = 0.01, p = 0.03), but these were no longer statistically significant after applying Bonferroni correction.
Table 2:
Results of Mann-Whitney U-tests comparing texture features between patients who developed liver metastases and controls.
| Staging CT | Surveillance CT | |||
|---|---|---|---|---|
| Feature | Spatial Scaling Factor | p-value | Spatial Scaling Factor | p-value |
| Skewness | 0 | 0.68 | 0 | 0.90 |
| 2 | 0.96 | 2 | 0.54 | |
| 3 | 0.41 | 3 | 0.38 | |
| 4 | 0.49 | 4 | 0.34 | |
| 5 | 0.36 | 5 | 0.49 | |
| 6 | 0.20 | 6 | 0.73 | |
| Standard deviation | 0 | 0.86 | 0 | 0.65 |
| 2 | 0.7 | 2 | 0.38 | |
| 3 | 0.47 | 3 | 0.61 | |
| 4 | 0.67 | 4 | 0.65 | |
| 5 | 0.94 | 5 | 0.5 | |
| 6 | 0.74 | 6 | 0.26 | |
| Kurtosis | 0 | 0.44 | 0 | 0.73 |
| 2 | 0.14 | 2 | 0.36 | |
| 3 | 0.01* | 3 | 0.33 | |
| 4 | 0.03* | 4 | 0.35 | |
| 5 | 0.75 | 5 | 0.46 | |
| 6 | 0.22 | 6 | 0.57 | |
| Entropy | 0 | 0.82 | 0 | 0.68 |
| 2 | 0.99 | 2 | 0.42 | |
| 3 | 0.74 | 3 | 0.51 | |
| 4 | 0.74 | 4 | 0.87 | |
| 5 | 0.88 | 5 | 0.86 | |
| 6 | 0.92 | 6 | 0.72 | |
These results were no longer statistically significant after applying Bonferroni correction for type 1 error (multiple hypothesis testing).
For models using entropy values as predictors, cross-validated Kaplan-Meier curves and corresponding log-rank statistics are shown in Figure 4. At staging CT, using linear predictor values to categorize patients into groups at high or low risk for liver metastases resulted in inaccurate prognostic predictions; patients categorized as high risk had a significantly greater survival function than those categorized as low risk (p = 0.02). This suggests that leaving out a single patient’s hepatic entropy values during model LOOCV significantly affected the parameter estimates of the Cox model. Incorrect prognostic categorization was also seen when using entropy values at surveillance (pre-metastasis) CT to predict liver metastasis-free survival and overall survival.
Figure 4:
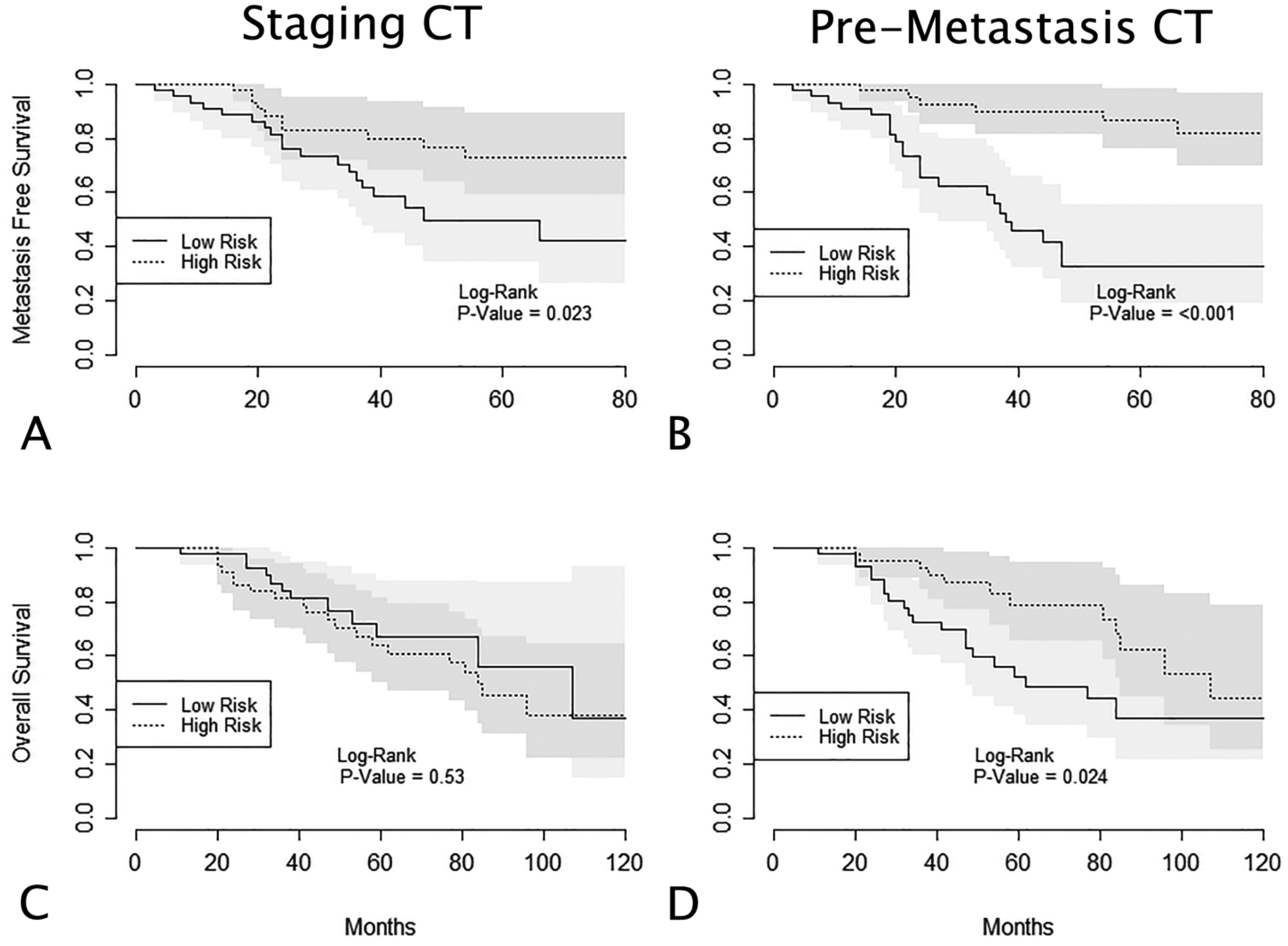
(A) Cross-validated Kaplan-Meier curves displaying liver metastasis-free survival for patients split into high and low risk groups based on linear predictor values at staging CT. (B) Liver metastasis-free survival for patients split into high and low risk groups based on linear predictor values at the “pre-metastasis” surveillance CT. (C, D) Overall survival for patients split into high and low risk groups based on linear predictor values at staging and pre-metastasis surveillance CT, respectively. Linear predictor values are calculated through leave-one-out cross-validation (LOOCV) of Cox survival models, using hepatic entropy values as covariates. In (A), (B), and (D), using linear predictor values to categorize patients into high and low risk groups resulted in incorrect predictions of patient prognoses (ie, “low risk” groups show decreased metastasis free survival).
For LASSO models using all 24 texture features as predictors, the partial likelihood deviance was still decreasing when the value for λ shrunk all coefficients to 0, which suggests that none of the calculated hepatic texture features from either staging or pre-metastasis CT scans are predictive of future occurrence of liver metastasis or overall survival.
Discussion:
In this study of patients with colorectal cancer, we have shown that CT texture features of the hepatic parenchyma shortly before the development of liver metastases are similar to those of matched colorectal cancer patients who do not develop liver metastases. We compared these CT texture features at both the initial cancer staging CT and at similar time intervals after biopsy-confirmed diagnosis (and immediately before CT-detectable metastases in the case cohort). Using survival analysis methods developed for high-dimensional data sets, we showed that these texture features were poor predictors of the occurrence of liver metastases and overall survival. Lastly, we tested the hypothesis that hepatic entropy values are predictive of liver metastases when measured on CT scans just before their development.
We were not able to predict overall survival or time to liver metastasis using survival models based on hepatic entropy values at staging and surveillance pre-metastatic CT. In some cases, using the models to assign patients to high and low risk groups resulted in categorizations that were worse than random assignment to those groups.
Prior studies using CT texture analysis have identified either uniformity or entropy of the liver parenchyma as potential predictors for clinical outcomes in patients with colorectal cancer [8, 12]. One study has suggested that entropy values of the liver parenchyma at staging CT may differ between patients who eventually develop liver metastases and those who do not, but this study had a limited sample size (total n = 29) [19]. We sought to test these intriguing hypotheses that entropy values at CT texture analysis might be predictive of either the development of hepatic metastases or overall survival. To improve our investigation, we not only included the initial staging CT, but also identified the last nominally normal CT (in terms of hepatic evaluation) before the development of liver metastases. Our results differ from these studies, and this difference may be due to a variety of reasons. Most notably, we used a more robust statistical methodology for feature selection and cross-validation for estimating model test error rates. These methods reduce data overfitting, leading to more conservative estimates of prediction accuracy. They are also less affected by the problems associated with multiple hypothesis testing, such as false positive associations between covariates and outcomes [20].
The statistical problems of multiple comparisons are inherent when performing analyses with large numbers of potential predictor variables, and independent validation studies of previously generated hypotheses are imperative before large-scale prospective studies of texture features’ clinical utility can be carried out [21]. This issue has caused criticism of texture analysis studies in the recently published literature. For example, one group conducted a systematic review of CT texture analysis studies and applied p-value correction to their results using the Benjamini-Hochberg method; none of the included studies’ results remained statistically significant after the corrections were applied [22]. Furthermore, they simulated 100 quantitative random variables in place of the original image-derived indices from one of the included studies and found that 10% of these variables were associated with clinical outcomes. The statistical methodologies of future texture analysis studies can be improved and false positive associations reduced by utilizing machine learning and data mining techniques developed for analyzing data in other ‘-omics’ disciplines [23, 24].
The results of texture analysis studies have been difficult to compare due to variability in acquisition, pre-processing, and reconstruction of images. For example, a recent phantom study demonstrated that texture features significantly varied across scanner models, and the authors suggested that researchers should develop a standardized acquisition technique when collecting images to be used in texture analysis studies [21]. Another study involving patients with non-small cell lung cancer tumors showed significant variations in over half (13/23) of the calculated CT texture features after simulating a decrease in tube current (mA), and also when comparing feature values from whole-tumor or largest cross-sectional slices of tumors [25]. Finally, the reconstruction algorithm most likely varied within our sample since our scans were obtained over a wide range of time, which also could have affected the texture parameters used in the study. Overall, these sources of variation make it difficult to validate results of texture studies performed at other institutions. The development of a texture-specific protocol that leads to minimal feature variation is necessary if predictive models are to be validated across different healthcare settings and translated into clinical practice.
There have been increasing efforts to produce guidelines and software that facilitate standardization, reproducibility, and collaboration between radiomics research groups. A recent review has proposed a set of potential guidelines for future texture analysis studies to facilitate reproducibility and pooling of results [21]. Additionally, open-source software platforms have been developed specifically for reproducible feature calculation [26]. These radiomics software platforms were developed with an emphasis on transparency and reproducibility of the computational methods used to perform texture analysis. It is paramount that future radiomics studies are as transparent as possible when describing the methods used to acquire, process, and analyze texture features from images.
There are several limitations to our study. Partly due to the retrospective method of data collection, CT scans included were performed at multiple institutions on a wide variety of scanner manufacturers and models in order to reach an adequate sample size. This could have biased the results and potentially could even mask actual differences between cases and controls. Also, we included patients with stage I-IV disease, although the majority of the patients in this study that developed liver metastases were diagnosed with stage III disease. Three patients had metastases at sites other than the liver at diagnosis, and this may have affected liver texture measurements in the liver which could conceivably bias the texture measurements within the case group. However, by design, there was a similar distribution of disease stage at diagnosis within the control group, and this may have sufficiently reduced the bias of the parameter confidence intervals.
In summary, we compared select CT texture features of the liver parenchyma in patients with colorectal cancer who developed metachronous liver metastases against those who did not develop liver metastases. These features were derived from staging CT images and from CT images just before the development of liver metastases. We were unable to demonstrate the predictive utility of the 24 calculated texture features using a machine learning algorithm developed for high-dimensional survival analysis. We also were unable to replicate the utility of entropy alone for predicting overall survival or development of liver metastases. Despite these findings, CT texture analysis still holds great promise for other clinical applications and is being investigated in the setting of response to therapeutics and evolution of the molecular profile of cancers. To advance the field of radiomics into clinical practice, future texture analysis studies must make efforts to reduce feature variation during image acquisition and computation so that generated models are easily replicable across research settings.
Key points:
Hepatic texture features on CT scans performed immediately prior to the development of identifiable colorectal liver metastases were unable to predict their imminent development.
CT texture features of the liver did not differ between cases and controls.
Although CT texture analysis can provide important prognostic information in oncology patients, this particular application failed to show benefit.
Acknowledgments
Funding provided by the Institute for Clinical and Translation Research (ICTR) at the University of Wisconsin - Madison. Patient consent was waived for this IRB-approved and HIPAA-compliant retrospective matched case-control study.
Dr. Pickhardt is co-founder of VirtuoCTC, consultant to Bracco, and shareholder in SHINE, Elucent, and Cellectar Biosciences; all other authors have no relevant financial disclosures
References
- 1.Steele CB, Rim SH, Joseph DA, et al. (2013) Colorectal cancer incidence and screening - United States, 2008 and 2010. Morb Mortal Wkly Rep Surveill Summ (Washington, DC 2002) 62 Suppl 3:53–60. [PubMed] [Google Scholar]
- 2.Report MW (2011) Vital signs: Colorectal cancer screening, incidence, and mortality--United States, 2002–2010. MMWR Morb Mortal Wkly Rep 60:884–9. [PubMed] [Google Scholar]
- 3.Manfredi S, Bouvier AM, Lepage C, et al. Incidence and patterns of recurrence after resection for cure of colonic cancer in a well defined population. [DOI] [PubMed]
- 4.Desch CE, Benson AB, Somerfield MR, et al. (2005) Colorectal cancer surveillance: 2005 update of an American Society of Clinical Oncology practice guideline. J Clin Oncol 23:8512–9. [DOI] [PubMed] [Google Scholar]
- 5.Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: Images Are More than Pictures, They Are Data. Radiology 278:563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganeshan B, Miles KA, Young RCD, Chatwin CR (2007) In Search of Biologic Correlates for Liver Texture on Portal-Phase CT. Acad Radiol 14:1058–1068. [DOI] [PubMed] [Google Scholar]
- 7.Miles KA, Hayball MP, Dixon AK (1993) Functional images of hepatic perfusion obtained with dynamic CT. Radiology 188:405–411. [DOI] [PubMed] [Google Scholar]
- 8.Ganeshan B, Miles KA, Young RCD, Chatwin CR (2007) Hepatic Enhancement in Colorectal Cancer. Texture Analysis Correlates with Hepatic Hemodynamics and Patient Survival. Acad Radiol 14:1520–1530. [DOI] [PubMed] [Google Scholar]
- 9.Haralick RM, Dinstein I, Shanmugam K (1973) Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. SMC-3: [Google Scholar]
- 10.Kruskal JB, Thomas P, Kane RA, Goldberg SN (2004) Hepatic perfusion changes in mice livers with developing colorectal cancer metastases. Radiology 231:482–490. [DOI] [PubMed] [Google Scholar]
- 11.Cuenod C, Leconte I, Siauve N, et al. (2001) Early changes in liver perfusion caused by occult metastases in rats: detection with quantitative CT. Radiology 218:556–561. [DOI] [PubMed] [Google Scholar]
- 12.Miles KA, Ganeshan B, Griffiths MR, et al. (2009) Colorectal cancer: texture analysis of portal phase hepatic CT images as a potential marker of survival. Radiology 250:444–52. [DOI] [PubMed] [Google Scholar]
- 13.Royston P, Altman DG (2013) External validation of a Cox prognostic model: principles and methods. BMC Med Res Methodol 13:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon RM, Subramanian J, Li M-C, Menezes S (2011) Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Brief Bioinform 12:203–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman J, Hastie T, Tibshirani R (2010) Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw 33: [PMC free article] [PubMed] [Google Scholar]
- 16.Hastie T, Tibshirani R, Friedman J (2001) The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer US [Google Scholar]
- 17.Witten DM, Tibshirani R (2010) Survival analysis with high-dimensional covariates. Stat Methods Med Res 19:29–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Team (2016) R: A Language and Environment for Statistical Computing. R Found. Stat. Comput [Google Scholar]
- 19.Rao S-X, Lambregts DM, Schnerr RS, et al. (2014) Whole-liver CT texture analysis in colorectal cancer: Does the presence of liver metastases affect the texture of the remaining liver? United Eur Gastroenterol J 2:530–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tello R, Crewson PE (2003) Hypothesis Testing II: Means. Radiology 227:1–4. [DOI] [PubMed] [Google Scholar]
- 21.Hatt M, Tixier F, Pierce L, et al. (2017) Characterization of PET/CT images using texture analysis: the past, the present… any future? Eur J Nucl Med Mol Imaging 44:151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chalkidou A, O’Doherty MJ, Marsden PK (2015) False Discovery Rates in PET and CT Studies with Texture Features: A Systematic Review. PLoS One 10:e0124165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Court LE, Fave X, Mackin D, et al. (2016) Computational resources for radiomics. Transl Cancer Res 5:340–348. [Google Scholar]
- 24.Kumar V, Gu Y, Basu S, et al. (2012) Radiomics: the process and the challenges. Magn Reson Imaging 30:1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackin D, Fave X, Zhang L, et al. (2015) Measuring Computed Tomography Scanner Variability of Radiomics Features. Invest Radiol 50:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Fried DV, Fave XJ, et al. (2015) ibex : An open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys 42:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]


