Abstract
Background and purpose:
Lonidamine is a hexokinase II inhibitor, works as an anticancer molecule, and is extensively explored in clinical trials. Limited information prevails about the stability-indicating methods which could determine the forced degradation of lonidamine under stressed conditions. Hence, we report the use of a rapid, sensitive, reproducible, and highly accurate liquid chromatography and mass spectrometry method to analyze lonidamine degradation.
Experimental approach:
The Xbridge BEH shield reverse phase C18 column (2.5 μm, 4.6 × 75 mm) using isocratic 50:50 water: acetonitrile with 0.1% formic acid can detect lonidamine with help of mass spectrometer in tandem with an ultraviolet (UV) detector at 260 nm wavelength.
Findings/ Results:
A linear curve with r2 > 0.99 was obtained for tandem liquid chromatography-mass spectrometry (LC-MS)-UV based detections. This study demonstrated (in the present set up of isocratic elution) that LC-MS based detection has a relatively high sensitivity (S/N (10 ng/mL): 220 and S/N (20 ng/mL): 945) and accuracy at lower detection and quantitation levels, respectively. In addition to developing the LC-MS method, we also report that the current method is stability-indicating and shows that lonidamine gets degraded over time under all three stress conditions; acidic, basic, and oxidative.
Conclusion and implications:
LC-MS based quantitation of lonidamine proved to be a better method compared to high-performance liquid chromatography (HPLC)-UV detections for mapping lonidamine degradation. This is the first report on the stability-indicating method for studying the forced degradation of lonidamine using LC-MS method.
Keywords: Forced degradation, LC-MS, Lonidamine, Stability indicating
INTRODUCTION
Approximately 1.7 million U.S. individuals are diagnosed with cancer each year and nearly half will die from the disease, according to data from the National Institute of Health (1,2). The rapidly growing nature of cancer cells indicates that cancer is an energy-dependent phenomenon. In the late 1990s, it was suggested that this rapid proliferation resulted from increased expression of genes coding for glucose transporters and glycolytic enzymes in tumor cells (3).
The glycolysis hypothesis provided an opportunity for cancer researchers to explore the use of novel and targeted therapies (4,5). In particular, they targeted hexokinase II (HKII), a protein that performs catalysis of the first step of glycolysis. Along with porin (a mitochondrial protein), it prevents the release of cytochrome C during apoptosis cascade. Research showed that the depletion of HKII in cancer cells increases the concentration of reactive oxygen species (ROS), which leads to cell death (6).
Furthermore, the presence of certain HKII inhibitors causes detachment of HKII enzyme from mitochondria, which leads to cell death by a drastic reduction in ATP (7,8). This makes HKII protein an important drug target for next- generation anticancer therapy. Three molecules that inhibit HKII have been studied for their anticancer effects including 2-deoxyglucose, 3-bromopyruvate, and clotrimazole (5).
Researchers have also studied the anticancer potential of lonidamine (2-indole-carboxylic acid) as HKII inhibitor. It selectively reduces glycolysis of tumor cells, possibly by inhibiting mitochondrial-bound hexokinase, which is absent in normal differentiated cells (9,10). Lonidamine also reduces lactate production in B-cell chronic leukemia, possibly due to reduced cellular ATP (which is required for glycolysis) (11). Furthermore, lonidamine has been shown to improve the sensitivity of cancer cells to known anticancer drugs such as doxorubicin, cisplatin, melphalan, and others (12,13,14,15). As a result, this molecule was explored in phase I and II clinical trials in combination therapy against breast cancer, glioblastoma multiforme, ovarian cancer, and lung cancer (16). Lonidamine was well-tolerated but limited by poor bioavailability after oral administration.
In light of this poor bioavailability, lonidamine is currently been explored in microencapsulation and targeted nanoparticle- based strategies to improve its delivery (17,18). Current methods for quantifying lonidamine are limited, particularly at the lower concentrations employed in preclinical experiments. Lonidamine is usually quantified using conventional high-performance liquid chromatography (HPLC) assays in biological fluids and samples (19). Most HPLC methods have relatively high limits of detection and quantitation levels, indicating relatively low sensitivities at lower concentrations (20,21,22). Newer high-resolution mass spectrometers and ultra HPLC systems can aid in developing highly sensitive assays for the quantitation of lonidamine. To the best of our knowledge, LC-MS method has not been used to quantify lonidamine for analytical applications. Furthermore, the best-known method for studying the stability study of lonidamine was conducted with HPLC with limited information about the degradation profile of lonidamine. Our research indicates that LC-MS based methods for quantitation of pharmaceutical active ingredients and their metabolites can prove to be significantly sensitive (23,24). In the current work, we demonstrated a highly sensitive LC-MS method for quantitating lonidamine using single or selected ion monitoring (SIM) mode. Results showed improved sensitivity of LC-MS over HPLC- ultraviolet (UV) detection. Finally, the assay was used to quantitatively assess the degradation profile of lonidamine under strong acidic, basic, oxidative stress conditions.
MATERIALS AND METHODS
Chemicals and reagents
Lonidamine was procured from Sigma Aldrich (St. Louis, MO, USA). LC-MS grade acetonitrile, water, and formic acid were purchased from Fischer Scientific (Fair Lawn, NJ, USA). XBridge C18 reverse phase (RP) HPLC column was procured from Waters Corp. (Milford, MA, USA). Sodium hydroxide pellets NF, hydrochloric acid, and hydrogen peroxide were purchased from Fisher Chemicals (Fair Lawn, NJ, USA).
Liquid chromatography conditions
A lonidamine quantitation assay was established using an isocratic elution method with 50:50 acetonitrile with 0.1% formic acid and water with 0.1% formic acid as a mobile phase. The sample injection was set to 20 μL with a flow rate of 0.350 mL/min. The temperature of the column was maintained at 30 °C with constant data acquisition for 20 min of run time. Ultra-high-performance LC-MS (UHPLC-MS) in tandem with UV (260 nm wavelength) was used for developing the quantitation method. Here, a Dionex 3000 UHPLC system (USA) was used for developing the tandem UHPLC-MS-UV method. The HPLC was a quaternary pump system with autosampler, column oven, and temperature-controlled sample tray. Exactive (v 1.1SP6) software was used for MS method development and data acquisition. Thermo Excaliber (v 3.0.63) was used for the integration of Chromeleon and Exactive for sample injections and LC-MS data acquisition in .raw format.
Mass spectrometry conditions
For identification of analyte (lonidamine) in solution eluting out of the HPLC column, a mass scanning range of 100 to 322 m/z was used. All scans were performed under positive ion mode. The sheath gas flow rate was maintained at 30 psi with an electron spray ionization voltage of 3.50 kV and a capillary temperature of 380 °C. Capillary, tube lens, and skimmer voltages were maintained at 45, 85, and 18 V, respectively. These parameters were adjusted using the auto-tune function of Thermo Exactive (v. 1.1 Sp6) software by direct infusion of lonidamine in the mass spectrometer. Using this function, the software automatically adjusts the parameters mentioned above to optimize the signal-to-noise (S/N) ratio (around 107 units) for lonidamine. Hence, an offline MS method was developed for the identification of lonidamine, which was then integrated with LC to develop quantitation and stability-indicating methods.
Sample preparation
Primary stocks of lonidamine were prepared in methanol (LC-MS grade) at a concentration of 1 mg/mL and stored at -20 °C. This aliquot was used for making secondary stock of 10 μg/mL. Later, the secondary stock solution was serially diluted in the concentration range of 10 to 10,000 ng/mL. All the serially diluted samples were stored at 4 °C for inter-day runs. All inter- and intra-day runs were performed at 20 °C. All the serially diluted samples were subjected to LC-MS-UV tandem runs.
Linearity
A linearity curve (for LC-MS and HPLC-UV) was prepared for the calibration standard samples (10 to 10,000 ng/mL) using values from three independent day runs, with two repeated injection for each set, n = 6 (3 × 2) injections. While developing the quantitation assay, the acceptable mean value of estimated concentration (% accuracy or recovery) for calibration and validation standards were kept within ± 15% to ± 20% of the theoretical value (25). The mean value of the lower limit of quantitation (LLOQ) was set within ± 10% coefficient of variance (CV). For other (higher) concentrations, this limit was set to ± 5% CV. According to Moorthy et al. at least 75% of the non-zero standards should meet these criteria and the linear coefficient of determination (r2) should be ≥ 0.99 (26). A linear regression (Y = mX + C) equation was determined for both data sets from concentration vs area of calibration standards. A linear regression equation from LC-MS standard runs was used to determine the time-dependent degradation of lonidamine under various forced degradation conditions.
Validation and stability
The quantitative method for LC-MS was validated for inter- and intra-day accuracy, %CV, and stability. Validation of the method was carried out using quality control (QC) standards of four random concentrations (80, 200, 3000, and 8000 ng/mL). Measurements were made for inter- and intra-day %CV and % accuracy. Furthermore, to check the stability of solutions, LC-MS analysis for lonidamine at various concentrations (20, 40, 500, and 10,000 ng/mL) were performed after freeze (-20°C)- thaw cycles.
Acid degradation studies of lonidamine
Preliminary studies have shown that lonidamine degrades at 1 μg/mL under 1 N HCl at 40 °C (28,29). Hence to assess the rate of degradation and probable degradation peaks of lonidamine, it was subjected to forced degradation with 1 N HCl under 40 and 80 °C with a final volume of solution as 10 mL. Samples (100 μL) were withdrawn at specific time (0, 6, 24, 48, and 72 h) points, centrifuged at 15,700 g for 10 min, and subjected to LC-MS runs to identify probable degradation chromatograms.
Alkali degradation studies of lonidamine
Similar to acidic degradation, lonidamine can also degrade under alkaline conditions. In order to assess the degradation process, lonidamine was treated with 1 N NaOH under 40 and 80 °C with a final volume (of solution) of 10 mL. Samples (100 μL) were withdrawn at specific time (0, 6, 24, 48, and 72 h) points, centrifuged at 15,700 g for 10 min, and subjected to LC-MS runs to identify probable degradation peaks in the chromatogram.
Oxidative degradation studies
Finally, lonidamine was subjected to oxidizing degradation using concentrated H2O2 with a sample volume of 10 mL (27,28). Samples were subjected to temperature treatment at 40 °C. Samples (100 μL) were withdrawn at specific time (0, 6, 24, and 48 h) points, centrifuged at 15,700 g for 10 min, and subjected to LC-MS runs to identify degradation profile.
Statistical analysis
Two tailed student’s t-test was used for the comparative analysis between data of interest. Wherever possible data is presented as mean ± standard deviation along with the respective P values.
RESULTS
Method development for the quantitative determination of lonidamine
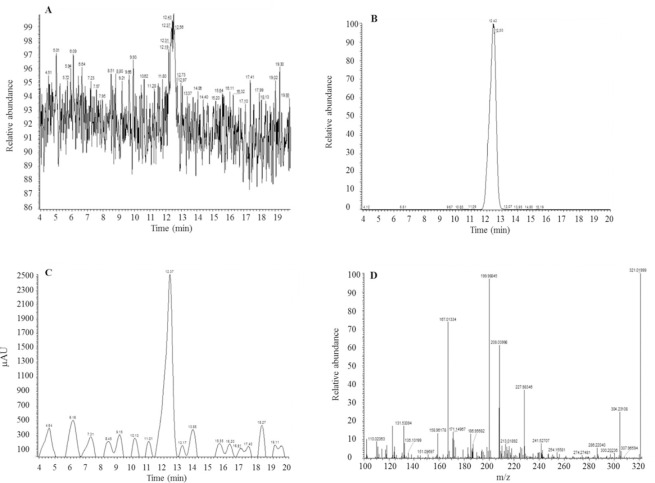
The retention time of standard lonidamine was found to be 12.34 ± 0.4 min with selected or targeted mass (M+H) + ion as 321.02 m/z, as shown in Fig. 1. The linearity equation (Y = 32792X) was established with correlation coefficient (r2) 0.9959. The average accuracy of inter- and intra-day run was found to be 88.14 ± 0.568% to 115.577 ± 0.469%, respectively. In the case of LC-MS method, although the signal intensity was detectable at 20 ng/mL, the % accuracy dropped below 100 ± 20%. Hence, for LC-MS assay, 10 ng/mL and 20 ng/mL were considered as LLOD and LLOQ, respectively. The tandem UV absorbance linearity equation (Y = 162.86X) was also found to have r2 = 0.9999 with average accuracy in the range of 90.84 ± 6.60% to 100.90 ± 0.31%.
Fig. 1.
Representative (A) TIC of lonidamine, (B) XIC-LC-MS chromatogram of lonidamine, (C) tandem HPLC-UV chromatogram, and (D) corresponding mass spectra of lonidamine with target mass ion (M+H)+ as 321.02 m/z. TIC, Total Ion chromatogram; XIC, extracted Ion chromatogram; LC-MS, liquid chromatography-mass spectrometry; HPLC, high-performance liquid chromatography; UV, ultraviolet.
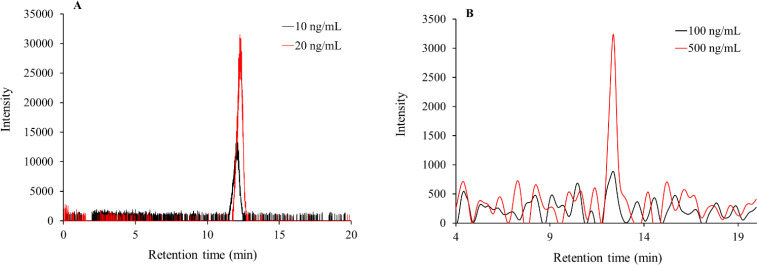
The accuracy% and CV% data for the linearity curve from LC-MS-UV tandem runs are summarized in Table 1. In case of HPLC-UV assay, the % accuracy dropped below 100 ± 20 % at concentrations below 500 ng/mL. Hence, 100 ng/mL (49.32 % accuracy) and 500 ng/mL (99.15% accuracy) were considered as LLOD and LLOQ for the assay. The relative sensitivity of LC-MS (Fig. 2A) was found to be higher compared to HPLC-UV (Fig. 2B) detections. Hence, looking at the % accuracy values for both LC-MS and HPLC-UV runs, linearity curve was established in the range of 20 to 10,000 ng/mL and 500 to 1000 ng/mL, respectively.
Table 1.
The CV and accuracy for the calibration standards used for plotting calibration curves for lonidamine using LC-MS runs, and tandem HPLC runs of lonidamine (with UV base detection) for its quantitation. Detection for the drug molecule was carried out at 230 nm wavelength.
| LC-MS | ||||||
|---|---|---|---|---|---|---|
| Con. (ng/mL) | Inter-day, n = 6 | Intra-day, n = 2 | Accuracy (%) Inter-day, n = 6 |
Accuracy (%) Intra-day, n = 2 |
||
| CV (%) | Area ± SD | CV (%) | Area ± SD | |||
| 20 | 1.61 | 412178 ± 6618 | 3.56 | 416858 ±14826 | 62.85 ± 1.01 | 63.56 ± 2.26 |
| 40 | 2.28 | 1314465 ± 29937 | 2.23 | 1335634 ± 29745 | 100.21 ± 2.28 | 101.82 ± 2.27 |
| 50 | 1.28 | 1693037 ± 21619 | 1.01 | 1708324 ± 17188 | 103.30 ± 1.32 | 104.19 ± 1.05 |
| 100 | 1.30 | 2877082 ± 37260 | 3.15 | 2903429 ± 91380 | 87.74 ± 1.14 | 88.54 ± 2.79 |
| 500 | 0.74 | 18570382 ± 138042 | 2.10 | 18667992 ± 392568 | 113.26 ± 0.84 | 113.86 ± 2.39 |
| 1000 | 1.27 | 37098195 ± 471480 | 1.81 | 37431582 ± 678901 | 113.13 ± 1.44 | 114.15 ± 2.07 |
| 2000 | 0.81 | 75582289 ± 615255 | 1.36 | 76017341 ± 1030986 | 115.24 ± 0.94 | 115.91 ± 1.57 |
| 5000 | 1.25 | 177493941 ± 2220627 | 1.68 | 179064161 ± 3005901 | 108.25 ± 1.35 | 109.21 ± 1.38 |
| 10000 | 0.87 | 318617153 ± 2779019 | 4.16 | 320582216 ± 13341608 | 97.16 ± 0.85 | 97.76 ± 4.07 |
| HPLC-UV | ||||||
| 100 | 124.7 | 8032 ± 10020 | 141.4 | 4836 ± 6840 | 49.32 ± 61.57 | 29.67 ± 42.00 |
| 500 | 7.12 | 80743 ± 5746 | 0.41 | 81475 ± 338 | 99.16 ± 7.05 | 100.05 ± 0.41 |
| 1000 | 8.70 | 155543 ± 13533 | 8.13 | 140343 ± 11411 | 95.51 ± 8.31 | 86.17 ± 7.01 |
| 2000 | 0.39 | 325148 ± 1266 | 2.34 | 324296 ± 7596 | 99.82 ± 0.38 | 99.56 ± 2.33 |
| 5000 | 1.75 | 823416 ± 14412 | 1.61 | 819876 ± 13223 | 101.1 ± 1.76 | 100.68 ± 1.62 |
| 10000 | 1.08 | 1624961 ± 17589 | 1.09 | 1623112 ± 17635 | 99.78 ± 1.07 | 99.66 ± 1.08 |
Con., Concentration; SD, standard deviation; CV, Coefficient of variance; LC-MS, chromatography-mass spectrometry; HPLC, high-performance liquid chromatography; UV, ultraviolet.
Fig. 2.
Representative XIC-LC-MS for (A) LLOD (S/N: 220) as 10 ng/mL and LLOQ (S/N: 945) as 20 ng/mL for lonidamine and (B) shows representative LLOD (S/N: 1) as 100 ng/mL and LLOQ (S/N:4) as 500 ng/mL for the HPLC-UV quantitation method. XIC, Extracted Ion chromatogram; LC-MS, liquid chromatography-mass spectrometry; LLOD, lower limit of detection; LLOQ, lower limit of quantitation; HPLC, high-performance liquid chromatography; UV, ultraviolet.
Validation and stability testing of LC-MS method for the quantitation of lonidamine
The LC-MS method and the calibration curve were validated using random known concentrations of lonidamine. Table 2 shows the data (accuracy% and CV%) for the validation runs. The accuracy for inter- and intra-day runs was found to be in the range of 93.36 ± 1.07% and 115.13 ± 2.50%, respectively. The intra- and inter-day validation results for random concentrations showed CV (< 5%) in the range of 0.99 ± 1.16% to 3.68 ± 1.14%. Furthermore, stability testing (for freeze-thaw cycles) of lonidamine solution using four additional random concentrations showed accuracy around 100 ± 20% and CV < 10% shown in Table 3. This indicates that the present method of LC-MS may serve to be optimum for the quantitation of degradation peaks in the stability-indicating method for lonidamine.
Table 2.
The CV and accuracy for random concentrations of lonidamine used for the validation of LC-MS based calibration curve.
| Con. (ng/mL) | Inter-day, n = 3 | Intra-day, n = 2 | Accuracy (%) Inter-day (n = 3) |
Accuracy (%) Intra-day (n = 2) |
||
|---|---|---|---|---|---|---|
| CV (%) | Area ± SD | CV (%) | Area ± SD | |||
| 80 | 2.871 | 2742049 ± 78827 | 0.17 | 2679149 ± 4575 | 104.52 ± 3.00 | 102.13 ± 0.17 |
| 200 | 2.26 | 7360566 ± 166086 | 4.48 | 7257893 ± 324949 | 112.23 ± 2.53 | 110.67 ± 4.95 |
| 3000 | 2.371 | 104886880 ± 2482132 | 1.75 | 102973246 ±1801549 | 106.62 ± 2.52 | 104.67 ± 1.83 |
| 8000 | 1.82 | 246914062 ± 4488524 | 2.41 | 242932821 ± 5858593 | 94.12 ± 1.71 | 92.60 ± 2.23 |
Con., Concentration; SD, standard deviation, CV, Coefficient of variance; LC-MS, chromatography-mass spectrometry.
Table 3.
The CV and accuracy (from LC-MS runs) for random concentration for lonidamine after freezethaw cycles (n = 2).
| Concentrations (ng/mL) | CV (%) | Accuracy (%) |
|---|---|---|
| 20 | 3.41 ± 0.60 | 62.43 ± 0.89 |
| 40 | 0.78 ± 0.20 | 99.19 ± 0.30 |
| 500 | 1.59 ± 0.20 | 108.73 ± 3.68 |
| 10000 | 4.89 ± 0.22 | 88.59 ± 1.59 |
| CV, Coefficient of variance; chromatography-mass spectrometry. | LC-MS, | liquid |
CV, Coefficient of variance; LC-MS, liquid chromatography-mass spectrometry.
Forced degradation of lonidamine in the presence of strong acid.
Due to exposure of lonidamine to 1 N HCl caused a time-dependent reduction in the concentration of the parent ion peak. This reduction was increased on a temperature- dependent basis as shown in Fig. 3.
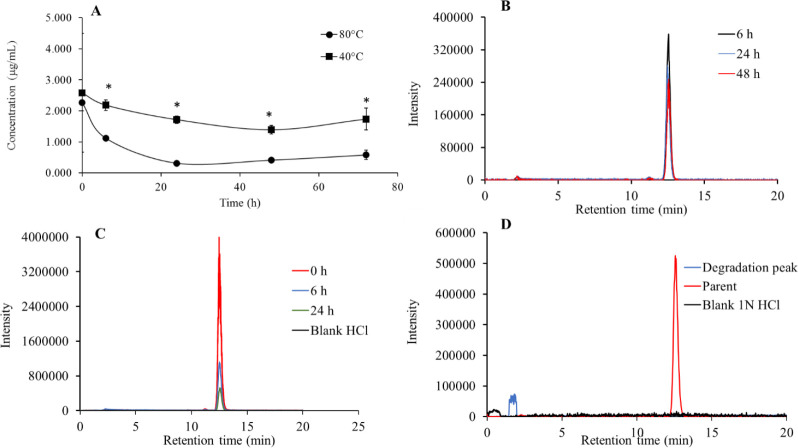
Fig. 3.
(A) Degradation of lonidamine in 1 N HCl at 40 and 80 °C, respectively. *P < 0.05 indicates significant differences between every two points regarding their corresponding time pint. Overall data suggest that lonidamine was degraded significantly at 80°C under 1N HCl; (B and C) chromatograms of lonidamine at 40 and 80 °C, respectively, at specific time points showing the relative degradation pattern in 1 N HCl; and (D) chromatograms of probable degradation peak vs standard and blank 1 N HCl.
The degradation of lonidamine was relatively higher at 80 °C (P < 0.05) after 20 h of incubation. The overall degradation of lonidamine was significantly (P < 0.05) higher at 80 °C in comparison to 40 °C in 1 N HCl. Degradation peak was also identified in the mass range 276 to 277 m/z (Fig. 3) at the retention time of 1.88 min.
Forced degradation of lonidamine in the presence of a strong base
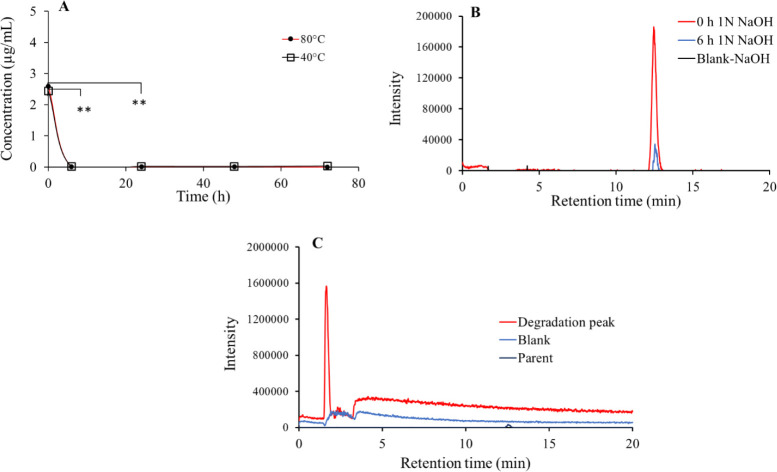
As shown in Fig. 4, it was observed that lonidamine gets degraded in basic (1N NaOH) condition showed a temperature-independent effect. It was detected that parent ion peak quantitatively reduced upon incubation of lonidamine in 1N NaOH. Later, it got significantly degraded within the first 6 h (P < 0.05) of incubation at both 40 and 80 °C. Insignificant (P > 0.05), temperature-dependent difference was observed in the degradation profile of lonidamine in presence of 1 N NaOH. There was a subsequent rise in the degradation peak (Fig. 4) around 1.66 min for a mass range of 172 to173 m/z.
Fig. 4.
(A) Time- and temperature-dependent degradation of lonidamine in the presence of 1 N NaOH. Although it shows complete degradation within first 6 to 24 h of incubation (**P < 0.01), no temperature-dependent effect was observed; (B) representative chromatograms of lonidamine showing time-dependent degradation of drug molecule in 1 N NaOH; and (C) degradation peak of lonidamine vs small standard peak for the drug and blank.
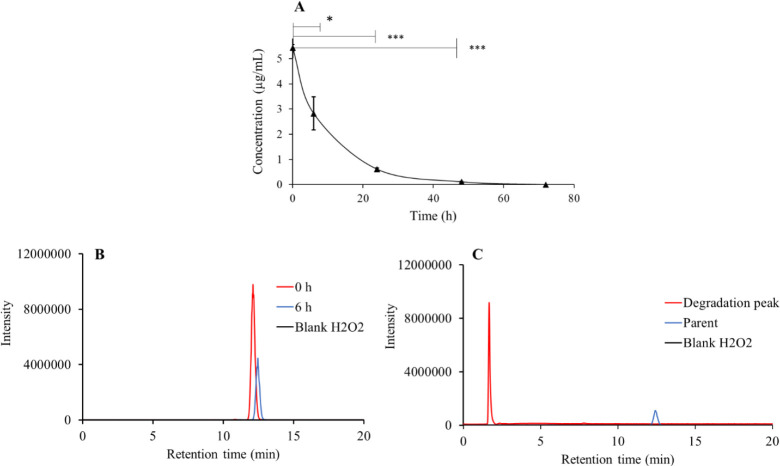
Forced degradation of lonidamine in the presence of a strong oxidizing agent
As shown in Fig. 5, lonidamine gets forced degraded at 40 °C temperature in a time-dependent manner with P < 0.05. It was noted that lonidamine gets degraded by the end of a 48-h (P < 0.05) time point. The lonidamine peak gets significantly reduced at a 48-h time point and consequently the presence of production (171 to 172 m/z) peak increases around Rt = 1.60 min.
Fig. 5.
(A) Significant time-dependent degradation of lonidamine in H2O2 at 40°C with (B) representative chromatogram of lonidamine showing time-dependent degradation, and (C) chromatogram of probable degradation product of lonidamine vs standard and blank H2O2. *P ≤ 0.05 and ***P ≤ 0.001 indicate significant differences.
DISCUSSION
Lonidamine is a hydrophobic anticancer molecule. A quantitation assay for lonidamine was developed using reverse phase chromatography. Lonidamine standards were subjected to LC-MS-UV runs using conventional isocratic elution chromatography with 50:50 water: acetonitrile with 0.1% formic acid. The present LC-MS based quantifiable method for lonidamine is sensitive for detections and quantitation in the nanogram concentration range (21). A UV based detection of eluting lonidamine (10 to 10000 ng/mL) standards were also used in tandem with mass spectrometer detection. Our results indicated that UV based HPLC detection could only provide accurate outcomes in the concentration range of 500 to 10000 ng/mL with a CV < 10% (Table 1). This observation and results are consistent with previously published work with low injection volumes (21). The result suggested that tandem HPLC-UV had higher values for lower limit of detection (LLOD), and the lower limit of quantitation (LLOQ) (Fig. 2B) as compared to LC-MS (Fig. 2A). This also confirms that the current LC-MS method provided a significant improvement over previous HPLC based UV detection methods by detecting and quantitating lower concentrations (19,22). Ioele et al. (21) provided an HPLC method with a linear calibration range of 0.5 to 50 μg/mL for lonidamine that was used for the qualitative impurity analysis which had relatively, higher LLOD and LLOQ values compared to the present study. Whereas, this study provided an online LC-MS method using high-resolution mass spectrometers systems with high sensitivity in mass detection with < 0.5 ppm error as compared to previously reported runs on HPLC and followed by separate GC-MS based confirmation of masses. Milane et al. (22) reported HPLC method for simultaneous estimation of lonidamine and paclitaxel from plasma and tissue samples of animals, where LLOD and LLOQ of 64 and 80 ng/mL was reported. Whereas, the present study data shows that with LC-MS method can go as low to 10 and 20 ng/mL for LLOD and LLOQ values. The previously published HPLC methods were not stability-indicating in nature. However, it showed that simple isocratic HPLC methods could be used for the efficient estimation of lonidamine in a complex matrix. To the best of our knowledge, this is the first reported LC-MS method for studying the degradation of lonidamine. This indicated that the present LC-MS method provides a simple robust and high sensitivity-based edge for mapping force degradation rates of lonidamine under various stress conditions.
The LC-MS (without UV detections) based linearity curve obtained for lonidamine described above were subjected to validation study using random (low, medium, high, and ultra-high) concentration ranges, as shown in Table 2. Intra- and inter-day validation results for random concentrations showed CV < 5%. The accuracy% for these validation standards for inter- and intra-day was found to be around 100 ± 20%. These studies indicated that the calibration curves are robust and with high accuracy as per the ICH guidelines and previously published reports (24,29,30,31). Linearity and validation study of linearity provides a necessary baseline for the stability of drug under ambient room temperature conditions. Further, the stability test for four random concentrations (Table 3) also supported the validation data performed by carrying out freeze-thaw cycles. It was observed that the lonidamine molecule has excellent stability at freezing and ambient room temperature at low and high concentrations, which is consistent with the previous report (21). Hence, the LC- MS based sensitive quantitation assay could be used for further analysis of the degradation of lonidamine under various forced degradation conditions.
Previous studies have shown that lonidamine (solid and solution) degrades in the presence of heat and light. It provides the necessary information to show that this molecule in the solution state is thermally stable at 21 and 40 °C. Lonidamine thermally stressed at 70 °C shows the degradation peak of the drug. Whereas, the solid lonidamine showed high stability at 60 and 100 °C and authors were able to perform only a small part of forced degradation studies (17,27). The detailed understanding of the stability of the lonidamine solution state in the presence of an external stress inducer becomes important. Hence, we performed temperature-dependent degradation of lonidamine in the presence of strong acidic (1 N HCl), basic (1 N NaOH), and oxidizing (H2O2) conditions.
To quantitatively estimate the degradation profile of lonidamine in the presence of a strong acid, we placed two vials containing 10 mL of acidified (treated with 1 N HCl) lonidamine solution in a water bath maintained at 40 and 80 °C. A time-dependent decrease in the concentration of lonidamine could be observed in Fig. 3A. There was a significant increase in the degradation rate due to a temperature increase to 80 °C. This could also be confirmed from the stepwise reduction in lonidamine LC- MS chromatogram peaks under two temperature conditions, as shown in Fig. 3B and C. This indicates that the degradation rate of lonidamine could get affected by high temperatures and strong acid. Lonidamine is degraded under temperature-dependent acid hydrolysis, and degradation peaks were also identified. The degradation peaks for lonidamine after 24 h is shown in Fig. 3D.
Further, a strong degradation of the lonidamine solution was observed with 1 N NaOH. It was observed that a strong base could cause significant damage to the structural integrity of lonidamine (Fig. 4A and B). Later, there was a dramatic reduction in the peak of lonidamine and a subsequent rise in the degradation peak (Fig. 4C). However, all the values are lesser then quantitation limits, hence we can consider complete degradation of the lonidamine.
Lonidamine degraded at both 40 and 80 °C under acidic and basic conditions. Taking this factor into consideration, lonidamine was treated with H2O2 and incubated at 40 °C in a water bath. The degradation rate of lonidamine was assessed at predetermined time points as shown in Fig. 5A and B. There was a significant degradation by the end of the 48-h time point. A degradation peak could be mapped as shown in Fig. 5C. The present LC-MS method could detect both the parent and degradation peaks (in the presence of 1 N HCl, 1 N NaOH, and H2O2) in a single run. The peaks of the parent ion remained uninterrupted by degradation.
The estimation of degradation chromatogram was carried out in an extremely accurate manner with an error of < 0.5 ppm. These results showed that lonidamine gets degraded in the presence of an acid, base, and strong oxidation conditions. Moreover, it was possible to detect the degradation peak of the parent molecule, which indicates that the LC-MS method presented in this study is stability indicating for lonidamine.
CONCLUSION
A highly sensitive, robust, and reproducible LC-MS method was developed with quantitation and detection limits in nanogram concentrations. The LLOD and LLOQ for LC-MS detection of lonidamine were found to be significantly better than the conventional HPLC-UV based detections and quantitation. Our results suggest that the current method is stability-indicating as it can map the degradation profile for lonidamine, with order degradation as 1 N NaOH > H2O2 > 1 N HCl. The stability-indicating method can help in studying the stability of formulations of lonidamine; currently being compounded for various clinical trials and future preclinical investigations.
CONFLICT OF INTEREST STATEMENT
All authors declare no conflict of interest in this study.
AUTHORS’ CONTRIBUTION
G. Kaushal, J. Eisenbery, B.E. Oeffinger, and M. Wheatley contributed to the development of the concept of this work. They provided necessary materials for conducting experiments for the work and helped to edit the manuscript; G. Kaushal and J. Eisenbery were supervisor and co-supervisor for this work, respectively. A.K. Rochani and G. Kaushal helped in designing experiments, data collection, analysis, writing, and editing the manuscript.
ACKNOWLEDGEMENTS
This work was financially supported by NIH under Grant No. R01EB023926. We would like to sincerely thank Ms. Jennifer Wilson, Office for Professional Writing, Publishing, & Communication, Thomas Jefferson University for thoroughly reading and correcting the manuscript.
REFERENCES
- 1.American Cancer Society. 2019. Available from: https://www cancer gov/about- cancer/understanding/statistics .
- 2.National Cancer Statistics. 2019. Available from: https://wwwcancergov/about- cancer/understanding/statistics .
- 3.Semenza GL, Artemov D, Bedi A, Bhuwalla Z, Chiles K, Feldser D, et al. ‘The metabolism of tumours’: 70 years later. Novartis Found Symp. 2001;240:251–260. [PubMed] [Google Scholar]
- 4.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis. Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. DOI: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Yang JM. Altered energy metabolism in cancer: a unique opportunity for therapeutic intervention. Cancer Biol Ther. 2013;14(2):81–89. doi: 10.4161/cbt.22958. DOI: 10.4161/cbt.22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JS, Ahn KJ, Kim JA, Kim HM, Lee JD, Lee JM, et al. Role of reactive oxygen species-mediated mitochondrial dysregulation in 3-bromopyruvate induced cell death in hepatoma cells: ROS-mediated cell death by 3-BrPA. J Bioenerg Biomembr. 2008;40(6):607–618. doi: 10.1007/s10863-008-9188-0. DOI: 10.1007/s10863-008-9188-0. [DOI] [PubMed] [Google Scholar]
- 7.Liemburg-Apers DC, Willems PH, Koopman WJ, Grefte S. Interactions between mitochondrial reactive oxygen species and cellular glucose metabolism. Arch Toxicol. 2015;89(8):1209–1226. doi: 10.1007/s00204-015-1520-y. DOI: 10.1007/s00204-015-1520-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da-Silva WS, Gomez-Puyou A, de Gomez-Puyou MT, Moreno-Sanchez R, De Felice FG, de Meis L, et al. Mitochondrial bound hexokinase activity as a preventive antioxidant defense: steady-state ADP formation as a regulatory mechanism of membrane potential and reactive oxygen species generation in mitochondria. J Biol Chem. 2004;279(38):39846–39855. doi: 10.1074/jbc.M403835200. DOI: 10.1074/jbc.M403835200. [DOI] [PubMed] [Google Scholar]
- 9.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25(34):4633–4646. doi: 10.1038/sj.onc.1209597. DOI: 10.1038/sj .onc. 1209597. [DOI] [PubMed] [Google Scholar]
- 10.Nath K, Guo L, Nancolas B, Nelson DS, Shestov AA, Lee SC, et al. Mechanism of antineoplastic activity of lonidamine. Biochim Biophys Acta. 2016;1866(2):151–162. doi: 10.1016/j.bbcan.2016.08.001. DOI: 10.1016/j.bbcan.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cervantes-Madrid D, Romero Y, Duenas-Gonzalez A. Reviving lonidamine and 6-Diazo-5-oxo-L-norleucine to be used in combination for metabolic cancer therapy. Biomed Res Int. 2015;2015:690492,1–13. doi: 10.1155/2015/690492. DOI: 10.1155/2015/690492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nath K, Guo L, Nancolas B, Nelson DS, Shestov AA, Lee SC, et al. Mechanism of antineoplastic activity of lonidamine. Biochim Biophys Acta. 2016;1866(2):151–162. doi: 10.1016/j.bbcan.2016.08.001. DOI: 10.1016/j.bbcan.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nath K, Nelson DS, Ho AM, Lee SC, Darpolor MM, Pickup S, et al. (31) P and (1) H MRS of DB-1 melanoma xenografts: lonidamine selectively decreases tumor intracellular pH and energy status and sensitizes tumors to melphalan. NMR Biomed. 2013;26(1):98–105. doi: 10.1002/nbm.2824. DOI: 10.1002/nbm.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nath K, Nelson DS, Heitjan D, Leeper DB, Zhou R, Glickson JD. Lonidamine induces intracellular tumor acidification and ATP depletion in breast, prostate and ovarian cancer xenografts and potentiates response to doxorubicin. NMR Biomed. 2015;28(3):281–290. doi: 10.1002/nbm.3240. DOI: 10.1002/nbm.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Lena M, Lorusso V, Latorre A, Fanizza G, Gargano G, Caporusso L, et al. Paclitaxel, cisplatin and lonidamine in advanced ovarian cancer A phase II study. Eur J Cancer. 2001;37(3):364–368. doi: 10.1016/s0959-8049(00)00400-7. DOI: 101016/s0959-8049(00)00400-7. [DOI] [PubMed] [Google Scholar]
- 16.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25(34):4633–4646. doi: 10.1038/sj.onc.1209597. DOI: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 17.Zhang BF, Xing L, Qiao JB, Cui PF, Wang FZ, Zhang JL, et al. In vivo synergistic antitumor effect and safety of siRNA and lonidamine dual-loaded hierarchical targeted nanoparticles. Int J Pharm. 2016;506(1-2):207–213. doi: 10.1016/j.ijpharm.2016.04.056. DOI: 10.1016/j.ijpharm.2016.04.056. [DOI] [PubMed] [Google Scholar]
- 18.Ruttala HB, Ramasamy T, Poudel BK, Ruttala RRT, Jin SG, Choi HG, et al. Multi-responsive albumin- lonidamine conjugated hybridized gold nanoparticle as a combined photothermal-chemotherapy for synergistic tumor ablation. Acta Biomater. 2020;101:531–543. doi: 10.1016/j.actbio.2019.11.003. DOI: 10.1016/j.actbio.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Grippa E, Gatto MT, Leone MG, Tita B, Adbel-Haq H, Vitalone A, et al. Analysis of lonidamine in rat serum and testis by high performance liquid chromatography. Biomed Chromatogr. 2001;15(1):1–8. doi: 10.1002/bmc.14. DOI: 10.1002/bmc.14. [DOI] [PubMed] [Google Scholar]
- 20.Chen BQ, Kankala RK, Wang SB, Chen AZ. Continous nanonization of lonidamine by modified- rapid expansion of supercritical solution process. J of Superitical fluids. 2018;133(1):486–493. DOI: 10.1016/j.supflu.2017.11.016. [Google Scholar]
- 21.Ioele G, De Luca M, Ragno G. Lonidamine and related impurities: HPLC analysis, stability profile and degradation pathways. Anal Methods. 2013;5(7):1715–1720. DOI: 10.1039/C3AY26467J. [Google Scholar]
- 22.Milane L, Duan ZF, Amiji M. Pharmacokinetics and biodistribution of lonidamine/paclitaxel loaded, EGFR-targeted nanoparticles in an orthotopic animal model of multi-drug resistant breast cancer. Nanomedicine. 2011;7(4):435–444. doi: 10.1016/j.nano.2010.12.009. DOI: 10.1016/j.nano.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam E, Rochani A, Kaushal G, Thoma BN, Tanjuakio J, West FM, et al. Pharmacokinetics of ketamine at dissociative doses in an adult patient with refractory status asthmaticus receiving extracorporeal membrane oxygenation therapy. Clin Ther. 2019;41(5):994–999. doi: 10.1016/j.clinthera.2019.03.005. DOI: 10.1016/j.clinthera.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rochani A, Lam E, Tanjuakio J, Hirose H, Kraft WK, Kaushal G. Simultaneous quantitative LC-MS method of ketamine, midazolam and their metabolites (dehydronorketamine, norketamine and 1hydroxymidazolam) for its application in patients on extracorporeal membrane oxygenation (ECMO) therapy. J Pharm Biomed Anal. 2020;178:112947,1–9. doi: 10.1016/j.jpba.2019.112947. DOI: 10.1016/j.jpba.2019.112947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.US Department of Health and Human Services, FDA, CDER, CVM. Bioanalytical Method Validation Guidance for Industry. 2018. Available from: https://wwwfdagov/regulatory-information/search- fda-guidance-documents/bioanalytical-method- validation-guidance-industry .
- 26.Moorthy GS, Jogiraju H, Vedar C, Zuppa AF. Development and validation of a sensitive assay for analysis of midazolam, free and conjugated 1- hydroxymidazolam and 4-hydroxymidazolam in pediatric plasma: application to pediatric pharmacokinetic study. J Chromatogr B Analyt Technol Biomed Life Sci. 2017;1067:1–9. doi: 10.1016/j.jchromb.2017.09.030. DOI: 10.1016/j.jchromb.2017.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blessy M, Patel RD, Prajapati PN, Agrawal YK. Development of forced degradation and stability indicating studies of drugs-a review. J Pharm Anal. 2014;4(3):159–165. doi: 10.1016/j.jpha.2013.09.003. DOI: 10.1016/j.jpha.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohammadi A, Rezanour N, Ansari Dogaheh M, Ghorbani Bidkorbeh F, Hashem M, Walker RB. A stability-indicating high performance liquid chromatographic (HPLC) assay for the simultaneous determination of atorvastatin and amlodipine in commercial tablets. J Chromatogr B Analyt Technol B iomed Life Sci. 2007;846(1-2):215–221. doi: 10.1016/j.jchromb.2006.09.007. DOI : 10.1016/j.jchromb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Duraisamy K, Jaganathan KS, Krishna MV. Method development and validation of HPLC tandem/mass spectrometry for quantification of perindopril arginine and amlodipine besylate combination in bulk and pharmaceutical formulations. Res Pharm Sci. 2017;12(4):307–314. doi: 10.4103/1735-5362.212048. DOI: 10.4103/1735-5362.212048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rezazadeh M, Emami J. A simple and sensitive HPLC method for analysis of imipramine in human plasma with UV detection and liquid-liquid extraction: application in bioequivalence studies. Res Pharm Sci. 2016;11(2):168–176. [PMC free article] [PubMed] [Google Scholar]
- 31.Varshosaz J, Emami J, Tavakoli N, Minaiyan M, Rahmani N, Ahmadi F, et al. Development and validation of a rapid HPLC method for simultaneous analysis of budesonide and its novel synthesized hemiesters in colon specific formulations. Res Pharm Sci. 2011;6(2):107–116. [PMC free article] [PubMed] [Google Scholar]