Abstract
Many infectious diseases are thought to have emerged in humans after the Neolithic revolution. While it is broadly accepted that this also applies to measles, the exact date of emergence for this disease is controversial. Here, we sequenced the genome of a 1912 measles virus and used selection-aware molecular clock modeling to determine the divergence date of measles virus and rinderpest virus. This divergence date represents the earliest possible date for the establishment of measles in human populations. Our analyses show that the measles virus potentially arose as early as the 6th century BCE, possibly coinciding with the rise of large cities.
One Sentence Summary:
Measles virus diverged from rinderpest virus in the 6th century BCE, which is compatible with an ancient emergence of measles.
Measles is a highly contagious viral disease that presents with rash, fever and respiratory symptoms. Before a live-attenuated vaccine was developed in the 1960s, the disease affected the vast majority of children (1, 2). Global vaccination campaigns resulted in a marked reduction of measles transmission and fatal cases and WHO has proclaimed an elimination goal. However, the disease still caused an estimated 110,000 deaths in 2017 (3) and incidence has recently been on the rise (4). Measles is caused by Measles morbillivirus (MeV), a negative sense single-stranded RNA virus from the family Paramyxoviridae (Order: Mononegavirales). MeV is an exclusively human pathogen whose closest relative was the now eradicated Rinderpest morbillivirus (RPV), a devastating cattle pathogen (5). It is generally accepted that measles emergence resulted from a spill-over from cattle to humans, although the directionality of this cross-species transmission event has never been formally established (supplementary text S1; 6).
It is unclear when measles first became endemic in human populations, but assuming an origin in cattle, the earliest possible date of MeV emergence is defined by the MeV-RPV divergence time. Several studies have provided estimates for this date using molecular clock analyses (7–10), with the most reliable (and oldest) estimate falling at the end of the 9th century CE (mean: 899 CE [95% highest posterior density (HPD) interval: 597 – 1144 CE]; (8). Here, we reassess the MeV-RPV divergence time using advanced, selection-aware Bayesian molecular clock modelling (11) on a dataset of heterochronous MeV genomes including the oldest human RNA virus genome sequenced to date, and show that a considerably earlier emergence can no longer be excluded.
Our re-examination was prompted by the broadly accepted view that molecular dating based on tip date calibration, i.e. the method used in previous efforts to estimate the timing of MeV-RPV divergence, underestimates deep divergence times (8). Rapid short-term substitution rates captured by tip calibration can often not be applied over long evolutionary timescales, because of the effects of long-term purifying selection and substitution saturation. This causes a discrepancy between short- and long-term substitution rates, which is referred to as the time-dependent rate phenomenon (12, 13). Since measurement timescales matter, a first step to arrive at accurate estimates is to maximize the time depth of tip calibration, for example through the use of ancient viral sequences (14, 15).
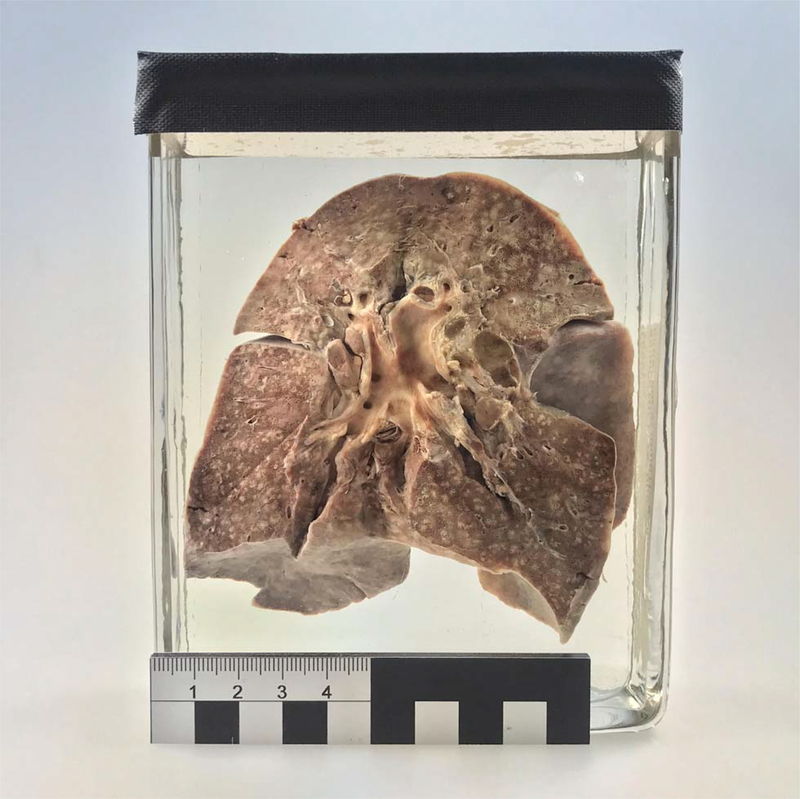
RNA tends to be much less stable in the environment than DNA, making the recovery of MeV genetic material from archeological remains unlikely (16). Pathology collections represent a more realistic source of MeV sequences that predate the oldest MeV genome – the genome of the Edmonston strain that was isolated in 1954 and attenuated to become the first measles vaccine. We examined a collection of lung specimens gathered by Rudolf Virchow and his successors between the 1870s and 1930s and preserved by the Berlin Museum of Medical History at the Charité (Berlin, Germany), and identified a 1912 case diagnosed with fatal measles-related bronchopneumonia (Fig. 1, fig. S1, supplementary texts S2 and S3). To retrieve MeV genetic material from this specimen, we first heat-treated 200mg of the formalin-fixed lung tissue to reverse macromolecule cross-links induced by formalin and subsequently performed nucleic acid extraction (17). Following DNase treatment and ribosomal RNA depletion, we built high-throughput sequencing libraries and shotgun sequenced them on Illumina® platforms. We generated 27,328,219 high quality reads, of which 0.46% were mapped to a MeV genome. Median insert size varied between 95 and 136 nucleotides and little damage was observed, suggesting good preservation of RNA molecules (fig. S2, table S1, supplementary text S4). The resulting 10,960 unique MeV reads allowed us to reconstruct an almost complete 1912 MeV genome: 15,257 of the 15,894 nucleotides in the MeV strain Edmonston (AF266288) were covered by at least 3 unique reads (11,988 nucleotides by at least 20 reads; mean coverage 54x).
Fig. 1.
Formalin-fixed lung specimen collected in 1912 in Berlin from a 2-year old girl diagnosed with measles-related bronchopneumonia (museum object ID: BMM 655/1912).
In addition to the 1912 genome and the 1954 Edmonston genome, only 2 genomes have been determined from MeV isolated prior to 1990 (Mvi/Lyon.FRA/77: HM562899; T11wild: AB481087). We therefore searched the strain collection of the German National Reference Laboratory (Robert Koch Institute, Berlin, Germany) for pre-1990 isolates. We found two strains from the pre-vaccine era isolated in 1960 by the National Reference Laboratory of former Czechoslovakia in Prague (MVi/Prague.CZE/60/1 and MVi/Prague.CZE/60/2; 18). We performed serial passages of these strains and determined their genome sequences at a mean coverage of 109x and 70x, respectively. The two genomes were nearly identical, differing at only four sites.
We performed Bayesian and maximum likelihood (ML) phylogenetic analyses to investigate the phylogenetic placement of the 1912 and 1960 genomes with respect to 127 available MeV genomes. Tip-dated Bayesian phylogenetic trees placed the 1912 genome as a sister lineage to all modern genomes while the two genomes from 1960 clustered together with the Edmonston strain (genotype A; fig. S3). The placement of the 1912 genome in the dated-tip tree was consistent with its placement in a non-clock ML tree reconstruction and with the rooting of a dated-tip tree excluding the 1912 genome (fig. S4 A and B). The relatedness of the 1912 and 1960 genomes to now extinct MeV lineages is in line with a marked reduction of MeV genetic diversity during the 20th century as a product of massive vaccination efforts.
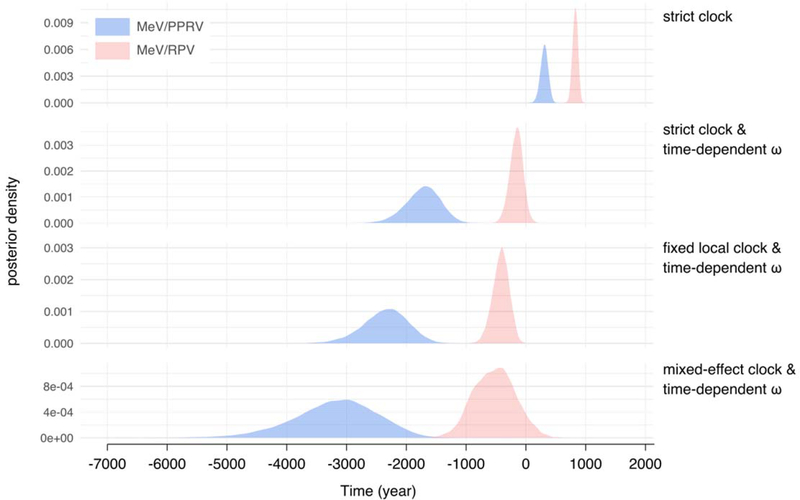
Having extended the time depth of MeV tip calibration, we subsequently focused our attention on estimating the timing of MeV-RPV divergence. We assembled a dataset of 51 genomes comprising MeV (including one of the 1960 genomes and the 1912 genome), RPV and Peste des petits ruminants virus (PPRV, the closest relative to MeV-RPV) sequences, ensuring they represented the known genetic diversity of these viruses (table S2). Prior to inferring a time-scaled evolutionary history for this dataset using a Bayesian phylogenetic framework, we assessed its temporal signal and tested it for substitution saturation. We confirmed a strong temporal signal (fig. S5, table S3) and did not identify strong substitution saturation (table S4). We constructed a series of increasingly complex evolutionary models to accommodate various sources of rate heterogeneity. Models ranged from a standard codon substitution model with a strict molecular clock assumption to a codon substitution model with time-varying selection combined with a clade-specific rate for PPRV and additional branch-specific random effects on the substitution rate. Adequately accommodating different sources of rate heterogeneity is known to provide a better correction for multiple hits in genetic distance estimation and the potential of codon substitution modelling in recovering deep viral divergence has specifically been demonstrated (8). This was reflected in the increasingly older estimates of MeV-RPV and PPRV-MeV-RPV divergence times and wider credible intervals for increasingly complex models (Fig. 2 and table S5). Parameter estimates of the substitution and clock models also provided evidence for a significant contribution of these different sources of rate heterogeneity to model fit improvement (table S5). We found a significantly negative coefficient for the time-dependent nonsynonymous/synonymous substitution rate ratio (ω) (11), indicating strong long-term purifying selection, a significantly positive coefficient for the fixed effect on the PPRV rate, indicating a faster evolutionary rate in this clade (as suggested by temporal signal analyses, fig. S5), and significant additional unexplained variation as modelled by the random effects (table S5). Our most complex model therefore provided the best description of the evolutionary process and significantly pushed back the divergence date of MeV and RPV, with a mean estimate at 528 BCE [95% HPD interval: 1174 BCE - 165 CE] (Fig. 3). These estimates were robust to (i) including or excluding the 1912 genome in the analyses (table S6), (ii) using a more conservative consensus genome for the 1912 sample (table S7), (iii) the prior specification on the age of the RPV genome or the inclusion of an additional RPV genome (table S7), and (iv) the coalescent prior specification (table. S7). A comparison of the four models in their ability to recover the age of the 1912 genome indicated that the most complex model yielded the best estimate (1929 CE [95% HPD interval: 1889 – 1961 CE]; fig. S6).
Fig. 2. Divergence time estimates for MeV and RPV (red) and for MeV/RPV and PPRV (blue) under increasingly complex evolutionary models.
Estimates for parameters of interest (posterior mean and 95% highest posterior density interval) under each model are provided in table S5.
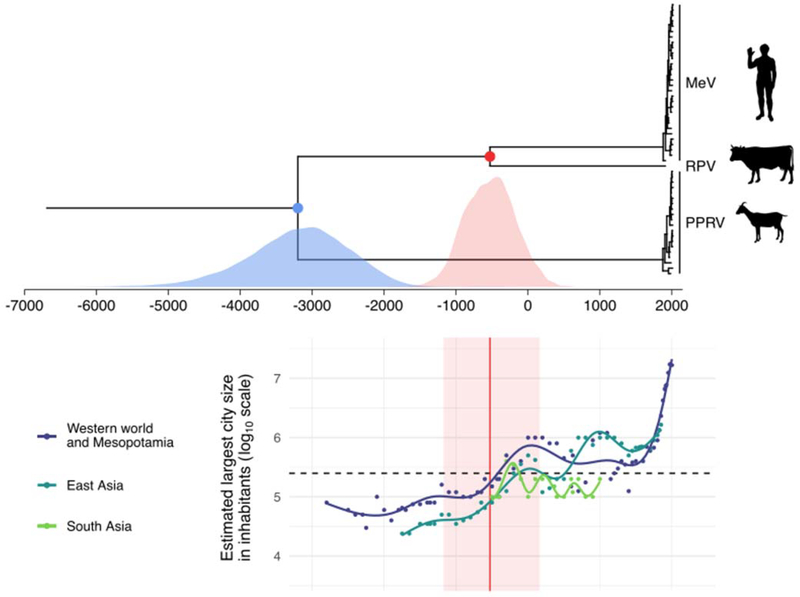
Fig. 3. Time-measured evolutionary history for MeV, RPV and PPRV, and largest city size over time in three well-studied regions of the world.
Upper figure displays maximum clade credibility (MCC) tree summarized from a Bayesian time-measured inference using tip-dating and accounting for long-term purifying selection. The red and blue points represent the mean estimates for the divergence times between MeV and RPV and MeV/RPV and PPRV, respectively; the corresponding divergence date estimates are depicted below as marginal posterior distributions. The lower figure represents the estimated size (log10 scale) of the largest city in the western world including Mesopotamia (dark blue), East Asia (teal) and South Asia (green) over time. The red vertical line represents the mean divergence time estimate between MeV and RPV and the red area its 95% highest posterior density interval. The dashed horizontal line represents the classical threshold for MeV maintenance in a population (i.e. 250,000 individuals). Dots show data points according to Morris (34) and Inoue et al. (26). Each line represents the fit of a generalized additive model with a cubic spline smoothing function.
The MeV/RPV divergence time provides the earliest possible date for measles emergence in humans, which is now compatible with the emergence of this disease more than 2,500 years ago. It seems plausible that the divergence of these lineages was closely followed by the cattle-to-human host jump and subsequent evolution into two distinct pathogens. However, the spill-over could have occurred at any time between the MeV/RPV divergence and the time to the most recent common ancestor of all MeV known to infect humans (1880 CE [95% HPD interval: 1865 – 1893 CE]). This raises the question of whether other sources of information can narrow down this timeframe and agree with an earlier timing of measles emergence.
The earliest clear clinical description of measles is often attributed to the Persian physician Rhazes, writing in the 10th century CE (19). But Rhazes was extremely familiar with all available medical literature at his time, and made use of earlier sources. Indian medical texts possibly describe measles several centuries prior to Rhazes (20). While clear descriptions of measles are missing in the Hippocratic corpus and the Greek medical tradition (at least through the prolific second-century writer Galen), such absence alone cannot be decisive. Retrospective diagnosis from pre-modern medical texts is notoriously fraught, especially for diseases like measles whose symptoms were easily confused with a variety of other conditions. Measles differential diagnosis remained a challenge well into more recent times (21). Therefore, any number of the large-scale “pestilences” described in ancient sources from Europe or China could reflect MeV outbreaks.
An ancient origin of measles seems all the more plausible in the light of demographic changes that are compatible with our understanding of (contemporary) MeV epidemiology. Populations large enough to support continuous MeV transmission, i.e. larger than the MeV critical community size (CCS) of 250,000–500,000 individuals (22–24), could not exist in Neolithic, Bronze Age, and early Iron Age settlements, which lacked both economic and political means to allow such numbers. Even if connectivity between such settlements may have created a larger pool of susceptible individuals, given the speed with which measles epidemics occur, and the efficacy of acquired immunity, epidemiologists have held that MeV could not have become endemic in urban populations below the CCS (25). In the late first millennium BCE, technologies (both economic and political) crossed a threshold promoting an upsurge in population sizes in Eurasia and South and East Asia. Although considerable uncertainty exists around population size estimates derived from ancient documents (e.g. literary observations, travelers’ reports, censuses, or references to the amount of food distributed in a city) or archaeological proxies (e.g. size of city walls, built-up area of settlement), there is broad agreement that a number of settlements in North Africa, India, China, Europe, and the Near East began to surpass the CCS for MeV by around 300 BCE, presumably for the first time in human history (Fig. 3; 26). From this period onward, there were consistently urban populations above the CCS for MeV.
Based on these considerations, our substantially older MeV/RPV divergence estimate provides grounds for sketching a new model of MeV’s evolutionary history. Under this scenario, a bovine virus, the common ancestor of modern strains of RPV and MeV, circulated in large populations of cattle (and possibly wild ungulates) since its divergence from PPRV around the 4th millennium BCE (3199 BCE [95% HPD interval: 4632 – 1900 BCE]; Fig. 3). As a fast-evolving RNA virus, it may have produced variants that were able to cross the species barrier on several occasions, but small human populations could only serve as dead-end hosts. Then, almost as soon as contiguous settlements reached sufficient sizes to maintain the virus’ continuous transmission (Fig. 3), it emerged as a human pathogen, the progenitor of modern-day MeV. It has been suggested that numerous concurrent human-bovine epidemics in the early medieval period (here 6th-10th centuries CE) were caused by an immediate ancestor of MeV and RPV that was pathogenic to both cattle and humans (27). The new RPV-MeV divergence date allows for the same inference to be made for earlier concurrent human-bovine mortality events well attested in e.g. Roman sources from the 5th century BCE on (28). During the following centuries, introduction of MeV into naive human populations and/or flare-ups of the disease might have caused some ancient epidemics whose etiology remains uncertain.
While our findings shed new light on the origin of measles, formally proving that the virus emerged soon after its divergence from RPV would require archeological genomic evidence. Most studies on ancient viruses have thus far focused on viruses with a double-stranded DNA genome (14, 29–32). However, genetic material of parvovirus B19 was also detected in early Neolithic skeletal remains, despite the relatively unstable nature of its single-stranded DNA genome (15). It remains to be determined if viral RNA recovery from such ancient specimens is feasible. Recently, RNA was extracted from the remains of a 14,300-year-old Pleistocene canid preserved in permafrost (33). While the majority of RNA fragments were extremely short (<30 nt), the authenticity of the sequences could be validated (33). Such advances highlight that it may not be completely impossible for ancient remains to still contain MeV RNA, especially if preserved under favorable circumstances, including natural mummification or preservation in cold environments (16). While awaiting such direct evidence, we believe that the proposed model of MeV evolution constitutes a compelling working hypothesis.
Supplementary Material
Acknowledgments:
The 1912 MeV genome was generated from a formalin-fixed lung specimen (museum object ID: BMM 655/1912) from the collection of the Berlin Museum of Medical History at the Charité (Berlin, Germany). Ethics approval was obtained from the ethics committee of the Charité (Berlin, Germany) under the reference number EA4/212/19. We thank Oliver Smith and Joel Wertheim for helpful suggestions. The Titan V GPU used for this research was donated by the NVIDIA Corporation.
Funding:
The research leading to these results has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 725422-ReservoirDOCS). PL acknowledges support by the Research Foundation -- Flanders (`Fonds voor Wetenschappelijk Onderzoek -- Vlaanderen’, FWO: G066215N, G0D5117N and G0B9317N). S.L. and B.V. are postdoctoral research fellows funded by the FWO. M.A.S. was partially supported through National Institutes of Health grant U19 AI135995.
Footnotes
Competing interests: All authors declare no competing interests.
Data and materials availability:
The sequencing data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB36265 (https://www.ebi.ac.uk/ena/data/view/PRJEB36265). Human reads have been removed from 1912 sequencing files prior to uploading (supplementary materials). Alignments, trees, and BEAST xml files are available at https://github.com/slequime/measles-history.
References and Notes:
- 1.Langmuir AD, Medical importance of measles. American journal of diseases of children (1960) 103, 224–226 (1962). [DOI] [PubMed] [Google Scholar]
- 2.Moss WJ, Measles. Lancet 390, 2490–2502 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Dabbagh A, Laws RL, Steulet C, Dumolard L, Mulders MN, Kretsinger K, Alexander JP, Rota PA, Goodson JL, Progress toward regional measles elimination—worldwide, 2000–2017. Morbidity and Mortality Weekly Report 67, 1323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO, Provisional data based on monthly data reported to WHO (Geneva) as of January 2020. https://www.who.int/immunization/monitoring_surveillance/burden/vpd/surveillance_type/active/measles_monthlydata/en/.
- 5.Roeder P, Mariner J, Kock R, Rinderpest: the veterinary perspective on eradication. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 368, 20120139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfe ND, Dunavan CP, Diamond J, Origins of major human infectious diseases. Nature 447, 279–283 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuse Y, Suzuki A, Oshitani H, Origin of measles virus: divergence from rinderpest virus between the 11th and 12th centuries. Virology journal 7, 52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wertheim JO, Kosakovsky Pond SL, Purifying selection can obscure the ancient age of viral lineages. Mol Biol Evol 28, 3355–3365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muniraju M, Munir M, Parthiban AR, Banyard AC, Bao J, Wang Z, Ayebazibwe C, Ayelet G, El Harrak M, Mahapatra M, Libeau G, Batten C, Parida S, Molecular evolution of peste des petits ruminants virus. Emerging infectious diseases 20, 2023–2033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura H, Saitoh M, Kobayashi M, Ishii H, Saraya T, Kurai D, Tsukagoshi H, Shirabe K, Nishina A, Kozawa K, Kuroda M, Takeuchi F, Sekizuka T, Minakami H, Ryo A, Takeda M, Molecular evolution of haemagglutinin (H) gene in measles virus. Scientific reports 5, 11648 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Membrebe JV, Suchard MA, Rambaut A, Baele G, Lemey P, Bayesian Inference of Evolutionary Histories under Time-Dependent Substitution Rates. Mol Biol Evol 36, 1793–1803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho SY, Duchene S, Molak M, Shapiro B, Time-dependent estimates of molecular evolutionary rates: evidence and causes. Molecular ecology 24, 6007–6012 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Aiewsakun P, Katzourakis A, Time-Dependent Rate Phenomenon in Viruses. Journal of virology 90, 7184–7195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mühlemann B, Jones TC, Damgaard PB, Allentoft ME, Shevnina I, Logvin A, Usmanova E, Panyushkina IP, Boldgiv B, Bazartseren T, Tashbaeva K, Merz V, Lau N, Smrcka V, Voyakin D, Kitov E, Epimakhov A, Pokutta D, Vicze M, Price TD, Moiseyev V, Hansen AJ, Orlando L, Rasmussen S, Sikora M, Vinner L, Osterhaus A, Smith DJ, Glebe D, Fouchier RAM, Drosten C, Sjogren KG, Kristiansen K, Willerslev E, Ancient hepatitis B viruses from the Bronze Age to the Medieval period. Nature 557, 418–423 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Mühlemann B, Margaryan A, Damgaard PB, Allentoft ME, Vinner L, Hansen AJ, Weber A, Bazaliiskii VI, Molak M, Arneborg J, Bogdanowicz W, Falys C, Sablin M, Smrcka V, Sten S, Tashbaeva K, Lynnerup N, Sikora M, Smith DJ, Fouchier RAM, Drosten C, Sjogren KG, Kristiansen K, Willerslev E, Jones TC, Ancient human parvovirus B19 in Eurasia reveals its long-term association with humans. Proceedings of the National Academy of Sciences of the United States of America 115, 7557–7562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith O, Gilbert MT, “Ancient RNA” in Paleogenomics: Genome-Scale Analysis of Ancient DNA, Lindqvist C, Rajora OP, Eds. (Springer, Cham, 2019). [Google Scholar]
- 17.Gilbert MT, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, Van Marck E, Worobey M, The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PloS one 2, e537 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santibanez S, Heider A, Gerike E, Agafonov A, Schreier E, Genotyping of measles virus isolates from central Europe and Russia. Journal of medical virology 58, 313–320 (1999). [PubMed] [Google Scholar]
- 19.Kim-Farley R, “Measles” in The Cambridge World History of Human Disease, Kiple KF, Ed. (Cambridge University Press, Cambridge, 1993). [Google Scholar]
- 20.Gupta KRL (translator), Madhava nidana: ayurvedic system of pathology (Sri Satguru Publications, Delhi, 1987). [Google Scholar]
- 21.Cunha BA, Smallpox and measles: historical aspects and clinical differentiation. Infectious disease clinics of North America 18, 79–100 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Bartlett MS, Measles periodicity and community size. Journal of the Royal Statistical Society. Series A (General) 120, 48–70 (1957). [Google Scholar]
- 23.Black FL, Measles endemicity in insular populations: critical community size and its evolutionary implication. Journal of theoretical biology 11, 207–211 (1966). [DOI] [PubMed] [Google Scholar]
- 24.Keeling MJ, Grenfell BT, Disease extinction and community size: modeling the persistence of measles. Science 275, 65–67 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Cliff AD, Haggett P, Smallman-Raynor M, Measles: an historical geography of a major human viral disease: from global expansion to local retreat, 1840–1990 (Blackwell, Oxford, 1993). [Google Scholar]
- 26.Inoue H, Álvarez A, Anderson EN, Owen A, Álvarez R, Lawrence K, Chase-Dunn C, Urban scale shifts since the bronze age: upsweeps, collapses, and semiperipheral development. Social Science History 39, 175–200 (2015). [Google Scholar]
- 27.Newfield TP, Human–bovine plagues in the early middle ages. Journal of Interdisciplinary History 46, 1–38 (2015). [Google Scholar]
- 28.Spinage CA, Cattle Plague: A History (Springer, Boston, 2003). [Google Scholar]
- 29.Biagini P, Theves C, Balaresque P, Geraut A, Cannet C, Keyser C, Nikolaeva D, Gerard P, Duchesne S, Orlando L, Willerslev E, Alekseev AN, de Micco P, Ludes B, Crubezy E, Variola virus in a 300-year-old Siberian mummy. The New England journal of medicine 367, 2057–2059 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Duggan AT, Perdomo MF, Piombino-Mascali D, Marciniak S, Poinar D, Emery MV, Buchmann JP, Duchene S, Jankauskas R, Humphreys M, Golding GB, Southon J, Devault A, Rouillard JM, Sahl JW, Dutour O, Hedman K, Sajantila A, Smith GL, Holmes EC, Poinar HN, 17th Century Variola Virus Reveals the Recent History of Smallpox. Current biology : CB 26, 3407–3412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patterson Ross Z, Klunk J, Fornaciari G, Giuffra V, Duchene S, Duggan AT, Poinar D, Douglas MW, Eden J-S, Holmes EC, The paradox of HBV evolution as revealed from a 16 th century mummy. PLoS pathogens 14, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krause-Kyora B, Susat J, Key FM, Kuhnert D, Bosse E, Immel A, Rinne C, Kornell SC, Yepes D, Franzenburg S, Heyne HO, Meier T, Losch S, Meller H, Friederich S, Nicklisch N, Alt KW, Schreiber S, Tholey A, Herbig A, Nebel A, Krause J, Neolithic and medieval virus genomes reveal complex evolution of hepatitis B. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith O, Dunshea G, Sinding MS, Fedorov S, Germonpre M, Bocherens H, Gilbert MTP, Ancient RNA from Late Pleistocene permafrost and historical canids shows tissue-specific transcriptome survival. PLoS biology 17, e3000166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris I, The measure of civilization: how social development decides the fate of nations (Princeton University Press, Princeton, 2013). [Google Scholar]
- 35.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA, metaSPAdes: a new versatile metagenomic assembler. Genome research 27, 824–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, Basic local alignment search tool. Journal of molecular biology 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 39.Jäger G, ClipAndMerge. https://github.com/apeltzer/ClipAndMerge.
- 40.Broad Institute, Picard. http://broadinstitute.github.io/picard.
- 41.Peltzer A, DeDup. https://github.com/apeltzer/DeDup.
- 42.Jonsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L, mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geneious 11.1.5. https://www.geneious.com.
- 44.Woelk CH, Pybus OG, Jin L, Brown DW, Holmes EC, Increased positive selection pressure in persistent (SSPE) versus acute measles virus infections. Journal of General Virology 83, 1419–1430 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Fukai K, Morioka K, Sakamoto K, Yoshida K, Characterization of the complete genomic sequence of the rinderpest virus Fusan strain cattle type, which is the most classical isolate in Asia and comparison with its lapinized strain. Virus genes 43, 249–253 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Katoh K, Standley DM, MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B, RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol 1, vev003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ, IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rambaut A, Lam TT, Max Carvalho L, Pybus OG, Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2, vew007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duchene S, Stadler T, Ho SY, Duchene DA, Dhanasekaran V, Baele G, Bayesian Evaluation of Temporal Signal in Measurably Evolving Populations. bioRxiv, 810697 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia X, Xie Z, Salemi M, Chen L, Wang Y, An index of substitution saturation and its application. Molecular phylogenetics and evolution 26, 1–7 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Xia X, Lemey P, “Assessing substitution saturation with DAMBE“ in The phylogenetic handbook: a practical approach to DNA and protein phylogeny, Lemey P, Salemi M, Vandamme AM, Eds. (Cambridge University Press, Cambridge, 2009). [Google Scholar]
- 53.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A, Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4, vey016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA, Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Systematic biology 67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chernomor O, Minh BQ, Forest F, Klaere S, Ingram T, Henzinger M, von Haeseler A, Split diversity in constrained conservation prioritization using integer linear programming. Methods in ecology and evolution 6, 83–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ayres DL, Cummings MP, Baele G, Darling AE, Lewis PO, Swofford DL, Huelsenbeck JP, Lemey P, Rambaut A, Suchard MA, BEAGLE 3: Improved Performance, Scaling, and Usability for a High-Performance Computing Library for Statistical Phylogenetics. Systematic biology 68, 1052–1061 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goldman N, Yang Z, A codon-based model of nucleotide substitution for protein-coding DNA sequences. Molecular biology and evolution 11, 725–736 (1994). [DOI] [PubMed] [Google Scholar]
- 58.Yoder AD, Yang Z, Estimation of primate speciation dates using local molecular clocks. Mol Biol Evol 17, 1081–1090 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Vrancken B, Rambaut A, Suchard MA, Drummond A, Baele G, Derdelinckx I, Van Wijngaerden E, Vandamme AM, Van Laethem K, Lemey P, The genealogical population dynamics of HIV-1 in a large transmission chain: bridging within and among host evolutionary rates. PLoS computational biology 10, e1003505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baron MD, Barrett T, Rescue of rinderpest virus from cloned cDNA. Journal of virology 71, 1265–1271 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, seqtk. https://github.com/lh3/seqtk. [Google Scholar]
- 63.Chandler T, Four thousand years of urban growth: An historical census (Mellen, Lewiston, 1987). [Google Scholar]
- 64.Modelski G, World cities: −3000 to 2000. (Faros, Washington D.C., 2000, 2003). [Google Scholar]
- 65.Bairoch P, Cities and economic development: from the dawn of history to the present (University of Chicago Press, Chicago, 1988). [Google Scholar]
- 66.Pasciuti D, Chase-Dunn C, Estimating the population sizes of cities. https://irows.ucr.edu/research/citemp/estcit/estcit.htm (Accessed 10/25/2019) [Google Scholar]
- 67.Pitt D, Sevane N, Nicolazzi EL, MacHugh DE, Park SDE, Colli L, Martinez R, Bruford MW, Orozco-terWengel P, Domestication of cattle: Two or three events? Evolutionary applications 12, 123–136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morand S, McIntyre KM, Baylis M, Domesticated animals and human infectious diseases of zoonotic origins: domestication time matters. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases 24, 76–81 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Mariner JC, McDermott J, Heesterbeek JA, Catley A, Roeder P, A model of lineage-1 and lineage-2 rinderpest virus transmission in pastoral areas of East Africa. Preventive veterinary medicine 69, 245–263 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Barrett T, Rossiter PB, Rinderpest: the disease and its impact on humans and animals. Advances in virus research 53, 89–110 (1999). [DOI] [PubMed] [Google Scholar]
- 71.McNeill WH, Plagues and Peoples. (Anchor Press, Norwell, 1976). [Google Scholar]
- 72.Patoret P-P, Yamanouchi K, Mueller-Doblies U, Rweyemamu MM, Horzinek M, Barrett T, “Rinderpest – an old and worldwide story: history to c.1902” in Rinderpest and Peste des Petits Ruminants: Virus Plagues of Large and Small Ruminants, Barrett T, Pastoret P-P, Taylor W, Eds. (Elsevier, Amsterdam, 2005). [Google Scholar]
- 73.Blancou J, “Old prophylactic methods“ in Rinderpest and Peste des Petits Ruminants: Virus Plagues of Large and Small Ruminants, Barrett T, Pastoret P-P, Taylor W, Eds. (Elsevier, Amsterdam, 2005). [Google Scholar]
- 74.OIE, Peste des Petits Ruminants. https://www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/Disease_cards/PESTE_DES_PETITS_RUMINANTS.pdf.
- 75.Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, Rupprecht CE, Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 329, 676–679 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Lembo T, Oura C, Parida S, Hoare R, Frost L, Fyumagwa R, Kivaria F, Chubwa C, Kock R, Cleaveland S, Batten C, Peste des petits ruminants infection among cattle and wildlife in northern Tanzania. Emerging infectious diseases 19, 2037–2040 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Couacy-Hymann E, Koffi MY, Kouadio VK, Mossoum A, Kouadio L, Kouassi A, Assemian K, Godji PH, Nana P, Experimental infection of cattle with wild type peste-des-petits-ruminants virus - Their role in its maintenance and spread. Research in veterinary science 124, 118–122 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Cottontail VM, Rasche A, Yordanov S, Seebens A, Knornschild M, Oppong S, Adu Sarkodie Y, Pongombo C, Lukashev AN, Schmidt-Chanasit J, Stocker A, Carneiro AJ, Erbar S, Maisner A, Fronhoffs F, Buettner R, Kalko EK, Kruppa T, Franke CR, Kallies R, Yandoko ER, Herrler G, Reusken C, Hassanin A, Kruger DH, Matthee S, Ulrich RG, Leroy EM, Drosten C, Bats host major mammalian paramyxoviruses. Nat Commun 3, 796 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ghawar W, Pascalis H, Bettaieb J, Melade J, Gharbi A, Snoussi MA, Laouini D, Goodman SM, Ben Salah A, Dellagi K, Insight into the global evolution of Rodentia associated Morbilli-related paramyxoviruses. Scientific reports 7, 1974 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pulliam JR, Epstein JH, Dushoff J, Rahman SA, Bunning M, Jamaluddin AA, Hyatt AD, Field HE, Dobson AP, Daszak P, Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis. Journal of the Royal Society, Interface 9, 89–101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Halpin K, Young PL, Field HE, Mackenzie JS, Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. The Journal of general virology 81, 1927–1932 (2000). [DOI] [PubMed] [Google Scholar]
- 82.Mina MJ, Kula T, Leng Y, Li M, De Vries RD, Knip M, Siljander H, Rewers M, Choy DF, Wilson MS, Measles virus infection diminishes preexisting antibodies that offer protection from other pathogens. Science 366, 599–606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB36265 (https://www.ebi.ac.uk/ena/data/view/PRJEB36265). Human reads have been removed from 1912 sequencing files prior to uploading (supplementary materials). Alignments, trees, and BEAST xml files are available at https://github.com/slequime/measles-history.